Abstract
Detection of low-affinity or transient interactions can be a bottleneck in our understanding of signaling networks. To address this problem, we developed an arrayed screening strategy based on protein complementation to systematically investigate protein-protein interactions in live human cells, and performed a large-scale screen for regulators of telomeres. Maintenance of vertebrate telomeres requires the concerted action of members of the Telomere Interactome, built upon the six core telomeric proteins TRF1, TRF2, RAP1, TIN2, TPP1, and POT1. Of the ∼12,000 human proteins examined, we identified over 300 proteins that associated with the six core telomeric proteins. The majority of the identified proteins have not been previously linked to telomere biology, including regulators of post-translational modifications such as protein kinases and ubiquitin E3 ligases. Results from this study shed light on the molecular niche that is fundamental to telomere regulation in humans, and provide a valuable tool to investigate signaling pathways in mammalian cells.
During mammalian DNA replication, linear chromosomal ends will gradually erode because of the inability of the DNA replication machinery to replicate the extreme 5′ terminus of a linear DNA sequence (1, 2). This inherent “end replication problem” is circumvented through specialized chromosomal end structures (telomeres) and the action of the RNA-containing DNA polymerase - telomerase (3–9). Telomere homeostasis is essential for genome stability, cell survival, and growth. Telomeres and telomerase help to ensure genome integrity in eukaryotes by enabling complete replication of the ends of linear DNA molecules, and preventing chromosomal rearrangements or fusion. For dividing cells such as stem cells and the majority of cancer cells, the telomerase is an essential positive regulator of their telomere length and ultimately determines the proliferative potential of these cells.
Mammalian telomeres consist of a series of (TTAGGG)n sequence repeats and terminate in 3′ single-stranded DNA overhangs that are extendable by the telomerase (10). Exposed linear chromosome ends or naturally occurring double-stranded breaks pose additional risks including activation of DNA damage responses. The ends of telomeres in mammalian cells appear to fold back in a T-loop structure, with the 3′ G-rich single-stranded overhang invading into the double-stranded telomere regions to form the d-loop (11). The structure of the telomeres, coupled with the coordinated action of a collection of proteins that protect the ends of chromosomes (12–15), contributes to the maintenance of telomere integrity, genome stability, and proper cell cycle progression.
In mammals, the most widely studied telomere-associated proteins include the double-stranded DNA binding proteins TRF1 and TRF2 (16, 17), the single-stranded telomeric DNA binding protein POT1 (18), and three associated factors (RAP1, TIN2, and TPP1) (19–23). Work from our lab and others suggest that TPP1, along with POT1, TIN2, TRF1, TRF2, and RAP1, form a higher order complex (the telosome/shelterin) at the telomeres (24–27). Information regarding the state of the telomere ends can be transmitted from TRF1 and TRF2 to POT1, through TPP1 and the other subunits (28). Furthermore, TRF1 and TRF2 function as bona fide protein hubs and interact with a diverse array of factors/complexes that are involved in cell cycle, DNA repair, and recombination to maintain telomere structure and length (12, 13, 27, 29–34). Consistent with the end protection function of this complex, many factors that are known to participate in DNA damage responses are recruited to the telomeres, such as the Mre11/Rad50/NBS1 complex, PARP-1, Ku70/80, DNA helicases BLM and WRN, Rad51D, nucleotide excision repair protein ERCC1/XPF, DNA nuclease Apollo, and the BRCT domain-containing protein MCPH1 (3, 4, 10, 20, 35–53). To date, much has been learned regarding the core telomere binding components, factors that constitutively associate with the telomeres. However, much remains unknown regarding the factors that are recruited to the telomeres upon damage or other signaling events, as well as the signaling cascades that must take place on or near the telomeres. In other words, the micro-environment—the complex regulatory network of protein-protein interactions—within which telomere homeostasis is achieved remains to be elucidated. Signaling regulators are often of low abundance, and their association with the targets may be transient or weak. Although conventional proteomic methods such as immunoprecipitation (IP) and mass spectrometry have been particularly informative in identifying core interacting proteins, regulatory components may be below the detection threshold.
We have developed a high-throughput protein-protein interaction screening strategy based on the principle of the yellow fluorescent protein (YFP)1-based protein complementation assay (PCA/bimolecular fluorescent complementation (BiFC)) (52, 54–56). In the PCA/BiFC assay, protein-protein interactions bring the two fragments (YFPn and YFPc) of YFP (tagged to two separate proteins) to close proximity and allow for their cofolding into a functional fluorescent protein (57, 58). PCA/BiFC enables the examination of interactions in live cells, providing spatial information about protein-protein interactions. Here we report the identification of over 300 telosome/shelterin associating proteins that mediate diverse signaling pathways. Many of these proteins regulate post-translational modifications including protein phosphorylation and ubiquitination. Our findings provide a high-resolution map of the telomere interactome (19, 59, 60), and should greatly facilitate further studies of telomere signaling.
EXPERIMENTAL PROCEDURES
Establishment of Stable Cell Lines
pDONR223 vectors encoding SOX2, TRF1, RAP1, TIN2, and POT1 came from human ORFeome v3.1 (Open Biosystems). Sequences encoding human TRF2 and TPP1 were PCR amplified and cloned into pENTR/d-TOPO vector (Invitrogen). Through Gateway recombination, the six telomere open reading frames (ORFs) were transferred individually into either pBabe-CMV-YFPn-DEST-neo or pBabe-CMV-DEST-YFPn-neo vectors (56). The resulting mammalian expression constructs enable tagging of Venus YFPn fragments at either the N- or C terminus of a protein. The vectors were then transfected individually into Phoenix cells for retrovirus production and subsequent infection of HTC75 cells. Infected cells were selected with 300 μg/ml G418 for up to 10 days to obtain cells stably expressing YFPn-tagged bait proteins. For each bait, cells that expressed either N- or C-terminally tagged YFPn-fusion proteins were mixed (1:1) for subsequent screens.
Establishment of Retroviral Array Libraries
Individual Gateway recombination reactions (in 96-well plates) were performed for all 12,212 ORFs from hORFeome with a mixture of pCl-CMV-YFPc-DEST-puro and pCL-CMV-DEST-YFPc-puro vectors (1:1) (56), to generate ORFs tagged with YFPc at either the N- or C terminus. The reaction products were subsequently used to transform DH5α and selected by ampicilin. Plasmid extractions were then carried out in 96-well plates for each pool of bacterial transformants using PureLink HQ 96 plasmid purification kit (Invitrogen) and Biomek FX Laboratory Automation Work station (Beckman Coulter). 11,880 ORFs were successfully cloned into the YFPc vectors. The YFPc-prey collections were then transfected into Phoenix cells to generate retroviruses for infection of bait cells. Retroviral supernatant was collected at 48 h or 72 h post-transfection, and stored in −80 °C before use. All transfection steps were done in 96-well formats using the Biomek 3000 Laboratory Automation Work station (Beckman Coulter).
High-Throughput Protein Complementation Array Screen Strategy
Cells from each bait cell line were seeded onto 96-well plates and infected with the arrayed YFPc-tagged prey library. At 2 days following the infection, cells were selected with 1 μg/ml of puromycin for 5–10 days. We were able to obtain 10058 SOX2, 11,685 TRF1, 11,006 TRF2, 11,724 RAP1, 11,398 TIN2, 11,385 TPP1, and 11,330 POT1 cell lines infected by the YFPc-tagged prey library. All work was performed using the Biomek3000 Laboratory Automation Work station. Cells were then harvested for high-throughput flow cytometric analysis using the LSRII flow cytometer equipped with a HTS sampler (BD Biosciences).
CytoArray
CytoArray is a data analysis platform custom designed for processing the large amount of flow cytometry data from the arrayed screen. Data processing is roughly divided into four major steps: defining positive regions, calculating weighted positive ratios (WPR), determining statistically significant cutoff values, and removing common contaminants (Supplemental Fig. S2).
Data from each well are plotted with green fluorescent protein (GFP) on the x axis and phycoerythrin on the y axis. First, any samples with <200 data points are automatically discarded. The remaining profiles are processed plate-by-plate for each bait. All the profiles within a plate are compiled to create a composite profile. It is assumed that the majority of these flow cytometry profiles would be negative for PCA/BiFC signals and closely resemble each other. Therefore, the composite profile can be used as an internal control to gate for negative versus positive regions when superimposed onto individual profiles within the same plate (Supplemental Fig. S3). For the composite profile, CytoArray determines the adjusted vertex and weight center of data point distribution, and uses the y value of the adjusted vertex and x value of the weight center to define the top and left boundary of the positive region. Starting from here, the remaining boundaries are defined by scanning across the distribution profile until this positive region contains 5% of data points.
CytoArray calculates weighted positive ratios (WPR) rather than positive percentages (PP: positive cell number/total live cell number) to measure PCA/BiFC signals, because PP ignores signal intensity (ratio of YFP/phycoerythrin) and does not differentiate between marginal versus significant data points. WPR increases the signal to noise ratio and improves the sensitivity of detection (Supplemental Fig. S3). WPR ranks all positive data points (on an arbitrary scale of 1–5) according to their distance from the leftmost boundary of the positive region, with the data points furthest away from that boundary assigned the highest value.
Next, CytoArray calculates the WPR cutoff. When the distribution of the WPR values from all the proteins that were defined as extracellular (based on cellular component annotation in Gene Ontology database) is compared with that of the WPR values from the entire screen for each bait, the trend is clear that the higher the WPR value, the fewer of these proteins can be found. Using these two distribution curves, CytoArray calculates the WPR ratios between them. For example, when the ratio is 90%, the corresponding WPR value is the threshold cutoff with a 90% positive ratio (Supplemental Fig. S4). Finally, common contaminants and nonspecific binding proteins are filtered out using data from similar screens of the unrelated bait SOX2.
Coprecipitation
To validate the interactions between each bait and prey protein pair, coprecipitation bythe glutathione S-transferase (GST) pull-down was performed in 96-well format using 96-well filter plates from Invitrogen. Sequences encoding telomeric proteins and candidate prey proteins were cloned into pDEST-27 (Invitrogen) for tagging with GST and pCl-2xFLAG for tagging with FLAG, respectively. Each bait-prey pair was cotransfected into 293T cells. The transfected cells were harvested after 2 days, and lysed with 1×NETN buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 0.5% Nonidet P-40, and 1 mm EDTA). The whole cell extract was then transferred to a 96-well lysate clearing filter plate (25 μm, Phenix), and centrifuged at 500 × g for 2 min (4 °C). The cleared lysate was then transferred into a 96-well binding plate (Purelink Clarificaiton plate, Invitrogen) preloaded with 100 μl/well of glutathione Sepharose 4B beads (10% slurry) (GE Healthcare Bio-Sciences AB), and incubated for 2 h at 4 °C with gentle agitation. The binding plate was then washed three times with 1× NETN, and the bound proteins eluted with elution buffer (50 mm Tris-HCl, pH 8.0, 20% glycerol, 20 mm reduced glutathione), centrifuged at 500 × g for 2 min and blotted onto polyvinylidene fluoride membranes using the Bio-Dot apparatus (Bio-Rad). The membrane was then probed with anti-FLAG-HRP (Sigma) or anti-GST-HRP (GE Healthcare Bio-Sciences AB) antibodies.
RESULTS
Examining Pair-Wise Interactions Among the Six Core Telomeric Proteins
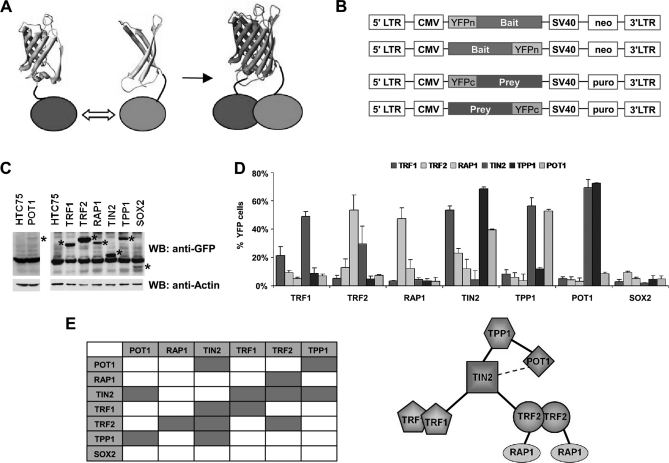
To investigate protein-protein interactions in human cells, we adopted and modified the YFP-based PCA/BiFC method (52, 56, 58). The N-terminal fragment (residues 1–155) of Venus YFP (YFPn) or the C-terminal fragment (residues 156–239) of YFP (YFPc) were tethered to either N- or C-terminal ends of candidate proteins and expressed in human cells for fluorescence complementation (Figs. 1A and 1B). A flexible linker of ∼30 amino acids, which covers a distance of ∼10 nm, was engineered to maximize complementation. Tagging protein pairs on either end with the two YFP fragments would cover all possible interaction configurations. Testing all these combinations individually, however, would be impractical on a large scale. Here, we performed PCA/BiFC assays using pools of cells expressing N- or C-terminally tagged fusion proteins (Fig. 1B). For example, bait-expressing cells were a mixture of cells that individually expressed bait proteins tagged with YFPn at either the N- or C terminus. Likewise, each prey protein was represented by a mixture of retroviruses encoding YFPc- fusion proteins tagged at either the N- or C terminus. As a result, four tagging combinations were simultaneously analyzed for each bait-prey pair.
Fig. 1.
PCA/BiFC reveals complex interactions among the six telomeric proteins. A, Principle of the PCA/BiFC assay. The N-terminal fragment (residues 1–155) of Venus YFP (YFPn) and C-terminal fragment (residues 156–239) of YFP (YFPc) were tethered to the N- or C-terminal ends of candidate proteins and expressed in human cells for fluorescence complementation. B, Schematic representation of the retroviral expression vectors for BiFC-based screening. LTR, long terminal repeat. Puro, puromycin selection marker. Neo, neomycin selection marker. YFPn and YFPc, N- or C-terminal fragment of YFP. C, Whole cell extracts from HTC75 cells expressing different Venus YFPn-tagged proteins were Western blotted with anti-GFP antibodies. Fusion proteins are indicated by *. Cells expressing YFPn-tagged SOX2 were used as a control for subsequent analysis. D, Flow cytometry analysis of the 6 × 6 PCA/BiFC assays of core telomeric protein pairs. YFPn-tagged SOX2 served as a negative control. E, Structural arrangement of the subunits within the six telomeric protein complex as determined by PCA/BiFC assays as in (D).
Interactions among the six core telomere proteins, TRF1, TRF2, RAP1, TPP1, TIN2, and POT1 have been studied extensively (19–21, 23–26, 61). For example, TIN2 is known to bind TRF1, TRF2, and TPP1, whereas TPP1 and TRF2 interact with POT1 and RAP1 respectively. In addition, TRF1 and TRF2 may form homodimers. We first utilized the modified PCA/BiFC strategy to analyze the pair-wise interactions within these six proteins (Figs. 1C and 1D). As shown in Fig. 1D, we found the pool/mixture strategy could accurately predict pair-wise interactions among these proteins, allowing us to construct the structural organization of the complex formed by the six telomeric proteins (Fig. 1E). Importantly, tagging YFP fragments onto either protein of the interacting pair resulted in similar percentages of YFP-positive cells (Fig. 1D). We also detected positive YFP signals for the TIN2-POT1 pair, suggesting a close proximity of these proteins in vivo. These findings indicate that the modified PCA/BiFC method worked well in detecting protein-protein interactions among the core telomeric proteins, as well as predicting their relative positions within the complex.
It is important to note that variations in expression levels of bait or prey proteins appeared not to affect fluorescence complementation or the sensitivity of signal detection (Fig. 1C), underlining the robustness of the assay. For example, POT1 exhibited fluorescence complementation comparable to others despite having the lowest expression level among the six telomeric proteins, suggesting that the initial level of YFP fusion proteins is not a major factor in this assay. It is possible that once the correct interacting partners meet, their interaction may help to stabilize both the prey and bait proteins, allowing for the detection of fluorescence complementation.
Genome-wide screening of proteins that interact with the six telomeric proteins
On the basis of our pilot studies, we developed an arrayed screening strategy based on protein complementation for genome-wide screening of protein-protein interactions and performed a screen for proteins that may be involved in telomere signaling in human cells (Supplemental Fig. S1). We first generated individual bait HTC75 cell lines that expressed various Venus YFPn-tagged telomeric proteins (and a negative control bait protein SOX2). Then, a YFPc-tagged expression prey library was constructed using the human ORFeome library (62). This ORFeome library, which contained 12,212 human full-length cDNAs in the pDNOR vector, was transferred well-by-well into YFPc-tagged retroviral destination vectors using the Gateway recombination system, resulting in a YFPc-tagged prey library arrayed in 134 96-well plates (Supplemental Fig. S1). Each reaction contained equal amount of YFPc-tagged retroviral destination vectors for either N- or C-terminal tagging. The success rate for cloning was estimated at 97%.
The YFPc prey library was transfected into packaging cell lines to produce retroviruses, which were subsequently used to infect bait cells (Supplemental Fig. S1). YFPc vectors encoding the six telomeric proteins were also added in this step to serve as positive controls for the screen. The infected cells were then selected in puromycin to enrich for prey expressing cells, and analyzed by a high-throughput flow cytometer to assess fluorescence complementation. A typical round of the arrayed screen for the six telomeric proteins included about 7 × 12,000 individual flow cytometry measurements.
CytoArray: An Automated Data Analysis Platform
To analyze the large amount of data generated from the screens, we developed the CytoArray program that takes into consideration various factors that affect the determination of negative versus positive populations, including variability in sample size and data plots. In Supplemental Fig. S2, we illustrated the workflow for data processing using CytoArray, and the steps taken to determine positive populations and reduce false positives (please see Experimental Procedures for a detailed description).
First, samples with <200 cells are discarded. The program then assumes that the majority of samples should exhibit no fluorescence complementation and largely resemble each other in the overall profile. Consequently, the data files for each plate are compiled into a single composite profile, which is then superimposed onto individual data files within that plate to gate for negative populations. Positive regions are thus defined and a weighed positive ratio (WPR) is calculated for each prey-bait pair (Supplemental Fig. S3). WPR takes into consideration both the fluorescence signal strength and number of YFP positive cells. We reasoned that in addition to the percentage of YFP positive cells, fluorescence intensity might also correlate with the frequency of protein-protein interactions and provide another indicator of the proximity of candidate interacting proteins. WPR values therefore can be used to rank interactions revealed by PCA/BiFC.
One concern with the PCA/BiFC assay is potential false-positives that result from nonspecific interactions or spontaneous cofolding of the YFP fragments (57, 58, 63). To address this, we established a bait-specific threshold using WPR values from all the genes and a subset of ∼700 extracellular proteins. The latter may be secreted or targeted to the membrane, therefore unlikely to interact with telomeric proteins. This internal control helped us to determine the cutoff values for positive interactions (Supplemental Fig. S4 and Supplemental Fig. S5A). We then selected data for the six telomere proteins from the arrayed screen to generate a 6 × 6 interaction matrix to test various thresholds (Supplemental Fig. S5B). At the 90% confidence level, our calculated thresholds predicted most of the interactions except for the TIN2-TRF2 pair (the weakest interaction as determined experimentally), indicating that our statistical process for determining cutoffs is stringent in scoring potential interacting proteins. To further exclude possible hits because of spontaneous cofolding of the YFP fragments, we performed a control screen using the transcription factor SOX2 that is not known to play a role in telomere biology (data not shown). The telomeric protein screen data were further processed to remove cross-reactive proteins found in the SOX2 screen, allowing us to eliminate 30 promiscuous proteins. An additional 21 proteins were discarded because they failed secondary screens. Through CytoArray, we obtained a high-quality data set containing 320 candidate human proteins that represent about 600 interactions (Supplemental Table S1).
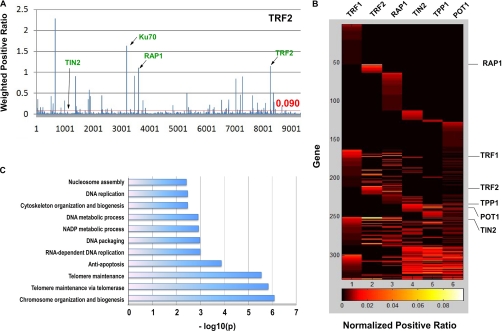
The list of candidate proteins identified from the arrayed screens includes all six telomeric proteins (Supplemental Table S1). RAP1 and POT1 emerged respectively as the top interactors of TRF2 and TPP1 (Fig. 2A and Supplemental Table S1), consistent with previous findings that TRF2-RAP1 and POT1-TPP1 form stable heterodimers (20, 64, 65). Within the human ORFeome library, a total of 14 full-length proteins have been reported to associate with the core telomere complex through 20 interactions. On the basis of these results (six missed interactions), we calculated the false-negative rate to be ∼30% (6/20). Two other known interactors, XRCC6/Ku70 (35) and PINX1 (66), were also among the best interacting proteins for TRF2 and TRF1 respectively (Fig. 2A and Supplemental Table S1).
Fig. 2.
Identification of proteins that associate with the six telomeric proteins through our BiFC-based screens. A, Screening data for TRF2. On the basis of CytoArray analysis, a cutoff value of 0.090 (90% confidence level) for the weighted positive ratio (WPR) was applied (indicated by the red line). X axis, prey proteins scored in the screen. Y axis, WPR values. Arrows indicate known interacting prey proteins. B, A heat-map of the identified interaction partners of the core telomere associated proteins. C, Enriched biological processes identified by the screens.
Construction of a High-Resolution Map of the Telomere Interactome
In addition to known interactors, the screens revealed ∼300 new putative regulators of telomeres (Supplemental Table S1). Approximately half of the identified proteins (161) interact with a single bait protein, suggesting that their association with the telomeric proteins did not result from spontaneous cofolding of YFP fragments. Some proteins appear to interact with up to five bait proteins, which may be partly explained by the fact that fluorescence complementation can occur between proteins that are close to each other, without direct contact. Therefore, positive PCA/BiFC signals may indicate close proximity rather than direct interactions. We further examined the interaction data set by analyzing the clustering of the identified proteins from all six baits (Fig. 2B). In theory, bait telomeric proteins that can interact with each other should give rise to targets that display significant overlap. Indeed, TRF2 interacting proteins are more closely clustered with RAP1-interacting proteins (p = 1.7E-05), whereas the TIN2-, TPP1-, and POT1- interacting proteins are clustered together (p < 2.6E-05) (Fig. 2B), consistent with our small-scale six-protein screen (Fig. 1D).
On the basis of our data and CytoArray analysis, we constructed a high-resolution map of the telomere interacting network -Telomere Interactome (Supplemental Fig. S6). Notably, >80% of the interactions in our data set have not been previously reported (28, 67). This may reflect one of the major advantages of PCA/BiFC - its ability to capture transient, infrequent, or low affinity interactions often missed by approaches such as IP/mass spectrometry. Using stringent cutoff criteria, we found 11 Gene Ontology biological processes that are statistically enriched in the network (Fig. 2C). Consistent with current knowledge about telomere biology, the top categories include telomere maintenance, chromosome organization and biogenesis, and anti-apoptosis.
Secondary Analysis of Candidate Proteins
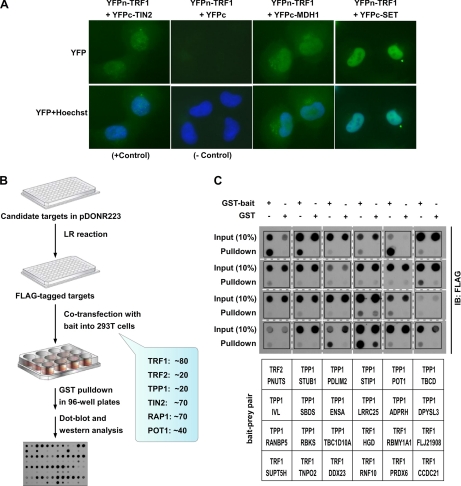
To confirm the results from our screens, we carried out secondary PCA/BiFC screens and found that the ∼300 proteins identified remained positive in individual PCA/BiFC assays (Fig. 3A). Next, we tagged the ∼300 genes with FLAG tag and coexpressed them individually with their respective GST-tagged bait proteins (Fig. 3B, Supplemental Fig. S7). Binding of these proteins to telomeric proteins was then determined by GST pull down and dot blotting (Fig. 3C, Supplemental Fig. S8A–E). On the basis of our analysis of the GST pulldown experiments (Supplemental Fig. S8F and Supplemental Table S2), ∼72% of the interactions could be verified by coprecipitation experiments. Both weak and strong binding signals could be observed. Such differences may reflect the range of interactions between bait and prey, both in their affinity and frequency, and lend more support to using PCA/BiFC to identify low affinity and transient interactions. These results further demonstrate the quality and reliability of our screen data. The candidate proteins that failed to bind in the coprecipitation assay may represent false positives, or require bridging factors or different conditions for the association to occur.
Fig. 3.
Confirmation of protein-protein interactions identified by the screens. A, Fluorescent microscopy images of bait-prey protein pairs tagged with YFP fragments. HTC75 cells stably coexpressing YFPn-tagged TRF1 and YFPc alone (negative control), YFPc-tagged TIN2 (positive control), YFPc-MDH1, or YFPc-SET were visualized live under a fluorescence microscope. Hoechst33342 was added to visualize the nuclei. B, A flow chart of secondary coprecipitation screens that were used to confirm the identified protein-protein interactions. C, Examples of coprecipitation screens. Cell extracts from 293T cells coexpressing FLAG-tagged candidate proteins with GST alone or GST-tagged telomeric bait proteins were incubated with GSH-agarose beads. The proteins bound to GSH beads were then dot-blotted and probed with anti-FLAG antibodies.
Identification of Regulatory Pathways in Telomere Signaling
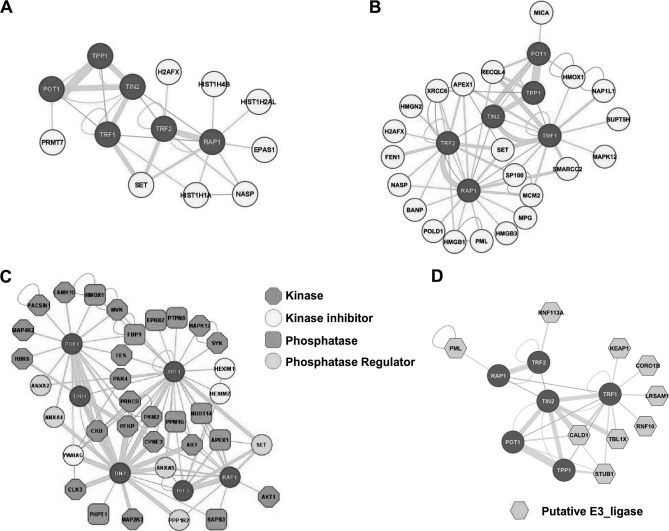
Telomere DNA and telomere binding proteins form the telomere chromatin. It is therefore expected that core telomere proteins will be in the vicinity of other DNA and chromatin associating factors. Indeed, 44 DNA binding proteins and chromatin regulators were identified in the Telomere Interactome (Supplemental Table S1). Among them are several RNA/DNA helicases including DDX19B, 21, 23, 24, and 38, as well as several DNA and protein methyltransferases including DNMT3A and PRMT7. An interesting pattern of association between histone proteins and the core telomeric proteins also emerges (Fig. 4A). In particular, the TRF2-RAP1 heterodimer is in the vicinity of histone H2A, H4, and H1. Whether this network represents the organization of telomere nucleosome warrants future investigation. Finally, the six telomeric proteins are known to communicate with the DNA damage repair pathways for telomere maintenance. Our screen has added additional players including the DNA base excision enzyme APEX1, endonuclease FEN1, helicase RECQL4, and DNA polymerase POLD1 (Fig. 4B).
Fig. 4.
Identification of signaling networks and regulatory components in the Telomere Interactome. A, Interaction of telomeric proteins with histone proteins. B, DNA repair and chromatin regulators that associate with the six telomeric proteins. C, Protein kinases, phosphatases, and their regulators are identified from the screen. D, Putative ubiquitin E3 ligases were found in the screen.
Among the ∼300 proteins identified, many are regulators of protein phosphorylation and ubiquitination (Fig. 4C and D). These include serine/threonine kinases (Akt1, CAMK1D, CLK3, MAP2K3, MAP4K2, MAPK12, and PAK4), and protein phosphatase catalytic and regulatory subunits (PPM1G, PHPT1, PTPN5, SAPS3, and PPP1R2). These proteins may form the circuitry that controls the phosphorylation of the six core telomeric proteins. Several RING finger or U-box containing proteins have also been found (Fig. 4D). These proteins could function as ubiquitin E3 ligases or stability regulators for telomere-associated proteins. Taken together, our study has demonstrated that regulatory components of the telomere interactome can be readily identified by our protein complementation based array screen. The results presented here should act as a catalyst for future investigations into the multitude of regulatory pathways at work for maintaining mammalian telomeres.
DISCUSSION
The importance of mapping protein-protein interaction networks and elucidating regulatory components that are integral to biological pathways cannot be overstated. Signaling regulators are often of low abundance, and their association with the targets may be transient or weak. Such characteristics make it difficult to capture and study key regulatory interactions, despite the wide array of tools that have been developed over the years for protein-protein interactions. Recent studies have demonstrated the benefit of detecting protein-protein interactions using protein complementation assay (PCA) systems (68). For example, a dihydrofolate reductase-based PCA method was used to investigate the yeast interactome, where thousands of interactions were identified (69). Given that many of the yeast genetic tools are not yet available in mammals, we decided to employ the PCA method that utilizes split Venus YFP to analyze the interactomes in human cells (52, 54, 56, 58). Compared with approaches such as yeast-two-hybrid and co-IP/mass spectrometry, it offers distinct advantages. Pair-wise protein-protein interactions are analyzed in live human cells, providing spatial information about protein-protein interactions. As a nontranscription based approach, it avoids bait self-activation and nonnuclear localization issues that frequently plague two-hybrid methods. PCA/BiFC is ideal for live or single cell experiments, circumventing the need for large numbers of cells and lengthy in vitro purification steps as is the case for IP/mass spectrometry. Most importantly, transient or weak interactions as well as low abundance regulators that may be lost during in vitro purification steps, can be “trapped” thanks to the cofolding of YFP fragments. This attribute really sets it apart from other screening methods including PCA approaches that utilize dihydrofolate reductase or luciferase (68, 70), and enables it to more readily identify regulatory interactions such as those between enzymes and their substrates. In support of this notion, we recently used a BiFC-based screening strategy and identified a rac-GDI protein that binds CARD9 in macrophages in a bacterial infection-dependent manner (71). We anticipate that our screen strategy will make major impact in this area and help to facilitate the process of identifying signal dependent interactions.
The screen strategy described here detects pair-wise interactions in the human proteome in a systematic manner, enables the identification of regulatory interactions such as those between enzymes and their substrates, and creates a high-resolution map of the interactome. Here we report our work on elucidating the interaction networks centering around the six core telomeric proteins. Screens utilizing split GFP or YFP have been performed previously (72, 73), however, split GFP or YFP cofolding is much less efficient than Venus YFP (74). In addition, previous screens were not systematic and the resulting interactions were not further characterized, and nonspecific interactions because of spontaneous cofolding of the YFP fragments were not eliminated. This is the first time an array-based high-throughput protein complementation screen technology has been used to map interaction networks in live human cells. Some of the identified proteins did not appear to localize to the telomeres in our secondary screens. Although we cannot rule out the possibility that they are false positives, it is possible that these proteins may be targeted to telomeres under specific conditions. Furthermore, telomeric proteins travel through different cellular compartments following their synthesis and have been implicated in nontelomeric activities (75, 76). Therefore, these proteins may have novel function outside of the telomeres. Nevertheless, the large number of newly identified and confirmed telomere interacting proteins is a testimony to the power of the screens. We believe that this technology will be an extremely valuable tool to study protein networks and signaling transduction in general, and help to relieve the bottleneck in our understanding of signaling pathways in human cells.
Our current library should detect the majority of interacting proteins within a radius of ∼20 nm. Differentiating between constitutive versus induced interactions (in response to signaling cues) can be easily achieved by our screening strategy. One limitation is the need to coexpress YFP-tagged bait and prey proteins in the same cells in order to achieve fluorescence complementation. YFP tags may alter the conformation or activity of the tagged proteins and lead to false negatives. In theory, high-affinity, direct interactions are likely to lead to higher percentage of PCA/BiFC positive cells and stronger fluorescence complementation. Although PCA/BiFC does appear to be more tolerant of differences in expression, high expression levels of bait and prey may still result in false positives. Therefore, secondary screens are needed to validate the identified interactions. The method described here may be further improved with an inducible expression system, particularly in cases where prolonged stable interactions between two proteins might be detrimental to cells. This will help to reduce the rate of false negatives as well. Our current PCA/BiFC array library contains ∼12,000 genes, approximately half of the genes in the human genome. Expanding the array library will certainly facilitate more complete interactome mapping.
Protein-protein interaction networks in human cells are much more complex than we originally anticipated (77). Our screens offer a multitude of candidates for further analysis of their potential role in telomere biology or other pathways. Over 80% of the identified proteins have been validated by secondary screens, which translates into an average of 40 binding partners for a given core telomere protein. It is unlikely that the identified ∼300 proteins bind simultaneously with the six telomeric proteins. Instead, they may form a variety of subcomplexes and associate with the six core telomeric proteins in different cellular compartments or at different times to mediate diverse biological processes. It is equally possible that some interactions are cell-type dependent or developmentally regulated. Further studies are needed to unravel how these processes contribute to the maintenance of telomere homeostasis.
Consistent with the major roles of the six telomeric proteins, the top category of biological processes scored in our screen belongs to chromatin organization, biogenesis, and telomere maintenance (Fig. 2C). In this category, we found many enzymes including helicases, methyltransferases, acetyl-transferases, kinases, and phosphatases. Whether these enzymes associate with the telomeres constitutively or transiently remain to be investigated. Understanding how these enzymes are regulated and their roles in telomere capping will be an intriguing area of research in the near future. Another interesting finding from our screen is the closeness of the TRF2-RAP1 heterodimer, but not TIN2-TPP1-POT1 subcomplex, to core histone subunits. This structural organization of the telomere chromatin is consistent with the model that the TPP1-POT1 complex is primarily involved in telomere ssDNA protection, and thus positioned more distal from the histone-coated dsDNA region.
It is intriguing to find apoptosis as one of the top biological processes mediated by the Telomere Interactome. Several proteins that are involved in stress response pathways and mitochondria function were found in our screens. It is possible that telomere dysfunction may trigger stress and apoptosis signaling through these proteins. In support of this hypothesis, it has been shown that disregulation of telomeric proteins in mammalian cells renders these cells sensitive to apoptosis in both p53-dependent and independent manner (78–82). Our result suggests that the six telomeric proteins may have a more direct role in connecting telomere dysfunction to apoptosis.
In addition, we found a collection of E3 ligases that likely control the stability and/or function of the six core telomeric proteins. More than one E3 ligase may associate with a given telomeric protein, perhaps to respond to distinct cellular signaling cues or function in different cellular compartments. As we begin to address the relationship between ubiquitination and telomere homeostatsis, such findings offer important clues to the participants of these biological events. We demonstrate here that our arrayed screen strategy can identify regulatory components of the telomere interactome, which makes it an invaluable tool for signal transduction and mechanistic studies.
Acknowledgments
We would like to thank Dr. Yun Kyoung Kang for technical support.
Footnotes
* This work is supported by NCI CA133249, NIGMS GM081627, the Welch Foundation Q-1673, and 2010CB945400. Z.S. is a Leukemia and Lymphoma Society Scholar. We would also like to acknowledge the support of the Dan L. Duncan Cancer Center (P30CA125123), and the Administrative and Genome-wide RNAi Screens Cores (IDDRC P30HD024064).
 This article contains supplemental Figs. 1–8 and Tables 1–2.
This article contains supplemental Figs. 1–8 and Tables 1–2.
1 The abbreviations used are:
- YFP
- yellow fluorescence protein
- PCA
- protein complementation assay
- BiFC
- bimolecular fluorescence complementation
- IP
- immunoprecipitation
- ORF
- open reading frame
- WPR
- weighted positive ratios
- GST
- glutathione S-transferase.
REFERENCES
- 1. Watson J. D. (1971) The regulation of DNA synthesis in eukaryotes. Adv. Cell Biol. 2, 1–46 [DOI] [PubMed] [Google Scholar]
- 2. Olovnikov A. M. (1973) A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 41, 181–190 [DOI] [PubMed] [Google Scholar]
- 3. Autexier C., Greider C. W. (1996) Telomerase and cancer: revisiting the telomere hypothesis. Trends Biochem. Sci. 21, 387–391 [PubMed] [Google Scholar]
- 4. McEachern M. J., Krauskopf A., Blackburn E. H. (2000) Telomeres and their control. Annu. Rev. Genet. 34, 331–358 [DOI] [PubMed] [Google Scholar]
- 5. Cech T. R. (2000) Life at the End of the Chromosome: Telomeres and Telomerase. Angew Chem. Int Ed. Engl. 39, 34–43 [DOI] [PubMed] [Google Scholar]
- 6. Harley C. B. (2002) Telomerase is not an oncogene. Oncogene 21, 494–502 [DOI] [PubMed] [Google Scholar]
- 7. Cong Y. S., Wright W. E., Shay J. W. (2002). Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 66, 407–425, table of contents [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Lange T. (2002) Protection of mammalian telomeres. Oncogene 21, 532–540 [DOI] [PubMed] [Google Scholar]
- 9. Wong J. M., Collins K. (2003) Telomere maintenance and disease. Lancet 362, 983–988 [DOI] [PubMed] [Google Scholar]
- 10. Cech T. R., Nakamura T. M., Lingner J. (1997) Telomerase is a true reverse transcriptase. A review. Biochemistry 62, 1202–1205 [PubMed] [Google Scholar]
- 11. Griffith J. D., Comeau L., Rosenfield S., Stansel R. M., Bianchi A., Moss H., de Lange T. (1999) Mammalian telomeres end in a large duplex loop. Cell 97, 503–514 [DOI] [PubMed] [Google Scholar]
- 12. Blackburn E. H. (2001) Switching and signaling at the telomere. Cell 106, 661–673 [DOI] [PubMed] [Google Scholar]
- 13. Maser R. S., DePinho R. A. (2002) Connecting chromosomes, crisis, and cancer. Science 297, 565–569 [DOI] [PubMed] [Google Scholar]
- 14. Smogorzewska A., de Lange T. (2004) Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 73, 177–208 [DOI] [PubMed] [Google Scholar]
- 15. Wright W. E., Shay J. W. (2005) Telomere-binding factors and general DNA repair. Nat. Genet. 37, 116–118 [DOI] [PubMed] [Google Scholar]
- 16. Bianchi A., Smith S., Chong L., Elias P., de Lange T. (1997) TRF1 is a dimer and bends telomeric DNA. EMBO J. 16, 1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Broccoli D., Smogorzewska A., Chong L., de Lange T. (1997) Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 17, 231–235 [DOI] [PubMed] [Google Scholar]
- 18. Baumann P., Cech T. R. (2001) Pot1, the putative telomere end-binding protein in fission yeast and humans. Science 292, 1171–1175 [DOI] [PubMed] [Google Scholar]
- 19. Kim S. H., Kaminker P., Campisi J. (1999) TIN2, a new regulator of telomere length in human cells. Nat. Genet. 23, 405–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li B., Oestreich S., de Lange T. (2000) Identification of human Rap1: implications for telomere evolution. Cell 101, 471–483 [DOI] [PubMed] [Google Scholar]
- 21. Liu D., Safari A., O'Connor M. S., Chan D. W., Laegeler A., Qin J., Songyang Z. (2004). PTOP interacts with POT1 and regulates its localization to telomeres. Nat. Cell Biol. 6, 673–680 [DOI] [PubMed] [Google Scholar]
- 22. Ye J. Z., Hockemeyer D., Krutchinsky A. N., Loayza D., Hooper S. M., Chait B. T., de Lange T. (2004) POT1-interacting protein PIP1: a telomere length regulator that recruits POT1 to the TIN2/TRF1 complex. Genes Dev. 18, 1649–1654 Epub 2004 Jul 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Houghtaling B. R., Cuttonaro L., Chang W., Smith S. (2004) A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr. Biol. 14, 1621–1631 [DOI] [PubMed] [Google Scholar]
- 24. Liu D., O'Connor M. S., Qin J., Songyang Z. (2004). Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 279, 51338–51342 [DOI] [PubMed] [Google Scholar]
- 25. Ye J. Z., Donigian J. R., Van Overbeek M., Loayza D., Luo Y., Krutchinsky A. N., Chait B. T., De Lange T. (2004) TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 16, 16. [DOI] [PubMed] [Google Scholar]
- 26. Kim S. H., Beausejour C., Davalos A. R., Kaminker P., Heo S. J., Campisi J. (2004) TIN2 mediates functions of TRF2 at human telomeres. J. Biol. Chem. 279, 43799–43804 [DOI] [PubMed] [Google Scholar]
- 27. de Lange T. (2005) Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 [DOI] [PubMed] [Google Scholar]
- 28. Xin H., Liu D., Songyang Z. (2008) The telosome/shelterin complex and its functions. Genome Biol. 9, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wright W. E., Shay J. W. (2001) Cellular senescence as a tumor-protection mechanism: the essential role of counting. Curr. Opin. Genet. Dev. 11, 98–103 [DOI] [PubMed] [Google Scholar]
- 30. Kim S. H., Kaminker P., Campisi J. (2002) Telomeres, aging and cancer: in search of a happy ending. Oncogene 21, 503–511 [DOI] [PubMed] [Google Scholar]
- 31. Baumann P. (2006) Are mouse telomeres going to pot? Cell 126, 33–36 [DOI] [PubMed] [Google Scholar]
- 32. Blasco M. A. (2007) The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 8, 299–309 [DOI] [PubMed] [Google Scholar]
- 33. Verdun R. E., Karlseder J. (2007) Replication and protection of telomeres. Nature 447, 924–931 [DOI] [PubMed] [Google Scholar]
- 34. Longhese M. P. (2008) DNA damage response at functional and dysfunctional telomeres. Genes Dev. 22, 125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Song K., Jung D., Jung Y., Lee S. G., Lee I. (2000) Interaction of human Ku70 with TRF2. FEBS Lett. 481, 81–85 [DOI] [PubMed] [Google Scholar]
- 36. Li B., Comai L. (2000) Functional interaction between Ku and the werner syndrome protein in DNA end processing. J. Biol. Chem. 275, 28349–28352 [DOI] [PubMed] [Google Scholar]
- 37. Zhu X. D., Küster B., Mann M., Petrini J. H., de, Lange T. (2000) Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25, 347–352 [DOI] [PubMed] [Google Scholar]
- 38. Opresko P. L., von Kobbe C., Laine J. P., Harrigan J., Hickson I. D., Bohr V. A. (2002). Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 277, 41110–41119 Epub 42002 Aug 41113 [DOI] [PubMed] [Google Scholar]
- 39. Takai H., Smogorzewska A., de Lange T. (2003) DNA damage foci at dysfunctional telomeres. Curr. Biol. 13, 1549–1556 [DOI] [PubMed] [Google Scholar]
- 40. Du X., Shen J., Kugan N., Furth E. E., Lombard D. B., Cheung C., Pak S., Luo G., Pignolo R. J., DePinho R. A., Guarente L., Johnson F. B. (2004) Telomere shortening exposes functions for the mouse Werner and Bloom syndrome genes. Mol. Cell. Biol. 24, 8437–8446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chang S., Multani A. S., Cabrera N. G., Naylor M. L., Laud P., Lombard D., Pathak S., Guarente L., DePinho R. A. (2004). Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat. Genet. 36, 877–882 Epub 2004 Jul 2004 [DOI] [PubMed] [Google Scholar]
- 42. Li B., Navarro S., Kasahara N., Comai L. (2004). Identification and biochemical characterization of a Werner's syndrome protein complex with Ku70/80 and poly(ADP-ribose) polymerase-1. J. Biol. Chem. 279, 13659–13667 Epub 12004 Jan 13620 [DOI] [PubMed] [Google Scholar]
- 43. Tarsounas M., Muñoz P., Claas A., Smiraldo P. G., Pittman D. L., Blasco M. A., West S. C. (2004) Telomere maintenance requires the RAD51D recombination/repair protein. Cell 117, 337–347 [DOI] [PubMed] [Google Scholar]
- 44. d'Adda di Fagagna F., Teo S. H., Jackson S. P. (2004) Functional links between telomeres and proteins of the DNA-damage response. Genes Dev. 18, 1781–1799 [DOI] [PubMed] [Google Scholar]
- 45. Stavropoulos D. J., Bradshaw P. S., Li X., Pasic I., Truong K., Ikura M., Ungrin M., Meyn M. S. (2002) The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum. Mol. Genet. 11, 3135–3144 [DOI] [PubMed] [Google Scholar]
- 46. Zhu X. D., Niedernhofer L., Kuster B., Mann M., Hoeijmakers J. H., de Lange T. (2003) ERCC1/XPF removes the 3′ overhang from uncapped telomeres and represses formation of telomeric DNA-containing double minute chromosomes. Mol. Cell 12, 1489–1498 [DOI] [PubMed] [Google Scholar]
- 47. O'Connor M. S., Safari A., Liu D., Qin J., Songyang Z. (2004). The human Rap1 protein complex and modulation of telomere length. J. Biol. Chem. 279, 28585–28591 Epub 22004 Apr 28520 [DOI] [PubMed] [Google Scholar]
- 48. Lillard-Wetherell K., Machwe A., Langland G. T., Combs K. A., Behbehani G. K., Schonberg S. A., German J., Turchi J. J., Orren D. K., Groden J. (2004). Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 13, 1919–1932 Epub 2004 Jun 30 [DOI] [PubMed] [Google Scholar]
- 49. Opresko P. L., Otterlei M., Graakjaer J., Bruheim P., Dawut L., Kølvraa S., May A., Seidman M. M., Bohr V. A. (2004) The Werner syndrome helicase and exonuclease cooperate to resolve telomeric D loops in a manner regulated by TRF1 and TRF2. Mol. Cell 14, 763–774 [DOI] [PubMed] [Google Scholar]
- 50. Collins K., Mitchell J. R. (2002) Telomerase in the human organism. Oncogene 21, 564–579 [DOI] [PubMed] [Google Scholar]
- 51. Granger M. P., Wright W. E., Shay J. W. (2002) Telomerase in cancer and aging. Crit. Rev. Oncol. Hematol. 41, 29–40 [DOI] [PubMed] [Google Scholar]
- 52. Kim H., Lee O. H., Xin H., Chen L. Y., Qin J., Chae H. K., Lin S. Y., Safari A., Liu D., Songyang Z. (2009) TRF2 functions as a protein hub and regulates telomere maintenance by recognizing specific peptide motifs. Nat. Struct. Mol. Biol. 16, 372–379 [DOI] [PubMed] [Google Scholar]
- 53. Peng G., Lin S. Y. (2009). BRIT1/MCPH1 is a multifunctional DNA damage responsive protein mediating DNA repair-associated chromatin remodeling. Cell Cycle 8, 3071–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ghosh I., Hamilton A. D., Regan L. (2000) antiparallel leucine zipper-directed protein reassembly: application to the green fluorescent protein. J. Am. Chem. Soc. 122, 5658 [Google Scholar]
- 55. Hu C. D., Chinenov Y., Kerppola T. K. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 [DOI] [PubMed] [Google Scholar]
- 56. Chen L. Y., Liu D., Songyang Z. (2007) Telomere maintenance through spatial control of telomeric proteins. Mol. Cell. Biol. 27, 5898–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wilson C. G., Magliery T. J., Regan L. (2004) Detecting protein-protein interactions with GFP-fragment reassembly. Nat. Methods 1, 255–262 [DOI] [PubMed] [Google Scholar]
- 58. Hu C. D., Kerppola T. K. (2003). Simultaneous visualization of multiple protein interactions in living cells using multicolor fluorescence complementation analysis. Nat. Biotechnol. 21, 539–545 Epub 2003 Apr 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. O'Connor M. S., Safari A., Xin H., Liu D., Songyang Z. (2006) A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc. Natl. Acad. Sci. U.S.A. 103, 11874–11879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Songyang Z., Liu D. (2006) Inside the mammalian telomere interactome: regulation and regulatory activities of telomeres. Crit. Rev. Eukaryot Gene Expr. 16, 103–118 [DOI] [PubMed] [Google Scholar]
- 61. Yang Q., Zheng Y. L., Harris C. C. (2005) POT1 and TRF2 Cooperate To Maintain Telomeric Integrity. Mol. Cell. Biol. 25, 1070–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lamesch P., Li N., Milstein S., Fan C., Hao T., Szabo G., Hu Z., Venkatesan K., Bethel G., Martin P., Rogers J., Lawlor S., McLaren S., Dricot A., Borick H., Cusick M. E., Vandenhaute J., Dunham I., Hill D. E., Vidal M. (2007) hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes. Genomics 89, 307–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. MacDonald M. L., Lamerdin J., Owens S., Keon B. H., Bilter G. K., Shang Z., Huang Z., Yu H., Dias J., Minami T., Michnick S. W., Westwick J. K. (2006) Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat. Chem. Biol. 2, 329–337 [DOI] [PubMed] [Google Scholar]
- 64. Xin H., Liu D., Wan M., Safari A., Kim H., Sun W., O'Connor M.S., Songyang Z. (2007). TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature 445, 559–562 [DOI] [PubMed] [Google Scholar]
- 65. Wang F., Podell E. R., Zaug A. J., Yang Y., Baciu P., Cech T. R., Lei M. (2007) The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 445, 506–510 [DOI] [PubMed] [Google Scholar]
- 66. Zhou X. Z., Lu K. P. (2001) The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell 107, 347–359 [DOI] [PubMed] [Google Scholar]
- 67. Déjardin J., Kingston R. E. (2009) Purification of proteins associated with specific genomic Loci. Cell 136, 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Michnick S. W., Ear P. H., Manderson E. N., Remy I., Stefan E. (2007) Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nat Rev Drug Discov 6, 569–582 [DOI] [PubMed] [Google Scholar]
- 69. Tarassov K., Messier V., Landry C. R., Radinovic S., Serna Molina M. M., Shames I., Malitskaya Y., Vogel J., Bussey H., Michnick S. W. (2008) An in vivo map of the yeast protein interactome. Science 320, 1465–1470 [DOI] [PubMed] [Google Scholar]
- 70. Paulmurugan R., Umezawa Y., Gambhir S. S. (2002) Noninvasive imaging of protein-protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc. Natl. Acad. Sci. U.S.A. 99, 15608–15613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wu W., Hsu Y. M., Bi L., Songyang Z., Lin X. (2009). CARD9 facilitates microbe-elicited production of reactive oxygen species by regulating the LyGDI-Rac1 complex. Nat. Immunol. 10, 1208–1214 [DOI] [PubMed] [Google Scholar]
- 72. Remy I., Michnick S. W. (2004) Regulation of apoptosis by the Ft1 protein, a new modulator of protein kinase B/Akt. Mol. Cell. Biol. 24, 1493–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ding Z., Liang J., Lu Y., Yu Q., Songyang Z., Lin S. Y., Mills G. B. (2006) A retrovirus-based protein complementation assay screen reveals functional AKT1-binding partners. Proc. Natl. Acad. Sci. U.S.A. 103, 15014–15019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Shyu Y. J., Liu H., Deng X., Hu C. D. (2006) Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques 40, 61–66 [DOI] [PubMed] [Google Scholar]
- 75. Kaminker P., Plachot C., Kim S. H., Chung P., Crippen D., Petersen O. W., Bissell M. J., Campisi J., Lelièvre S. A. (2005) Higher-order nuclear organization in growth arrest of human mammary epithelial cells: a novel role for telomere-associated protein TIN2. J. Cell Sci. 118, 1321–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Canudas S., Houghtaling B. R., Kim J. Y., Dynek J. N., Chang W. G., Smith S. (2007) Protein requirements for sister telomere association in human cells. EMBO J. 26, 4867–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., Klitgord N., Simon C., Boxem M., Milstein S., Rosenberg J., Goldberg D. S., Zhang L. V., Wong S. L., Franklin G., Li S., Albala J. S., Lim J., Fraughton C., Llamosas E., Cevik S., Bex C., Lamesch P., Sikorski R. S., Vandenhaute J., Zoghbi H. Y., Smolyar A., Bosak S., Sequerra R., Doucette-Stamm L., Cusick M. E., Hill D. E., Roth F. P., Vidal M. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 78. Karlseder J., Broccoli D., Dai Y., Hardy S., de Lange T. (1999) p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2. Science 283, 1321–1325 [DOI] [PubMed] [Google Scholar]
- 79. Karlseder J., Kachatrian L., Takai H., Mercer K., Hingorani S., Jacks T., de Lange T. (2003) Targeted deletion reveals an essential function for the telomere length regulator Trf1. Mol. Cell. Biol. 23, 6533–6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Biroccio A., Rizzo A., Elli R., Koering C. E., Belleville A., Benassi B., Leonetti C., Stevens M. F., D'Incalci M., Zupi G., Gilson E. (2006) TRF2 inhibition triggers apoptosis and reduces tumourigenicity of human melanoma cells. Eur. J. Cancer 42, 1881–1888 [DOI] [PubMed] [Google Scholar]
- 81. Guo X., Deng Y., Lin Y., Cosme-Blanco W., Chan S., He H., Yuan G., Brown E.J., Chang S. (2007) Dysfunctional telomeres activate an ATM-ATR-dependent DNA damage response to suppress tumorigenesis. EMBO J. 26, 4709–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kim S. H., Davalos A. R., Heo S. J., Rodier F., Zou Y., Beausejour C., Kaminker P., Yannone S. M., Campisi J. (2008) Telomere dysfunction and cell survival: roles for distinct TIN2-containing complexes. J. Cell Biol. 181, 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]