Abstract
Integrated top-down bottom-up proteomics combined with on-line digestion has great potential to improve the characterization of protein isoforms in biological systems and is amendable to high throughput proteomics experiments. Bottom-up proteomics ultimately provides the peptide sequences derived from the tandem MS analyses of peptides after the proteome has been digested. Top-down proteomics conversely entails the MS analyses of intact proteins for more effective characterization of genetic variations and/or post-translational modifications. Herein, we describe recent efforts toward efficient integration of bottom-up and top-down LC-MS-based proteomics strategies. Since most proteomics separations utilize acidic conditions, we exploited the compatibility of pepsin (where the optimal digestion conditions are at low pH) for integration into bottom-up and top-down proteomics work flows. Pressure-enhanced pepsin digestions were successfully performed and characterized with several standard proteins in either an off-line mode using a Barocycler or an on-line mode using a modified high pressure LC system referred to as a fast on-line digestion system (FOLDS). FOLDS was tested using pepsin and a whole microbial proteome, and the results were compared against traditional trypsin digestions on the same platform. Additionally, FOLDS was integrated with a RePlay configuration to demonstrate an ultrarapid integrated bottom-up top-down proteomics strategy using a standard mixture of proteins and a monkey pox virus proteome.
In-depth characterization and quantitation of protein isoforms, including post-translationally modified proteins, are challenging goals of contemporary proteomics. Traditionally, top-down (1, 2) and bottom-up (3, 4) proteomics have been two distinct analytical paths for liquid-based proteomics analysis. Top-down proteomics is the mass spectrometry (MS)-based characterization of intact proteins, whereas bottom-up proteomics requires a chemical or enzymatic proteolytic digestion of all proteins into peptides prior to MS analysis. Both strategies have their own strengths and challenges and can be thought of as complementary rather than competing analytical techniques.
In a top-down proteomics approach, proteins are usually separated by one- or two-dimensional liquid chromatography (LC) and identified using high performance MS (5, 6). This approach is very attractive because it allows the identification of protein isoforms arising from various amino acid modifications, genetic variants (e.g. single nucleotide polymorphisms), mRNA splice variants, and multisite modifications (7) (e.g. specific histone modifications) as well as characterization of proteolytic processing events. However, there are several challenges that have limited the broad application of the approach. Typically, intact proteins are less soluble than their peptide complement, which effectively results in greater losses during various stages of sample handling (i.e. limited sensitivity). Similarly, proteins above ∼40–50 kDa in size are more difficult to ionize, detect, and dissociate in most high throughput MS work flows. Additionally, major challenges associated with MS data interpretation and sensitivity, especially for higher molecular mass proteins (>100 kDa) and highly hydrophobic proteins (e.g. integral membrane proteins), remain largely unsolved, thus limiting the applicability of top-down proteomics on a large scale.
Bottom-up proteomics approaches have broad application because peptides are easier to separate and analyze via LC coupled with tandem mass spectrometry (MS/MS), offering a basis for more comprehensive protein identification. As this method relies on protein digestion (which produces multiple peptides for each protein), the sample complexity can become exceedingly large, requiring several dimensions of chromatographic separations (e.g. strong cation exchange and/or high pH reversed phase) prior to the final LC separation (typically reversed phase (RP)1 C18), which is oftentimes directly coupled with the mass spectrometer (3, 8). In general, the bottom-up analysis rarely achieves 100% sequence coverage of the original proteins, which can result in an incorrect/incomplete assessment of protein isoforms and combinatorial PTMs. Additionally, the digested peptides are not detected with uniform efficiency, which challenges and distorts protein quantification efforts.
Because the data obtained from top-down and bottom-up work flows are complementary, several attempts have been made to integrate the two strategies (9, 10). Typically, these efforts have utilized extensive fractionation of the intact protein separation followed by bottom-up analysis of the collected fractions. Results so far have encouraged us to consider on-line digestion methods for integrating top-down and bottom-up proteomics in a higher throughput fashion. Such an on-line digestion approach would not only benefit in terms of higher sample throughput and improved overall sensitivity but would also allow a better correlation between the observed intact protein and its peptide digestion products, greatly aiding data analysis and protein characterization efforts.
So far, however, none of the on-line integrated methods have proven robust enough for routine high throughput analyses. One of the reasons for this limited success relates to the choice of the proteolytic enzyme used for the bottom-up segment. Trypsin is by far the most widely used enzyme for proteome analyses because it is affordable (relative to other proteases), it has been well characterized for proteome research, and it offers a nice array of detectable peptides due to a fairly even distribution of lysines and arginines across most proteins. However, protein/peptide RPLC separations (optimal at low pH) are fundamentally incompatible with on-line trypsin digestion (optimal at pH ∼ 8) (11, 12). Therefore, on-line coupling of trypsin digestion and RPLC separations is fraught with technological challenges, and proposed solutions (12) have not proven to be robust enough for integration into demanding high throughput platforms.
Our approach to this challenge was to investigate alternative proteases that may be more compatible with automated on-line digestion, peptide separation, and MS detection. Pepsin, which is acid-compatible (i.e. it acts in the stomach to initially aid in the digestion of food) (13), is a particularly promising candidate. This protease has previously been successfully used for the targeted analyses of protein complexes, hydrogen/deuterium exchange experiments (14, 15), and characterization of biopharmaceuticals (16, 17). Generally, pepsin preferentially cleaves the peptide bond located on the N-terminal side of hydrophobic amino acids, such as leucine and phenylalanine, although with less specificity than the preferential cleavage observed for trypsin at arginine and lysine. The compatibility of pepsin with typical LC-MS operation makes it an ideal choice for the development of novel approaches combining protein digestion, protein/peptide separation, and MS-based protein/peptide identification.
To develop an automated system capable of simultaneously capturing top-down and bottom-up data, enzyme kinetics of the chosen protease must be extremely fast (because one cannot wait hours as is typical when performing off-line proteolysis). Another requirement is the use of immobilized enzyme or a low enough concentration of the enzyme such that autolysis products do not obscure the detection of substrate peptides. The latter was a concern when using pepsin because prior hydrogen/deuterium exchange experiments used enzyme:substrate ratios up to 1:2 (18, 19). To test whether or not such a large concentration of pepsin was necessary, we performed pepsin digestion at ratios of 1:20. Many alternative energy inputs into the system were considered for speeding up the digestion. For instance, it has been shown that an input of ultrasonic energy could accelerate the reaction rate of a typical trypsin digestion while using small amounts of a protease (20). Because ultrasonic energy results in an increase of temperature and microenvironments of high pressure, it has been hypothesized that the higher temperature was the component responsible for the enhanced enzyme activity (21). López-Ferrer et al. (22, 23), however, have demonstrated that application of higher pressure with incorporation of a Barocycler alone can make trypsin display faster enzyme kinetics. This phenomenon can easily be integrated with an LC separation (which already operates at elevated pressure) to enable an automatable ultrarapid on-line digestion LC-MS proteomics platform. Herein, we refer to this platform as the fast on-line digestion system (FOLDS) (23). Although FOLDS has been described before using trypsin, here the system is characterized with pepsin, and the results obtained are compared with results attainable with trypsin. Like trypsin, pepsin produced efficient protein digestion in just a few minutes when placed under pressure. Because of the natural maximal activity of pepsin at low pH, the FOLDS can be incorporated with a RePlay (Advion Biosciences, Ithaca, NY) system, and this powerful combination is what ultimately makes the integration of top-down and bottom-up proteomics analyses possible. The integrated analysis begins with a chromatographic separation of intact proteins. The separated proteins are then split into two streams. One stream proceeds directly to the mass spectrometer for MS and/or tandem MS analysis. The second stream is split into a long capillary where the chromatographic separation of the proteins is maintained, but their arrival to the mass spectrometer for detection is delayed. This is in essence the concept of RePlay (24, 25). Herein, we have taken the RePlay a step further by implementing our FOLDS technology into the second split delayed stream of proteins. While these delayed proteins travel down the long and narrow capillary, we exposed them to pepsin where, in combination with the pressure, the proteins are quickly and reproducibly digested. These peptide fragments are subsequently subjected to MS and/or tandem MS analysis. The FOLDS RePlay system allows the rapid and robust incorporation of the integrated top-down bottom-up proteomics work flow with the ability to not only identify proteins but also to sequence multisite/combinatorial PTMs because all detected peptides (from the FOLDS analysis) are confined to the original chromatographic peak of the protein they were derived from. The analysis of protein mixtures using this integrated strategy reduces the total amount of samples required to obtain both the top-down and bottom-up data, increases throughput, and improves protein sequence coverage.
EXPERIMENTAL PROCEDURES
Materials
Sequencing grade trypsin was obtained from Promega (Madison, WI). Protein standards, pepsin, iodoacetamide, ammonium bicarbonate, formic acid, and HPLC grade solvents were purchased from Sigma-Aldrich. Tris(2-carboxyethyl)phosphine was purchased from Pierce.
Apparatus
The Barocycler NEP-3229 instrument, MicroTube, and a pressure cycling technology MicroTube adapter kit were obtained from Pressure BioSciences (West Bridgewater, MA). A UTR200 Sonoreactor was purchase from Hischler (Teltow, Germany).
Off-line Pressure-assisted Protein Digestion
Standard proteins and a Shewanella oneidensis proteome were digested as described previously (22). Briefly, 150 μl of protein solution at 1 mg/ml protein concentration was reduced and alkylated with tris(2-carboxyethyl)phosphine and iodoacetamide at a final concentration of 5 and 50 mm, respectively, and subjected to ultrasound irradiation for 3 min at 50% power using a Sonoreactor. For the samples that were to be digested with pepsin, samples were diluted 10× with 1% formic acid in water, and pepsin was added in a 1:20 pepsin:sample (w/w) ratio. For the samples that were to be digested with trypsin, the samples were diluted 10× with 50 mm ammonium bicarbonate, pH 8.0, and trypsin was added at a 1:20 trypsin:sample (w/w) ratio. 150 μl of resultant peptide solution was transferred to a MicroTube, and digestion was accomplished in the Barocycler. The pressure program cycle used in the Barocycler experiments consisted of 20 s at 25,000 p.s.i. and 5 s at ambient pressure. The number of cycles was carried out to obtain a total time under pressure of 1, 2, or 5 min. Once the samples were digested, the reaction was quenched either by raising the pH with NH4OH to about pH 8 in the case of the pepsin digestions or by lowering the pH of the solution with formic acid to about pH 3 for the trypsin digestions. The samples were then analyzed by LC-MS/MS as described elsewhere (26, 27).
FOLDS on Dual On-line Solid Phase Extraction/Reversed Phase Liquid Chromatography (DO-SPE/RPLC)
We have incorporated the FOLDS into a DO-SPE/RPLC system, which has been reported previously (28). Briefly, this system incorporated the following fluidic components: a six-port injection valve with a 5-μl sample loop, three four-port valves, and two six-port valves (VICI Valco, Houston, TX) rated to 15,000 p.s.i. Enolase was used to evaluate the pepsin digestion efficiency in experiments using the on-line two-column setup. 100 ng of enolase was loaded into the 5-μl sample loop with the pepsin in an enzyme:substrate ratio of 1:50. The sample loop was pressurized to 10,000 p.s.i. using a syringe pump (Teledyne ISCO, Lincoln, NE), which delivered mobile phase A and was used to digest and load the sample onto the SPE column. The FOLDS was operated at pressure ranging from 0 up to 10,000 p.s.i. Mobile phase A consisted of nanopure water with 0.1% formic acid, pH < 3. LC experiments were performed on a Nano-HPLC 1200 system (Agilent Technologies, Santa Clara, CA) equipped with two 1-cm-long SPE columns and a 15-cm-long 75-μm-inner diameter analytical column; all of the columns were packed in house with 3-μm diameter C18 bonded particles (Phenomenex, Torrance, CA). The system was coupled to an LTQ (Thermo Scientific, San Jose, CA) mass spectrometer.
Postcolumn On-line Digestion
The system consisted of the standard LC system described above with a 15-cm-long 75-μm-inner diameter column. A three-way tee was connected to the end of the column, allowing introduction of pepsin into the system using a syringe pump (Harvard Apparatus, South Natick, MA). The output was connected to a 10-μm-inner diameter × 10-m-long capillary with an integrated nano-ESI emitter. The LC system was coupled with an LTQ (Thermo Scientific) mass spectrometer.
Integrated Top-down Bottom-up Proteomics Analysis Using a Modified “RePlay” Digestion System
The integration of top-down and bottom-up proteomics was demonstrated using both a mixture of standard proteins and an extract of monkey pox proteins. The standard protein mixture consisted of carbonic anhydrase, lactoglobulins A and B, cytochrome c, ubiquitin, and myoglobin with 2.5 μg of total protein injected onto the capillary column for intact protein separation. The intact protein HPLC utilized an exponential gradient at 8,000-p.s.i. constant pressure with solvent A consisting of 20% acetonitrile, 5% isopropanol, 0.6% acetic acid, and 0.01% trifluoroacetic acid and solvent B consisting of 45% acetonitrile, 45% isopropanol, 0.6% acetic acid, and 0.01% trifluoroacetic acid. The capillary column (75-μm inner diameter × 75 cm long) was packed in house with 5-μm Jupiter C5 particles (Phenomenex, Torrance, CA). The column flow rate was ∼400 nl/min at the beginning of the gradient.
The intact protein HPLC eluent was split with half of the flow (∼200 nl/min) directed to a TriVersa NanoMate (Advion Biosciences) for ESI and introduction of the intact protein ions into the mass spectrometer. The remaining intact protein HPLC eluent (∼200 nl/min) was directed into the Advion RePlay device (Advion Biosciences) and captured in a 50-μm-inner diameter × 22-m-long RePlay capture capillary. A 0.1 μg/μl solution of active pepsin protease was mixed with the split LC eluent prior to capture in the capillary column. The flow rate of the pepsin solution was 50 nl/min. Proteins and pepsin were in contact for ∼220 min (the length of the initial HPLC separation) in the RePlay capture capillary at 1,000 p.s.i.
Intact protein ions generated by ESI using the TriVersa NanoMate were introduced into a modified Bruker 12-T FTICR mass spectrometer. A compensated open cylindrical cell with improved mass measurement accuracy and extended dynamic range (5, 29) was used to acquire all mass spectra. Tandem MS was carried out utilizing external precursor ion selection and accumulation followed by external CID with nitrogen as the collision gas. Nitrogen gas was leaked into the external accumulation cell at a constant pressure with the source vacuum gauge reading 5.5 × 10−6 millibar. Preselection of precursor ions and calculation of appropriate collision energy were performed using apexControl software (Bruker Daltonics, Billerica, MA) with the AMS_EnergyFile parameter set to 0.01053, 12.78947 for all charge states.
During column equilibration after the first HPLC separation, the Advion RePlay device was switched to allow the captured protein-pepsin mixture to flow (∼200 nl/min) to the TriVersa NanoMate for ESI-MS characterization of the peptic peptides in the 12-T FTICR mass spectrometer. Tandem MS of selected peptides was carried out as described previously (26, 27).
Data Analysis
For IT-MS/MS data, we used either a SEQUEST (30) database search engine or SpectrumMill to identify the peptides. A previously published method (31, 32) was used to calculate and control the error rates associated with the peptide identifications. For the corresponding RePlay digestion analyses acquired on the 12-T FTICR mass spectrometer proteins and peptides were identified using an in-house software that deisotoped the data (ICR2LS, Decon2ls) (33) and matched the theoretically generated protein/peptide masses to the masses obtained experimentally. A subset of identifications was also analyzed by comparison of the observed and calculated isotope distributions. Protein and peptide MS/MS data sets were analyzed by de novo reconstruction of short amino acid sequences based on the observed MS/MS fragmentation patterns (34, 35).
RESULTS AND DISCUSSION
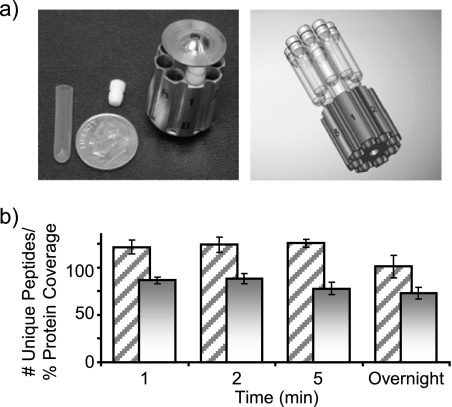
It has been recently demonstrated that an increase in pressure can significantly accelerate the reaction rate of a trypsin digestion (22). Herein, pepsin was evaluated as a candidate proteolytic enzyme displaying the same capacity of enhanced kinetics under pressure. The experiments were performed using the Barocycler, an apparatus that provides precise time control over the application of pressure and enables automated and rapid cycling between a given pressure and ambient pressure. The samples contained in small silicon tubes (MicroTubes) (Fig. 1a) were directly subjected to the pressure changes that were transduced through the sample tube onto the enclosed sample solution. One of the main advantages of this system is its ability to handle up to 48 small samples (with sample sizes of 50–150 μl); thus, it is possible to carry out rapid and thorough digestions of 48 samples simultaneously in only a few minutes. Rapid digestions under high pressure have been demonstrated with trypsin, but it was not known whether this phenomenon was limited to trypsin or whether it could be applied to other proteolytic enzymes. Hence, we first characterized bovine serum albumin (BSA) pepsin digestion in the controlled off-line setting using the Barocycler. After reduction and alkylation, the mixture of BSA and pepsin was subjected to 25,000 p.s.i. for 1, 2, or 5 min. These experiments were performed in duplicate to validate the results. Fig. 1 shows the average number of unique peptides identified and the overall proteome coverage obtained for each condition. In agreement with previous experiments performed with trypsin (22), the results were similar for all conditions studied with a slightly higher number of peptides identified for the pressurized digestions than for the overnight digestion (Fig. 1b).
Fig. 1.
a, the left panel shows a photograph of the MicroTube insert, and MicroTube holder used for off-line pressure digestion experiments with a dime for size comparison. The right panel shows how MicroTubes are inserted into the holder. b, the histogram depicts the average number of unique identified peptides and average protein coverage obtained from two duplicate LC-MS/MS runs of bovine serum albumin digested with pepsin for three different lengths of time at 25,000 p.s.i. and an overnight digestion at 37 °C and ambient pressure. The percentage of protein coverage is depicted as a solid bar, and uniquely identified peptides are depicted as diagonal lines. Error bars represent one standard deviation.
We previously hypothesized that the increase in enzyme kinetics (and consequentially coverage) is due to a denaturing effect of pressure, which acts to force (most of the) proteins into a more linear or open conformation (23). In other words, the pressure input provides enough energy to overcome any decreased solvent entropy effects associated with the linear conformation, and this opening makes the proteins more accessible to the enzyme. What is curious is that the proteases (which are also proteins) maintain activity despite being under the same influence of the pressure. A possible explanation might be that perhaps the active sites of the enzymes possess a higher degree of stability, which may translate into the maintenance of these highly conserved amino acid sequence regions across many species because the preservation of these sites is critical for the function of the enzyme (36). This higher degree of stability from an evolution perspective could then also explain why the active enzyme pocket of the proteases remains active even under high pressure (37). The overall agreement between the chromatograms (supplemental Fig. 1), number of proteins, and peptide coverage nicely illustrates minimal variations in the extent and the observed high degree of reproducibility of the digestions (Fig. 1).
We further explored 1-min pressurized pepsin digestions with a mixture of standard proteins to ensure that our observations were not limited to BSA. This protein mixture was also subjected to digestion with trypsin (instead of pepsin) under the same conditions (with the exception of the buffer pH). As shown in Fig. 2, results for the two enzymes were similar in terms of protein coverage, normalized peptide counts, and reproducibility. Fig. 2b shows the protein coverage attained by each protease alone and when the results were combined. A significant gain in protein coverage was realized when data from two digestions were combined as previously demonstrated by Coon and co-workers (38). This strategy could be particularly useful for characterization of combinatorial PTMs because of the higher sequence coverage. Fig. 2c shows the overlap between replicate analyses attained for pepsin and trypsin digestions, indicating a similar degree of reproducibility for the two digestion procedures. For the pepsin digestion, each technical replicate had on average 64 ± 4% of the total identified peptides. The technical replicates of the trypsin digestion each had on average 70 ± 6% of the total identified peptides.
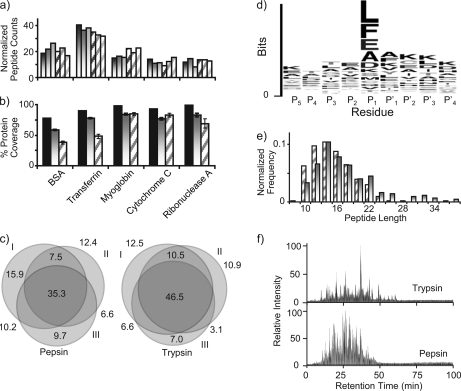
Fig. 2.
Comparison of enzyme performance for pepsin and trypsin pressure-assisted digestions using a five-standard protein mixture consisting of BSA, transferrin, myoglobin, cytochrome c, and ribonuclease A. a and b, histograms comparing normalized peptide counts and percentage of protein coverage for three technical replicates of pressurized digestions using either trypsin (shaded gradation bars) or pepsin (diagonal striped bars) for 1 min. Solid black bars indicate the total unique protein coverage when data from both digestions were combined. c, Venn diagrams showing the peptide overlap between three technical digestion replicates (I–III), including the percentage of identified peptides in each region. d, sequence logo for the amino acid sequence motif of pepsin-generated peptides identified by MS/MS analysis. e, histogram depicting the frequency of peptide length identified from the peptides identified either by trypsin (shaded gradation bars) or pepsin (diagonal striped bars). f, representative chromatograms corresponding to the LC-MS/MS of a pressure-assisted digestion for trypsin (top) and pepsin (bottom).
Although the reproducibility and experimental peptide coverage was on par with that obtained from a trypsin digestion, peptides obtained from a pepsin digestion (supplemental Table 1) were observed to exhibit less amino acid cleavage specificity as expected. For bottom-up approaches, this lack of specificity would require downstream peptide MS/MS data analyses to be conducted with no enzyme restrictions, naturally leading to higher false discovery rates. Although pepsin in general is considered to be a nonspecific protease, in replicate analyses, pepsin does appear to reproducibly cleave at the specific positions within a protein. This suggests that there are predictable pepsin cleavage rules, but these rules are not presently understood, nor are they as straightforward as they are for trypsin. In an attempt to better characterize expected cleavage specificity patterns of pepsin, a list of peptide sequences containing the P4, P3, P2, P1, P1′, P2′, P3′, and P4′ residues for all identified peptides was compiled (with cleavage occurring between P1 and P1′). These sequences were then submitted to WebLogo (http://weblogo.berkeley.edu/) to obtain a graphical representation of the amino acid sequence preferences around the cleavage sites. The WebLogo graph (Fig. 2d) displaying the amino acid positional preference and frequency of cleavage sites from the data collected in this study shows that the P1 position had an overall preference for the expected amino acids of Leu and Phe but also (to a lesser extent for) Glu, Ala, Asp, and Lys. P2 and P1′ appear to be somewhat conserved, signifying the need for further research into protease specificity (39) to truly understand and predict pepsin cleavage preferences.
When looking at the peptide lengths, there were no great differences between the detected peptides from the pepsin and trypsin digestions (Fig. 2e). Although the detectable peptides did not appear to have much difference in the distribution of their lengths, there were substantial differences in their RPLC (C18) retention behavior (Fig. 2f). Overall, peptic peptides appear to elute sooner and with a more normal (bell-shaped) distribution, suggesting that they have a more common level of hydrophobicity in comparison with the tryptic peptides, which appear to cover a larger range of peptides with more diverse hydrophobicities. The fact that these two proteases generate peptides that show different chromatographic behavior in addition to the combinatorial gain in sequence coverage illustrated in Fig. 2b demonstrates how multiple digestion strategies could be used to improve proteome coverage in terms of total proteins identified and number of peptides identified for each protein.
Because the ultimate goal of high throughput proteomics is to analyze complex protein mixtures, we applied the high pressure digestion approach to S. oneidensis whole cell lysate. Using either trypsin or pepsin, triplicate global proteome digestions were performed. The six protein digests were analyzed by LC-MS/MS, and the resulting data were processed using a SEQUEST search without enzyme restrictions. A total of 11,105 spectra passed a 1% false discovery rate filter cutoff (27). This translates to an average of 1,385 and 1,124 unique peptides obtained per data set for the trypsin and pepsin digests, respectively. The total number of unique peptide identifications (summed over three replicates for each digestion) was in the range of ∼2,150 in both cases. Fig. 3 shows the number of unique peptides and proteins and their overall distribution obtained in each replicate experiment. A significant overlap (>40%) in identified peptides was obtained between the trypsin and pepsin digestion methods. Of all peptides that did not overlap, nearly half were evenly divided between those uniquely identified in either the pepsin digestion or the trypsin digestion.
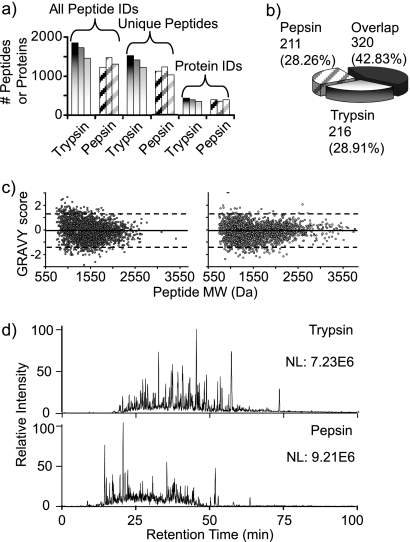
Fig. 3.
Comparison of enzyme performance and peptide properties for pepsin and trypsin pressure-assisted digestions using S. oneidensis protein extract as model proteome. a, histograms comparing the performance of 1-min pressure-assisted digestion with trypsin (shaded gradation bars) and pepsin (diagonal striped bars) across three technical replicates. b, pie chart representing the overlap of enzyme exclusive peptide identifications. c, GRAVY values plotted against the calculated molecular masses for the identified peptic (left) and tryptic (right) peptides. d, representative chromatograms corresponding to the LC-MS/MS of pressure-assisted digestion for trypsin (top) and pepsin (bottom) after a 2-min digestion.
The chromatograms reconstructed from the LC-MS data acquired from the S. oneidensis pepsin and trypsin digests differed significantly as was demonstrated earlier with the standard protein mixture (Fig. 3d) with peptic peptides in general eluting earlier in the gradient than tryptic peptides (again in agreement with standard protein mixture results). Over the last decade, there have been many efforts in the area of developing models for peptide retention time prediction (40, 41). Findings published by Petritis et al. (41) suggested that hydrophobicity and peptide length contribute to its interaction with a C18 stationary phase less than the N and C termini. Our data are in agreement with these earlier results as shown in supplemental Fig. 2 where the significant differences in the chromatographic profiles cannot be explained by small variations observed between peptide lengths and the grand average hydropathicity (GRAVY) scores. The sequence logo for pepsin digests depicted in Fig. 2d shows that the majority of the amino acids in positions P5 to P2 as well as P1′ to P4′ are hydrophilic. We thus hypothesize that for pepsin digestion these intermediate amino acids, which are mostly polar, may have a greater influence than in the case of tryptic peptides to explain their early elution times. We have to note, however, that the limited number of observations makes this conclusion preliminary.
GRAVY plots can be a useful way to look at hydrophobic domains and even the overall hydrophobicity of many peptides (42). (Positive GRAVY scores indicate that the sequence is hydrophobic, and negative GRAVY scores indicate that the sequence is hydrophilic.) By carefully looking at scatter plots of molecular mass versus the GRAVY scores for each identified peptide (Fig. 3c), it becomes evident that peptic peptides cover a smaller molecular mass range than the tryptic peptides. The opposite trend is observed when the hydropathicity dimension is considered. Approximately 8% of the identified peptic peptides have a GRAVY score above 1.5 (versus 2% for the tryptic peptides). Interestingly, the pepsin plot has a pattern similar to that of the plot published by Poetsch and co-workers (43) in their report on the advantages of using elastase digestion for membrane proteomics. The authors claimed that small peptides (i.e. below ∼700 Da) with higher GRAVY scores represent potential transmembrane peptides. Our data suggest that a pepsin digestion also could be more effective in cleaving the membrane proteins and producing peptides more compatible with LC-MS analysis. However, a more targeted study is needed to validate this hypothesis as the data presented here are derived from a global cell lysate and not from a purified membrane fraction.
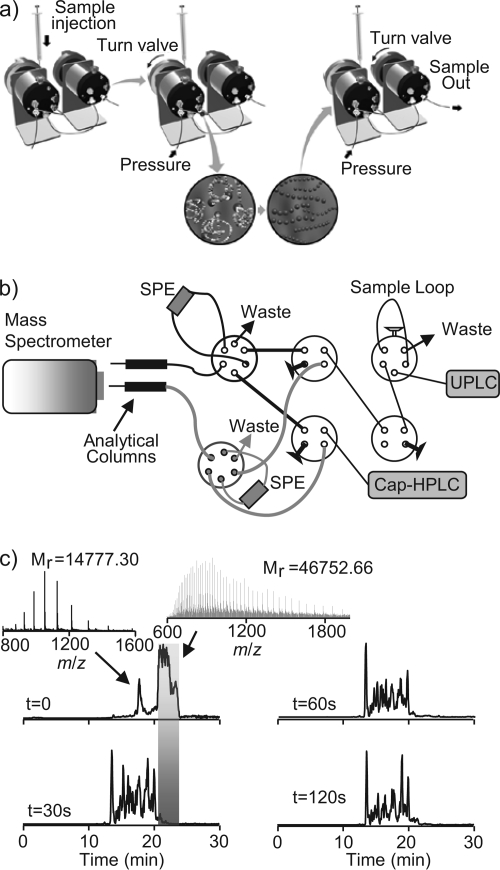
We have previously published a FOLDS that utilized the high pressure capability of an LC system to rapidly digest target proteins and complex protein mixtures under pressure in the presence of trypsin (23) (Fig. 4a). The system used ultrahigh performance LC pumps pressurizing water up to 10,000 p.s.i. to accelerate the enzyme kinetics of trypsin. Herein, we coupled the FOLDS with a DO-SPE-RPLC system that uses two on-line SPE columns and two separation columns (Fig. 4b). The major advantage of a dual column system is a higher duty cycle because one column can be regenerated and loaded with the next sample while the other column is conducting the separation. We modified the FOLDS system to accommodate pepsin digestions (all the parameters remain the same as in the case of trypsin with the exception of a digestion buffer, which consists of 1% formic acid to make it compatible with the pepsin). Enolase was used to evaluate the pepsin digestion efficiency using the on-line two-column setup (Fig. 4b). The sample loop was pressurized to 10,000 p.s.i., then the secondary valve was opened, and the sample was released and loaded onto the SPE column (Fig. 4c). For time zero, the enzyme was mixed with the protein and immediately loaded onto the SPE column. Under this condition, we detected the intact protein at 22 min in the gradient with a few additional large fragments (with masses over 10 kDa) eluting earlier. These large fragments were found to be degradation products of the enolase generated during the brief exposure of the protein to pepsin under pressure, but most of the protein remained undigested as expected. When the protein and the enzyme were pressurized together for 30 s in the loop, complete digestion was observed (Table I). Prolonging the reaction time up to 2 min resulted in similar LC-MS chromatograms, indicating that fast pepsin digestion in small capillaries under 10,000-p.s.i. pressure is feasible. Gross and co-workers (44) recently explored a similar setup for hydrogen/deuterium exchange experiments.
Fig. 4.
Setup and validation of FOLDS for pepsin digestions. a, illustration showing the FOLDS platform. b, schematic diagram of the two-column FOLDS. The components include three six-port injection valves and four four-port valves. The system is coupled to a two-column RPLC system, and an LTQ mass spectrometer was used for MS/MS analysis. c, comparison of the chromatograms (and representative spectra) obtained from the two-column FOLDS pepsin digestion of enolase as a function of the digestion time. UPLC, ultraperformance LC; Cap-HPLC, capillary HPLC.
Table I. MS/MS results for time course digestion experiment performed with two-column FOLDS.
| Digestion time | Protein identified | SpectrumMill protein score | Protein coveragea | No. unique peptides |
|---|---|---|---|---|
| s | % | |||
| 30 | Enolase | 823.66 | 81 | 60 |
| 45 | Enolase | 854.36 | 70 | 58 |
| 60 | Enolase | 800.95 | 87 | 54 |
a Percentage of the amino acid sequence of the protein covered by the identified peptides.
In efforts to provide a greater depth of information from proteome analyses, several methods of coupling protein-level (i.e. top-down) and peptide-level (i.e. bottom-up) LC-MS analyses after digestion into an integrated platform have been attempted with varying degrees of success. Many approaches have been developed, and most of them use an integrated on-column microreactor based on some form of enzyme immobilization for the rapid digestion of protein fractions into peptides (11, 45, 46). Although many of these approaches were successful in digesting proteins on line, their application to demanding proteomics experiments proved to be limited because of rapid inactivation of the immobilized enzyme presumably by endogenous proteases, mobile phase additives and impurities, organic solvents, endogenous inhibitors, and other factors (47). Immobilized enzyme inactivation limits the number of times the reactor can be used, preventing the automation and market viability of on-line digestion platforms.
To circumvent these challenges, we propose an approach similar to that of Chen and co-workers (18) developed with the triaxial probe for on-line proteolysis integral to their hydrogen/deuterium exchange experiments but with a design modification to our reactor to handle low flow rates, low sample concentrations, and incorporation of a RePlay system (24). As the name suggests, in RePlay mode of operation, flow from the initial RPLC intact protein separation is split into two streams; one stream is directed into the mass spectrometer, whereas the second stream is diverted into a long capture capillary, allowing the separated constituents to be analyzed via MS again in any desired way. In its original implementation, this approach allowed the sample (mixture of peptides) to be reanalyzed after the first analysis had finished to improve the overall quality of the analysis (e.g. increase the number of identified species or enable a more targeted analysis the second time around) (24). For our purposes, we utilized the storage capillary as a pressurized digestion reactor. This was accomplished using a three-way zero-volume tee to continually introduce the pepsin into the stream of separated proteins as they pass through the 10-m-long, 15-μm-inner diameter capillary at a concentration of 0.1 μg/μl. After an appropriate residence/digestion time, the stream was directed to the mass spectrometer for analysis (via TriVersa NanoMate). One of the key advantages of this approach (besides the compatibility between RPLC and pepsin digestion conditions) is a continuous introduction of a constant stream of fresh pepsin, which significantly reduces the inactivation problems described above.
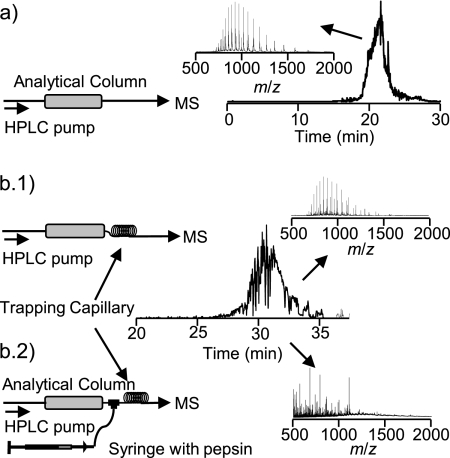
Fig. 5 shows the results of a RePlay-based integrated top-down bottom-up platform using pepsin to digest myoglobin in a proof-of-principle experiment. In a control experiment (Fig. 5a), the system was operated without pepsin, and the characteristic charge state distribution of intact myoglobin was detected in the mass spectrometer. Because the pumped-in pepsin is introduced as a second stream into the protein sample stream, lateral diffusion of the enzyme into the sample was required, and a minimum residence time of about 10 min was needed for complete digestion (Fig. 5b). No noticeable peak broadening effect was observed.
Fig. 5.
LC-MS setup schematics (left) and total ion chromatogram (right) for on-line digestion of myoglobin. Insets show mass spectra extracted at the indicated LC elution times. a, control experiment (no digestion). b, on-line digestion. b.1 and b.2, effect of the insertion of the delay/trapping capillary without (b.1) and with (b.2) pepsin infusion.
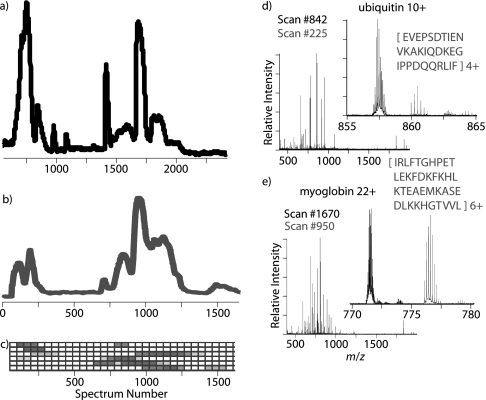
Next, a standard protein mixture consisting of carbonic anhydrase, lactoglobulins A and B, cytochrome c, ubiquitin, and myoglobin was evaluated using this coupled top-down bottom-up analysis approach (Fig. 6). The eluent of an HPLC separation of a 2.5-μg injection of a protein mixture was split with half of the flow introduced into a 12-T FTICR mass spectrometer for analysis (Fig. 6a) and the other half mixed with an active pepsin digestion solution and stored in a capture capillary utilizing an Advion RePlay device. After finishing analysis of the intact proteins, the RePlay device was switched from capture to replay mode, and the digested proteins were introduced to the 12-T FTICR mass spectrometer for analysis (Fig. 6b). The capture capillary was maintained at a minimum pressure of 1,000 p.s.i. to ensure complete digestion of the proteins by the time the first intact protein RPLC run finished (∼220 min). Complete digestion was verified by the lack of detected intact protein in mass spectra of the pepsin RePlay analysis and the appearance of multiple lower charge state peaks corresponding to digested peptides (Fig. 6, insets). Digested peptides were typically in the 400–4,000-Da range. The number of peptides originating from each standard protein was calculated in 50 spectra bins. This number of observed peptides was then normalized relative to each protein and plotted graphically. As seen in Fig. 6c, digested peptides from each protein appeared to be localized into discrete segments of the RePlay analysis. This setup eliminated the typical need to collect fractions (9), thus decreasing the sample requirements and increasing the throughput. Because the resulting peptic peptides elute in the same order as the separated intact proteins, this approach preserves a direct link between the intact protein and its corresponding peptides, providing a greater confidence in the sequence determination and characterization of (multisite) PTMs. Complete protein digestion was achieved, and the high mass accuracy offered by FTMS allowed the matching of proteins with their resulting peptic peptides (i.e. classical peptide fingerprinting). Our data also agree with the recent results published from other groups (44, 48) where the application of as little as ∼100 p.s.i. produced an effective digestion in 30 min or less. In our case, the digestion time depends on the time required for the first chromatographic separation, which is longer than 30 min. It is important to note that with more complex mixtures of proteins (i.e. a whole cell proteome) this approach may require further modification and/or downstream peptide separation because with the low specificity of pepsin the number of theoretical peptide candidates may become overwhelmingly large, making the data analysis exceptionally challenging.
Fig. 6.
LC-MS and RePlay pepsin digestion analysis of protein standard mixture containing carbonic anhydrase, lactoglobulins A and B, cytochrome c, ubiquitin, and myoglobin. a, a total ion chromatogram of the initial intact protein RPLC analysis. b, the corresponding total ion chromatogram for the RePlay pepsin digestion analysis. c, a plot of the normalized number of digested peptides corresponding to a particular protein as a function of elution time. The darker boxes indicate more peptides were observed. Each row contains data corresponding to a different protein. From top to bottom, these proteins are cytochrome c, ubiquitin, lactoglobulin A, lactoglobulin B, myoglobin, and carbonic anhydrase. d and e, mass spectra of both the initial intact RPLC separation plotted in black and the RePlay pepsin digestion analysis plotted in grey. A significant number of intact ubiquitin (d) and myoglobin (e) peaks were only detected in the initial RPLC run, and digestion products were only observed in the RePlay run as illustrated in the zoomed-in regions.
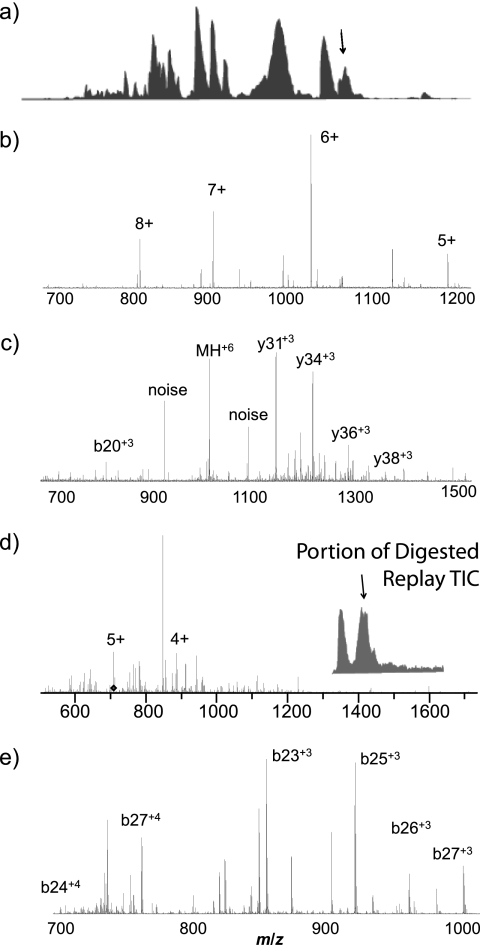
Fortunately, tandem MS results of both the intact proteins and digested peptides can aid in protein and PTM identification and characterization and mitigate the data analysis challenge. Initial results demonstrating the effectiveness of the integrated top-down bottom-up strategy that incorporates tandem MS of intact and on line-digested proteins from a single intact protein RPLC separation are shown in Fig. 7. The tandem MS spectra of both the initial RPLC separation of a 2-μg sample of monkey pox viral proteins (Fig. 7c) and the digested RePlay analysis (Fig. 7d) were processed using in-house de novo sequencing tools (34, 35), and several peptides corresponding to viral protein fragments were identified. For example, de novo MS/MS analysis identified GFTNKNKLEKLSTNKELESYSSSPLQEPIRLNDFLGLLECVKKNIPLTDIPTKD, a protein fragment from the monkey pox structural protein VP8 (Fig. 7c), 73 min into the gradient of the original protein RPLC-MS experiment. At the corresponding elution time in a RePlay run, similar de novo analysis based on the observed MS/MS fragment ions identified the sequence GFTNKNKLEKLSTNKELESYSSSPLQEPIRL as a digestion product of the original larger protein fragment (Fig. 7d). Although detailed analysis of detected/identified monkey pox proteins in this series of experiments will be published separately, the results presented in Fig. 7 are sufficient to illustrate the power and the potential of this comprehensive integrated top-down bottom-up approach featuring high performance tandem MS capabilities.
Fig. 7.
LC-MS/MS of isolated monkey pox viral proteins with RePlay pepsin digestion MS/MS. a, the total ion chromatogram of the initial intact protein RPLC separation. b, a typical MS scan. The arrow in a indicates when in the RPLC this spectrum was acquired. c, the 6+ peak near 1,021 m/z in b was selected for isolation and MS/MS. De novo MS/MS analysis using in-house tools identified a fragment from the monkey pox structural protein VP8 based on the observed fragment peptides. d, representative mass spectrum from the RePlay pepsin digestion corresponding to a. e, tandem mass spectrum acquired for the 5+ peak in d, which corresponds to a pepsin digestion product of the original larger protein fragment based on de novo analysis of the observed fragmentation pattern.
CONCLUSIONS
The combined use of pepsin and pressure has been demonstrated to be an effective approach to the ultrarapid characterization of proteins and peptides. High pressure accelerated the rate of proteolysis, thus eliminating the need for long incubation times, and the use of pepsin allowed a straightforward coupling with LC-MS because of its acid-compatible activity. These developments taken altogether have enabled FOLDS, which is a significant step toward creating a robust and fully automated on-line proteomics platform with integrated protein separation, digestion, and MS detection. Additionally, FOLDS was combined with the RePlay technology into a fully integrated top-down bottom-up proteomics platform. Integrated top-down bottom-up proteomics utilizing on-line pepsin digestion proved to be a feasible, facile, and robust platform, which combined with enhancements in the HPLC and tandem MS of proteins to enable high throughput analysis of complex biological systems.
Acknowledgments
We thank Rui Zhao and Ron Moore for helpful suggestions and technical support. Portions of this work were supported by the William R. Wiley Environmental Molecular Sciences Laboratory (EMSL) Intramural Research and Capability Development Program, the U.S. Department of Energy (DOE) Office of Biological and Environmental Research (OBER), the National Institutes of Health (NIH) Grant RR018522 (to R. D. S.) from the National Center for Research Resources and Grant R21 CA12619-01 from the NCI. Portions of this work were also supported by the Laboratory Directed Research and Development Program of the Pacific Northwest National Laboratory. Work was performed using EMSL, a national user facility sponsored by DOE-OBER.
Footnotes
 This article contains supplemental Table 1 and Figs. 1 and 2.
This article contains supplemental Table 1 and Figs. 1 and 2.
1 The abbreviations used are:
- RP
- reversed phase
- FOLDS
- fast on-line digestion system
- PTM
- post-translational modification
- SPE
- solid phase extraction
- T
- tesla
- DO
- dual on-line.
REFERENCES
- 1. Kelleher N. L., Taylor S. V., Grannis D., Kinsland C., Chiu H. J., Begley T. P., McLafferty F. W. (1998) Efficient sequence analysis of the six gene products (7–74 kDa) from the Escherichia coli thiamin biosynthetic operon by tandem high-resolution mass spectrometry. Protein Sci. 7, 1796–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zubarev R. A., Horn D. M., Fridriksson E. K., Kelleher N. L., Kruger N. A., Lewis M. A., Carpenter B. K., McLafferty F. W. (2000) Electron capture dissociation for structural characterization of multiply charged protein cations. Anal. Chem. 72, 563–573 [DOI] [PubMed] [Google Scholar]
- 3. Washburn M. P., Wolters D., Yates J. R., 3rd (2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19, 242–247 [DOI] [PubMed] [Google Scholar]
- 4. Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G. R., Vandekerckhove J. (2003) Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol. 21, 566–569 [DOI] [PubMed] [Google Scholar]
- 5. Parks B. A., Jiang L., Thomas P. M., Wenger C. D., Roth M. J., Boyne M. T., 2nd, Burke P. V., Kwast K. E., Kelleher N. L. (2007) Top-down proteomics on a chromatographic time scale using linear ion trap Fourier transform hybrid mass spectrometers. Anal. Chem. 79, 7984–7991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sharma S., Simpson D. C., Tolić N., Jaitly N., Mayampurath A. M., Smith R. D., Paša-Tolić L. (2007) Proteomic profiling of intact proteins using WAX-RPLC 2-D separations and FTICR mass spectrometry. J. Proteome Res. 6, 602–610 [DOI] [PubMed] [Google Scholar]
- 7. Lee S. W., Berger S. J., Martinović S., Paša-Tolić L., Anderson G. A., Shen Y., Zhao R., Smith R. D. (2002) Direct mass spectrometric analysis of intact proteins of the yeast large ribosomal subunit using capillary LC/FTICR. Proc. Natl. Acad. Sci. U.S.A. 99, 5942–5947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wolters D. A., Washburn M. P., Yates J. R., 3rd (2001) An automated multidimensional protein identification technology for shotgun proteomics. Anal. Chem. 73, 5683–5690 [DOI] [PubMed] [Google Scholar]
- 9. Wu S., Lourette N. M., Tolić N., Zhao R., Robinson E. W., Tolmachev A. V., Smith R. D., Paša-Tolić L. (2009) An integrated top-down and bottom-up strategy for broadly characterizing protein isoforms and modifications. J. Proteome Res. 8, 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu S., Yang F., Zhao R., Tolíc N., Robinson E. W., Camp D. G., 2nd, Smith R. D., Pasa-Tolíc L. (2009) Integrated workflow for characterizing intact phosphoproteins from complex mixtures. Anal. Chem. 81, 4210–4219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Slysz G. W., Schriemer D. C. (2003) On-column digestion of proteins in aqueous-organic solvents. Rapid Commun. Mass Spectrom. 17, 1044–1050 [DOI] [PubMed] [Google Scholar]
- 12. Slysz G. W., Schriemer D. C. (2005) Blending protein separation and peptide analysis through real-time proteolytic digestion. Anal. Chem. 77, 1572–1579 [DOI] [PubMed] [Google Scholar]
- 13. Cox M. N., David R. (2008) Lehninger Principles of Biochemistry, 5th Ed., W. H. Freeman, New York [Google Scholar]
- 14. Zhang H. M., Bou-Assaf G. M., Emmett M. R., Marshall A. G. (2009) Fast reversed-phase liquid chromatography to reduce back exchange and increase throughput in H/D exchange monitored by FT-ICR mass spectrometry. J. Am. Soc. Mass Spectrom. 20, 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang H. M., Kazazić S., Schaub T. M., Tipton J. D., Emmett M. R., Marshall A. G. (2008) Enhanced digestion efficiency, peptide ionization efficiency, and sequence resolution for protein hydrogen/deuterium exchange monitored by Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 80, 9034–9041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun S., Mo W., Ji Y., Liu S. (2001) Use of nitrocellulose films for affinity-directed mass spectrometry for the analysis of antibody/antigen interactions. Rapid Commun. Mass Spectrom. 15, 1743–1746 [DOI] [PubMed] [Google Scholar]
- 17. Sun S., Mo W., Ji Y., Liu S. (2001) Preparation and mass spectrometric study of egg yolk antibody (IgY) against rabies virus. Rapid Commun. Mass Spectrom. 15, 708–712 [DOI] [PubMed] [Google Scholar]
- 18. Chen M., Cook K. D., Kheterpal I., Wetzel R. (2007) A triaxial probe for on-line proteolysis coupled with hydrogen/deuterium exchange-electrospray mass spectrometry. J. Am. Soc. Mass Spectrom. 18, 208–217 [DOI] [PubMed] [Google Scholar]
- 19. Kheterpal I., Williams A., Murphy C., Bledsoe B., Wetzel R. (2001) Structural features of the Abeta amyloid fibril elucidated by limited proteolysis. Biochemistry 40, 11757–11767 [DOI] [PubMed] [Google Scholar]
- 20. López-Ferrer D., Capelo J. L., Vázquez J. (2005) Ultra fast trypsin digestion of proteins by high intensity focused ultrasound. J. Proteome Res. 4, 1569–1574 [DOI] [PubMed] [Google Scholar]
- 21. Havlis J., Thomas H., Sebela M., Shevchenko A. (2003) Fast-response proteomics by accelerated in-gel digestion of proteins. Anal. Chem. 75, 1300–1306 [DOI] [PubMed] [Google Scholar]
- 22. López-Ferrer D., Petritis K., Hixson K. K., Heibeck T. H., Moore R. J., Belov M. E., Camp D. G., 2nd, Smith R. D. (2008) Application of pressurized solvents for ultrafast trypsin hydrolysis in proteomics: proteomics on the fly. J. Proteome Res. 7, 3276–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. López-Ferrer D., Petritis K., Lourette N. M., Clowers B., Hixson K. K., Heibeck T., Prior D. C., Paša-Tolić L., Camp D. G., 2nd, Belov M. E., Smith R. D. (2008) On-line digestion system for protein characterization and proteome analysis. Anal. Chem. 80, 8930–8936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Waanders L. F., Almeida R., Prosser S., Cox J., Eikel D., Allen M. H., Schultz G. A., Mann M. (2008) A novel chromatographic method allows on-line reanalysis of the proteome. Mol. Cell. Proteomics 7, 1452–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waanders L. F., Chwalek K., Monetti M., Kumar C., Lammert E., Mann M. (2009) Quantitative proteomic analysis of single pancreatic islets. Proc. Natl. Acad. Sci. U.S.A. 106, 18902–18907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shen Y., Tolić N., Masselon C., Paša-Tolić L., Camp D. G., 2nd, Hixson K. K., Zhao R., Anderson G. A., Smith R. D. (2004) Ultrasensitive proteomics using high-efficiency on-line micro-SPE-nanoLC-nanoESI MS and MS/MS. Anal. Chem. 76, 144–154 [DOI] [PubMed] [Google Scholar]
- 27. López-Ferrer D., Heibeck T. H., Petritis K., Hixson K. K., Qian W., Monroe M. E., Mayampurath A., Moore R. J., Belov M. E., Camp D. G., 2nd, Smith R. D. (2008) Rapid sample processing for LC-MS-based quantitative proteomics using high intensity focused ultrasound. J. Proteome Res. 7, 3860–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Min H. K., Hyung S. W., Shin J. W., Nam H. S., Ahn S. H., Jung H. J., Lee S. W. (2007) Ultrahigh-pressure dual online solid phase extraction/capillary reverse-phase liquid chromatography/tandem mass spectrometry (DO-SPE/cRPLC/MS/MS): a versatile separation platform for high-throughput and highly sensitive proteomic analyses. Electrophoresis 28, 1012–1021 [DOI] [PubMed] [Google Scholar]
- 29. Tolmachev A. V., Robinson E. W., Wu S., Paša-Tolić L., Smith R. D. (2009) FT-ICR MS optimization for the analysis of intact proteins. Int. J. Mass Spectrom. 281, 32–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eng J. K., McCormack A. L., Yates J. R., 3rd (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 31. López-Ferrer D., Martínez-Bartolomé S., Villar M., Campillos M., Martín-Maroto F., Vázquez J. (2004) Statistical model for large-scale peptide identification in databases from tandem mass spectra using SEQUEST. Anal. Chem. 76, 6853–6860 [DOI] [PubMed] [Google Scholar]
- 32. Kim B. C., Lopez-Ferrer D., Lee S. M., Ahn H. K., Nair S., Kim S. H., Kim B. S., Petritis K., Camp D. G., Grate J. W., Smith R. D., Koo Y. M., Gu M. B., Kim J. (2009) Highly stable trypsin-aggregate coatings on polymer nanofibers for repeated protein digestion. Proteomics 9, 1893–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jaitly N., Mayampurath A., Littlefield K., Adkins J. N., Anderson G. A., Smith R. D. (2009) Decon2LS: an open-source software package for automated processing and visualization of high resolution mass spectrometry data. BMC Bioinformatics 10, 87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shen Y., Tolić N., Hixson K. K., Purvine S. O., Anderson G. A., Smith R. D. (2008) De novo sequencing of unique sequence tags for discovery of post-translational modifications of proteins. Anal. Chem. 80, 7742–7754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shen Y., Tolić N., Hixson K. K., Purvine S. O., Pasa-Toliæ L., Qian W. J., Adkins J. N., Moore R. J., Smith R. D. (2008) Proteome-wide identification of proteins and their modifications with decreased ambiguities and improved false discovery rates using unique sequence tags. Anal. Chem. 80, 1871–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang K., Horst J. A., Cheng G., Nickle D. C., Samudrala R. (2008) Protein meta-functional signatures from combining sequence, structure, evolution, and amino acid property information. PLoS Comput. Biol. 4, e1000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shoichet B. K., Baase W. A., Kuroki R., Matthews B. W. (1995) A relationship between protein stability and protein function. Proc. Natl. Acad. Sci. U.S.A. 92, 452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Swaney D. L., Wenger C. D., Coon J. J. (2010) Value of using multiple proteases for large-scale mass spectrometry-based proteomics. J. Proteome Res. 9, 1323–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rodriguez J., Gupta N., Smith R. D., Pevzner P. A. (2008) Does trypsin cut before proline? J. Proteome Res. 7, 300–305 [DOI] [PubMed] [Google Scholar]
- 40. Petritis K., Kangas L. J., Ferguson P. L., Anderson G. A., Pasa-Toliæ L., Lipton M. S., Auberry K. J., Strittmatter E. F., Shen Y., Zhao R., Smith R. D. (2003) Use of artificial neural networks for the accurate prediction of peptide liquid chromatography elution times in proteome analyses. Anal. Chem. 75, 1039–1048 [DOI] [PubMed] [Google Scholar]
- 41. Petritis K., Kangas L. J., Yan B., Monroe M. E., Strittmatter E. F., Qian W. J., Adkins J. N., Moore R. J., Xu Y., Lipton M. S., Camp D. G., 2nd, Smith R. D. (2006) Improved peptide elution time prediction for reversed-phase liquid chromatography-MS by incorporating peptide sequence information. Anal. Chem. 78, 5026–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kyte J., Doolittle R. F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 [DOI] [PubMed] [Google Scholar]
- 43. Rietschel B., Arrey T. N., Meyer B., Bornemann S., Schuerken M., Karas M., Poetsch A. (2009) Elastase digests: new ammunition for shotgun membrane proteomics. Mol. Cell. Proteomics 8, 1029–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jones L. M., Zhang H., Vidavsky I., Gross M. L. (2010) Online, high-pressure digestion system for protein characterization by hydrogen/deuterium exchange and mass spectrometry. Anal. Chem. 82, 1171–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shui W., Fan J., Yang P., Liu C., Zhai J., Lei J., Yan Y., Zhao D., Chen X. (2006) Nanopore-based proteolytic reactor for sensitive and comprehensive proteomic analyses. Anal. Chem. 78, 4811–4819 [DOI] [PubMed] [Google Scholar]
- 46. Slysz G. W., Lewis D. F., Schriemer D. C. (2006) Detection and identification of sub-nanogram levels of protein in a nanoLC-trypsin-MS system. J. Proteome Res. 5, 1959–1966 [DOI] [PubMed] [Google Scholar]
- 47. Percy A. J., Schriemer D. C. (2010) Rheostatic control of tryptic digestion in a microscale fluidic system. Anal. Chim. Acta 657, 53–59 [DOI] [PubMed] [Google Scholar]
- 48. Yang H. J., Hong J., Lee S., Shin S., Kim J., Kim J. (2010) Pressure-assisted tryptic digestion using a syringe. Rapid Commun. Mass Spectrom. 24, 901–908 [DOI] [PubMed] [Google Scholar]