Abstract
The molecular basis governing the differentiation of human embryonic stem cells (hESCs) remains largely unknown. Systems-level analysis by proteomics provides a unique approach to tackle this question. However, the requirement of a large number of cells for proteomics analysis (i.e. 106–107 cells) makes this assay challenging, especially for the study of rare events during hESCs lineage specification. Here, a fully integrated proteomics sample processing and analysis platform, termed rare cell proteomic reactor (RCPR), was developed for large scale quantitative proteomics analysis of hESCs with ∼50,000 cells. hESCs were completely extracted by a defined lysis buffer, and all of the proteomics sample processing procedures, including protein preconcentration, reduction, alkylation, and digestion, were integrated into one single capillary column with a strong cation exchange monolith matrix. Furthermore, on-line two-dimensional LC-MS/MS analysis was performed directly using RCPR as the first dimension strong cation exchange column. 2,281 unique proteins were identified on this system using only 50,000 hESCs. For stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative study, a ready-to-use and chemically defined medium and an in situ differentiation procedure were developed for complete SILAC labeling of hESCs with well characterized self-renewal and differentiation properties. Mesoderm-enriched differentiation was studied by RCPR using 50,000 hESCs, and 1,086 proteins were quantified with a minimum of two peptides per protein. Of these, 56 proteins exhibited significant changes during mesoderm-enriched differentiation, and eight proteins were demonstrated for the first time to be overexpressed during early mesoderm development. This work provides a new platform for the study of rare cells and in particular for further elucidating proteins that govern the mesoderm lineage specification of human pluripotent stem cells.
hESCs1 provide a novel means to study early human development and also hold strong therapeutic potentials (1). However, our understanding of the biological mechanisms involved in hESC developmental processes, such as differentiation into mesoderm and its progenies, blood, endothelial cells, bone, heart, and skeletal muscles, is limited. Mass spectrometry-based proteomics has been developed as a systems biology approach to explore gene functions at the post-transcriptional level (2, 3).
There are two main challenges with the stable isotope labeling by amino acids in cell culture (SILAC)-based quantitative proteomics study of hESCs. The first challenge is the number of cells required for proteomics analysis, and the second challenge is the maintenance of hESCs necessitated for SILAC labeling. Recently, two research groups have reported SILAC protocols to study the self-renewal and differentiation of hESCs (4, 5). In these methods, up to 10 million hESCs per sample were cultured in modified mouse embryonic fibroblast-conditioned medium (MEF-CM). The large number of cell required per analysis hampers the study of lineage specification of hESCs. To put it in context, to obtain 10 millions hESCs requires at least 15 days of culture, representing a culture medium cost of ∼$250 per sample, not considering the cost of SILAC reagent. Moreover, MEF-CM used for SILAC labeling of hESCs contains numerous known and unknown factors (6, 7), which raises a serious issue for the elucidation of a given factor affecting hESC differentiation. It would be prohibitively expensive and difficult to simultaneously examine and compare multiple factors that impact lineage specification during hESC differentiation using current methodology. As well, it would be difficult to study a small number of ancestor cells during developmental processes, such as early precursors of blood, bone, or muscles. These ancestor cells can be <5% of the total cell population after fractionation for proteomics analysis. Therefore, new technologies that can handle a limited number of cells and that can provide suitable growth and SILAC labeling conditions are needed.
In recent years, we and others have developed novel analytical techniques that improve the processing of proteomics samples (8–11). We have previously shown that a device, called the proteomic reactor, can improve the processing of proteomics samples over conventional approaches (12–15). We have also shown that a smaller version of this device can be used to handle a limited number of cells (12). Here, we report the development of the rare cell proteomic reactor (RCPR) that integrates 1) a monolith-based proteomic reactor that allows the processing of the proteome of hESCs and 2) the on-line coupling of the proteomic reactor in a two-dimensional HPLC-ESI-MS/MS analysis for the identification and quantitation of proteins. Using this approach, we have shown that we can analyze hESCs down to only 500 cells introduced on the proteomic reactor. We also report that 2,281 unique proteins can be readily identified on this system using only 50,000 hESCs. In addition, based on previous studies (4, 5, 16–18), we developed a new SILAC medium with a chemically defined background using readily available reagents for hESC maintenance and differentiation. Furthermore, we report a new in situ differentiation procedure that generates more differentiated cells and offers a simple and efficient way to study cellular commitment of hESCs. We studied the mesoderm differentiation of hESCs using the RCPR and were able to quantify 1,086 proteins (excluding one-peptide results) with an FPR rate of 0.45%. Altogether, the RCPR provides a new platform to allow simultaneous study of multiple factors and rare ancestor cells during different stages of hESC development. This work paves the way toward further elucidating crucial proteins controlling cellular commitment, lineage restriction, and terminal differentiation of human pluripotent stem cells.
EXPERIMENTAL PROCEDURES
Human Embryonic Stem Cell Culture, SILAC Labeling, BIO Treatment, and Harvest
H1, H9, and CA1 hESC lines were maintained under feeder-free culture conditions as described previously (16, 19, 20) (refer to supplemental experimental methods for details). For SILAC labeling, hESCs were cultured on Matrigel-coated plates in either heavy or light SILAC labeling medium for 8 days to achieve complete SILAC incorporation. Media were changed daily, and cells were passaged every 4–5 days.
A new chemically defined SILAC medium was developed and used in this study. The medium was prepared using commercially available agents devoid of MEF conditioning and dialyzing. It is formulated with the advanced DMEM/F12-Flex medium supplemented with 100 mg/liter l-arginine, 200g/liter glucose, 1 mm nonessential amino acids, 2 mm l-glutamine, 0.1 mm 2-mercaptoethanol, 1× N2 supplements, 1× B27 supplements, and 100 mg/liter l-[13C6]lysine HCl or 100 mg/liter non-labeled l-lysine HCl. All reagents were purchased from Invitrogen. The media were sterile filtered, and basic FGF (120 ng/ml; R&D Systems) was added immediately before use. The high concentration of basic FGF was important for hESC maintenance in the chemically defined SILAC medium.
To differentiate hESCs, 0.8 μm GSK-3 inhibitor 6-bromoindirubin-3′-oxime (BIO; Calbiochem) or vehicle control was added into the heavy or light SILAC labeling medium for 3–4 days. After 8 days, hESCs were subjected to immunocytochemistry, alkaline phosphatase staining, Q-PCR, and MS analyses.
To harvest hESCs, hESC colonies were dissociated into single cells with collagenase IV and cell dissociation buffer (Invitrogen), and the cell number was counted using a hemocytometer. hESCs were then aliquoted at a defined cell number into different tubes. The cells were lysed immediately with RCPR lysis buffer (10 mm HEPES, pH 7.4, 150 mm NaCl, 2 mm CaCl2, 2 mm MgCl2, 0.6 m guanidine HCl, 1% (v/v) Triton X-100, protease inhibitor mixture) before proteomics analysis.
RT-PCR, Q-PCR, Alkaline Phosphatase Staining, Immunofluorescence Microscopy, and Quantitative Image Analysis
The assays were conducted as described previously (16, 19–21) and detailed in the supplemental experimental methods and supplemental Table 1.
Rare Cell Proteomic Reactor
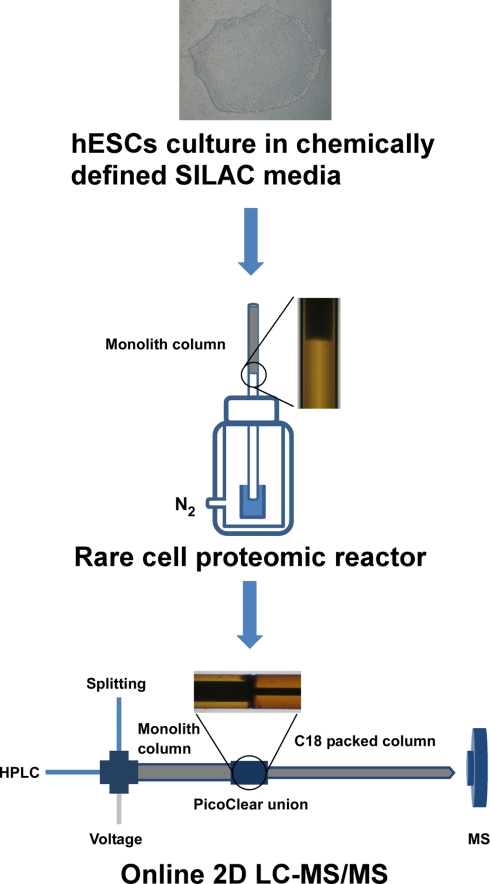
As shown in Fig. 1, the RCPR consists of a capillary with integrated strong cation exchange (SCX) monolith matrix and a custom-made vessel pressurized with nitrogen for parallel sample loading. The SCX monolith column was prepared by modifying a method reported previously (22). Briefly, a fused silica capillary (Polymicro Technologies, Phoenix, AZ) was pretreated with 0.1 m NaOH for 1 h, washed with water and methanol, and then dried by nitrogen. 3-(Trimethoxysilyl)propyl methacrylate mixed with an equal amount of methanol was introduced into the pretreated capillary and incubated at 60 °C overnight. After the reaction, the capillary was washed with methanol and dried by nitrogen. The polymerization mixture containing 100 mg 2-acrylamido-2-methyl-1-propanesulfonic acid, 60 mg of bisacrylamide, 270 μl of DMSO, 200 μl of dodecanol, 50 μl of N,N-dimethylformamide, and 2 mg of 1,1′-azobis(cyclohexanecarbonitrile) was sonicated on ice for 5 min. The resulting transparent solution was infused into the pretreated capillary (200-μm inner diameter × 15 cm) to fill 5 cm of the capillary. Then, the capillary was sealed at both ends and incubated at 60 °C for 12 h. The prepared monolith column was extensively washed with methanol prior to use.
Fig. 1.
Schematic overview of typical procedure for SILAC-based quantitative proteomics analysis of hESCs using RCPR platform. hESCs were cultured in chemically defined SILAC media first, limited amounts of cells were collected for RCPR operation on a nitrogen-pressurized vessel, and then the RCPR was connected directly with a C18 analytical column for on-line two-dimensional LC-MS/MS analysis.
For RCPR operation, the monolith column was preconditioned with 10 mm potassium phosphate buffer, pH 3 before sample loading, and the flow rate was measured on the pressurized vessel. Cell lysate was loaded directly onto the preconditioned SCX monolith column at 62 p.s.i. The SCX monolith column was then washed with wash buffer (8 mm potassium phosphate buffer, 20% (v/v) ACN). For protein reduction, 100 mm DTT dissolved in 10 mm ammonium bicarbonate (ABC) was loaded onto the column and incubated for 30 min at room temperature. After that, the monolith column was washed with 10 mm ABC briefly. Finally, 10 mm iodoacetamide and 2 μg/μl trypsin (Promega) dissolved in 20 mm ABC, pH 8 were loaded for simultaneously alkylation and digestion. After 2 h of incubation at room temperature, the monolith column was disconnected from the pressurized vessel, sealed at both ends, and stored at 4 °C for on-line two-dimensional LC-MS/MS analysis.
On-line Two-dimensional LC-MS/MS Analysis
The on-line two-dimensional LC-MS/MS analysis was performed on a standard LC-MS system consisting of an Agilent 1200 capillary pump, Agilent microautosampler (Agilent Technologies, Waldbronn, Germany), and LTQ (for method optimization) or LTQ-Orbitrap XL (for SILAC) mass spectrometers equipped with a nanospray source (Thermo, San Jose, CA).
For on-line two-dimensional LC separation (see Fig. 1), the RCPR was cut right after the monolith matrix to remove the blank capillary part using a diamond scribe and connected in tandem to a C18 tip column by a PicoClear union (New Objective, Woburn, MA). When used with the LTQ mass spectrometer, the RCPR was connected to a 120-mm × 75-μm-inner diameter C18 tip column packed with Magic C18 AQ resins (5 μm, 200 Å; Michrom Bioresources, Auburn, CA), and the system flow rate was restricted to 200 nl/min. When used with the LTQ-Orbitrap XL mass spectrometer, the RCPR was connected to a 200-mm × 50-μm-inner diameter C18 tip column packed with ReproSil-Pur C18 resins (3 μm, 200 Å; Dr. Maisch GmbH, Ammerbuch, Germany), and the system flow rate was restricted to 100 nl/min. The trapped peptides on RCPR were eluted onto C18 analytical column by 13 stepwise elutions with different concentrations of NH4OAc, pH 2.7. After each elution, the tandem column system was washed with 0.1% acetic acid in water, and a 60- or 90-min ACN gradient elution from 5 to 35% was performed using 0.1% acetic acid in ACN for the reversed phase LC-MS analysis.
The LTQ and LTQ-Orbitrap XL mass spectrometers were operated in a positive ionization mode. A voltage of 1.8 kV was applied to generate the electrospray ionization. All MS and MS/MS spectra were acquired in a data-dependent mode. The instrument was set so that one full MS scan was followed by 10 MS/MS scans. For the LTQ-Orbitrap XL, the full-scan MS spectra (from m/z 400 to 2,000) were acquired with a resolution of 60,000 at m/z 400 after accumulation to a target value of 500,000. The 10 most intense ions at a threshold above 500 counts were selected for fragmentation by CID at a normalized collision energy of 35%.
MS Data Analysis
Peak lists were generated from the raw file using Mascot Distiller (version 2.0.0.0, Matrix Science, London, UK) for LTQ data and DTASupercharge (version 2.0a7, SourceForge) for LTQ-Orbitrap XL data. The acquired MS/MS spectra were searched against the human International Protein Index (IPI) protein sequence database (version 3.66, 86,845 protein entries; European Bioinformatics Institute). A decoy database with the reversed sequence of each entry in the forward database was also searched for evaluation of the false positive rate. The database searching was performed using Mascot (version 2.2.02, Matrix Science) with the following parameters: trypsin as digestion enzyme, carbamidomethyl (Cys) as a fixed modification, and oxidation (Met) as a variable modification. The SILAC label ([13C6]lysine) was set as an extra variable modification for Orbitrap XL data. The number of allowed missed cleavages was set to 2. The precursor and fragment mass tolerances were set at 2.0 and 0.8 Da for the LTQ data and 7 ppm and 0.5 Da for the Orbitrap XL data, respectively. The significance threshold was set to 0.05. A protein hit required at least one “bold red peptide,” i.e. the most logical assignment of the peptide in the database selected, to be reported. Mascot cutoff scores were set to 30. All of the raw files were searched by Mascot separately. The false positive rate was controlled to be less than 1% using the equation FPR = Number of false peptides/(Number of true peptides + Number of false peptides) × 100 (23). MSQuant (version 2.0a81, SourceForge) was used for calculating the relative peptide abundance with a default setting. The exported files from MSQuant were further integrated, and results were normalized by StatQuant (version 1.2.2, GForge). Manual validation was performed to assign shared peptides belonging to multiple protein groups to the protein group with the highest number of identified peptides (24). Molecular functions and protein networks were analyzed using the Ingenuity pathways analysis (IPA) software (version 8.5, Ingenuity® Systems).
RESULTS AND DISCUSSION
We are interested in the study of the proteome of rare cells. In these instances, the amount of starting material is limited, and the analytical techniques need to be adapted accordingly. The recovery from every step in the process needs to be maximized with a particular attention to cell lysis, the recovery and digestion of proteins, and the recovery and separation of peptides for identification by mass spectrometry. In this study, we report a fully integrated proteomics platform, termed RCPR, that minimizes sample loss and maximizes the mass spectrometry analysis efficiency. The RCPR allowed efficient capture and processing of proteins on a capillary proteomic reactor composed of an SCX monolith matrix. As well, the direct coupling of the capillary proteomic reactor with a nanoflow reversed phase column on line with an ESI mass spectrometer allowed the efficient transfer of the analytes from the proteomic reactor to the nanoflow reversed phase column and the on-line analysis of the peptides by mass spectrometry. Here, we also describe the application of the RCPR to the analysis of hESCs and their differentiation.
Optimization of Lysis Buffer
First, we developed a well defined cell lysis buffer compatible with the RCPR to completely solubilize proteins from the rare cell sample in as little volume as possible. The typical procedure in proteomics analysis from cell culture is to add cell lysis buffer into a cell pellet and remove the unsolubilized fraction by centrifugation prior to protein digestion. Unfortunately, this causes protein loss, which is an issue when dealing with a limited amount of samples. Instead, we optimized the cell lysis buffer composition and volume according to the number of cells to maintain a cell-to-buffer volume ratio lower than 2,500 cells/μl. After adding the RCPR lysis buffer and pipetting several times, the cell pellet disappears completely, and the solution becomes transparent. Because the high concentration of guanidine HCl will possibly influence the binding of proteins onto the SCX monolith column, we next optimized the proper concentration of guanidine HCl in RCPR lysis buffer. As shown in supplemental Fig. 1, a concentration of guanidine HCl lower than 1 m does not cause protein loss. We selected to set the composition of the RCPR lysis buffer to 0.6 m guanidine HCl, which allows extracting 50% more proteins in comparison with RCPR lysis buffer without guanidine HCl (data not shown).
Monolithic Proteomic Reactor Design
Second, we optimized the protein recovery and processing techniques to maximize recovery when dealing with a limited number of cells. We previously demonstrated that the proteomic reactor is an efficient approach to process proteomics samples (12). This technology greatly simplifies the handling of proteins, reduces the volumes required for analysis, and shortens the processing times. Here, we introduce an SCX monolith column for proteomic reactor operation to further reduce sample loss and dead volume. The SCX monolith column has several advantages over our previously reported proteomic reactor based on SCX beads. First, the monolith matrix is polymerized directly inside the capillary (see Fig. 1, inset photo), and the whole proteomic reactor operation is performed in one single capillary column. A frit, SCX beads, and a union are not needed to prepare the reactor column, and surface loss is reduced. Second, the binding capacity of the monolith column is ∼4 μg of protein/cm for a 200-μm-inner diameter column, which doubles the capacity of the previous proteomic reactor. This allows more proteins to be captured on a smaller volume of the column. Our comparison based on the previous off-line protocol indicates that 50% more proteins can be identified by the monolith proteomic reactor than by the bead proteomic reactor when as few as 220 and 4,200 cells are used (supplemental Table 2).
The separation ability of the analytical technique is also a critical factor for large scale proteomics. Here, to enhance the separation ability of the RCPR platform we coupled the proteomic reactor with an on-line two-dimensional LC-MS/MS analysis system. As shown in Fig. 1, after trypsin digestion, the SCX monolith proteomic reactor was connected directly with the LC-MS/MS system for on-line two-dimensional separation. To reduce the possible dead volume between the SCX monolith column and C18 analytical column, the capillary columns were cleaved and carefully connected using a PicoClear union. As shown in Fig. 1, inset, the two columns are perfectly aligned. SCX-based peptide separation has been shown previously to be well suited as a first dimensional separation in on-line two-dimensional LC-MS/MS analysis because of good orthogonality in the separation and solvents utilized in SCX- and reversed phase-based separation (25). Here, the monolithic proteomic reactor plays a dual role as a bed for extraction and chemical/biochemical reactions and as the first dimension in two-dimensional LC separation. This key design makes the whole RCPR platform a fully integrated proteomics sample processing and mass spectrometry analysis system.
Minimum Number of hESCs That Can Be Processed by RCPR
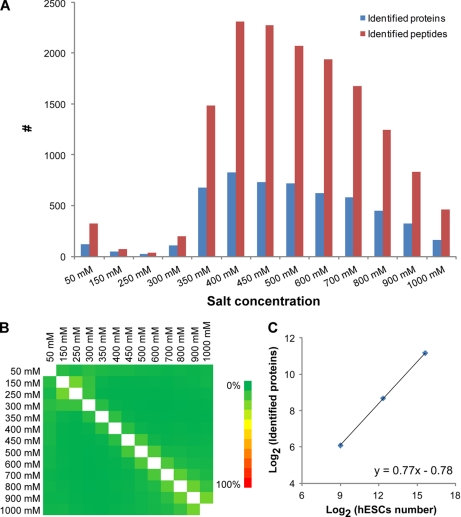
To test the applicability of RCPR for large scale proteomics study with limited starting material, the default number of hESCs was loaded, processed, and analyzed by RCPR. As an example, Fig. 2, A and B, shows the results for processing and analyzing 50,000 hESCs. To fully explore the complexity of the digested peptides, the on-line two-dimensional LC-MS/MS in the RCPR was performed with 13 subsequent salt step elutions from 50 mm to 1 m salt. After each salt step elution, peptides were eluted from the SCX proteomic reactor to the reversed phase column, separated by reversed phase chromatography, and analyzed by ESI-MS/MS. Fig. 2A illustrates the number of peptides and proteins identified from each salt step, and Fig. 2B illustrates the overlaps for peptides identified between the different fractions. The majority of the peptides were eluted at a salt concentration above 350 mm, indicating a strong retention of the peptides on the proteomic reactor. Moreover, the carryover between neighboring fractions and distal fractions is minimal. We analyzed 500, 5,000, and 50,000 hESCs on the RCPR, and 68, 409, and 2,281 unique proteins were identified, respectively. An excellent linearity between the number of unique proteins identified and the number of cells processed was obtained (Fig. 2C). This clearly demonstrates that the RCPR is compatible with the analysis of a limited number of cells, such as fractionated precursors from differentiated hESCs, and the distribution of the identified proteins is generally consistent with a previous report (26).
Fig. 2.
Performance of RCPR applied to large scale proteome analysis of hESCs. A, the distribution of identified proteins and peptides by on-line two-dimensional LC-MS/MS analysis with 13 salt fractions. B, heat map of unique peptide overlap between two salt fractions. C, the distribution of proteins identified by RCPR using different amounts of hESCs (n = 2).
Quantitative Proteomics Study of hESCs Using Novel Chemically Defined SILAC Media
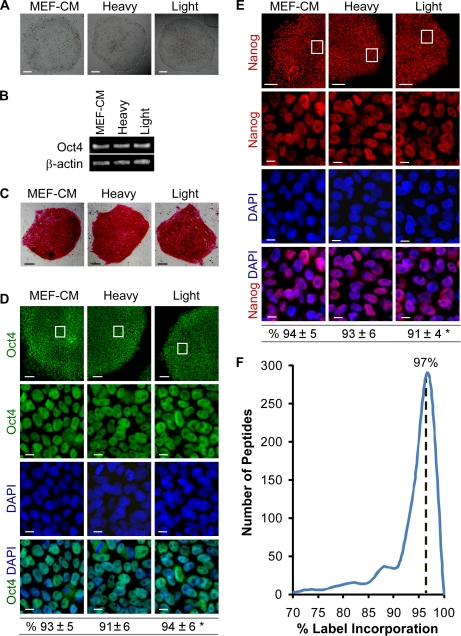
We are interested in studying the quantitative changes that occur in the proteome of hESCs using a limited number of cells. It has been reported that SILAC labeling of hESCs is not a trivial task because of the specific conditions required for hESC maintenance (4, 5, 27). Recently, Blagoev and co-workers (4) developed a SILAC labeling protocol by cultivating and SILAC labeling hESCs in a dialyzed MEF-CM. Although they reported excellent results, the generation of the medium is not straightforward. Another recipe reported by Bendall et al. (5) is to prepare the SILAC medium first followed by MEF conditioning. Notably, MEF-conditioned medium contains numerous anonymous xenogenic factors from MEF cells that may complicate the data interpretation when studying a given factor that impacts the self-renewal and differentiation of hESCs. Instead, we report a new SILAC medium with a chemically defined background for the SILAC-based quantitative proteomics study of hESCs. It can be simply made using commercially available reagents directly without MEF conditioning (for details see “Experimental Procedures”). Importantly, this new chemically defined SILAC medium is capable of maintaining hESCs in an undifferentiated state for over five cell doublings (an average doubling time for hESCs is ∼36 h (28–30)), fulfilling the requirement for complete SILAC incorporation. As shown in Fig. 3, hESCs cultured over 8 days in both heavy and light SILAC labeling media maintained their undifferentiated characteristics and were indistinguishable from hESCs maintained in MEF-CM. Moreover, the hESCs cultured in both heavy and light SILAC labeling media exhibited typical colony morphology (Fig. 3A), expressed OCT4 pluripotency gene (Fig. 3B), and were stained positively for pluripotency markers alkaline phosphatase, OCT4, and NANOG (Fig. 3, C–E). These results demonstrate that the simplified SILAC medium is capable of maintaining hESCs in an undifferentiated state. Similar results were obtained from three different hESC lines (H1, H9, and CA1), suggesting that the chemically defined SILAC media used in this study are not cell line-specific.
Fig. 3.
Chemically defined SILAC media are capable of maintaining hESCs in undifferentiated state. A, hESCs (CA1 line) cultured in the chemically defined SILAC medium (heavy or light) for 12 days exhibit typical hESC colony morphology indistinguishable from those maintained in MEF-CM. Bright field images are shown. Scale bars, 200 μm. B, hESCs cultured in the three different media express similar levels of OCT4 gene as determined by RT-PCR at day 12. C–E, similar to the cells maintained in MEF-CM, CA1 hESCs cultured in the chemically defined SILAC media for 12 days express pluripotency markers alkaline phosphatase (C), OCT4 (D), and NANOG (E) as determined by immunofluorescence microscopy. Boxed areas in the top panels are enlarged in the bottom panels (D and E). Scale bars, 100 μm (C and D and E, top panels) and 10 μm (the enlarged bottom panels of D and E). The nuclei counterstained with the DNA-specific fluorophore DAPI (blue) show the total number of cells. The percentages of OCT4- and NANOG-positive cells (CA1 and H1 lines) were quantitatively assessed and are shown at the bottom of D and E. Data are mean ± S.D. from three independent experiments counting the number of positive cells within the total population of eight fields of view per experiment (>1,500 cells/group). Eight fields of view were randomly selected. *, p > 0.05 among groups. F, the incorporation efficiency of undifferentiated hESCs after 8 days of SILAC labeling. The median ratio is indicated.
We next examined the incorporation efficiency of isotope-labeled lysine using the SILAC labeling medium containing 100 mg/liter l-[13C6]lysine HCl. The analysis was based on 578 peptides that were identified repeatedly (supplemental Table 3). The efficiency of labeling on lysine residue was estimated to be 97% after culture of hESCs for 8 days in the SILAC labeling medium (Fig. 3F). Based on the atom purity of the stable isotope-labeled lysine, this incorporation rate almost reaches the maximal achievable labeling efficiency.
Application of RCPR in Study of hESC Differentiation
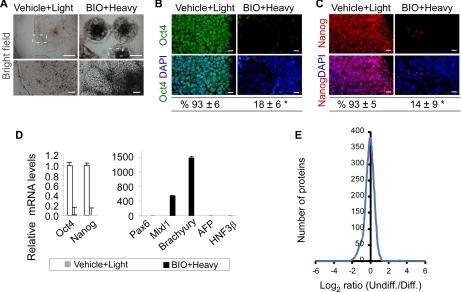
The conventional approach of cellular aggregates (formation of embryoid body) in hESC differentiation sometimes produces a limited number of differentiated cells (31). Instead, we developed an in situ differentiation protocol to improve cell yields after hESC differentiation. Briefly, hESCs were cultured in the chemically defined SILAC media with the addition of a GSK-3 kinase inhibitor, BIO (0.8 μm), from days 4 to 8. GSK-3 kinase is one of the key components in the canonical Wnt signaling pathways (32). Inhibition of GSK-3 kinase leads to Wnt activation (32), which facilitates the mesoderm differentiation of hESCs (33, 34). Our results showed that the in situ differentiation procedure produced up to 5-fold greater numbers of live cells than the formation of embryoid body and drove hESCs to differentiate efficiently. In the absence of BIO, hESCs were maintained in an undifferentiated state as characterized by colony morphology and positive staining for pluripotency markers alkaline phosphatase (data not shown), OCT4, and NANOG (Fig. 4, B and C). In contrast, exposing hESCs to BIO for 4 days resulted in a marked change in colony morphology (Fig. 4A) and a significant decrease in OCT4 and NANOG gene expression (Fig. 4D). Consistently, the majority of cells lost OCT4 and NANOG protein expression as determined by fluorescence immunocytochemistry and quantitative image analysis (Fig. 4, B and C). Furthermore, as opposed to the undifferentiated hESCs, the differentiated hESCs expressed a higher level of genes known to be expressed during mesoderm differentiation, such as BRACHYURY (1,600-fold increase) and MIXL1 (600-fold increase), whereas three other genes representing ectoderm and endoderm differentiation were not significantly up-regulated (Fig. 4D). These results demonstrate that undifferentiated and differentiated hESCs (mesoderm-enriched specification) can be obtained in the chemically defined SILAC media using vehicle or BIO treatment.
Fig. 4.
Application of RCPR in study of hESCs differentiation. A–C, in contrast to the vehicle control, exposing hESCs to 0.8 μm GSK-3 inhibitor BIO for 4 days results in cell differentiation, which is characterized by a marked change in colony morphology (A, bright field; H1 line; boxed areas are enlarged in the bottom panels) and a negative staining for OCT4 (B, green; CA1 line) and NANOG (C, red; H1 line). The nuclei were counterstained with the DNA-specific fluorophore DAPI (blue). Similar results were obtained when hESCs were cultured in light SILAC medium and treated with BIO in comparison with those cultured in heavy SILAC medium and treated with vehicle. Scale bars, 500 μm (A, top panels); 50 μm (A, enlarged bottom panels); 20 μm (B and C, immunofluorescence images). The percentages of OCT4- and NANOG-positive hESCs were quantified and are shown at the bottom of B and C. S.D. is displayed (>1,500 cells/group per experiment). *, p < 0.01. D, exposing hESCs to BIO (0.8 μm) for 3–4 days also results in a marked down-regulation of pluripotency genes OCT4 and NANOG and an up-regulation of BRACHYURY and MIXL1 (mesoderm genes) as determined by Q-PCR. PAX6 is an ectoderm gene; AFP and HNF3β are endoderm genes. Q-PCR data are expressed relative to the vehicle control after being normalized to β-ACTIN. Error bars represent S.D. (n = 4). E, histogram of the SILAC ratio of undifferentiated (Undiff.) hESCs and differentiated (Diff.) hESCs treated with BIO (n = 1,086, S.D. = 0.55).
We next mixed the cell lysates of 25,000 cells from both undifferentiated hESCs and BIO-differentiated hESCs and applied them onto the RCPR for sample processing and on-line two-dimensional LC-MS/MS analysis with optimized salt fractions. Three replicate experiments with the same amount of starting material were performed. 2,347 proteins were identified, and 1,086 proteins with a minimum of two peptides per protein were kept for further quantitative analysis (FPR of 0.45% and SILAC ratio S.D. of less than 2; supplemental Table 4). Quantified peptide information and MS/MS spectra for one peptide identifications were included in supplemental information. 95% of the proteins could be quantified in at least two experiments, and only 57 proteins were quantified in one experiment. The relative quantitation of the 57 proteins was manually validated by inspecting the raw files, and the S.D. of the quantitation was less than 0.76. The quantified proteins are involved in a variety of cellular and molecular functions (supplemental Fig. 2), suggesting that the RCPR can be used for a global quantitative study of the hESC proteome at great depth. Fig. 4E shows that the SILAC ratio (a log-transformed distribution) for the most quantified proteins is nearly centered on 0 with overall S.D. of 0.55, which demonstrates the high accuracy of the SILAC-based quantitation. Of these quantified proteins, 1,030 proteins fell within the 95% confidence interval, whereas 56 proteins exhibited a significant SILAC ratio change (supplemental Table 5). The cutoff ratios for proteins with significant changes are 2.62 and 0.58 for up-regulation and down-regulation, respectively. Of the 56 proteins, 45 proteins were up-regulated, whereas only 11 proteins were down-regulated after differentiation of hESCs into an early mesoderm-enriched lineage (Fig. 4), representing the possible contribution of mesoderm differentiation to significant protein up-regulation. Interestingly, 84% of these proteins are either known to directly interact or linked by one other protein in the Ingenuity Knowledge Base (supplemental Fig. 3). For example, the up-regulated proteins APR2/3, ARPC3, ARPC1B, and ARPC2 are part of the ARP2/3 complex, whereas CORO1B is known to interact with the ARP2/3 complex. The ARP2/3 complex and CORO1B are involved in the regulation of the actin cytoskeleton (35). Furthermore, comparison with recent transcriptome analyses of hESC self-renewal and differentiation indicated that 52% of the matching entries (protein-RNA entries) have the same expression trend at the RNA and protein levels (supplemental Table 6) (36–40).
To further explore potential proteins associated with self-renewal and early mesoderm differentiation of hESCs, identified proteins showing significant change after treatment with BIO (a mesoderm inducer) were grouped and extracted for validation analysis. As shown in Table I, there were three down-regulated proteins; SOX2 and isoform 1 of DNMT3B are well known self-renewal markers (41, 42), and L1TD1 has been demonstrated to be enriched in undifferentiated hESCs in a recent transcriptome study (43). Our recent experiments suggest that L1TD1 is one of the most important factors during hESC self-renewal,2 consistent with our proteomics results. Notably, of the eight up-regulated proteins, two proteins are associated with hematopoiesis and angiogenesis (MEST and PTK7) (44–46), two are associated with chondrogenesis (PLOD2 and P4HA2) (47, 48), two are associated with myogenesis (ERO1L and CAMK2D) (49, 50), and one is associated with mesoderm commitment (PRTG) (51). SARS, which is associated with angiogenesis (52), was up-regulated after mesoderm-enriched differentiation and further validated by Q-PCR. Consistently, three down-regulated proteins also exhibited a significant decrease in mRNA expression, whereas eight up-regulated proteins showed an increase in mRNA expression as determined subsequently by Q-PCR assays (Table I).
Table I. Proteins identified with significant change that are associated with self-renewal and mesoderm differentiation after 4 days of BIO treatment.
| Accession no. (IPI) | Protein description | Gene symbol | No. of peptides (unique/quantified) | BIO/control ratio |
Ref. | |
|---|---|---|---|---|---|---|
| SILACa | Q-PCRb | |||||
| IPI00298947 | Isoform 1 of mesoderm-specific transcript homolog protein | MEST | 2/2 | 5.27 ± 0.09 | 11.88 ± 0.14 | 44 |
| IPI00168813 | Isoform 3 of tyrosine-protein kinase-like 7 | PTK7 | 2/2 | 2.66 ± 0.04 | 1.29 ± 0.03 | 45, 46 |
| IPI00003128 | Isoform IIb of prolyl 4-hydroxylase subunit α-2 | P4HA2 | 2/3 | 3.33 ± 0.21 | 2.35 ± 0.04 | 47 |
| IPI00337495 | Isoform 2 of procollagen-lysine,2-oxoglutarate 5-dioxygenase 2 | PLOD2 | 2/2 | 3.73 ± 0.23 | 2.12 ± 0.03 | 48 |
| IPI00386755 | ERO1-like protein α | ERO1L | 1/2 | 3.04 ± 0.16 | 1.39 ± 0.06 | 49 |
| IPI00172636 | Isoform δ6 of calcium/calmodulin-dependent protein kinase type II δ chain | CAMK2D | 4/4 | 4.35 ± 0.83 | >4c | 50 |
| IPI00658004 | Protogenin | PRTG | 1/2 | 7.69 ± 0.9 | 14.67 ± 0.04 | 51 |
| IPI00220637 | Seryl-tRNA synthetase, cytoplasmic | SARS | 7/2 | 2.15 ± 0.23 | 1.59 ± 0.08 | 52 |
| IPI00009703 | Transcription factor SOX-2 | SOX2 | 4/3 | 0.11 ± 0.05 | 0.09 ± 0.08 | 41 |
| IPI00012593 | Isoform 1 of DNA (cytosine-5)-methyltransferase 3B | DNMT3B | 17/39 | 0.07 ± 0.04 | 0.08 ± 0.00 | 42 |
| IPI00253050 | LINE-1 type transposase domain-containing protein 1 | L1TD1 | 12/25 | 0.12 ± 0.16 | 0.05 ± 0.07 | 43 |
a SILAC ratio of BIO differentiation/self-renewal with S.D. The SILAC ratios were validated manually at a mass tolerance of 7 ppm.
b Q-PCR validation with S.D. (n = 2).
c Control sample is undetectable.
This work demonstrates that the RCPR is suitable for the quantitative analysis of the proteome changes that occur in as little as 50,000 hESCs after SILAC labeling. It also demonstrates that during hESC differentiation into a mesoderm-enriched lineage 56 proteins are significantly changed and that eight proteins previously associated with late mesoderm progenies are first shown to be expressed during early mesoderm commitment. The RCPR shown to be provides a new platform to allow us to further study these molecular details during different stages of hESC differentiation using rare ancestor cells.
Acknowledgments
We are grateful to Drs. A. Nagy, J. Rossant, M. Gertsenstein, K. Vinterstein, M. Mileikovsky, and J. Draper for providing the CA1 hESC line and to Dr. P. W. Zandstra's group for distributing the CA1 cell line.
Footnotes
* This work was supported by funds from Genome Canada through the Ontario Genomics Institute (Grant 2008-OGI-TD-01), funds from the Province of Ontario, and an Early Research Award from the Ontario Government (to D. F. and L. W.); by Operating Grant MOP-158235 from the Canadian Institutes of Health Research (CIHR) and CIHR New Investigator Award MSH-166732 (to L. W.); and by a Natural Sciences and Engineering Research Council (Canada) discovery grant (to D. F.).
 This article contains supplemental experimental methods, Figs. 1–3, Tables 1–6, quantified peptide information, and sequence information and MS/MS spectra for one-peptide identifications.
This article contains supplemental experimental methods, Figs. 1–3, Tables 1–6, quantified peptide information, and sequence information and MS/MS spectra for one-peptide identifications.
2 R. Tian, S. Wang, L. Wang, and D. Figeys, unpublished observations.
1 The abbreviations used are:
- hESCs
- human embryonic stem cells
- RCPR
- rare cell proteomic reactor
- SCX
- strong cation exchange
- SILAC
- stable isotope labeling by amino acids in cell culture
- MEF
- mouse embryonic fibroblast
- MEF-CM
- mouse embryonic fibroblast-conditioned medium
- FPR
- false positive rate
- BIO
- 6-bromoindirubin-3′-oxime
- Q-PCR
- quantitative PCR
- ABC
- ammonium bicarbonate
- IPI
- International Protein Index.
REFERENCES
- 1. Yu J., Thomson J. A. (2008) Pluripotent stem cell lines. Genes Dev. 22, 1987–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 3. Cravatt B. F., Simon G. M., Yates J. R., 3rd (2007) The biological impact of mass-spectrometry-based proteomics. Nature 450, 991–1000 [DOI] [PubMed] [Google Scholar]
- 4. Prokhorova T. A., Rigbolt K. T., Johansen P. T., Henningsen J., Kratchmarova I., Kassem M., Blagoev B. (2009) Stable isotope labeling by amino acids in cell culture (SILAC) and quantitative comparison of the membrane proteomes of self-renewing and differentiating human embryonic stem cells. Mol. Cell. Proteomics 8, 959–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bendall S. C., Hughes C., Stewart M. H., Doble B., Bhatia M., Lajoie G. A. (2008) Prevention of amino acid conversion in SILAC experiments with embryonic stem cells. Mol. Cell. Proteomics 7, 1587–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prowse A. B., McQuade L. R., Bryant K. J., Marcal H., Gray P. P. (2007) Identification of potential pluripotency determinants for human embryonic stem cells following proteomic analysis of human and mouse fibroblast conditioned media. J. Proteome Res. 6, 3796–3807 [DOI] [PubMed] [Google Scholar]
- 7. Lim J. W., Bodnar A. (2002) Proteome analysis of conditioned medium from mouse embryonic fibroblast feeder layers which support the growth of human embryonic stem cells. Proteomics 2, 1187–1203 [DOI] [PubMed] [Google Scholar]
- 8. Waanders L. F., Chwalek K., Monetti M., Kumar C., Lammert E., Mann M. (2009) Quantitative proteomic analysis of single pancreatic islets. Proc. Natl. Acad. Sci. U.S.A. 106, 18902–18907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. López-Ferrer D., Petritis K., Lourette N. M., Clowers B., Hixson K. K., Heibeck T., Prior D. C., Pasa-Toliæ L., Camp D. G., 2nd, Belov M. E., Smith R. D. (2008) On-line digestion system for protein characterization and proteome analysis. Anal. Chem. 80, 8930–8936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krenkova J., Lacher N. A., Svec F. (2009) Highly efficient enzyme reactors containing trypsin and endoproteinase LysC immobilized on porous polymer monolith coupled to MS suitable for analysis of antibodies. Anal. Chem. 81, 2004–2012 [DOI] [PubMed] [Google Scholar]
- 11. Wiœniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 12. Ethier M., Hou W., Duewel H. S., Figeys D. (2006) The proteomic reactor: a microfluidic device for processing minute amounts of protein prior to mass spectrometry analysis. J. Proteome Res. 5, 2754–2759 [DOI] [PubMed] [Google Scholar]
- 13. Vasilescu J., Zweitzig D. R., Denis N. J., Smith J. C., Ethier M., Haines D. S., Figeys D. (2007) The proteomic reactor facilitates the analysis of affinity-purified proteins by mass spectrometry: application for identifying ubiquitinated proteins in human cells. J. Proteome Res. 6, 298–305 [DOI] [PubMed] [Google Scholar]
- 14. Hou W., Ethier M., Smith J. C., Sheng Y., Figeys D. (2007) Multiplexed proteomic reactor for the processing of proteomic samples. Anal. Chem. 79, 39–44 [DOI] [PubMed] [Google Scholar]
- 15. Zhou H., Hou W., Denis N. J., Zhou H., Vasilescu J., Zou H., Figeys D. (2009) Glycoproteomic reactor for human plasma. J. Proteome Res. 8, 556–566 [DOI] [PubMed] [Google Scholar]
- 16. Wang L., Li L., Menendez P., Cerdan C., Bhatia M. (2005) Human embryonic stem cells maintained in the absence of mouse embryonic fibroblasts or conditioned media are capable of hematopoietic development. Blood 105, 4598–4603 [DOI] [PubMed] [Google Scholar]
- 17. Yao S., Chen S., Clark J., Hao E., Beattie G. M., Hayek A., Ding S. (2006) Long-term self-renewal and directed differentiation of human embryonic stem cells in chemically defined conditions. Proc. Natl. Acad. Sci. U.S.A. 103, 6907–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 19. Wang L., Li L., Shojaei F., Levac K., Cerdan C., Menendez P., Martin T., Rouleau A., Bhatia M. (2004) Endothelial and hematopoietic cell fate of human embryonic stem cells originates from primitive endothelium with hemangioblastic properties. Immunity 21, 31–41 [DOI] [PubMed] [Google Scholar]
- 20. Li L., Wang S., Jezierski A., Moalim-Nour L., Mohib K., Parks R. J., Retta S. F., Wang L. (2010) A unique interplay between Rap1 and E-cadherin in the endocytic pathway regulates self-renewal of human embryonic stem cells. Stem Cells 28, 247–257 [DOI] [PubMed] [Google Scholar]
- 21. Chadwick K., Wang L., Li L., Menendez P., Murdoch B., Rouleau A., Bhatia M. (2003) Cytokines and BMP-4 promote hematopoietic differentiation of human embryonic stem cells. Blood 102, 906–915 [DOI] [PubMed] [Google Scholar]
- 22. Wang F., Dong J., Jiang X., Ye M., Zou H. (2007) Capillary trap column with strong cation-exchange monolith for automated shotgun proteome analysis. Anal. Chem. 79, 6599–6606 [DOI] [PubMed] [Google Scholar]
- 23. Elias J. E., Gygi S. P. (2007) Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 [DOI] [PubMed] [Google Scholar]
- 24. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 25. Fournier M. L., Gilmore J. M., Martin-Brown S. A., Washburn M. P. (2007) Multidimensional separations-based shotgun proteomics. Chem. Rev. 107, 3654–3686 [DOI] [PubMed] [Google Scholar]
- 26. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 27. Van Hoof D., Muñoz J., Braam S. R., Pinkse M. W., Linding R., Heck A. J., Mummery C. L., Krijgsveld J. (2009) Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5, 214–226 [DOI] [PubMed] [Google Scholar]
- 28. Hoffman L. M., Carpenter M. K. (2005) Characterization and culture of human embryonic stem cells. Nat. Biotechnol. 23, 699–708 [DOI] [PubMed] [Google Scholar]
- 29. Richards M., Fong C. Y., Chan W. K., Wong P. C., Bongso A. (2002) Human feeders support prolonged undifferentiated growth of human inner cell masses and embryonic stem cells. Nat. Biotechnol. 20, 933–936 [DOI] [PubMed] [Google Scholar]
- 30. Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998) Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- 31. Wang L., Schulz T. C., Sherrer E. S., Dauphin D. S., Shin S., Nelson A. M., Ware C. B., Zhan M., Song C. Z., Chen X., Brimble S. N., McLean A., Galeano M. J., Uhl E. W., D'Amour K. A., Chesnut J. D., Rao M. S., Blau C. A., Robins A. J. (2007) Self-renewal of human embryonic stem cells requires insulin-like growth factor-1 receptor and ERBB2 receptor signaling. Blood 110, 4111–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nelson W. J., Nusse R. (2004) Convergence of Wnt, beta-catenin, and cadherin pathways. Science 303, 1483–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dravid G., Ye Z., Hammond H., Chen G., Pyle A., Donovan P., Yu X., Cheng L. (2005) Defining the role of Wnt/beta-catenin signaling in the survival, proliferation, and self-renewal of human embryonic stem cells. Stem Cells 23, 1489–1501 [DOI] [PubMed] [Google Scholar]
- 34. Sumi T., Tsuneyoshi N., Nakatsuji N., Suemori H. (2008) Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development 135, 2969–2979 [DOI] [PubMed] [Google Scholar]
- 35. Cai L., Holoweckyj N., Schaller M. D., Bear J. E. (2005) Phosphorylation of coronin 1B by protein kinase C regulates interaction with Arp2/3 and cell motility. J. Biol. Chem. 280, 31913–31923 [DOI] [PubMed] [Google Scholar]
- 36. Cai J., Chen J., Liu Y., Miura T., Luo Y., Loring J. F., Freed W. J., Rao M. S., Zeng X. (2006) Assessing self-renewal and differentiation in human embryonic stem cell lines. Stem Cells 24, 516–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Desbordes S. C., Placantonakis D. G., Ciro A., Socci N. D., Lee G., Djaballah H., Studer L. (2008) High-throughput screening assay for the identification of compounds regulating self-renewal and differentiation in human embryonic stem cells. Cell Stem Cell 2, 602–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sperger J. M., Chen X., Draper J. S., Antosiewicz J. E., Chon C. H., Jones S. B., Brooks J. D., Andrews P. W., Brown P. O., Thomson J. A. (2003) Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc. Natl. Acad. Sci. U.S.A. 100, 13350–13355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brandenberger R., Khrebtukova I., Thies R. S., Miura T., Jingli C., Puri R., Vasicek T., Lebkowski J., Rao M. (2004) MPSS profiling of human embryonic stem cells. BMC Dev. Biol. 4, 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bhattacharya B., Miura T., Brandenberger R., Mejido J., Luo Y., Yang A. X., Joshi B. H., Ginis I., Thies R. S., Amit M., Lyons I., Condie B. G., Itskovitz-Eldor J., Rao M. S., Puri R. K. (2004) Gene expression in human embryonic stem cell lines: unique molecular signature. Blood 103, 2956–2964 [DOI] [PubMed] [Google Scholar]
- 41. Fong H., Hohenstein K. A., Donovan P. J. (2008) Regulation of self-renewal and pluripotency by Sox2 in human embryonic stem cells. Stem Cells 26, 1931–1938 [DOI] [PubMed] [Google Scholar]
- 42. Adewumi O., Aflatoonian B., Ahrlund-Richter L., Amit M., Andrews P. W., Beighton G., Bello P. A., Benvenisty N., Berry L. S., Bevan S., Blum B., Brooking J., Chen K. G., Choo A. B., Churchill G. A., Corbel M., Damjanov I., Draper J. S., Dvorak P., Emanuelsson K., Fleck R. A., Ford A., Gertow K., Gertsenstein M., Gokhale P. J., Hamilton R. S., Hampl A., Healy L. E., Hovatta O., Hyllner J., Imreh M. P., Itskovitz-Eldor J., Jackson J., Johnson J. L., Jones M., Kee K., King B. L., Knowles B. B., Lako M., Lebrin F., Mallon B. S., Manning D., Mayshar Y., McKay R. D., Michalska A. E., Mikkola M., Mileikovsky M., Minger S. L., Moore H. D., Mummery C. L., Nagy A., Nakatsuji N., O'Brien C. M., Oh S. K., Olsson C., Otonkoski T., Park K. Y., Passier R., Patel H., Patel M., Pedersen R., Pera M. F., Piekarczyk M. S., Pera R. A., Reubinoff B. E., Robins A. J., Rossant J., Rugg-Gunn P., Schulz T. C., Semb H., Sherrer E. S., Siemen H., Stacey G. N., Stojkovic M., Suemori H., Szatkiewicz J., Turetsky T., Tuuri T., van den Brink S., Vintersten K., Vuoristo S., Ward D., Weaver T. A., Young L. A., Zhang W. (2007) Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat. Biotechnol. 25, 803–816 [DOI] [PubMed] [Google Scholar]
- 43. Brandenberger R., Wei H., Zhang S., Lei S., Murage J., Fisk G. J., Li Y., Xu C., Fang R., Guegler K., Rao M. S., Mandalam R., Lebkowski J., Stanton L. W. (2004) Transcriptome characterization elucidates signaling networks that control human ES cell growth and differentiation. Nat. Biotechnol. 22, 707–716 [DOI] [PubMed] [Google Scholar]
- 44. Huang T. S., Hsieh J. Y., Wu Y. H., Jen C. H., Tsuang Y. H., Chiou S. H., Partanen J., Anderson H., Jaatinen T., Yu Y. H., Wang H. W. (2008) Functional network reconstruction reveals somatic stemness genetic maps and dedifferentiation-like transcriptome reprogramming induced by GATA2. Stem Cells 26, 1186–1201 [DOI] [PubMed] [Google Scholar]
- 45. Chang S. J., Huang T. S., Wang K. L., Wang T. Y., Yang Y. C., Chang M. D., Wu Y. H., Wang H. W. (2008) Genetic network analysis of human CD34+ hematopoietic stem/precursor cells. Taiwan J. Obstet. Gynecol. 47, 422–430 [DOI] [PubMed] [Google Scholar]
- 46. Müller-Tidow C., Schwäble J., Steffen B., Tidow N., Brandt B., Becker K., Schulze-Bahr E., Halfter H., Vogt U., Metzger R., Schneider P. M., Büchner T., Brandts C., Berdel W. E., Serve H. (2004) High-throughput analysis of genome-wide receptor tyrosine kinase expression in human cancers identifies potential novel drug targets. Clin. Cancer Res. 10, 1241–1249 [DOI] [PubMed] [Google Scholar]
- 47. Grimmer C., Balbus N., Lang U., Aigner T., Cramer T., Müller L., Swoboda B., Pfander D. (2006) Regulation of type II collagen synthesis during osteoarthritis by prolyl-4-hydroxylases: possible influence of low oxygen levels. Am. J. Pathol. 169, 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bock O., Höftmann J., Theophile K., Hussein K., Wiese B., Schlué J., Kreipe H. (2008) Bone morphogenetic proteins are overexpressed in the bone marrow of primary myelofibrosis and are apparently induced by fibrogenic cytokines. Am. J. Pathol. 172, 951–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Patel B. B., Li X. M., Dixon M. P., Blagoi E. L., Seeholzer S. H., Chen Y., Miller C. G., He Y. A., Tetruashvily M., Chaudhry A. H., Ke E., Xie J., Cooper H., Bellacosa A., Clapper M. L., Boman B. M., Zhang T., Litwin S., Ross E. A., Conrad P., Crowell J. A., Kopelovich L., Knudson A., Yeung A. T. (2007) Searchable high-resolution 2D gel proteome of the human colon crypt. J. Proteome Res. 6, 2232–2238 [DOI] [PubMed] [Google Scholar]
- 50. Højlund K., Yi Z., Hwang H., Bowen B., Lefort N., Flynn C. R., Langlais P., Weintraub S. T., Mandarino L. J. (2008) Characterization of the human skeletal muscle proteome by one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. Mol. Cell. Proteomics 7, 257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Vesque C., Anselme I., Couvé E., Charnay P., Schneider-Maunoury S. (2006) Cloning of vertebrate Protogenin (Prtg) and comparative expression analysis during axis elongation. Dev. Dyn. 235, 2836–2844 [DOI] [PubMed] [Google Scholar]
- 52. Herzog W., Müller K., Huisken J., Stainier D. Y. (2009) Genetic evidence for a noncanonical function of seryl-tRNA synthetase in vascular development. Circ. Res. 104, 1260–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]