Abstract
Epithelial-mesenchymal transition (EMT) describes a process whereby polarized epithelial cells with restricted migration transform into elongated spindle-shaped mesenchymal cells with enhanced motility and invasiveness. Although there are some molecular markers for this process, including the down-regulation of E-cadherin, our understanding of plasma membrane (PM) and associated proteins involved in EMT is limited. To specifically explore molecular alterations occurring at the PM, we used the cationic colloidal silica isolation technique to purify PM fractions from epithelial Madin-Darby canine kidney cells during Ras/TGF-β-mediated EMT. Proteins in the isolated membrane fractions were separated by one-dimensional SDS-PAGE and subjected to nano-LC-MS/MS-based protein identification. In this study, the first membrane protein analysis of an EMT model, we identified 805 proteins and determined their differential expression using label-free spectral counting. These data reveal that Madin-Darby canine kidney cells switch from cadherin-mediated to integrin-mediated adhesion following Ras/TGF-β-mediated EMT. Thus, during the EMT process, E-cadherin, claudin 4, desmoplakin, desmoglein-2, and junctional adhesion molecule A were down-regulated, whereas integrins α6β1, α3β1, α2β1, α5β1, αVβ1, and αVβ3 along with their extracellular ligands collagens I and V and fibronectin had increased expression levels. Conspicuously, Wnt-5a expression was elevated in cells undergoing EMT, and transient Wnt-5a siRNA silencing attenuated both cell migration and invasion in these cells. Furthermore, Wnt-5a expression suppressed canonical Wnt signaling induced by Wnt-3a. Wnt-5a may act through the planar cell polarity pathway of the non-canonical Wnt signaling pathway as several of the components and modulators (Wnt-5a, -5b, frizzled 6, collagen triple helix repeat-containing protein 1, tyrosine-protein kinase 7, RhoA, Rac, and JNK) were found to be up-regulated during Ras/TGF-β-mediated EMT.
Epithelial-mesenchymal transition (EMT)1 is a process essential for morphogenesis during embryonic development but has more recently been implicated in tumor metastasis (1). Morphologically, EMT describes the conversion of epithelial cells with cobblestone-like morphology and restricted cell migration into elongated fibroblast-like mesenchymal cells with enhanced cell motility and invasiveness in vitro (2, 3). It is currently thought that a subset of cancerous cells undergo EMT to enable them to escape from surrounding cells, penetrate into neighboring lymph or blood vessels, and passage to distant sites to form secondary metastases (2, 4). The biochemical events by which cells undergo and maintain EMT, especially those involving membrane and extracellular environmental cues, remain poorly understood.
The complexity of EMT is exemplified in the cooperation of multiple signaling pathways, including Wnt, Notch, and Hedgehog (5, 6). Although the precise details and mechanisms of pathway cross-talk remain largely unknown (7), one of the best characterized examples is the interaction between the TGF-β and Ras signaling pathways (8–11). Oncogenic Ras is a well known molecular EMT effector that causes cell scattering via PI3K signaling and drives the autocrine production of TGF-β via Raf/MAPK signaling (12). Although TGF-β normally functions as a tumor suppressor during the early stages of cancer progression, it is known to be involved in later stages of carcinogenesis (13–15).
In mammalian tissues, epithelial cell polarity and intercellular adhesion is manifested through the assembly of tight junctions (TJs), adherens junctions (AJs), and desmosomes (16). Epithelial TJs and AJs are asymmetrically distributed at the apical region of the lateral cell membrane, forming a circumferential belt that separates the plasma membrane (PM) into apical and basolateral domains (17). TJs consist of occludins, claudins, and junctional adhesion molecules as well as cytoplasmic-plaque ZO proteins that associate with the actin cytoskeleton (18, 19). AJs are located directly below TJs and also encircle the apex of epithelial cells. E-cadherin is the best characterized AJ constituent and is juxtaposed to cytoplasmic catenins, actinins, and vinculin, which mediate attachment to actin filaments. Desmosomes are composed of desmosomal cadherins, proteins from the armadillo family, and members of the plakin family of cytolinkers (20). Desmosomes reside more basally than AJs and confer strong cell-cell adhesion through anchoring of the intermediate filament cytoskeleton (21). A hallmark of the EMT process is the loss of cell-cell adhesion that stems from the down-regulation of these cell junction proteins (6). Molecular mechanisms known to be involved include transcriptional repression mediated by transcription factors such as snail, slug, and members of the Zeb and basic helix-loop-helix families (22). Although the diminished expression of some PM proteins during EMT has been documented, knowledge of global PM protein expression remains much more limited.
The paucity of information on PM proteins is largely due to their low solubility, which presents technical challenges in their isolation, and their low copy number (23, 24). Proteomics isolation strategies, including ultracentrifugation, affinity capture, and solubility-based isolation methods, are commonly used to enrich for PM and peripherally associated proteins for the purpose of mass spectrometry-based identification and characterization (25). In the present study, a partitioning-type purification strategy was used to isolate adherent and non-adherent plasma membrane fractions from MDCK cells based on the cationic colloidal silica (CCS) method developed by Chaney and Jacobson (26). To gain insights into comprehensive membrane protein expression changes during EMT, we performed a comparative proteomics analysis of MDCK cell PMs following oncogenic Ras/TGF-β-mediated EMT (21D1 cells). For proteins with altered abundance based on label-free spectral counting, gene expression levels during EMT were examined via microarray analysis. Furthermore, siRNA was used to confirm the functional outcomes of modulators with up-regulated expression. Together, these studies revealed changes in levels of several adhesion receptors and their extracellular ligands as well as signaling molecules that regulate cell migration and invasion.
EXPERIMENTAL PROCEDURES
Cell Culture
MDCK cells (5 × 105 cells/150-mm culture dish) were maintained in Dulbecco's modified Eagle's medium (DMEM) with high glucose (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (Invitrogen), 60 μg/ml benzylpenicillin (CSL Ltd.), and 100 μg/ml streptomycin (Sigma-Aldrich) and incubated in 37 °C with 10% CO2-saturated humidity. The H-Ras-transformed derivative cell line 21D1 (27, 28) was maintained in 10% fetal bovine serum in DMEM plus 30% (v/v) conditioned medium from the previous culture. Cells were grown to 80–90% confluence and harvested for plasma membrane purification, Western immunoblotting analysis, or total RNA extraction.
Immunofluorescence Microscopy
Cells cultured on coverslips (25-mm diameter, 1.7-mm thickness) were washed twice with phosphate-buffered saline (PBS) (20 mm phosphate, pH 7.3, 120 mm NaCl), fixed with 3.7% (v/v) formaldehyde at 25 °C for 10 min, permeabilized using 0.2% (v/v) Triton X-100 in PBS at 25 °C for 10 min, and blocked by 10% (w/v) BSA in PBS with Tween 20 at 25 °C for 1 h with gentle shaking. Coverslips were subsequently incubated with either a mouse anti-E-cadherin (BD Biosciences) or rabbit anti-ZO-1 (Invitrogen) primary antibody at 25 °C for 2 h. Cells were washed three times with PBS followed by incubation with the corresponding fluorescent dye-labeled secondary antibody, Alexa Fluor 488 goat anti-mouse or Alexa Fluor 546 goat anti-rabbit antibody (Invitrogen), at 25 °C for 1 h. Imaging was performed on an inverted IX50 microscope (Olympus) equipped with a charge-coupled device camera (Model 11.3, Diagnostic Instruments).
Wound Healing Assay
The in vitro wound healing/scratch cell migration assay was performed as described previously (29). Briefly, cells were seeded onto 6-well plates and cultured until confluent upon which “wounds” were created using a pipette tip to scratch a straight line on the culture plate. Cells were washed, and fresh culture medium was replaced to remove detached cells. Cells were further cultured for 24 h, and the percentage of wound closure was assessed. Phase-contrast images were acquired at 0- and 24-h time points using an inverted IX50 microscope (Olympus) equipped with a charge-coupled device camera (Model 11.3, Diagnostic Instruments). SPOT advanced imaging software (version 4.0.4) was used to acquire and process the images.
Transwell Migration and Invasion Assays
The in vitro cell migration assay was performed by seeding 10,000 cells in the top chamber of an 8-μm polycarbonate membrane Transwell insert (Corning). Medium containing 15% (v/v) conditioned medium and 10% (v/v) FCS with or without 40 ng/ml recombinant mouse Wnt-5a (R&D Systems) was used as the chemoattractant in the lower chamber. Cells were incubated at 37 °C with 10% CO2 for 24 h, and the membrane was fixed with 100% methanol and stained with hematoxylin. Cells that migrated through the membrane were counted in 10 random high power fields. The in vitro cell invasion assay was performed as described previously (30). The assay was performed similarly to the migration assay except the Transwell insert was coated with 75 μl of matrix containing 20% Matrigel (BD Biosciences) and 1 mg/ml collagen I (BD Biosciences) in serum-free DMEM (Invitrogen). The matrix was solidified by incubation at 37 °C for 2 h. 10,000 cells were resuspended in serum-free DMEM (containing 0.1% BSA (w/v)) and plated in the top chamber on the solidified matrix. Both assays were performed in duplicate or triplicate as stated.
Cationic Colloidal Silica Plasma Membrane Purification
Purification of adherent cell membranes was performed as described previously (31). Briefly, MDCK and 21D1 cells were washed with 1× PBS plus 1 mm CaCl2 and 1 mm MgCl2 twice and equilibrated using PMCB buffer (20 mm MES, pH 5.3, 0.5 mm CaCl2, 1 mm MgCl2, 135 mm NaCl). The cell surface was then coated with 0.5% (w/v) cationic colloidal silica in PMCB buffer for 1 min. After excess silica was discarded and the cells were washed with PMCB buffer, 1% (v/v) polyacrylic acid (Sigma-Aldrich) in PMCB buffer was overlaid for 1 min and then washed with PMCB buffer. Dishes were washed with hypotonic buffer containing 2.5 mm imidazole solution and incubated in this buffer for 30 min. Cells were then disrupted with hypotonic buffer by shear force generated by a syringe with a modified 18-gauge needle (see Ref. 31 for details). The adherent membrane (attached to the culture dish) fraction was washed twice with 5 m NaCl and collected using detergent. To separate the non-adherent membrane (containing the cationic colloidal silica) from the cytosol and intracellular organelles, the non-adherent membrane fraction was overlaid on 60% (w/v) OptiPrep (Sigma-Aldrich) and centrifuged at 75,000 × g at 4 °C for 30 min. The resultant non-adherent membrane pellet was collected and washed twice with 5 m NaCl. Membrane fractions were precipitated using acetone and resolubilized in SDS-PAGE sample buffer. Membrane protein samples from 10 biological replicates were pooled prior to electrophoresis and proteomics analysis.
One-dimensional SDS-PAGE
Adherent and non-adherent membrane protein samples were resuspended in 1× NuPAGE lithium dodecyl sulfate sample buffer (Invitrogen), and the protein concentration was estimated by densitometry using BenchMark Protein Ladder (Invitrogen) as the standard. Adherent and non-adherent membrane protein samples containing 50 mm DTT were boiled at 95 °C for 10 min and loaded onto 4–12% NuPAGE Novex Bis-Tris gels (Invitrogen). Electrophoresis was performed in MES running buffer (Invitrogen) at 150 V (constant voltage) until the tracking dye reached the bottom of the gel. Proteins were visualized using Imperial Protein Stain (Pierce) or subjected to Western immunoblotting analysis.
Western Immunoblotting
Proteins were transferred onto nitrocellulose membranes using the iBlotTM dry blotting system (Invitrogen). Membranes were incubated in blocking solution (5% (w/v) milk powder in TBST (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.05% (v/v) Tween 20) for 1 h before being probed with anti-Ras (Millipore), anti-vimentin (Millipore), anti-E-cadherin (BD Biosciences), or anti-actin (Sigma-Aldrich) primary antibodies for 1 h. Membranes were washed three times for 5 min with TBST and then probed with the corresponding secondary antibody labeled with IRDye 700/800CW (LI-COR Biosciences, Inc.). Membranes were washed twice with TBST and scanned using the Odyssey infrared imaging system (LI-COR Biosciences, Inc.).
In-gel Protein Digestion
Individual gel lanes were excised into 24 gel slices (2–3 mm), reduced, alkylated, and subjected to in-gel tryptic digestion as described previously (32). Briefly, gel slices were reduced with 10 mm DTT for 30 min, alkylated with 25 mm iodoacetamide for 20 min, and trypsinized (6 ng/μl) (Worthington) for 5 h at 37 °C. Peptide digests were extracted and dried via centrifugal lyophilization (SpeedVac, Savant) to a volume of 10 μl. Digests were then subjected to MS/MS analysis on the LTQ-Orbitrap (Thermo Fischer Scientific).
Nano-LC-MS/MS
A 96-well plate containing peptide digests was loaded into the autosampler for injection and fractionation by nanoflow reversed-phase HPLC (Model 1200, Agilent). Fractionation was performed using a nanoACQUITY (C18) 150-mm × 0.15-mm-inner diameter reversed-phase ultraperformance LC column (Waters) developed with a linear 60-min gradient from 0 to 100% Buffer B (0.1% (v/v) aqueous formic acid, 60% (v/v) acetonitrile) with a flow rate of 0.8 μl/min at 45 °C where Buffer A was 0.1% (v/v) aqueous formic acid. The capillary HPLC system was coupled on line to the LTQ-Orbitrap mass spectrometer equipped with a nanoelectrospray ion source (Thermo Fisher Scientific). Positive ion mode was used for data-dependent acquisition. Survey MS scans were acquired with the resolution set to 30,000. Each scan was recalibrated in real time by co-injecting an internal standard from ambient air into the C-trap (33). The five most intense ions per cycle were fragmented and analyzed in the linear trap. Target ions already selected for MS/MS were dynamically excluded for 180 s.
Database Searching and Bioinformatics Analysis
Peak lists were generated using extract-msn with the following parameters: minimum mass, 700; maximum mass, 5,000; grouping tolerance, 0.01 Da; intermediate scans, 200; minimum group count, 1; minimum number of peaks, 10; and total ion current, 100. Peak lists for each LC-MS/MS run were merged into a single MGF file for Mascot searches. Database search parameters were as follows. Carboxymethylation of cysteine was set as a fixed modification (+58 Da) as were variable modifications such as NH2-terminal acetylation (+42 Da) and oxidation of methionine (+16 Da). Allowance was made for up to two missed tryptic cleavages. Peptide mass tolerance was ±20 ppm, and the number of 13C atoms was defined as 1. MGF files were searched against the LudwigNR_subset database (Q308)2 comprising 487,924 entries using the Mascot search algorithm (version 2.2.04, Matrix Science) (34).
An in-house software program (MSPro) was used for parsing and summarizing the output files from Mascot searches as described previously (35). Peptide identifications were deemed significant if the ion score was greater than or equal to the homology score (or identity score if there was no homology score). False positive protein identifications were estimated by searching MS/MS spectra against the corresponding reversed sequence (decoy) database (36). A false discovery rate of 1% (protein score of 46) was used with significant protein hits containing at least two peptides.
The BioMart data-mining tool (http://www.ensembl.org/biomart/index.html) was used to obtain Ensembl protein description, gene name, and microarray probe information. UniProt (http://www.uniprot.org) and Protein Information Resource (http://pir.georgetown.edu) were used to obtain gene ontology annotation. Transmembrane-spanning α-helices were predicted using the web-based prediction program TMHMM version 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0) (37, 38).
Significant relative spectral count -fold change ratios (Rsc) were determined using a modified formula from a previous serial analysis of gene expression study by Beissbarth et al. (39),
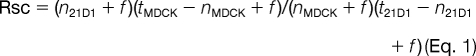 |
where n is the significant protein spectral count, t is the total number of significant spectra in the sample, and f is a correction factor set to 1.25 (40). The total number of spectra was counted only for the significant peptides identified (ion score greater than or equal to homology score). When Rsc was less than 1, the negative inverse Rsc value was used.
GeneChip Expression Analysis
Microarray expression profiling was performed as described previously (27). Briefly, total RNA was extracted from MDCK and Ras-transformed/TGF-β-induced MDCK cells using the RNeasy minikit (Qiagen). A T7 promoter-linked oligo(dT) primer was used to synthesize double-stranded cDNA using an Affymetrix One Cycle cDNA synthesis kit (Millennium Sciences). Double-stranded cDNA was then subjected to in vitro transcription to incorporate a biotinylated ribonucleotide analog (pseudouridine) into the newly synthesized cRNA using the Affymetrix IVT labeling kit (Millennium Sciences). Labeled cRNA was isolated, fragmented to the 50–200-bp size range, and then hybridized to the Canine Genome 2.0 GeneChip Array (Affymetrix) at 45 °C for 16 h with a 60-rpm rotating wheel. Chips were washed in the Affymetrix Fluidics Station 450 and scanned using the Affymetrix GeneChip Scanner 3000. GeneChip Operating Software (GCOS version 1.4) was used to process scanned data.
Bioinformatics Analysis of WNT5A Promoter
MatInspector (version 8.0) (41), a bioinformatics tool to identify transcription factor binding sites in DNA sequences, was used to locate sites in the promoter region of WNT5A. General core promoter elements (e.g. TATA box) and vertebrate matrix (Matrix Library 8.2) with a core similarity of 0.75 were used. Additionally, 1000 bp upstream of the WNT5A transcription initiation site was downloaded from the NCBI Map Viewer (42). The downloaded DNA sequence was input into PROMO (version 3.0.2) (43) and TFSEARCH (version 1.3) to identify transcription factor binding sites. Human and vertebrate transcription factors were searched in PROMO and TFSEARCH, respectively. Results from the three search algorithms were concatenated, and the gene list was filtered based on mRNA transcript expression obtained from the microarray analysis.
Luciferase Gene Reporter Assay
To determine canonical Wnt signaling activity, gene reporter assays were performed using the Steady-Glo Luciferase Assay System (Promega). 21D1 cells in a 24-well plate were transfected using Metafectine transfection reagent (Biontex) for 6 h with 250 ng of Super TopFlash (STF) plasmid containing firefly luciferase driven by TCF/LEF-responsive promoter (a kind gift from Dr. Randall Moon) (44). Following incubation, medium was replaced with serum-free DMEM containing 20 ng/ml recombinant mouse Wnt-3a (R&D Systems), 20 or 40 ng/ml recombinant mouse Wnt-5a (R&D Systems), or 30% conditioned medium from 21D1 cells. Cells were cultured for a further 48 h before being lysed with luciferase cell culture lysis reagent (Promega). Lysates were collected, and luciferase activity was measured in a 96-well plate using a Glomax 96 microplate luminometer (Promega) according to manufacturer's instructions. Each assay was performed in duplicate.
Semiquantitative RT-PCR
Total RNA from MDCK and 21D1 cells was extracted using TRIzol reagent (Invitrogen), and the amount of RNA was estimated by reading the OD at 260 nm. Primer oligo(dT), a component in the Superscript III first strand synthesis system (Invitrogen), was used to reverse transcribe cDNA. Primer pairs were as follows: Wnt-5a, 5′-CCAAGGGCTCGTACGAGAGC-3′ and 5′-CGAACTGGTCCACGATCTCC-3′; and an internal control GAPDH (glyceraldehyde 3-phosphate dehydrogenase), 5′-AACATCATCCCTGCTTCCAC-3′ and 5′-GGCAGGTCAGATCCACAACT-3′. The PCR was carried out using native Taq DNA polymerase (Invitrogen) in a GeneAmp PCR system 9700 (Applied Biosystems). PCR products were analyzed by electrophoresis on a 2% (w/v) agarose gel and visualized with ethidium bromide.
siRNA Transfection
Customized Stealth siRNA (Invitrogen) targeting the Canis familiaris Wnt-5a transcript was designed using the Invitrogen RNAi Designer with the Ensembl accession number ENSCAFT00000013003. The siRNA sequence 5′-GGGCAUCCAAGAGUGCCAGUAUCAA-3′ corresponding to residues 301–325 of Wnt-5a was selected from three siRNA duplexes to be the most effective in reducing Wnt-5a expression. 21D1 cells were seeded in a 6-well plate and grown to 50–60% confluence upon which 250 pmol of either Wnt-5a siRNA or a scrambled negative control was transfected into the cells using Lipofectamine 2000 transfection reagent (Invitrogen) in a ratio of 20 pmol of siRNA/1 μl of transfection reagent. After 6 h of incubation, the medium was replaced with 15% conditioned medium with or without recombinant mouse Wnt-5a (R&D Systems), and cells were further cultured for 48 h. Transfected cells were then subjected to RNA extraction for RT-PCR or used in the in vitro wound healing or cell invasion assay.
RESULTS AND DISCUSSION
Oncogenic H-Ras Expression and TGF-β Stimulation Drive EMT Progression in MDCK Cells
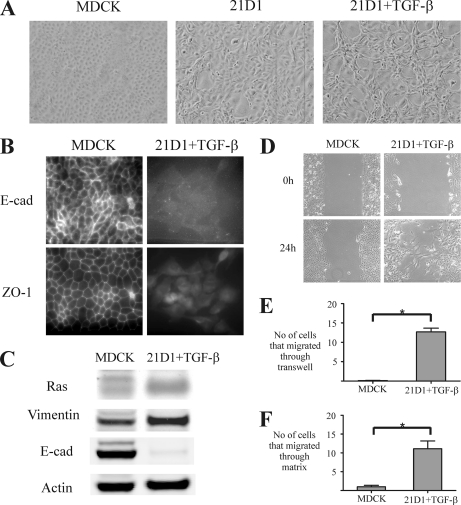
It is well documented that Ras and TGF-β act synergistically to promote EMT in MDCK cells as well as several other cell models, including mouse mammary EpH4 cells (8, 12, 45, 46). We have previously shown that MDCK cells stably transformed with constitutive oncogenic H-Ras (21D1 cells) undergo EMT (27, 28). In this study, 21D1 cells were treated with TGF-β to initiate an autocrine TGF-β loop that further enhances the EMT phenotype (8, 47). Noticeably, MDCK cells transformed with Ras undergo a distinct morphological alteration from round cobblestone-like to fibroblast-like cells, and stimulation with TGF-β further elongates cell shape and increases scattering (Fig. 1A). In addition, 21D1 cells exhibit loss of cell polarity as evidenced by reduced expression of tight junction proteins (ZO-1) and adherens junction proteins such as E-cadherin between adjacent cells (Fig. 1, B and C) and have increased expression of the mesenchymal marker vimentin compared with wild-type MDCK cells (Fig. 1C). 21D1 cells stimulated with TGF-β demonstrate increased migration and invasion compared with wild-type MDCK cells (Fig. 1, D–F) and unstimulated 21D1 cells (data not shown). The elevated motility of 21D1 cells is evident in the wound healing assay where individual cells migrated into the “wounded” gap area after 24 h (Fig. 1D). In contrast, MDCK cells show sheetlike migration, leaving the wound entirely unclosed (Fig. 1D). This finding was supported by the Transwell migration assay where a greater number of 21D1 cells migrated from the upper chamber through the membrane insert toward the lower chamber compared with MDCK cells (Fig. 1E). In a similar fashion, 21D1 cells showed enhanced invasiveness in the Transwell invasion assay (Fig. 1F). On average, only one MDCK cell penetrated through the collagen-Matrigel matrix compared with 11 21D1 cells.
Fig. 1.
Oncogenic H-Ras and TGF-β stimulation induces EMT in MDCK cells. A, MDCK cells transformed with oncogenic H-Ras undergo morphological alterations from round cobblestone-like cells with tight cell-cell adhesion to long spindle-shaped cells with reduced cell contact with neighboring cells. Stimulation with TGF-β further elongates cell shape and increases scattering. B, immunofluorescence microscopy reveals strong positive staining of epithelial cell junction proteins E-cadherin (E-cad) and ZO-1 between borders of MDCK cells. In contrast, 21D1 cells stimulated with TGF-β exhibit loss of E-cadherin and ZO-1 localization and protein expression. C, MDCK cells undergoing oncogenic Ras- and TGF-β-induced EMT show diminished expression of the epithelial marker E-cadherin and increased expression of the mesenchymal marker vimentin by Western immunoblotting. D, the wound healing assay reveals that 21D1 cells have increased cell migration compared with MDCK cells as individual cells leave the front to migrate into the wounded area. E, 21D1 cells exhibit elevated migration in the Transwell migration assay as a significant (*) number of 21D1 cells move through the membrane filter toward the lower chamber (n = 3). F, a significant number of 21D1 cells relative to MDCK cells penetrated through the collagen-Matrigel matrix, demonstrating their enhanced cell invasion during EMT (n = 2). Error bars represent mean ± S.D. Significance is determined by a p value ≤0.05 using the Student's t test.
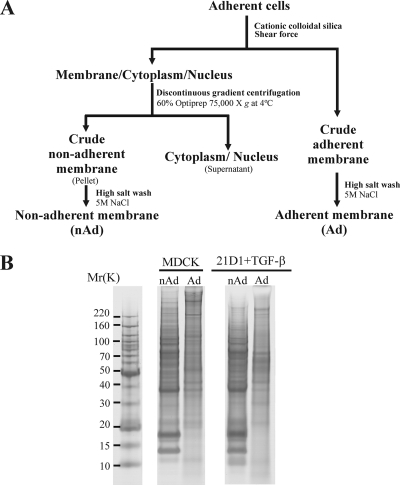
Proteomics Profiling of Plasma Membranes Isolated Using Cationic Colloidal Silica
To gain insights into proteins expressed at the PM during EMT, we performed a proteomics analysis of MDCK and Ras-transformed MDCK cells stimulated with TGF-β. A modified version of the CCS method (26, 31) was applied to purify PMs for proteomics analysis (Fig. 2A). This strategy yielded two PM fractions from each cell line, an adherent (Ad) fraction (still attached to the culture dish) and a non-adherent (nAd) fraction (coated with CCS and isolated by density-based ultracentrifugation). Proteins in PM preparations were fractionated by SDS-PAGE (Fig. 2B) and subjected to LC-MS/MS protein identification (48). In total, 805 proteins were identified (supplemental Table S1), encompassing 757 from MDCK cells and 724 from 21D1 cells. Of the 805 proteins identified, 81 were unique to MDCK cells, 48 were unique to 21D1 cells, and 676 were detected in both cell line PM fractions (supplemental Fig. S1A). A Venn diagram depicting proteins identified in each fraction (Ad and nAd) of both cell lines is given in supplemental Fig. S1B; complete protein and peptide lists can be found in supplemental Tables S2–S5.
Fig. 2.
Plasma membrane purification. A, experimental work flow outlining purification of Ad and nAd membranes from MDCK and 21D1 cells. Membrane protein isolation and purification are based on the cationic colloidal silica method (26) and a modified version (31). B, SDS-PAGE protein gel banding patterns of Ad and nAd isolated membrane fractions. Gel lanes were excised and trypsinized, and extracted peptides were subjected to LC-MS/MS protein identification.
Of the 805 proteins identified, 225 (28%) had membrane classification according to GO (gene ontology) Slim annotation with 104 of these (13%) predicted to contain at least one transmembrane-spanning helix using the hidden Markov model prediction algorithm (38). Although our proteome data sets show enrichment for PM proteins, basic proteins such as nuclear constituents may be present due to nonspecific binding with the negatively charged polyacrylic acid following cell disruption. At this stage, we cannot exclude the possibility that these non-membrane annotated proteins either interact with the membrane indirectly or are part of a membrane-associated complex.
Label-free Spectral Counting Reveals EMT Proteins with Altered Abundance
Relative protein abundance was determined using a modified version of protein Rsc described previously (28). For each protein, the Rsc formula generated by Beissbarth et al. (39) and modified by Old et al. (40) first normalizes the number of spectral counts identified by the total number of spectra identified in the sample and then compares it against another sample to give insights into protein enrichment. This semiquantitative calculation was used to rank the 805 proteins identified from most up-regulated to most down-regulated in 21D1 cells relative to MDCK cells (supplemental Table S1). The results using this approach are consistent with published literature for many well known EMT markers. For example, E-cadherin (Rsc, −18.29), claudin 4 (Rsc, −3.08), desmoplakin (Rsc, −29.64), and cytokeratin 18 (Rsc, −7.18) were down-regulated following Ras/TGF-β-mediated EMT, whereas integrin αV (Rsc, 11.76), fibronectin (Rsc, 13.48), and vimentin (Rsc, 2.05) were up-regulated (6, 49). In addition, several proteins involved in mediating cell-cell adhesion were down-regulated, including junctional adhesion molecule A (Rsc, −1.68), desmoglein-2 (Rsc, −3.57), and desmoglein-3 (Rsc, −8.15) (50, 51), whereas elevated levels of transmembrane proteins such as tetraspanin 8 (Rsc, 26.8) were detected. Increased expression of proteins involved in Wnt signaling (Wnt-5a (Rsc, 158.08) and Wnt-5b (Rsc, 28.41)) and Notch signaling pathways (jagged-1 (Rsc, 38.09)) were also observed. Wnt-5a was found to be the most up-regulated protein with 18 unique significant peptides and 194 spectral counts observed only in the Ad 21D1 PM fraction. The most significant differentially expressed proteins identified (i.e. top 5% dysregulated (−11.3 ≥ Rsc ≥ 11.3)) are listed in Table I with their corresponding transcript expression data and gene ontology annotation.
Table I. Selected proteins differentially expressed during Ras/TGF-β-induced EMT.
| Accession numbera | Geneb | Descriptionb | BLASTc | Proteomics Rscd | Microarray |
Signal transductiong | Structuralg | Peptidase or inhibitorg | Transportg | |
|---|---|---|---|---|---|---|---|---|---|---|
| Probe IDe | FCf | |||||||||
| Proteins with increased abundance following Ras transformation | ||||||||||
| ENSCAFP00000012033 | WNT5A | Protein Wnt-5a precursor | Q6DK41 | 158.08 | CfaAffx.13012.1.S1_at | 45.25 | ✓ | |||
| ENSCAFP00000000926 | CTHRC1 | Collagen triple helix repeat-containing protein 1 precursor | A2RRY6 | 139.37 | Cfa.15223.1.A1_at | 19.70 | ||||
| ENSCAFP00000000701 | SYNE1 | Nesprin-1 | Q8NF91 | 51.02 | Cfa.14004.1.A1_s_at | NC | ||||
| ENSCAFP00000002301 | ADAMTSL1 | ADAMTS-like protein 1 precursor | B1AMM2 | 43.75 | CfaAffx.3281.1.S1_s_at | NC | ||||
| ENSCAFP00000029402 | COL5A1 | Collagen α-1(V) chain precursor | O88207 | 42.94 | CfaAffx.30396.1.S1_s_at | 1.62 | ✓ | |||
| ENSCAFP00000008417 | JAG1 | Protein jagged-1 precursor | P78504 | 38.09 | CfaAffx.9396.1.S1_at | 5.28 | ✓ | |||
| ENSCAFP00000026045 | CLTC | Clathrin heavy chain 1 (CLH-17) | Q80U89 | 37.29 | CfaAffx.27024.1.S1_s_at | 1.23 | ✓ | |||
| ENSCAFP00000006943 | EMILIN1 | Elastin microfibril interface-located protein 1 | Q53SY9 | 33.25 | Cfa.16928.1.S1_s_at | NC | ✓ | |||
| ENSCAFP00000023434 | WNT5B | Protein Wnt-5b precursor | B1WBR9 | 28.41 | CfaAffx.24413.1.S1_at | 13.00 | ✓ | |||
| O46392 | COL1A2 | Collagen α-2(I) chain precursor | O46392 | 28.41 | Cfa.1262.1.S2_s_at | 1176.26 | ✓ | |||
| ENSCAFP00000000667 | TSPAN8 | Tetraspanin-8 | O55158 | 26.80 | Cfa.6237.1.S1_at | 1.62 | ||||
| ENSCAFP00000006423 | FOLH1 | FOLH1 protein (fragment) | Q2VPJ0 | 25.18 | Cfa.13491.1.A1_s_at | −2.14 | ✓ | |||
| ENSCAFP00000006067 | EHD2 | EH domain-containing protein 2 | Q9NZN4 | 21.96 | Cfa.16731.1.S1_s_at | 45.25 | ||||
| ENSCAFP00000019418 | PRPF6 | Pre-mRNA-processing factor 6 | Q91YR7 | 21.15 | Cfa.20738.1.S1_s_at | NC | ||||
| ENSCAFP00000021445 | HTRA3 | Probable serine protease HTRA3 precursor (EC 3.4.21.-) | P83110 | 20.34 | CfaAffx.22424.1.S1_at | 9.19 | ✓ | |||
| ENSCAFP00000003680 | COL15A1 | Collagen α-1(XV) chain precursor | P39059 | 19.54 | Cfa.18161.1.S1_s_at | NC | ✓ | |||
| ENSCAFP00000009742 | ITGA5 | Integrin α5 precursor | P08648 | 19.54 | CfaAffx.10710.1.S1_s_at | 4.92 | ✓ | |||
| A1YVW4 | PRNP | Major prion protein | A1YVW4 | 19.54 | Cfa.20442.2.S1_s_at | 6.50 | ||||
| O46605 | MDR1 | Multidrug resistance p-glycoprotein | O46605 | 18.07 | Cfa.14115.1.A1_at | NC | ✓ | |||
| ENSCAFP00000023923 | SERPINE2 | Glia-derived nexin precursor | P07093 | 17.93 | CfaAffx.24902.1.S1_s_at | 12.13 | ||||
| ENSCAFP00000027208 | ITGA2 | Integrin α2 precursor | P17301 | 17.29 | CfaAffx.28187.1.S1_at | 1.52 | ✓ | |||
| ENSCAFP00000002306 | ADAMTSL3 | ADAMTS-like protein 3 Precursor | A2A344 | 17.12 | CfaAffx.3277.1.S1_s_at | NC | ✓ | |||
| ENSCAFP00000003319 | GLIPR2 | Golgi-associated plant pathogenesis-related protein 1 | Q0VCH9 | 16.31 | CfaAffx.4299.1.S1_s_at | 25.99 | ||||
| ENSCAFP00000019213 | ITGA6 | Integrin α6 precursor | P23229 | 16.31 | Cfa.19343.1.S1_s_at | 4.29 | ✓ | |||
| ENSCAFP00000002677 | RscB4 | Multidrug resistance protein 3 (EC 3.6.3.44) | P21439 | 15.51 | CfaAffx.3656.1.S1_s_at | 2.14 | ✓ | |||
| ENSCAFP00000022384 | PLAU | Urokinase-type plasminogen activator (fragment) | Q8MHY7 | 14.70 | Cfa.127.1.S1_s_at | 4.92 | ✓ | |||
| ENSCAFP00000021176 | FN1 | Fibronectin (FN) (fragment) | P02751-15 | 13.48 | Cfa.3707.3.S1_s_at | 21.11 | ✓ | |||
| ENSCAFP00000023653 | EPHA2 | Ephrin type-A receptor 2 precursor | Q8N3Z2 | 12.15 | CfaAffx.24616.1.S1_s_at | 1.74 | ✓ | |||
| ENSCAFP00000012605 | DKFZP686E01144 | ADAMTS-1 precursor (EC 3.4.24.-) | Q5HYL0 | 11.95 | CfaAffx.13584.1.S1_s_at | 4.92 | ✓ | |||
| ENSCAFP00000021616 | ITGAV | Integrin αV precursor | P80746 | 11.76 | Cfa.18044.1.S1_at | 1.62 | ✓ | |||
| ENSCAFP00000002651 | PTK7 | Tyrosine-protein kinase-like 7 precursor | Q13308 | 11.48 | CfaAffx.3630.1.S1_s_at | NC | ✓ | |||
| ENSCAFP00000026360 | RAVER1 | Ribonucleoprotein PTB-binding 1 | Q8IY67-2 | 11.48 | CfaAffx.27339.1.S1_at | NC | ||||
| Proteins with decreased abundance following Ras transformation | ||||||||||
| ENSCAFP00000026847 | LAMA3 | Laminin subunit α3 precursor | Q6VU68 | −308.78 | Cfa.14.1.A1_at | −4.59 | ✓ | |||
| Q867A2 | LAMC2 | Laminin 5 γ2 | Q867A2 | −263.36 | Cfa.135.1.S1_at | −2.30 | ||||
| ENSCAFP00000017535 | LAMB3 | Laminin β3 (fragment) | Q13751 | −156.38 | CfaAffx.18514.1.S1_at | −6.06 | ✓ | |||
| P25473 | CLU | Clusterin precursor | P25473 | −95.11 | Cfa.1254.1.S1_s_at | NC | ||||
| ENSCAFP00000016115 | NPNT | Nephronectin precursor | Q91V88-4 | −63.13 | Cfa.17266.1.S1_s_at | −8.57 | ||||
| Q9XST1 | 3OST1 | Heparan sulfate 3-O-sulfotransferase-1 (fragment) | Q9XST1 | −41.59 | Cfa.3538.1.S1_s_at | −1.74 | ||||
| ENSCAFP00000003954 | COL12A1 | Collagen α-1(XII) chain precursor | Q99715 | −39.20 | CfaAffx.4940.1.S1_s_at | NC | ✓ | |||
| ENSCAFP00000019760 | ECHS1 | Enoyl-CoA hydratase, mitochondrial precursor (EC 4.2.1.17) | Q58DM8 | −38.40 | Cfa.15945.1.S1_at | −1.52 | ||||
| ENSCAFP00000014066 | DSP | Desmoplakin | P15924 | −29.64 | CfaAffx.15045.1.S1_at | −24.25 | ✓ | |||
| P13206 | EDN1 | Endothelin-1 precursor | P13206 | −28.84 | Cfa.125.1.S1_s_at | −119.43 | ||||
| ENSCAFP00000014095 | PCNP | PEST proteolytic signal-containing nuclear protein | Q32PF3 | −28.05 | Cfa.14096.1.S1_at | NC | ||||
| ENSCAFP00000003865 | EPCAM | Tumor-associated calcium signal transducer 1 precursor | Q3T0L5 | −27.66 | CfaAffx.4844.1.S1_at | −168.90 | ||||
| ENSCAFP00000007319 | ITGB4 | Integrin β4 precursor | P16144 | −26.45 | Cfa.6403.1.A1_at | −2.46 | ✓ | |||
| ENSCAFP00000007960 | EBNA1BP2 | Probable rRNA-processing protein EBP2 | Q3T0K6 | −24.86 | Cfa.3338.1.A1_s_at | NC | ||||
| Q29471-2 | ANXA13 | Annexin-13 Isoform B | Q29471-2 | −23.27 | Cfa.3796.1.A1_s_at | −24.25 | ||||
| P54714 | TPI1 | Triose-phosphate isomerase (EC 5.3.1.1) | P54714 | −22.47 | Cfa.6532.1.A1_at | NC | ||||
| ENSCAFP00000007234 | KCTD14 | BTB/POZ domain-containing protein KCTD14 | Q9BQ13 | −21.68 | CfaAffx.8213.1.S1_at | −22.63 | ✓ | |||
| Q95LE0 | CDH1 | E-cadherin (fragment) | Q95LE0 | −18.49 | Cfa.3488.1.S1_s_at | −78.79 | ||||
| ENSCAFP00000028105 | RPL17 | Rpl17 protein | Q6PHZ1 | −18.49 | CfaAffx.2842.1.S1_x_at | NC | ✓ | |||
| ENSCAFP00000033877 | RPL14 | 60 S ribosomal protein L14 | Q45RF0 | −16.90 | Cfa.1050.2.S1_s_at | 1.23 | ✓ | |||
| ENSCAFP00000025082 | MUC1 | Endometrial mucin-1 (fragment) | B1AVQ5 | −16.11 | CfaAffx.26061.1.S1_at | −12.13 | ||||
| ENSCAFP00000022387 | MRTO4 | mRNA turnover protein 4 homolog | Q9UKD2 | −16.11 | CfaAffx.23366.1.S1_s_at | NC | ||||
| ENSCAFP00000015542 | SNRPA1 | U2 small nuclear ribonucleoprotein A′ | P09661 | −16.11 | CfaAffx.16521.1.S1_at | NC | ||||
| ENSCAFP00000010012 | FLT1 | VEGFR-1 receptor (fragment) | P17948 | −15.31 | Cfa.19323.1.S1_s_at | −3.48 | ✓ | |||
| ENSCAFP00000010578 | BMS1 | Ribosome biogenesis protein BMS1 homolog | Q14692 | −14.51 | CfaAffx.11557.1.S1_s_at | −1.62 | ||||
| ENSCAFP00000023638 | TNS4 | Tensin-4 precursor (C-terminal tensin-like protein) | Q32PJ7 | −14.51 | CfaAffx.24618.1.S1_s_at | −2.83 | ||||
| ENSCAFP00000002355 | RPS6 | 40 S ribosomal protein S6 | Q8BT09 | −13.72 | Cfa.10190.1.S1_s_at | 1.23 | ✓ | |||
| ENSCAFP00000002431 | RBM28 | RNA-binding protein 28 | Q9NW13 | −13.72 | Cfa.3335.1.A1_at | −1.32 | ||||
| ENSCAFP00000028676 | DCI | 3,2-trans-Enoyl-CoA isomerase, mitochondrial precursor | P42126 | −13.72 | CfaAffx.29655.1.S1_at | −1.87 | ||||
| ENSCAFP00000019692 | SLC16A1 | Monocarboxylate transporter 1 (fragment) | P53985 | −12.92 | CfaAffx.20671.1.S1_at | −2.46 | ✓ | |||
| ENSCAFP00000002516 | RPL7L1 | Ribosomal protein L7-like 1 | Q6DKI1 | −12.92 | Cfa.18852.1.S1_s_at | −1.15 | ✓ | |||
| ENSCAFP00000011930 | HELLS | Lymphoid-specific helicase (EC 3.6.1.-) | Q9NRZ9 | −12.13 | CfaAffx.12909.1.S1_at | −3.25 | ||||
| ENSCAFP00000024805 | RAB25 | Ras-related protein Rab-25 | P57735 | −11.33 | CfaAffx.25784.1.S1_at | −55.72 | ||||
| ENSCAFP00000004160 | TMPRSS9 | Transmembrane protease, serine 9 (EC 3.4.21.-) | P69526 | −11.33 | CfaAffx.5135.1.S1_at | NC | ✓ | |||
| ENSCAFP00000004990 | FAM136A | Protein FAM136A | Q96C01 | −11.33 | CfaAffx.5969.1.S1_s_at | −1.74 | ||||
| ENSCAFP00000000121 | CDK2 | Cell division protein kinase 2; cyclin-dependent kinase (EC 2.7.11.22) | Q6P751 | −11.33 | CfaAffx.1100.1.S1_at | NC | ||||
a Protein accession numbers obtained from the Ensembl (http://www.ensembl.org/index.html) and UniProt (http://www.uniprot.org/) databases.
b Gene symbols and protein descriptions obtained from the Ensembl and UniProt databases.
c UniProt homolog obtained from the Ensembl database. When unavailable, the protein sequence was BLAST (basic local alignment search tool)-searched for the UniProt homolog.
d Relative protein spectral count ratios reflecting protein abundance (see Equation 1 under “Experimental Procedures”).
e Probe identifiers obtained from the Ensembl database.
f -Fold change differences calculated using Affymetrix GeneChip Operating Software (http://www.affymetrix.com). NC, no change.
g Gene ontology (GO Slim) annotation obtained from the Protein Information Resource (http://pir.georgetown.edu/) with classifiers (GO:0004871, GO:0005198, GO:0008233, and GO:0005215).
mRNA Transcript and Protein Expression Levels Are Concordantly Regulated during EMT
mRNA expression analysis was performed to ascertain the correlation between transcript and protein dysregulation during Ras/TGF-β-induced EMT. Selected probe information was extracted from the complete microarray data set (supplemental Table S6), and correlation analysis was performed. Of the significantly dysregulated proteins listed in Table I, 68% of the proteins exhibited consistent mRNA transcript regulation. Consistent with its elevated protein expression, Wnt-5a was found to have a 45-fold increase in mRNA expression in 21D1 cells relative to MDCK cells. In addition to confirming proteomics findings, we mined the microarray data sets to gain further insights into aspects of the EMT process that may explain changes in cell behavior. For example, membrane profiling revealed up-regulation of Wnt-5a, whereas microarray analysis shed light on which downstream signaling pathways may be active (see Wnt-5a section below).
Integrins and Extracellular Ligands Are Regulated Concomitantly during EMT
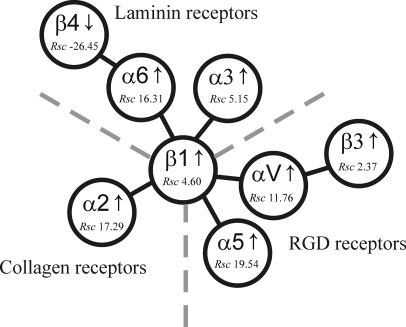
One striking feature identified in our PM proteomics study was that cells undergoing Ras/TGF-β EMT switch from cell-cell (e.g. E-cadherin) to cell-matrix adhesion as evidenced by increased integrin expression. Integrins are a large family of cell surface adhesion receptors that mediate interactions between glycoprotein ligands in the extracellular matrix (ECM) or on neighboring cells (52). The integrin family comprises 24 distinct heterodimers formed by the association of 18 α-subunits with eight β-subunits (53). It is currently thought that each integrin has a specific, non-redundant function that regulates many aspects of cell behavior, including cell adhesion, signaling, proliferation, differentiation, and migration (52). Integrins consist of a large extracellular domain, a single transmembrane domain, and typically a small cytoplasmic domain (52). The extracellular domains recognize the RGD recognition sequence in collagen, laminin, and ECM proteins (54), whereas the intracellular domain anchors to the cytoskeleton through linker proteins such as talin, α-actinin, vinculin, and paxillin. These intracellular links provide mechanical continuity between the outside and inside of the cell (55). Although crystal structure determination and null mutation studies in mice have provided insights into the diverse functions of integrins, the precise role of each individual member is still to be defined (56, 57). Of the eight integrin subunits identified in this study, α2, α3, α5, α6, αV, β1, and β3 (with the exception of β4) were found to have elevated expression levels in 21D1 cells (supplemental Table S1) and can complex together to form seven individual integrins (Fig. 3). It is interesting to note that for a subset of integrin extracellular ligands, including laminin (integrin α6β4), collagen (integrin α2β1), and fibronectin (integrins α5β1 and αVβ3), changes in expression are shown to correlate with that of its corresponding integrin receptor (Fig. 3). Reduced expression levels of integrin α6β4 were observed in 21D1 cells with virtually no detectable deposition of its ligand (laminin 5), and no peptide spectral counts were observed for either the α3 and γ2 chains of laminin 5, whereas only two peptide spectral counts were detected for the β3 chain.
Fig. 3.
Integrin regulation during Ras/TGF-β-mediated EMT. Integrin α6β4, which binds laminin 5, was down-regulated during EMT, whereas other laminin-binding integrins, including α6β1 and α3β1, had elevated expression levels. The major collagen-binding integrin, α2β1, was up-regulated during the EMT process. Increased expression levels of integrin RGD receptors that bind fibronectin and vitronectin, namely integrins α5β1, αVβ1, and αVβ3, were detected during plasma membrane proteomics profiling. Rsc values indicate relative abundance based on label-free spectral counting (see “Experimental Procedures”). Figure was adapted from Hynes (52).
Integrin regulation detected in our study concurs with that reported in other EMT studies. For example, up-regulation of α6β1 was recently reported by Colomiere et al. (58) in ovarian cancer cells; it promotes EMT cell migration via activation of the JAK2/STAT3 pathway. Furthermore, elevated integrin α3β1 observed in our study is consistent with increased expression levels reported in lung epithelial cells undergoing EMT where E-cadherin internalization and translocation of phosphorylated Smad2 and β-catenin initiates mesenchymal gene transcription (59). Our PM profiling data are also consistent with previous studies suggesting that cells undergoing EMT induce expression of integrins that interact with the ECM to facilitate cell movement and motility (60). The major collagen-binding integrin, α2β1, was significantly up-regulated in our study (Fig. 3), and this may enhance the interaction between fibrillar collagens including collagens I and V (53). Both collagens I (Rsc, 28.41) and V (Rsc, 42.94) were found to have increased expression following Ras/TGF-β-mediated EMT (supplemental Table S1). Although collagen I is currently regarded as an EMT marker (61–63), its precise involvement in EMT is unknown. However, collagen expression has been reported to induce integrin α2β1, which in turn facilitates cell scattering and increases the speed of cell locomotion (64).
The major fibronectin receptors, integrins α5β1 and αVβ3, exhibited elevated expression levels during the MDCK cell EMT process (supplemental Table S1). This result is in agreement with oncogenic Ras/TGF-β EMT studies in EpH4 cells (60) where integrins α5β1 and αVβ3 were found to be involved in cell migration and cell signaling (65). In addition, the well known EMT marker fibronectin (49, 66) was found to be up-regulated (Rsc, 13.48) in 21D1 cells (67). The contribution of the integrin α5β1 and fibronectin interaction during cell migration is exemplified in a study by White et al. (68) where tubular epithelial cells undergoing EMT demonstrate enhanced motility when cultured on fibronectin; when the α5 subunit was knocked down using siRNA, cell movement was attenuated. Likewise, the expression of the other fibronectin-binding integrin, αVβ3, found to be up-regulated in our study has been shown to increase in mammary epithelial cells undergoing TGF-β-induced EMT. In this case, it complexes with the TGF-β receptor to enhance cell invasion via MAPK- and Smad2-mediated gene transcription (69).
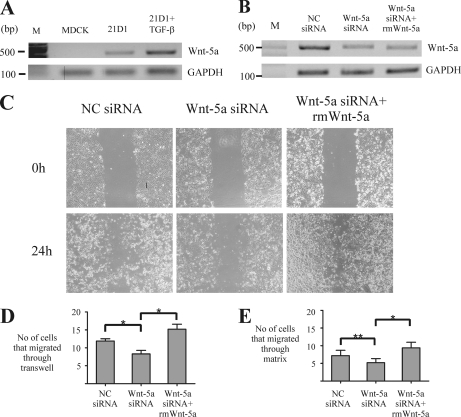
Wnt-5a Is Up-regulated in MDCK Cells Undergoing Ras/TGF-β-mediated EMT
A salient finding of the proteomics PM profiling study was the up-regulation of Wnt-5a during Ras/TGF-β-mediated EMT (Rsc, 158.08; microarray, 45-fold increase). To determine whether Ras or TGF-β induces Wnt-5a expression, RNA from MDCK, 21D1, and 21D1 + TGF-β cells was extracted and analyzed using semiquantitative RT-PCR (Fig. 4A). These data suggest that Wnt-5a expression is induced by Ras, and levels are further increased by TGF-β, which is consistent with the report that both molecules work synergistically to promote EMT (8). As some of the molecular mechanisms that regulate Wnt-5a expression have been revealed, we mined our microarray data set to see whether any known Wnt-5a transcription factors (TFs) had increased expression in 21D1 cells. However, we found no change in the transcriptional levels of STAT3, CUX1/CUTL1, NF-κB, or any of the FOX family members of TFs. Only SMAD-4 and -5 were up-regulated in our study (1.4- and 1.9-fold, respectively) (supplemental Table S6). Thus, we wanted to identify novel TFs that may induce Wnt-5a expression, so we performed a bioinformatics analysis using the WNT5A promoter region to identify potential TF binding sites and compared this with their expression in our microarray data set. Eleven TFs from this data set were up-regulated at the mRNA transcript level and might contribute to Wnt-5a expression in our Ras/TGF-β-mediated EMT model (Table II). An inspection of our microarray data set revealed that early B-cell factor 1/transcription factor COE1, transcription factor 7-like 2, and sex determining region Y-box 5/transcription factor SOX5 were among the TF genes most up-regulated in 21D1 cells. Interestingly, transcription factor 7-like 2, commonly known as TCF4, is known to play a key role in the Wnt signaling pathway, whereas nuclear factor of activated T-cells 5 and SOX5 are involved in cell migration and invasion (70–72). It is possible that these TFs induce the expression of mesenchymal genes, including WNT5A, to stimulate cell motility and invasiveness during Ras/TGF-β-mediated EMT.
Fig. 4.
siRNA silencing of Wnt-5a attenuates 21D1 cell migration and invasion. A, transcript expression of Wnt-5a was not detected in MDCK cells by semiquantitative RT-PCR; however, transformation with H-Ras and stimulation with TGF-β induces Wnt-5a expression. B, expression of Wnt-5a in 21D1 cells is attenuated by RNA interference using siRNA duplexes targeting Wnt-5a. The addition of scrambled negative control (NC) siRNA did not affect Wnt-5a expression. Expression of Wnt-5a was reduced in 21D1 cells using transient siRNA transfection, and cell migration was assessed using the wound healing (C) and Transwell migration (D) assays. Both assays show that cell migration is significantly impeded when expression of Wnt-5a is reduced. However, cell migration is restored and even elevated with the addition of rm Wnt-5a (n = 2). E, the ability of 21D1 cells to invade through the matrix is decreased when expression of Wnt-5a is reduced using siRNA duplexes, although this reduction is not statistically significant (**). The invasive nature of 21D1 cells is rescued with the addition of recombinant Wnt-5a (n = 2). Error bars represent mean ± S.D. Significance is determined by a p value ≤0.05 using the Student's t test.
Table II. Transcription factors up-regulated during EMT that are predicted to bind promoter of Wnt-5a.
HMG, high mobility group.
| Probe IDa | Gene symbolb | Gene nameb | Microarray FCc |
|---|---|---|---|
| Cfa.12330.1.A1_s_at | EBF1 | Early B-cell factor 1 | 32 |
| Cfa.3808.1.S1_s_at | TCF7L2 | Transcription factor 7-like 2 (T-cell-specific, HMG box) | 29.86 |
| CfaAffx.18082.1.S1_s_at | SOX5 | SRY (sex determining region Y) box 5 | 8 |
| CfaAffx.28854.1.S1_s_at | JUN | jun oncogene | 3.73 |
| Cfa.11535.1.A1_at | MZF1 | Myeloid zinc finger 1 | 2.64 |
| CfaAffx.2790.1.S1_s_at | EGR1 | Early growth response 1 | 2.3 |
| Cfa.10604.1.S1_at | GABPA | GA-binding protein transcription factor, α-subunit, 60 kDa | 2 |
| Cfa.19790.1.S1_at | NFAT5 | Nuclear factor of activated T-cells 5, tonicity-responsive | 1.87 |
| CfaAffx.23894.1.S1_at | STAT5B | Signal transducer and activator of transcription 5B | 1.52 |
| Cfa.174.1.S1_s_at | TFAP2B | Transcription factor AP-2β (activating enhancer-binding protein 2β) | 1.52 |
| Cfa.1648.1.S1_at | SMAD4 | SMAD family member 4 | 1.41 |
a Probe identifiers obtained from the Ensembl database.
b Gene symbol and gene name descriptions obtained from the Ensembl and UniProt databases.
c -Fold change differences calculated using Affymetrix GeneChip Operating Software (http://www.affymetrix.com).
Wnt-5a Enhances Cell Migration and Invasion during EMT
As Wnt-5a was the most up-regulated protein in the PM profiling study, we evaluated its involvement in 21D1 cell migration and invasion using siRNA-based RNA interference. Three siRNA duplexes targeting Wnt-5a were designed using the BLOCK-iTTM RNAi Designer (Invitrogen). 100 pmol of each duplex was transfected into 21D1 cells, Wnt-5a expression was measured using RT-PCR, and the concentration of the most effective duplex required to silence Wnt-5a expression was optimized (data not shown). 21D1 cells were transiently transfected with 250 pmol of either Wnt-5a siRNA or a scrambled negative control, and the expression of Wnt-5a was analyzed by semiquantitative RT-PCR (Fig. 4B). An approximately 2-fold reduction in Wnt-5a expression levels was observed in 21D1 cells following Wnt-5a silencing compared with cells transfected with the negative control (Fig. 4B). Wnt-5a expression could not be entirely abolished, i.e. restored to the levels in MDCK cells (Fig. 4A), as 21D1 cell transfection efficiency is only 70% (28), and higher amounts of transfection reagent were toxic for the cells. Thus, 250 pmol of Wnt-5a siRNA was transfected into 21D1 cells, and migration and invasion assays were performed (Fig. 4, C–E). Reduced expression levels of Wnt-5a impaired 21D1 cell motility in both migration assays with fewer cells migrating into the wounded area in the wound healing assay and through the Transwell filter in the Transwell migration assay (Fig. 4, C and D). In addition, the Transwell invasion assay revealed that the enhanced invasiveness of 21D1 cells was attenuated by knocking down expression of Wnt-5a with fewer cells able to penetrate through the matrix compared with the negative control (Fig. 4E), although this reduction was not statistically significant (p = 0.38). Noticeably, the migration and invasion capability of 21D1 cells was restored via the addition of recombinant Wnt-5a protein to the cell culture medium (Fig. 4, C–E). Although only a 2-fold knockdown of Wnt-5a was achieved using transient siRNA silencing, this reduction was sufficient to impair cell migration and invasion. These functional data unequivocally demonstrate that Wnt-5a is an important promoter of the EMT process (73). Furthermore, we suspect that increasing the level of Wnt-5a knockdown using either a combination of Wnt-5a siRNAs or Wnt-5a shRNAs would further revert the cells toward an MDCK/epithelial phenotype.
Wnt-5a Represses Canonical Wnt Signaling following Ras/TGF-β-mediated EMT
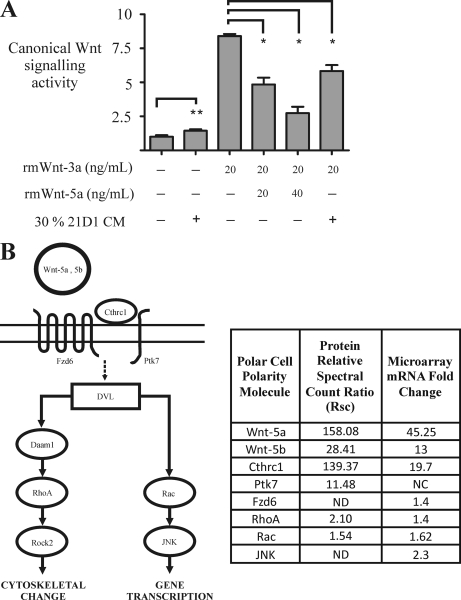
The Wnt family of secreted glycoproteins comprises 19 members that regulate proliferation, migration, polarity, and differentiation during embryogenesis (74). Several members, including Wnt-1, Wnt-2, and Wnt-3a, signal through the canonical Wnt pathway through GSK3β and activate β-catenin/TCF to induce gene transcription (74). In contrast, other Wnts, including Wnt-5a, Wnt-5b, and Wnt-11, activate the non-canonical Wnt pathways through the calcium-dependent/PKC or planar cell polarity (PCP)/JNK signaling pathways (75). Involvement of both canonical and non-canonical Wnt signaling pathways has been reported during EMT (7, 9, 76–78), so we wanted to establish which one was active in our cell model. Using the luciferase gene reporter assay (see “Experimental Procedures”), we examined canonical Wnt signaling in 21D1 cells using recombinant mouse (rm) Wnts and culture medium conditioned by 21D1 cells (Fig. 5A). When canonical Wnt signaling was stimulated in 21D1 cells by the addition of 20 ng/ml rm Wnt-3a, TCF/LEF gene reporter activity increased 8-fold. However, when 20 ng/ml rm Wnt-5a was added together with 20 ng/ml rm Wnt-3a, canonical Wnt activity decreased by 40%. It appears that the suppression of canonical Wnt signaling is dose-dependent as 40 ng/ml rm Wnt5-a reduced activity by 80%. Importantly, when 30% conditioned medium from 21D1 cells was added to 21D1 cells stimulated with 20 ng/ml rm Wnt-3a, canonical Wnt signaling activity was reduced by 25%. These results suggest that 21D1 cells may secrete Wnt-5a-like factors to repress canonical Wnt signaling during Ras/TGF-β-mediated EMT. Topol et al. (79) have previously demonstrated that Wnt-5a signaling antagonizes the canonical Wnt signaling pathway by promoting β-catenin degradation. In that case, the process is independent of GSK3β phosphorylation, and Siah2 ubiquitinates β-catenin for subsequent proteasomal degradation (80). Precisely how Wnt-5a represses canonical Wnt-3a signaling in this our model remains unknown.
Fig. 5.
Wnt-5a expression during EMT represses canonical Wnt signaling. A, canonical Wnt signaling activity was examined using the T-cell factor-luciferase reporter assay under different culture conditions. 21D1 cells cultured in 30% conditioned medium (CM) showed no significant change (**) in canonical Wnt signaling activity. Addition of rm Wnt-3a stimulates canonical Wnt signaling; however, this activity is significantly (*) repressed following the addition of 20 ng/ml and to a greater extent by 40 ng/ml recombinant Wnt-5a. In a similar fashion, addition of 30% conditioned medium from 21D1 cells significantly attenuates canonical Wnt signaling (n = 2). Error bars represent mean ± S.D. Significance is determined by a p value ≤0.05 using the Student's t test. B, the non-canonical Wnt signaling PCP pathway may be activated during Ras/TGF-β-induced EMT. Several upstream components, core components, downstream effectors, and PCP modulators were revealed to be up-regulated in our integrated proteomics and transcriptomics EMT studies. Cthrc1, collagen triple helix repeat-containing protein 1; Fzd6, frizzled 6; Ptk7, tyrosine-protein kinase 7; DVL, dishevelled; ND, not detected; NC, no change. Pathway interactions are based on information obtained from the Kyoto Encyclopedia of Genes and Genomes database (http://www.genome.jp/kegg/pathway.html).
Wnt-5a May Promote EMT via Non-canonical Planar Cell Polarity Wnt Signaling Pathway
Non-canonical Wnt signaling encompasses several Wnt-activated cellular pathways that do not promote β-catenin-mediated transcription. These include the calcium-dependent/PKC, Siah2/APC, and PCP/JNK pathways (73, 81, 82). Although calcium-dependent signaling leads to increased intracellular calcium levels and activates downstream effectors, including PKC, the PCP/JNK pathway is activated through the seven-transmembrane frizzled receptors, which in turn activate a suite of downstream signal transducers, including Daam1, Rho, Rac, Rho kinase, JNK, and profilin (83). PKC signaling has been reported to enhance EMT in melanoma (76); however, PCP signaling is yet to be described in the context of EMT, although it has been found to promote gastric cancer progression (77). A major finding of our proteomics and microarray studies was the up-regulation of several known components and modulators of PCP signaling, strongly suggesting the activation of this pathway during Ras/TGF-β-mediated EMT (Fig. 5B and supplemental Tables S1 and S6). For example, the extracellular non-canonical Wnt ligands Wnt-5a and Wnt-5b were significantly up-regulated at both the mRNA transcript and protein levels (Fig. 5B). The transmembrane receptor frizzled 6 was up-regulated 1.4-fold in the microarray analysis, and the co-receptor tyrosine-protein kinase 7 had an Rsc value of 11.48 (Fig. 5B). In addition, the secreted glycoprotein collagen triple helix repeat-containing protein 1 (Cthrc1), a known modulator/co-receptor of the PCP pathway (83), was significantly up-regulated at both the protein and mRNA transcript levels. Cthrc1 stabilizes the Wnt ligand/frizzled receptor interaction and specifically activates PCP signaling via the downstream effectors RhoA and Rac (84). In our study, RhoA, Rac, and JNK were also found to have elevated expression levels during Ras/TGF-β-mediated EMT. Delineation of the precise PCP Wnt signaling mechanism awaits further experimentation.
Concluding Remarks
Membrane protein receptors on epithelial cells mediate cell-cell and cell-matrix contacts to maintain epithelial polarity, structure, and function. Plasma membrane proteomics profiling indicates that MDCK cells undergoing Ras/TGF-β-mediated EMT switch from cadherin- to integrin-mediated adhesion and co-regulate the expression of their extracellular ligands. Expression of Wnt-5a is up-regulated during EMT, resulting in enhanced cell migration and invasion as confirmed by transient siRNA-based silencing. Wnt-5a expression represses canonical Wnt signaling and may promote Ras/TGF-β-mediated EMT through activation of the planar cell polarity pathway of the non-canonical Wnt signaling pathway.
Acknowledgments
Analysis of proteomics data described in this work was supported using the Australian Proteomics Computational Facility, which is funded by National Health and Medical Research Council of Australia Grant 381413. We thank Bo Wang for providing access to the MDCK and 21D1 cells lines, Dr. Randall Moon for providing the Super TopFlash (STF) plasmid, Robert Goode for assistance with membrane purification, and Donna Dorow for critical reading of the manuscript.
Footnotes
* This work was supported by National Health and Medical Research Council of Australia Program Grant 487922 (to R. J. S.) and Grants 280913 and 433619 (to H.-J. Z.), The Australian Government Endeavor International Postgraduate Research Scholarship and The University of Melbourne International Research Scholarship (to Y.-S. C.), and The University of Melbourne Research Scholarship (to R. A. M.).
 This article contains supplemental Fig. S1 and Tables S1–S6.
This article contains supplemental Fig. S1 and Tables S1–S6.
2 E. A. Kapp and C. Iseli, unpublished data.
1 The abbreviations used are:
- EMT
- epithelial-mesenchymal transition
- MDCK
- Madin-Darby canine kidney
- TJ
- tight junction
- AJ
- adherens junction
- CCS
- cationic colloidal silica
- PCP
- planar cell polarity
- Rsc
- relative spectral count -fold change ratio
- ZO
- zonula occludin
- TCF
- T-cell factor
- Ad
- adherent
- nAd
- non-adherent
- ECM
- extracellular matrix
- PM
- plasma membrane
- rm
- recombinant mouse
- TF
- transcription factor
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- MGF
- Mascot generic format
- LEF
- lymphoid enhancer factor
- ADAMTS
- A disintegrin and metalloproteinase with trombospondin motifs
- EM
- epis homology
- PTB
- polypyrimidine tract binding
- PEST
- protein domains rich in protein, glutamic acid, aspartic acid, serine or threonine
- BTB/POZ
- bric-a-brac, tramtrack, broad-complex, pox, poxvirus zinc finger
- SMAD
- mother is aganist decapentaplegic homolog
- APC
- adenomatous polyposis coli.
REFERENCES
- 1. Kang Y., Massagué J. (2004) Epithelial-mesenchymal transitions: twist in development and metastasis. Cell 118, 277–279 [DOI] [PubMed] [Google Scholar]
- 2. Thiery J. P. (2002) Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2, 442–454 [DOI] [PubMed] [Google Scholar]
- 3. Thiery J. P. (2003) Epithelial-mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 15, 740–746 [DOI] [PubMed] [Google Scholar]
- 4. Hugo H., Ackland M. L., Blick T., Lawrence M. G., Clements J. A., Williams E. D., Thompson E. W. (2007) Epithelial-mesenchymal and mesenchymal-epithelial transitions in carcinoma progression. J. Cell. Physiol. 213, 374–383 [DOI] [PubMed] [Google Scholar]
- 5. Bailey J. M., Singh P. K., Hollingsworth M. A. (2007) Cancer metastasis facilitated by developmental pathways: sonic hedgehog, Notch, and bone morphogenic proteins. J. Cell. Biochem. 102, 829–839 [DOI] [PubMed] [Google Scholar]
- 6. Thiery J. P., Sleeman J. P. (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 7, 131–142 [DOI] [PubMed] [Google Scholar]
- 7. Yang J., Weinberg R. A. (2008) Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell 14, 818–829 [DOI] [PubMed] [Google Scholar]
- 8. Oft M., Peli J., Rudaz C., Schwarz H., Beug H., Reichmann E. (1996) TGF-beta1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev. 10, 2462–2477 [DOI] [PubMed] [Google Scholar]
- 9. Huber M. A., Kraut N., Beug H. (2005) Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr. Opin. Cell Biol. 17, 548–558 [DOI] [PubMed] [Google Scholar]
- 10. Boyer B., Vallés A. M., Edme N. (2000) Induction and regulation of epithelial-mesenchymal transitions. Biochem. Pharmacol. 60, 1091–1099 [DOI] [PubMed] [Google Scholar]
- 11. Fujimoto K., Sheng H., Shao J., Beauchamp R. D. (2001) Transforming growth factor-beta1 promotes invasiveness after cellular transformation with activated Ras in intestinal epithelial cells. Exp. Cell Res. 266, 239–249 [DOI] [PubMed] [Google Scholar]
- 12. Janda E., Lehmann K., Killisch I., Jechlinger M., Herzig M., Downward J., Beug H., Grünert S. (2002) Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J. Cell Biol. 156, 299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Campanaro S., Picelli S., Torregrossa R., Colluto L., Ceol M., Del Prete D., D'Angelo A., Valle G., Anglani F. (2007) Genes involved in TGF beta1-driven epithelial-mesenchymal transition of renal epithelial cells are topologically related in the human interactome map. BMC Genomics 8, 383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Derynck R., Akhurst R. J., Balmain A. (2001) TGF-beta signaling in tumor suppression and cancer progression. Nat. Genet. 29, 117–129 [DOI] [PubMed] [Google Scholar]
- 15. Wakefield L. M., Roberts A. B. (2002) TGF-beta signaling: positive and negative effects on tumorigenesis. Curr. Opin. Genet. Dev. 12, 22–29 [DOI] [PubMed] [Google Scholar]
- 16. Perez-Moreno M., Jamora C., Fuchs E. (2003) Sticky business: orchestrating cellular signals at adherens junctions. Cell 112, 535–548 [DOI] [PubMed] [Google Scholar]
- 17. Lee D. B., Huang E., Ward H. J. (2006) Tight junction biology and kidney dysfunction. Am. J. Physiol. Renal Physiol. 290, F20–F34 [DOI] [PubMed] [Google Scholar]
- 18. Ebnet K., Suzuki A., Ohno S., Vestweber D. (2004) Junctional adhesion molecules (JAMs): more molecules with dual functions? J. Cell Sci. 117, 19–29 [DOI] [PubMed] [Google Scholar]
- 19. McNeil E., Capaldo C. T., Macara I. G. (2006) Zonula occludens-1 function in the assembly of tight junctions in Madin-Darby canine kidney epithelial cells. Mol. Biol. Cell 17, 1922–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chidgey M., Dawson C. (2007) Desmosomes: a role in cancer? Br. J. Cancer 96, 1783–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holthöfer B., Windoffer R., Troyanovsky S., Leube R. E. (2007) Structure and function of desmosomes. Int. Rev. Cytol. 264, 65–163 [DOI] [PubMed] [Google Scholar]
- 22. Peinado H., Olmeda D., Cano A. (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 23. Rabilloud T. (2003) Membrane proteins ride shotgun. Nat. Biotechnol. 21, 508–510 [DOI] [PubMed] [Google Scholar]
- 24. Yates J. R., 3rd, Gilchrist A., Howell K. E., Bergeron J. J. (2005) Proteomics of organelles and large cellular structures. Nat. Rev. Mol. Cell Biol. 6, 702–714 [DOI] [PubMed] [Google Scholar]
- 25. Macher B. A., Yen T. Y. (2007) Proteins at membrane surfaces-a review of approaches. Mol. Biosyst. 3, 705–713 [DOI] [PubMed] [Google Scholar]
- 26. Chaney L. K., Jacobson B. S. (1983) Coating cells with colloidal silica for high yield isolation of plasma membrane sheets and identification of transmembrane proteins. J. Biol. Chem. 258, 10062–10072 [PubMed] [Google Scholar]
- 27. Mathias R. A., Wang B., Ji H., Kapp E. A., Moritz R. L., Zhu H. J., Simpson R. J. (2009) Secretome-based proteomic profiling of Ras-transformed MDCK cells reveals extracellular modulators of epithelial-mesenchymal transition. J. Proteome Res. 8, 2827–2837 [DOI] [PubMed] [Google Scholar]
- 28. Mathias R. A., Chen Y. S., Wang B., Ji H., Kapp E. A., Moritz R. L., Zhu H. J., Simpson R. J. (2010) Extracellular remodelling during oncogenic Ras-induced epithelial-mesenchymal transition facilitates MDCK cell migration. J. Proteome Res. 9, 1007–1019 [DOI] [PubMed] [Google Scholar]
- 29. Liang C. C., Park A. Y., Guan J. L. (2007) In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2, 329–333 [DOI] [PubMed] [Google Scholar]
- 30. Albini A., Iwamoto Y., Kleinman H. K., Martin G. R., Aaronson S. A., Kozlowski J. M., McEwan R. N. (1987) A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 47, 3239–3245 [PubMed] [Google Scholar]
- 31. Goode R. J., Simpson R. J. (2009) Purification of basolateral integral membrane proteins by cationic colloidal silica-based apical membrane subtraction. Methods Mol. Biol. 528, 177–187 [DOI] [PubMed] [Google Scholar]
- 32. Moritz R. L., Eddes J. S., Reid G. E., Simpson R. J. (1996) S-Pyridylethylation of intact polyacrylamide gels and in situ digestion of electrophoretically separated proteins: a rapid mass spectrometric method for identifying cysteine-containing peptides. Electrophoresis 17, 907–917 [DOI] [PubMed] [Google Scholar]
- 33. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 34. Perkins D. N., Pappin D. J., Creasy D. M., Cottrell J. S. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 [DOI] [PubMed] [Google Scholar]
- 35. Greening D. W., Glenister K. M., Kapp E. A., Moritz R. L., Sparrow R. L., Lynch G. W., Simpson R. J. (2008) Comparison of human platelet membrane-cytoskeletal proteins with the plasma proteome: towards understanding the platelet-plasma nexus. Proteomics Clin. Appl. 2, 63–77 [DOI] [PubMed] [Google Scholar]
- 36. Kapp E. A., Schütz F., Connolly L. M., Chakel J. A., Meza J. E., Miller C. A., Fenyo D., Eng J. K., Adkins J. N., Omenn G. S., Simpson R. J. (2005) An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: sensitivity and specificity analysis. Proteomics 5, 3475–3490 [DOI] [PubMed] [Google Scholar]
- 37. Sonnhammer E. L., von Heijne G., Krogh A. (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6, 175–182 [PubMed] [Google Scholar]
- 38. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 39. Beissbarth T., Hyde L., Smyth G. K., Job C., Boon W. M., Tan S. S., Scott H. S., Speed T. P. (2004) Statistical modeling of sequencing errors in SAGE libraries. Bioinformatics 20, Suppl. 1, i31–i39 [DOI] [PubMed] [Google Scholar]
- 40. Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 41. Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 42. Sayers E. W., Barrett T., Benson D. A., Bolton E., Bryant S. H., Canese K., Chetvernin V., Church D. M., Dicuccio M., Federhen S., Feolo M., Geer L. Y., Helmberg W., Kapustin Y., Landsman D., Lipman D. J., Lu Z., Madden T. L., Madej T., Maglott D. R., Marchler-Bauer A., Miller V., Mizrachi I., Ostell J., Panchenko A., Pruitt K. D., Schuler G. D., Sequeira E., Sherry S. T., Shumway M., Sirotkin K., Slotta D., Souvorov A., Starchenko G., Tatusova T. A., Wagner L., Wang Y., John Wilbur W., Yaschenko E., Ye J. (2010) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 38, D5–D16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Messeguer X., Escudero R., Farré D., Núñez O., Martínez J., Albà M. M. (2002) PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18, 333–334 [DOI] [PubMed] [Google Scholar]
- 44. Kaykas A., Yang-Snyder J., Héroux M., Shah K. V., Bouvier M., Moon R. T. (2004) Mutant Frizzled 4 associated with vitreoretinopathy traps wild-type Frizzled in the endoplasmic reticulum by oligomerization. Nat. Cell Biol. 6, 52–58 [DOI] [PubMed] [Google Scholar]
- 45. Safina A. F., Varga A. E., Bianchi A., Zheng Q., Kunnev D., Liang P., Bakin A. V. (2009) Ras alters epithelial-mesenchymal transition in response to TGFbeta by reducing actin fibers and cell-matrix adhesion. Cell Cycle 8, 284–298 [DOI] [PubMed] [Google Scholar]
- 46. Gotzmann J., Mikula M., Eger A., Schulte-Hermann R., Foisner R., Beug H., Mikulits W. (2004) Molecular aspects of epithelial cell plasticity: implications for local tumor invasion and metastasis. Mutat. Res. 566, 9–20 [DOI] [PubMed] [Google Scholar]
- 47. Jakowlew S. B. (2006) Transforming growth factor-beta in cancer and metastasis. Cancer Metastasis Rev. 25, 435–457 [DOI] [PubMed] [Google Scholar]
- 48. Simpson R. J., Connolly L. M., Eddes J. S., Pereira J. J., Moritz R. L., Reid G. E. (2000) Proteomic analysis of the human colon carcinoma cell line (LIM 1215): development of a membrane protein database. Electrophoresis 21, 1707–1732 [DOI] [PubMed] [Google Scholar]
- 49. Lee J. M., Dedhar S., Kalluri R., Thompson E. W. (2006) The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J. Cell Biol. 172, 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kurrey N. K., K A., Bapat S. A. (2005) Snail and Slug are major determinants of ovarian cancer invasiveness at the transcription level. Gynecol. Oncol. 97, 155–165 [DOI] [PubMed] [Google Scholar]
- 51. Severson E. A., Parkos C. A. (2009) Structural determinants of junctional adhesion molecule A (JAM-A) function and mechanisms of intracellular signaling. Curr. Opin. Cell Biol. 21, 701–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hynes R. O. (2002) Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 53. Humphries J. D., Byron A., Humphries M. J. (2006) Integrin ligands at a glance. J. Cell Sci. 119, 3901–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ruoslahti E. (1996) RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 12, 697–715 [DOI] [PubMed] [Google Scholar]
- 55. Bosman F. T., Stamenkovic I. (2003) Functional structure and composition of the extracellular matrix. J. Pathol. 200, 423–428 [DOI] [PubMed] [Google Scholar]
- 56. Xiong J. P., Goodman S. L., Arnaout M. A. (2007) Purification, analysis, and crystal structure of integrins. Methods Enzymol. 426, 307–336 [DOI] [PubMed] [Google Scholar]
- 57. Chen C., Sheppard D. (2007) Identification and molecular characterization of multiple phenotypes in integrin knockout mice. Methods Enzymol. 426, 291–305 [DOI] [PubMed] [Google Scholar]
- 58. Colomiere M., Findlay J., Ackland L., Ahmed N. (2009) Epidermal growth factor-induced ovarian carcinoma cell migration is associated with JAK2/STAT3 signals and changes in the abundance and localization of alpha6beta1 integrin. Int. J. Biochem. Cell Biol. 41, 1034–1045 [DOI] [PubMed] [Google Scholar]
- 59. Kim Y., Kugler M. C., Wei Y., Kim K. K., Li X., Brumwell A. N., Chapman H. A. (2009) Integrin alpha3beta1-dependent beta-catenin phosphorylation links epithelial Smad signaling to cell contacts. J. Cell Biol. 184, 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Maschler S., Wirl G., Spring H., Bredow D. V., Sordat I., Beug H., Reichmann E. (2005) Tumor cell invasiveness correlates with changes in integrin expression and localization. Oncogene 24, 2032–2041 [DOI] [PubMed] [Google Scholar]
- 61. Zeisberg M., Neilson E. G. (2009) Biomarkers for epithelial-mesenchymal transitions. J. Clin. Investig. 119, 1429–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Thiery J. P., Chopin D. (1999) Epithelial cell plasticity in development and tumor progression. Cancer Metastasis Rev. 18, 31–42 [DOI] [PubMed] [Google Scholar]
- 63. Luo G. H., Lu Y. P., Yang L., Song J., Shi Y. J., Li Y. P. (2008) Epithelial to mesenchymal transformation in tubular epithelial cells undergoing anoxia. Transplant. Proc. 40, 2800–2803 [DOI] [PubMed] [Google Scholar]
- 64. Vallés A. M., Boyer B., Tarone G., Thiery J. P. (1996) Alpha 2 beta 1 integrin is required for the collagen and FGF-1 induced cell dispersion in a rat bladder carcinoma cell line. Cell Adhes. Commun. 4, 187–199 [DOI] [PubMed] [Google Scholar]
- 65. Morgan M. R., Byron A., Humphries M. J., Bass M. D. (2009) Giving off mixed signals—distinct functions of alpha5beta1 and alphavbeta3 integrins in regulating cell behaviour. IUBMB Life 61, 731–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang Z., Zhang X., Gang H., Li X., Li Z., Wang T., Han J., Luo T., Wen F., Wu X. (2007) Up-regulation of gastric cancer cell invasion by Twist is accompanied by N-cadherin and fibronectin expression. Biochem. Biophys. Res. Commun. 358, 925–930 [DOI] [PubMed] [Google Scholar]
- 67. Zuk A., Hay E. D. (1994) Expression of beta 1 integrins changes during transformation of avian lens epithelium to mesenchyme in collagen gels. Dev. Dyn. 201, 378–393 [DOI] [PubMed] [Google Scholar]
- 68. White L. R., Blanchette J. B., Ren L., Awn A., Trpkov K., Muruve D. A. (2007) The characterization of alpha5-integrin expression on tubular epithelium during renal injury. Am. J. Physiol. Renal Physiol. 292, F567–F576 [DOI] [PubMed] [Google Scholar]
- 69. Galliher A. J., Schiemann W. P. (2006) Beta3 integrin and Src facilitate transforming growth factor-beta mediated induction of epithelial-mesenchymal transition in mammary epithelial cells. Breast Cancer Res. 8, R42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Jauliac S., López-Rodriguez C., Shaw L. M., Brown L. F., Rao A., Toker A. (2002) The role of NFAT transcription factors in integrin-mediated carcinoma invasion. Nat. Cell Biol. 4, 540–544 [DOI] [PubMed] [Google Scholar]
- 71. Kwan K. Y., Lam M. M., Krsnik Z., Kawasawa Y. I., Lefebvre V., Sestan N. (2008) SOX5 postmitotically regulates migration, postmigratory differentiation, and projections of subplate and deep-layer neocortical neurons. Proc. Natl. Acad. Sci. U.S.A. 105, 16021–16026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. O'Connor R. S., Mills S. T., Jones K. A., Ho S. N., Pavlath G. K. (2007) A combinatorial role for NFAT5 in both myoblast migration and differentiation during skeletal muscle myogenesis. J. Cell Sci. 120, 149–159 [DOI] [PubMed] [Google Scholar]
- 73. McDonald S. L., Silver A. (2009) The opposing roles of Wnt-5a in cancer. Br. J. Cancer 101, 209–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Katoh M., Katoh M. (2007) WNT signaling pathway and stem cell signaling network. Clin. Cancer Res. 13, 4042–4045 [DOI] [PubMed] [Google Scholar]
- 75. Katoh M. (2005) WNT/PCP signaling pathway and human cancer (review). Oncol. Rep. 14, 1583–1588 [PubMed] [Google Scholar]
- 76. Dissanayake S. K., Wade M., Johnson C. E., O'Connell M. P., Leotlela P. D., French A. D., Shah K. V., Hewitt K. J., Rosenthal D. T., Indig F. E., Jiang Y., Nickoloff B. J., Taub D. D., Trent J. M., Moon R. T., Bittner M., Weeraratna A. T. (2007) The Wnt5A/protein kinase C pathway mediates motility in melanoma cells via the inhibition of metastasis suppressors and initiation of an epithelial to mesenchymal transition. J. Biol. Chem. 282, 17259–17271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kurayoshi M., Oue N., Yamamoto H., Kishida M., Inoue A., Asahara T., Yasui W., Kikuchi A. (2006) Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res. 66, 10439–10448 [DOI] [PubMed] [Google Scholar]
- 78. Taki M., Kamata N., Yokoyama K., Fujimoto R., Tsutsumi S., Nagayama M. (2003) Down-regulation of Wnt-4 and up-regulation of Wnt-5a expression by epithelial-mesenchymal transition in human squamous carcinoma cells. Cancer Sci. 94, 593–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Topol L., Jiang X., Choi H., Garrett-Beal L., Carolan P. J., Yang Y. (2003) Wnt-5a inhibits the canonical Wnt pathway by promoting GSK-3-independent beta-catenin degradation. J. Cell Biol. 162, 899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Germani A., Prabel A., Mourah S., Podgorniak M. P., Di Carlo A., Ehrlich R., Gisselbrecht S., Varin-Blank N., Calvo F., Bruzzoni-Giovanelli H. (2003) SIAH-1 interacts with CtIP and promotes its degradation by the proteasome pathway. Oncogene 22, 8845–8851 [DOI] [PubMed] [Google Scholar]
- 81. Semenov M. V., Habas R., Macdonald B. T., He X. (2007) SnapShot: noncanonical Wnt signaling pathways. Cell 131, 1378 [DOI] [PubMed] [Google Scholar]
- 82. MacLeod R. J., Hayes M., Pacheco I. (2007) Wnt5a secretion stimulated by the extracellular calcium-sensing receptor inhibits defective Wnt signaling in colon cancer cells. Am. J. Physiol. Gastrointest. Liver Physiol. 293, G403–G411 [DOI] [PubMed] [Google Scholar]
- 83. Wang Y. (2009) Wnt/Planar cell polarity signaling: a new paradigm for cancer therapy. Mol. Cancer Ther. 8, 2103–2109 [DOI] [PubMed] [Google Scholar]
- 84. Yamamoto S., Nishimura O., Misaki K., Nishita M., Minami Y., Yonemura S., Tarui H., Sasaki H. (2008) Cthrc1 selectively activates the planar cell polarity pathway of Wnt signaling by stabilizing the Wnt-receptor complex. Dev. Cell 15, 23–36 [DOI] [PubMed] [Google Scholar]