Abstract
Selected reaction monitoring mass spectrometry (SRM-MS) is playing an increasing role in quantitative proteomics and biomarker discovery studies as a method for high throughput candidate quantification and verification. Although SRM-MS offers advantages in sensitivity and quantification compared with other MS-based techniques, current SRM technologies are still challenged by detection and quantification of low abundance proteins (e.g. present at ∼10 ng/ml or lower levels in blood plasma). Here we report enhanced detection sensitivity and reproducibility for SRM-based targeted proteomics by coupling a nanospray ionization multicapillary inlet/dual electrodynamic ion funnel interface to a commercial triple quadrupole mass spectrometer. Because of the increased efficiency in ion transmission, significant enhancements in overall signal intensities and improved limits of detection were observed with the new interface compared with the original interface for SRM measurements of tryptic peptides from proteins spiked into non-depleted mouse plasma over a range of concentrations. Overall, average SRM peak intensities were increased by ∼70-fold. The average level of detection for peptides also improved by ∼10-fold with notably improved reproducibility of peptide measurements as indicated by the reduced coefficients of variance. The ability to detect proteins ranging from 40 to 80 ng/ml within mouse plasma was demonstrated for all spiked proteins without the application of front-end immunoaffinity depletion and fractionation. This significant improvement in detection sensitivity for low abundance proteins in complex matrices is expected to enhance a broad range of SRM-MS applications including targeted protein and metabolite validation.
Although mass spectrometry (MS)-based proteomics is a promising high throughput technology for biomarker discovery and validation (1–5), only a handful of cancer biomarkers have been approved by the United States Food and Drug Administration for clinical use in the last decade (6, 7). Assuming that low abundance biomarkers do exist in the biofluids to be studied, the success of biomarker discovery efforts primarily depends on the sensitivity, accuracy, and robustness of the measurement technologies; the quality and size of patient cohorts and clinical samples and execution within the context of an overall difficult and expensive path to clinical application that encompasses discovery, verification, and validation stages (1, 5, 8–10). A multiplexed assay platform increasingly considered for biomarker verification is selected reaction monitoring (SRM)1 by tandem mass spectrometry using e.g. a triple quadrupole (QqQ) mass spectrometer to attain high throughput quantitative measurements of targeted proteins in complex matrices (1, 11, 12).
SRM utilizes two stages of mass filtering by selecting a specific analyte ion of interest (precursor ion) in the first stage followed by a specific fragment ion derived from the precursor (fragment ion) filter in the second stage after collision-activated dissociation. Typically, several transitions (precursor/fragment ion pairs) are monitored for greater selectivity and confidence in a targeted peptide assay, and large numbers of peptides can be monitored during a single LC-MS/MS analysis. The two-stage mass selection by individual quadrupoles enables more rapid and continuous monitoring of specific ions derived from analytes of interest such as peptides and leads to significantly enhanced detection sensitivity and quantitative accuracy compared with broad (i.e. non-targeted) LC-MS or LC-MS/MS measurements (11, 12). Both the sensitivity and selectivity of SRM-MS make this technique well suited for the targeted detection and quantification of low abundance proteins in highly complex biofluids (13–16). The precision and reproducibility of SRM-based measurements of proteins in plasma across different laboratories have recently been assessed (17).
Despite its promise, present SRM measurements still do not provide sufficient sensitivity for reliable detection and quantification of low abundance proteins in biofluids (e.g. present in plasma at ∼10 ng/ml or lower levels) primarily because of factors related to high sample complexity and the large dynamic range of relative protein abundances (7, 18, 19). Given sufficient selectivity, the sensitivity achievable is generally related to the peptide MS and MS/MS signal intensities obtained. One of the key factors limiting peptide MS intensities is the significant ion losses encountered between the electrospray ionization (ESI) source and the interface to the mass spectrometer. In typical LC-ESI-MS interfaces, the mass spectrometer inlet (e.g. heated capillary followed by a skimmer) presently provides total ion utilization and ion transmission efficiencies on the order of ∼1% (20) due to a combination of limited ion sampling from the atmospheric pressure ion source into the inlet and inefficient transmission of ions entering the first reduced pressure stage of the mass spectrometer.
The electrodynamic ion funnel (21), which has been developed to efficiently capture, focus, and transmit ions to the high vacuum region of the mass spectrometer, is expected to provide a large benefit to SRM analyses. The original ion funnel interfaces, which operated at a maximum of ∼5 torr, were able to enhance signal intensities for a variety of MS analyzers (22–24) by replacing the inefficient skimmer interface. Although achieving near lossless ion transmission to high vacuum, losses at the atmospheric pressure interface went unmitigated. More recently, a high pressure ion funnel interface capable of operating at a pressure of ∼30 torr was introduced (25). The higher operating pressures accommodated greater gas loads and enabled more efficient ion sampling from atmospheric pressure through a multicapillary inlet. With a dual ion funnel interface comprising a high pressure ion funnel with a heated multicapillary inlet followed by a standard ion funnel operated at 1–2 torrs, highly efficient ion sampling from atmospheric pressure to high vacuum is readily achieved.
In this study, we report the enhanced sensitivity and reproducibility of SRM-based targeted proteomics measurements achieved by implementing a dual stage electrodynamic ion funnel interface that incorporates a multicapillary inlet with a triple quadrupole mass spectrometer. A series of LC-SRM-MS measurements were made using mouse plasma samples spiked with various concentrations of tryptic peptides from five standard proteins to evaluate the improvements in detection sensitivity and reproducibility attained by this modified interface relative to a standard Thermo (single capillary inlet/skimmer) interface. A ∼10-fold improvement in the limit of detection (LOD) as well as improved measurement reproducibility was achieved.
EXPERIMENTAL PROCEDURES
Materials and Chemicals
The standard proteins bovine carbonic anhydrase, Escherichia coli β-galactosidase, equine skeletal muscle myoglobin, chicken ovalbumin, and bovine cytochrome c and the standard peptides bradykinin fragment 1–7, kemptide, melittin, methionine enkephalin, renin substrate porcine, and [d-Ala2]-deltorphin II were purchased from Sigma. Ammonium bicarbonate, dithiothreitol (DTT), and iodoacetamide were from Sigma. Sequencing grade modified porcine trypsin was from Promega (Madison, WI), and the mouse plasma was purchased from Equitech Bio, Inc. (Kerrville, TX).
Sample Preparation
The initial mouse plasma protein concentration was ∼40 mg/ml as determined by BCA protein assay (Pierce). Individual stock solutions were prepared for each of the following five standard proteins: bovine carbonic anhydrase, E. coli β-galactosidase, equine skeletal muscle myoglobin, chicken ovalbumin, and bovine cytochrome c. First, the standard proteins were digested separately using the same protocol as that used to digest the mouse plasma. Briefly, ∼1 mg of proteins was denatured and reduced in 50 mm NH4HCO3 buffer (pH 8.0), 8 m urea, 10 mm DTT for 1 h at 37 °C. Protein cysteinyl residues were alkylated with 40 mm iodoacetamide for 90 min at room temperature. The resulting mixtures were diluted 6-fold with 50 mm NH4HCO3 (pH 8.0), and then trypsin was added at a trypsin-to-protein ratio of 1:50 (w/w). Each sample was incubated at 37 °C for 3 h, then loaded onto a 1-ml solid phase extraction C18 column (Supelco, Bellefonte, PA), and washed with 4 ml of 0.1% trifluoroacetic acid (TFA), 5% acetonitrile (ACN). Peptides were eluted from the solid phase extraction column with 1 ml of 0.1% TFA, 80% ACN; lyophilized; and redissolved in 200 μl of 50 mm NH4HCO3. The final peptide concentration in each of the stock solutions was determined by BCA protein assay (Pierce), and then aliquots of the five solutions were combined to form a new stock solution with equal concentrations of each of the proteins. Samples for SRM analysis were prepared by spiking different aliquots of the mixture of digested protein standards into a mouse plasma tryptic digest to desired concentrations (see Table I). The spiking values were calculated in relation to the protein concentration in the original mouse plasma. After spiking, mouse samples were adjusted to a final concentration of 0.5 μg/μl total protein. The samples were then split into aliquots, frozen in liquid nitrogen, and stored at −80 °C until use.
Table I. Standard protein spiking levels used are listed in ng/ml protein concentration in original plasma sample (40 mg/ml total protein) and mass to mass ratio (spiking level relative to total plasma protein).
| Sample | ng/ml | Mass to mass ratio |
|---|---|---|
| S9 | 40,000 | 1 × 10−3 |
| S8 | 4,000 | 1 × 10−4 |
| S7 | 400 | 1 × 10−5 |
| S6 | 80 | 2 × 10−6 |
| S5 | 40 | 1 × 10−6 |
| S4 | 8 | 2 × 10−7 |
| S3 | 4 | 1 × 10−7 |
| S2 | 0.8 | 2 × 10−8 |
| S1 | 0.2 | 5 × 10−9 |
| S0 | 0 | 0 |
LC-SRM-MS
Peptide samples were analyzed using a custom-built automated four-column high pressure capillary LC system (26) coupled on line to a triple quadrupole mass spectrometer (TSQ Quantum Ultra, Thermo Fisher Scientific). All LC separations were performed using the same capillary column, which was prepared by slurry-packing 3-μm Jupiter C18 bonded particles (Phenomenex, Torrance, CA) into a 65-cm-long, 75-μm-inner diameter fused silica capillary (Polymicro Technologies, Phoenix, AZ). This capillary column was connected via a Valco 100-μm-inner diameter stainless steel union to a chemically etched fused 20-μm-inner diameter silica emitter (27) produced in house. The mobile phase consisted of 0.1% formic acid in water (A) and 0.1% formic acid in ACN (B). After loading 5 μl of the peptide solution onto the column (2.5 μg nominally), an exponential gradient elution was performed by increasing the mobile phase composition from 0 to 70% B over 100 min. The inlet capillary of the mass spectrometer was maintained at 200 °C with an ESI voltage of 2.5 kV that was applied to the Valco union.
Dual Electrodynamic Ion Funnel Interface
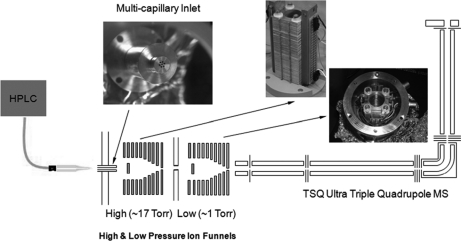
LC-SRM-MS analyses were performed using either the standard Thermo interface or the dual ion funnel interface. In the dual ion funnel configuration (25), one high pressure funnel (∼17 torr) and one low pressure funnel (∼1 torr) were used in tandem (Fig. 1). Both ion funnels consisted of 100 stainless steel electrodes; each electrode was 0.5 mm thick and separated by 0.5-mm-thick Teflon sheets. The inner diameter of each of the first 55 electrodes was 25.4 mm, and the inner diameters of the last 45 electrodes decreased linearly from 25.4 to 2.0 mm. The total capacitance of the high pressure ion funnel electrodes was 1.6 nanofarads, which resulted in higher radio frequency (RF) peak-to-peak voltages for effective ion transmission in the pressure regime of the high pressure ion funnel. A 180° out-of-phase RF field was applied to adjacent electrodes to create an effective potential well that confined the ions, whereas a DC gradient pushed the ions through the ion funnel. The high pressure ion funnel had an entrance DC bias of 425 V and a bias of 235 V on the last RF ion funnel electrode. The low pressure ion funnel had DC biases of 220 and 7 V on the first and last RF ion funnel electrodes, respectively.
Fig. 1.
Schematic of Thermo QqQ Quantum Ultra mass spectrometer equipped with dual ion funnel interface. The main features of this interface are the incorporation of a multicapillary inlet along with two consecutive ion funnels, one high pressure (∼17 torr) funnel and one low pressure (∼1 torr) funnel used in tandem.
The two ion funnels were separated by a 2.3-mm-inner diameter conductance-limiting DC-only orifice. The funnels also incorporated a jet disrupter disk with a diameter of 6.5 mm located ∼20 mm from the ion funnel entrance along the funnel axis (28). The DC-only electric potential of the jet disrupter and conductance limit orifice electrode were independently controlled at 390 and 230 V for the high pressure ion funnel and 175 and 2 V for the low pressure ion funnel, respectively.
The dual ion funnel configuration enabled incorporation of a multicapillary inlet, which significantly increased ion transmission into the mass spectrometer relative to the standard, single capillary inlet interface. The multicapillary inlet was constructed using an array of seven thin wall stainless tubes (600 μm inner diameter and 900 μm outer diameter) soldered into a central hole of cylindrical heating block. The vacuum arrangement of the instrument was also modified in the dual ion funnel configuration. Rather than using the standard two rotary vacuum pumps (Edwards 30, Tewksbury, MA) connected to the TSQ main vacuum line, one of the original pumps was used in conjugation with a second pump via a common header and throttled with a gate valve (Oerlikon Leybold) to maintain the desired pressure in the higher pressure ion funnel chamber. The common gate valve was throttled to achieve ∼1 torr in the low pressure ion funnel chamber.
Selection of Peptides and Transitions
Peptides and SRM transitions for each protein were identified, and collision energies were optimized by direct infusion of a 500 nm digested solution of each standard protein in a 1:1 mixture of solvents A and B at an infusion rate of 300 nl/min. Three transitions were targeted for each peptide, and the two best responding peptides were selected for each of the standard proteins. Similarly, two transitions for each quality control (QC) peptide were also selected. The selection of peptides and transitions was based on the intensity data in the acquired mass spectra. During SRM analysis, a dwell time of 10 ms and a scan window of 0.002 m/z were used. Both Q1 and Q3 were set to a peak width of 0.7 full-width at half-maximum. During LC-SRM analysis, centroid data were acquired in positive ion mode.
Data Analysis
Data were analyzed using Thermo Xcalibur 2.0.7, specifically with Qual Browser. Peak detection and integration were performed using the Genesis algorithm with automated determinations performed on the highest concentration spiked samples (S9, 40 μg/ml and S8, 4 μg/ml; see Table I) using the following parameters: 10% of highest peak, 2.0 minimum peak height, and a signal-to-noise threshold of 0.5. In addition, peaks were constrained (Constrain Peak Width option selected) to a peak height of 5% and tailing factor of 1.5 (2.0 for the bovine cytochrome c peptides). To aid more accurate determination of the retention time, peaks were initially smoothed (Gaussian, 15 points), and the distance from the nearest QC peptide peak was determined. Six standard peptides, bradykinin fragment 1–7, kemptide, melittin, methionine enkephalin, renin substrate porcine, and [d-Ala2]-deltorphin II, were used as QC peptides for LC retention time markers and for monitoring the overall performance of the platform. The LOD was determined by the lowest detectable sample concentration in this study having a signal-to-noise area ratio of at least 3 for the best transition. Peak smoothing was not used in determining the peak areas. The peak widths for each peptide were determined by averaging the data obtained using the two highest spiked concentrations (S8 and S9), and the elution time difference observed relative to the nearest QC marker peaks served as a peak integration window for the lower (S0–S7) spike concentrations. Analyte peak identification at the lower concentrations was determined based on three main criteria. 1) The difference of elution time from the nearest QC peaks was within ±10% of the average difference observed from the two highest concentrations. 2) Analyte peak widths were similar (i.e. within ±10%) to the average of peak widths from the two top concentrations. 3) Analyte peak area decreased with decreasing analyte concentration levels.
RESULTS AND DISCUSSION
This study evaluated the improvements in SRM-MS measurements achievable based upon the increased signal levels obtained using a much more efficient ESI-MS interface. Fig. 1 shows the configuration of the dual ion funnel interface coupled to the QqQ mass spectrometer. The high pressure ion funnel (∼17 torr) used a seven-capillary inlet instead of the single capillary inlet typically applied for a standard low pressure (∼1 torr) ion funnel configuration. The greater sampling efficiency achieved with the seven-capillary inlet allowed significantly more ions to be transmitted into the mass spectrometer from the ESI source, thus providing overall higher ion transmission efficiency to the mass spectrometer. To evaluate the dual ion funnel interface for SRM measurements, we selected five standard proteins that were spiked into a digest of (non-depleted) mouse plasma at nine concentration levels. Protein concentrations ranged from 0.2 to 40,000 ng/ml in mouse plasma as shown in Table I (S1–S9). A side-by-side comparison between the dual ion funnel interface and the standard Thermo interface was accomplished by performing triplicate analyses of these 10 samples using both interface configurations. Six standard peptides spiked into each sample at a level of 500 nm prior to LC-SRM analyses served as QC peptides. For each of the five standard proteins, two proteotypic peptides were selected from which three precursor-to-fragment transitions for each selected peptide were evaluated.
Improved SRM Peak Intensity
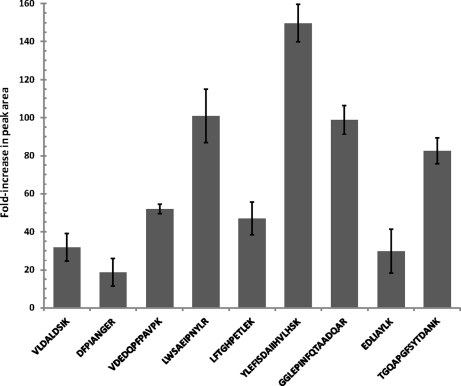
Fig. 2 shows the average increase in SRM peak areas attained with the multicapillary inlet/dual ion funnel interface relative to those observed with the standard interface. The data for the nine peptides in this figure originated from triplicate LC-SRM-MS analyses of S9 (Table I). On average, peak areas were enhanced ∼70-fold with the dual ion funnel interface with peak area improvements ranging from ∼20- to ∼150-fold for individual peptides. Significant interference from the plasma matrix was observed with one of the peptides, HIATNAVLFFGR, which made its peak area calculation ambiguous. Additional data for all of the SRM transitions are available in supplemental Table 1.
Fig. 2.
Average increase in SRM peak area for dual ion funnel configuration relative to Thermo interface for best transitions from nine peptides in S9 sample (see Table I). Error bars represent standard deviations of the measurements.
Overall, the observed increases in SRM peak intensities are in good agreement with expectations based on earlier observations for various MS platforms. For example, >10-fold signal gains were observed when a standard low pressure ion funnel interface was coupled to a triple quadrupole mass spectrometer (22), and an additional 5-fold enhancement in signal intensity relative to a low pressure ion funnel configuration was achieved using a dual ion funnel fitted with a multicapillary inlet in a time-of-flight (TOF) mass spectrometer (25). An overall gain of ∼40-fold was observed when a multiemitter/dual ion funnel configuration was compared with an unmodified TOF (29).
Enhanced Detection of Low Abundance Proteins
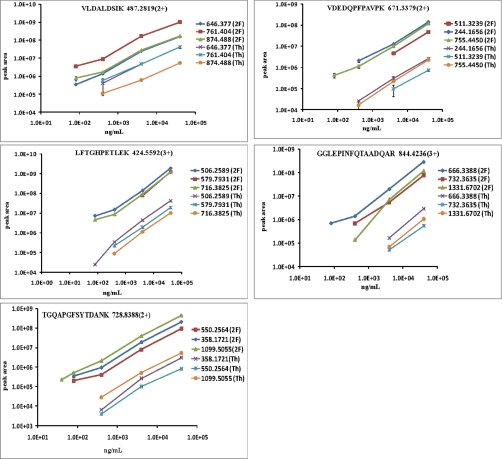
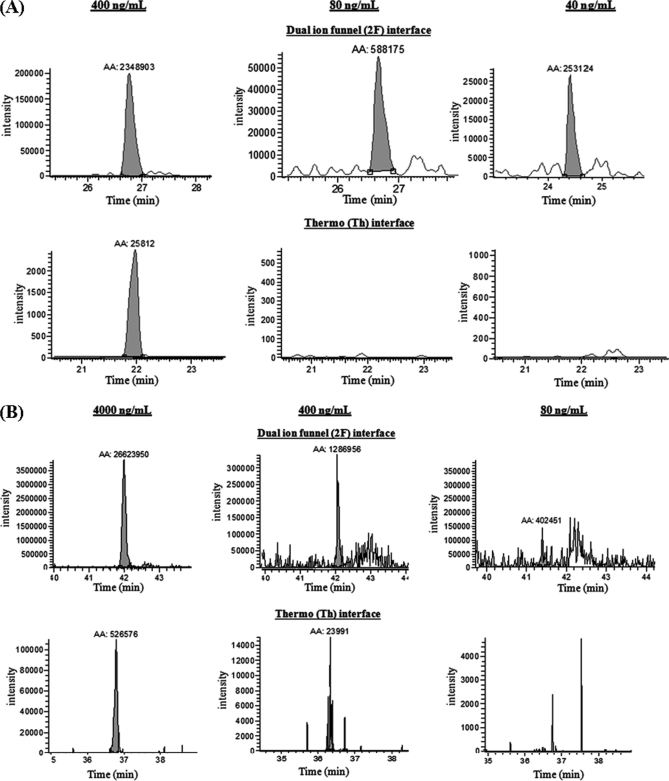
Our next step was to assess whether the increased signal intensities enhanced detection of the low abundance spiked proteins in mouse plasma, i.e. an improved lower LOD for targeted proteins. Fig. 3 shows the SRM peak areas observed for five peptides at different analyte concentrations and all transitions with both the standard and dual ion funnel interfaces; some examples of SRM chromatograms for the high intensity transitions, m/z 728.842+ > 1099.51+ for peptide TGQAPGFSYTDANK and m/z 482.772+ > 494.30+ for peptide EDLIAYLK of bovine cytochrome c at different concentration levels (40–400 ng/ml and 80–4000 ng/ml, respectively), are shown in Fig. 4. As shown, the enhanced peak intensities resulted in better LC peak shapes and improved signal-to-noise ratio, thus leading to lower LOD. Table II shows the LOD and coefficient of variance (CV) for the best transitions for each peptide with the two interface configurations. The best performing transition was defined as the transition that was observed with highest signal intensity. The data demonstrate that both sensitivity and detection limit are improved by using the dual ion funnel configuration. With the dual ion funnel interface, six of 10 peptides were detected at 40–80 ng/ml levels; with the standard interface, the LODs for the same six peptides were 400–4000 ng/ml with the exception of one peptide detected at 80 ng/ml. On average, the overall improvement in LOD for all targeted peptides using the dual ion funnel interface was ∼10-fold with only one exception for peptide LFTGHPETLEK (from myoglobin) because of interference between the peptide of interest and a matrix peak that confounded quantification of proteins present at <80 ng/ml. The LC-SRM chromatograms for all transitions listed in Table II are available as supplemental Figs. A–J.
Fig. 3.
Plots of peak area versus concentration curves for each of five selected peptides in mouse plasma are constructed (peak area versus ng/ml protein concentration) with dual ion funnel (2F; upper three bold traces) interface and standard Thermo (Th; lower three narrow traces) configurations. Peptide VLDALDSIK is from protein bovine carbonic anhydrase, peptide VDEDQPFPAVPK is from E. coli β-galactosidase, peptide LFTGHPETLEK is from equine skeletal muscle myoglobin, peptide GGLEPINFQTAADQAR is from chicken ovalbumin, and peptide TGQAPGFSYTDANK is from bovine cytochrome c. Three transitions are monitored for each of the peptides. Error bars represent standard deviations of the measurements.
Fig. 4.
Comparison of observed extracted ion chromatograms using dual ion funnel and standard Thermo interfaces for two intense transitions (m/z 728.842+ > 1099.51+ of peptide TGQAPGFSYTDANK (A) and m/z 482.772+ > 494.30+ of peptide EDLIAYLK (B)) from protein bovine cytochrome c. AA, peak area; 2F, dual ion funnel; Th, standard Thermo.
Table II. Summary of peptide data for best transitions studied.
| Protein peptide sequence | Precursor ion m/z | Fragment ion m/z | Standard interface |
Dual ion funnel interface |
|||
|---|---|---|---|---|---|---|---|
| LOD | CV | LOD | CV | CV at thermo LODa | |||
| ng/ml | % | ng/ml | % | % | |||
| Bovine carbonic anhydrase | |||||||
| VLDALDSIK | 487.28192+ | 874.4880+ (y8) | 400 | 20.4 | 80 | 23.5 | 9.7 |
| DFPIANGER | 509.75132+ | 378.7036+2 (y72+) | 80 | 38.3 | 40 | 36.6 | 11.6 |
| E. coli β-galactosidase | |||||||
| VDEDQPFPAVPK | 671.33792+ | 755.4450+ (y7) | 400 | 45.3 | 80 | 24.3 | 18.2 |
| LWSAEIPNLYR | 681.36422+ | 662.3620+ (y5) | 4,000b | 16.4 | 4,000b | 5.5 | 5.5 |
| Equine skeletal muscle myoglobin | |||||||
| LFTGHPETLEK | 424.55923+ | 506.25892+ (y92+) | 80 | 25.2 | 80b | 5.3 | 5.3 |
| YLEFISDAIIHVLHSK | 629.01213+ | 740.41962+ (y132+) | 40,000 | 9.9 | 4,000 | 4.2 | 10.2 |
| Chicken ovalbumin | |||||||
| HIATNAVLFFGR | 673.37242+ | 1,095.5946+ (y10) | 40,000b | 40.0 | 40,000b | 2.5 | 2.5 |
| GGLEPINFQTAADQAR | 844.42363+ | 666.33882+ (y122+) | 4,000 | 13.0 | 80 | 14.9 | 7.24 |
| Bovine cytochrome c | |||||||
| EDLIAYLK | 482.77112+ | 494.2973+ (y4) | 400 | 54.4 | 80 | 7.6 | 3.5 |
| TGQAPGFSYTDANK | 728.83882+ | 1,099.5055+ (y10) | 400 | 79.1 | 40 | 9.4 | 11.9 |
a Percent CV for selected best transitions at the concentration of the LOD for the standard Thermo interface obtained with the dual ion funnel interface.
b Interference peaks from the matrix were observed to limit the detection and quantification of the peaks of interest.
Improved Reproducibility for Quantification
In addition to enhanced sensitivity, we also observed improved reproducibility. Both interfaces were first compared for the reproducibility of their best transitions of each peptide using the same concentration levels. Here Thermo interface LOD levels were used for comparison. Then reproducibility was also calculated for only the dual ion funnel interface at its own LOD level. Table II shows that the CV values for the selected best transitions obtained with the dual ion funnel interface are lower than those obtained with the standard interface for the same set of samples at specific concentration levels. For the Thermo interface, the CV of the selected best transitions at its lower detection level ranges from 9.9% to as high as 79.1% with an average of 34.2%, whereas the CV values obtained with the dual ion funnel interface range from 2.5 to 11.9% with an average of 8.6% for various peptides at the same concentrations as the Thermo LOD levels. The CV values for nine of 10 peptides are less than 25%, ranging from 2.5 to 24.3%, with an average of 10.8% for peptides at the dual funnel interface lower detection levels.
Although the improvements in both sensitivity and reproducibility are anticipated because of the increased ion transmission through the new interface, such significant benefits observed are attributed to the unique selectivity enabled by the two-stage mass filtering in SRM measurements. In typical LC-MS measurements, the detection of analyte is largely limited by signal-to-noise ratios where chemical background noise is often dominant in these measurements. Because the ion funnel interface enhances overall ion transmission without bias, the signal-to-noise ratios in LC-MS measurements mostly do not improve; thus, they have a limited effect on LC-MS analyte detection sensitivity as observed previously (25). However, the unique two-stage mass filtering for the parent/fragment ions at different quadrupoles in SRM-MS measurements significantly reduces the background chemical noise to a very low level by allowing only analyte signal to pass. As a result, the detection of a given analyte by SRM is governed by whether there are sufficient analyte signals available to overcome the electronic noise of the detector. Therefore, SRM-MS constitutes an ideal platform for taking advantage of the increased ion transmission of the dual ion funnel interface. In the SRM measurement mode, the increased ion transmission results in enhanced signal intensities (Fig. 2) without a proportionally increased noise level due to the mass filtering, leading to a lower limit of detection observed (Fig. 3). Similarly, the increased signals improved measurement reproducibility (Table II) because the increased signal intensities reduce the contribution of detector electronic noise, which is often the main contributor of overall variations at low signal levels, thus improving measurement reproducibility.
Besides sensitivity, another challenge in LC-SRM-MS measurements is the degree of interference peaks eluted closely with the analytes of interest as clearly observed in our data (supplemental Figs. A–J). Such interference peaks often make it difficult to identify exact analyte peak for quantification. Although the use of stable isotope-labeled standard peptides is helpful to identify analyte peaks based on their elution time, developments in better LC separation and strategies that simplify the sample complexity will be necessary to further enhance the selectivity of overall measurements.
Conclusions
In this study, we demonstrated that the implementation of a new ESI-MS interface that incorporates multicapillary inlets and dual electrodynamic ion funnels on a triple quadrupole mass spectrometer significantly improves SRM sensitivity and reproducibility as assessed by the LOD and CV values of peptides spiked into non-depleted mouse plasma. Significant improvement in ion transmission efficiency to the mass spectrometer with the new interface was confirmed by the observation of a ∼70-fold average increase in SRM peak intensities for all peptides relative to the unmodified instrument interface. The increased ion signal intensities result in a lower analyte LOD and better reproducibility. The LOD improved on average by ∼10-fold for the selected peptides. For all selected peptides, better reproducibility was confirmed by coefficients of variance from triplicate measurements. 40–80 ng/ml proteins spiked into mouse plasma were detected without the application of front-end immunoaffinity depletion and fractionation using this new platform. We anticipate that the coupling of this dual ion funnel interface with a triple quadrupole mass spectrometer will enable broad applications involving targeted measurements of low abundance analytes (e.g. proteins or metabolites) in complex biological samples because of the enhanced sensitivity and reproducibility. When coupling with front-end single (30) or dual stage (19) immunoaffinity depletion and fractionation, SRM-MS with this new interface is expected to allow measurements at sub-ng/ml levels for proteins in human plasma.
Acknowledgments
Experimental work was performed in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the United States Department of Energy (DOE) and located at Pacific Northwest National Laboratory, which is operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL0 1830.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Director's New Innovator Award Program Grant 1-DP2OD006668-01 (to W.-J. Q.) and National Center of Proteomics Research Resource for Integrative Biology Grant RR018522 (to R. D. S.). Portions of this work were also supported by the Entertainment Industry Foundation (EIF) and the EIF Women's Cancer Research Fund.
 This article contains supplemental Table 1 and Figs. A–J.
This article contains supplemental Table 1 and Figs. A–J.
1 The abbreviations used are:
- SRM
- selected reaction monitoring
- LOD
- limit of detection
- CV
- coefficient of variance
- QqQ
- triple quadrupole
- RF
- radio frequency
- DC
- direct current
- QC
- quality control.
REFERENCES
- 1. Rifai N., Gillette M. A., Carr S. A. (2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 24, 971–983 [DOI] [PubMed] [Google Scholar]
- 2. Lu M., Faull K. F., Whitelegge J. P., He J., Shen D., Saxton R. E., Chang H. R. (2007) Proteomics and mass spectrometry for cancer biomarker discovery. Biomarker Insights 2, 347–360 [PMC free article] [PubMed] [Google Scholar]
- 3. Schiess R., Wollscheid B., Aebersold R. (2009) Targeted proteomic strategy for clinical biomarker discovery. Mol. Oncol. 3, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hanash S. M., Pitteri S. J., Faca V. M. (2008) Mining the plasma proteome for cancer biomarkers. Nature 452, 571–579 [DOI] [PubMed] [Google Scholar]
- 5. Qian W. J., Jacobs J. M., Liu T., Camp D. G., 2nd, Smith R. D. (2006) Advances and challenges in liquid chromatography-mass spectrometry-based proteomics profiling for clinical applications. Mol. Cell. Proteomics 5, 1727–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ludwig J. A., Weinstein J. N. (2005) Biomarkers in cancer staging, prognosis and treatment selection. Nat. Rev. Cancer 5, 845–856 [DOI] [PubMed] [Google Scholar]
- 7. Anderson N. L., Anderson N. G. (2002) The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics 1, 845–867 [DOI] [PubMed] [Google Scholar]
- 8. Polanski M., Anderson N. L. (2007) A list of candidate cancer biomarkers for targeted proteomics. Biomarker Insights 1, 1–48 [PMC free article] [PubMed] [Google Scholar]
- 9. Ye X., Blonder J., Veenstra T. D. (2009) Targeted proteomics for validation of biomarkers in clinical samples. Brief. Funct. Genomic. Proteomic. 8, 126–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carr S. A., Anderson L. (2008) Protein quantitation through targeted mass spectrometry: the way out of biomarker purgatory?. Clin. Chem. 54, 1749–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Anderson L., Hunter C. L. (2006) Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics 5, 573–588 [DOI] [PubMed] [Google Scholar]
- 12. Keshishian H., Addona T., Burgess M., Kuhn E., Carr S. A. (2007) Quantitative, multiplexed assays for low abundance proteins in plasma by targeted mass spectrometry and stable isotope dilution. Mol. Cell. Proteomics 6, 2212–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lange V., Picotti P., Domon B., Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuzyk M. A., Smith D., Yang J., Cross T. J., Jackson A. M., Hardie D. B., Anderson N. L., Borchers C. H. (2009) Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol. Cell. Proteomics 8, 1860–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kuhn E., Addona T., Keshishian H., Burgess M., Mani D. R., Lee R. T., Sabatine M. S., Gerszten R. E., Carr S. A. (2009) Developing multiplexed assays for troponin I and interleukin-33 in plasma by peptide immunoaffinity enrichment and targeted mass spectrometry. Clin. Chem. 55, 1108–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. DeSouza L. V., Romaschin A. D., Colgan T. J., Siu K. W. (2009) Absolute quantification of potential cancer markers in clinical tissue homogenates using multiple reaction monitoring on a hybrid triple quadrupole/linear ion trap tandem mass spectrometer. Anal. Chem. 81, 3462–3470 [DOI] [PubMed] [Google Scholar]
- 17. Addona T. A., Abbatiello S. E., Schilling B., Skates S. J., Mani D. R., Bunk D. M., Spiegelman C. H., Zimmerman L. J., Ham A. J., Keshishian H., Hall S. C., Allen S., Blackman R. K., Borchers C. H., Buck C., Cardasis H. L., Cusack M. P., Dodder N. G., Gibson B. W., Held J. M., Hiltke T., Jackson A., Johansen E. B., Kinsinger C. R., Li J., Mesri M., Neubert T. A., Niles R. K., Pulsipher T. C., Ransohoff D., Rodriguez H., Rudnick P. A., Smith D., Tabb D. L., Tegeler T. J., Variyath A. M., Vega-Montoto L. J., Wahlander A., Waldemarson S., Wang M., Whiteaker J. R., Zhao L., Anderson N. L., Fisher S. J., Liebler D. C., Paulovich A. G., Regnier F. E., Tempst P., Carr S. A. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 27, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu X., Veenstra T. D. (2008) Analysis of biofluids for biomarker research. Proteomics Clin. Appl. 2, 1403–1412 [DOI] [PubMed] [Google Scholar]
- 19. Qian W. J., Kaleta D. T., Petritis B. O., Jiang H., Liu T., Zhang X., Mottaz H. M., Varnum S. M., Camp D. G., 2nd, Huang L., Fang X., Zhang W. W., Smith R. D. (2008) Enhanced detection of low abundance human plasma proteins using a tandem IgY12-SuperMix immunoaffinity separation strategy. Mol. Cell. Proteomics 7, 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Page J. S., Kelly R. T., Tang K., Smith R. D. (2007) Ionization and transmission efficiency in an electrospray ionization-mass spectrometry interface. J. Am. Soc. Mass Spectrom. 18, 1582–1590 [DOI] [PubMed] [Google Scholar]
- 21. Kelly R. T., Tolmachev A. V., Page J. S., Tang K., Smith R. D. (2010) The ion funnel: theory, implementations, and applications. Mass Spectrom. Rev. 29, 294–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shaffer S. A., Tang K., Anderson G. A., Prior D. C., Udseth H. R., Smith R. D. (1997) A novel ion funnel for focusing ions at elevated pressure using electrospray ionization mass spectrometry. Rapid Commun. Mass Spectrom. 11, 1813–1817 [Google Scholar]
- 23. Shaffer S. A., Tolmachev A., Prior D. C., Anderson G. A., Udseth H. R., Smith R. D. (1999) Characterization of an improved electrodynamic ion funnel interface for electrospray ionization mass spectrometry. Anal. Chem. 71, 2957–2964 [DOI] [PubMed] [Google Scholar]
- 24. Kim T., Tolmachev A. V., Harkewicz R., Prior D. C., Anderson G., Udseth H. R., Smith R. D. (2000) Design and implementation of a new electrodynamic ion funnel. Anal. Chem. 72, 2247–2255 [DOI] [PubMed] [Google Scholar]
- 25. Ibrahim Y., Tang K., Tolmachev A. V., Shvartsburg A. A., Smith R. D. (2006) Improving mass spectrometer sensitivity using a high-pressure electrodynamic ion funnel interface. J. Am. Soc. Mass Spectrom. 17, 1299–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Livesay E. A., Tang K., Taylor B. K., Buschbach M. A., Hopkins D. F., LaMarche B. L., Zhao R., Shen Y., Orton D. J., Moore R. J., Kelly R. T., Udseth H. R., Smith R. D. (2008) Fully automated four-column capillary LC-MS system for maximizing throughput in proteomic analyses. Anal. Chem. 80, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kelly R. T., Page J. S., Luo Q., Moore R. J., Orton D. J., Tang K., Smith R. D. (2006) Chemically etched open tubular and monolithic emitters for nanoelectrospray ionization mass spectrometry. Anal. Chem. 78, 7796–7801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim T., Tang K., Udseth H. R., Smith R. D. (2001) A multicapillary inlet jet disruption electrodynamic ion funnel interface for improved sensitivity using atmospheric pressure ion sources. Anal. Chem. 73, 4162–4170 [DOI] [PubMed] [Google Scholar]
- 29. Kelly R. T., Page J. S., Zhao R., Qian W. J., Mottaz H. M., Tang K., Smith R. D. (2008) Capillary-based multi nanoelectrospray emitters: improvements in ion transmission efficiency and implementation with capillary reversed-phase LC-ESI-MS. Anal. Chem. 80, 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pieper R., Su Q., Gatlin C. L., Huang S. T., Anderson N. L., Steiner S. (2003) Multi-component immunoaffinity subtraction chromatography: an innovative step towards a comprehensive survey of the human plasma proteome. Proteomics 3, 422–432 [DOI] [PubMed] [Google Scholar]