Abstract
Several mass spectrometry-driven techniques allow to map the substrate repertoires and specificities of proteases. These techniques typically yield long lists of protease substrates and processed sites with (potential) physiological relevance, but in order to understand the primary function of a protease, it is important to discern bystander substrates from critical substrates. Because the former are generally processed with lower efficiency, data on the actual substrate cleavage efficiency could assist in categorizing protease substrates. In this study, quantitative mass spectrometry following metabolic proteome labeling (SILAC), combined with the isolation of N-terminal peptides by Combined Fractional Diagonal Chromatography, was used to monitor fluxes in the concentration of protease-generated neo-N-termini. In our experimental setup, a Jurkat cell lysate was treated with the human serine protease granzyme B (hGrB) for three different incubation periods. The extensive list of human granzyme B substrates previously catalogued by N-terminal Combined Fractional Diagonal Chromatography (1) was then used to assign 101 unique hGrB-specific neo-N-termini in 86 proteins. In this way, we were able to define several sites as getting efficiently cleaved in vitro and consequently recognize potential physiologically more relevant substrates. Among them the well-known hGrB substrate Bid was confirmed as being an efficient hGrB substrate next to several other potential regulators of hGrB induced apoptosis such as Bnip2 and Akap-8. Several of our proteomics results were further confirmed by substrate immunoblotting and by using peptide substrates incubated with human granzyme B.
Proteolysis is a process during which a target protein is either partially processed into (stable) fragments (often implicated in protease-mediated propagation of signaling events) or completely shred into its composite amino acids (protein catabolism) by proteolytic enzymes or proteases. Proteases have key effector roles in human homeostasis, pathogenesis, and autoimmunity, so not surprisingly, it is considered of utmost importance to elucidate their specific functions in these processes, among others by profiling their physiological substrate repertoires (2, 3). Because proteolytic cleavage results in the formation of new protein N- and C-termini, many efforts were recently devoted to the specific isolation of neo-terminal peptides. This resulted in the development of several proteomics technologies to study protease substrate specificities or catalogue substrate repertoires (e.g. recently reviewed in (4)).
Because of this highly evolving field of targeted proteomics, a wealth of protease-specific substrate cleavage data has emerged, which typically originate from ex vivo analyses; e.g. the hundreds of potential granzyme B substrates that were reported by our group (1). Several studies already indicated that the high sensitivity of mass spectrometers might also lead to the identification of very rare and inefficient cleavage events (e.g. (1, 5)). These are often believed to occur as the result of nonphysiological conditions of protease concentration and substrate localization (especially in ex vivo studies) and therefore affect proteins that are probably not of crucial importance to understand the physiological function of a protease. It has therefore become highly important to distinguish the key targets of proteases identified in such screens in order to reveal the primary signaling pathways or protein assemblies on which proteases act. Because it was also shown that substrate processing resulting in loss-of-function happens faster than processing resulting in gain-of-function (6), clear challenges to distinguish efficient from inefficient, and early from late substrate processing events in large-scale protease studies are emerging.
To discern bystander from critical cleavage events of human granzyme B (hGrB)1—a protease involved in granule-mediated cell death by cells from the innate and adaptive immune system—we here used a three-plexed (time-based) N-terminal Combined Fractional Diagonal Chromatography (COFRADIC) positional proteomics setup. N-terminal COFRADIC uses consecutive reverse phase high performance liquid chromatography (RP-HPLC) separations of peptide mixtures and sorts out N-terminal peptides from whole proteome digests (7, 8). Metabolic proteome labeling was introduced by isotope variants of arginine because sorted N-terminal peptides, by nature of the used chemical modification reactions, all end on arginine. Here, three-plexed proteomics was performed using 12C6, 13C6, and 13C615N4 arginine stable isotope labeling by amino acids in cell culture (SILAC) labeling (9), introducing labels that provide ample spacing and thus allow direct quantification of the individual peptide ions (see Results section). Our general assumption was the following. For each identified protein N-terminal peptide and hGrB-generated neo-N-terminal peptide, three possible isotopic variants may be expected. Protein N termini that are not affected by hGrB and of which the concentration remains constant, can be easily read from their MS spectra. On the other hand, in our time-based proteomics setup, different trends for neo-N-termini generated by hGrB are expected. We show that hGrB cleavage sites fully cleaved at the shortest time of incubation—and as such assigned as efficient cleavage sites—are readily distinguished from less efficient sites based on the pattern of their isotopically labeled peptide ions in MS spectra. Validation of the cleavage efficiency of several categorized hGrB sites was done by immunoblot detection following SDS-PAGE separation, in addition to monitoring in vitro peptide processing by RP-HPLC. Furthermore, kinetics of processing was analyzed for several representative substrates. The results of all these biochemical characterization experiments were very well in agreement with the data obtained from the proteomics screen, among others confirming the known human BH3-interacting domain death agonist as a crucial and efficiently cleaved hGrB substrate, thus validating our targeted proteomics approach for identifying prime targets of proteases.
EXPERIMENTAL PROCEDURES
Cell Culture
Human Jurkat T cells were from the ATCC (TIB-152, American Type Culture Collection, Manassas, VA) and were grown in RPMI 1640 medium supplemented with 10% dialyzed fetal bovine serum (Invitrogen, Carlsbad, CA), 100 units/ml penicillin (Invitrogen), and 100 μg/ml streptomycin (Invitrogen). Cells were grown in media containing either natural (12C6), 13C6, or 13C615N4 l-arginine (Cambridge Isotope Labs, Andover, MA) (9) at a concentration of 230 μm (i.e. 20% of the suggested concentration present in RPMI at which l-arginine to proline conversion was not detectable). Cells were cultured at 37 °C and in 5% CO2 for at least six population doublings to ensure complete incorporation of the labeled arginine.
Preparation of Cell Lysates for N-Terminal COFRADIC Analysis
SILAC-labeled cells were washed in d-PBS and resuspended at 7 × 106 cells per ml in 50 mm Tris-HCl and 100 mm NaCl at pH 8.0. Cells were pre-incubated with the pan-caspase inhibitor N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone at a final concentration of 50 μm for 15 min at 37 °C and then lysed by three rounds of freeze-thawing. Supernatants, cleared by centrifugation for 10 min at 16,000 × g (the protein concentration was determined to be 1.3 mg/ml), were treated with 200 nm of human recombinant granzyme B (hGrB) for different time-points at 37 °C. Recombinant human granzyme B was expressed and purified from Pichia pastoris, and its activity was determined as described previously (10, 11). Solid guanidinium hydrochloride was added to a final concentration of 4 m in order to inactivate hGrB and denature all proteins. Before mixing the samples in equal amounts, proteins were reduced and alkylated simultaneously, using tris(2-carboxyethyl)phospine.hydrochloride (TCEP.HCl; 1 mm final concentration) and iodoacetamide (2 mm final concentration) respectively, for 1 h at 30 °C. Subsequent steps of the N-terminal COFRADIC protocol were performed as described previously (7). The proteome was digested overnight at 37 °C with sequencing-grade, modified trypsin (Promega, Madison, WI) (enzyme/substrate of 1/100 w/w).
Liquid Chromatography (LC)-tandem MS (MS/MS) Analysis and Data Processing
Electrospray ionization (ESI) LC-MS/MS analysis was performed on an ESI-quadrupole (Q)-time-of-flight (TOF) (Waters Corporation) mass spectrometer as described before (12). ESI-Q-TOF MS/MS peptide fragmentation spectra were converted to pkl files using the Masslynx® software (version 4.1, Waters Corporation). N-terminal peptides were identified using a locally installed version of the MASCOT database search engine version 2.2.1 (www.matrixscience.com) and the Swiss-Prot database (version 57.11 of UniProtKB/Swiss-Prot protein database, containing 512,994 sequence entries of which 20,328 originated from human) was searched with restriction to human proteins. Truncated peptide databases made by DBToolkit (13), were searched in parallel to pick up protein processing events more efficiently (e.g. (14)). The following search parameters were used. Peptide mass tolerance was set at 0.2 Da and peptide fragment mass tolerance at 0.1 Da with the ESI-QUAD-TOF as selected instrument for peptide fragmentation rules for the Q-TOF Premier data. Endoproteinase Arg-C/P (= no restriction towards arginine-proline cleavage) was set as enzyme allowing one missed cleavage. Variable modifications were pyroglutamate formation of N-terminal glutamine, pyrocarbamidomethyl formation of N-terminal alkylated cysteine, deamidation of asparagine, acetylation and tri-deuteroacetylation of the alpha-N-terminus. Fixed modifications were methionine oxidation (sulfoxide), carbamidomethyl for cysteine, tri-deuteroacetylation of lysine and for identifying heavy labeled peptides, [13C6] or [13C615N4]-arginine were additionally set as fixed modifications. Only MS/MS-spectra and corresponding identifications that exceeded the corresponding MASCOT's threshold score of identity (at 95% confidence level) and that were ranked one, were withheld. In addition, spectra that received a low MASCOT ion score (five or less points above threshold for identity) were further interrogated and only spectra that contained b- and y-fragment ions covering a stretch of three consecutive amino acids were considered identified (see Supplemental Data listing identified MS/MS-spectra). The estimated false discovery rate by searching decoy databases was typically found to lie between 2% and 4% on the spectrum level (7). Whenever a peptide matched to multiple members of a protein family (redundancy), the protein entry reported (column “Protein description,” Supplemental Table 1) was according to its alphabetical ranking. In each case, other matching protein entries were reported under the column “Isoforms.” Corresponding orthologous mouse cleavage sites, as determined in (1), are reported in Supplemental Table 1.
The peptide intensity ratios were individually calculated from the MS data in the Masslynx® software version 4.1 environment. In order to correct for small variations in initial protein concentrations, the 12C6/13C6 and 12C6/13C615N4 ratios of the 1788 database-annotated protein N-terminal peptides (peptides starting at position 1 or 2) were first subjected to robust statistics using base-2 logarithm values of their ratios (15). The corresponding ratio means, being 0.0115 (12C6/13C6) and −0.0209 (12C6/13C615N4), were then used to correct for the calculated ratios of all peptides identified. Then, we used the R software package for statistical computing to calculate the probability distributions of log2 transformed N-terminal peptide ratios between the first and second time point (N(0.0115, 0.1416) (mean, standard deviation)) and between the second and third time point (N(-0.0209, 0.1890)). Protease substrate intensity ratios falling within the 99.5% probability interval of the protein N-terminal peptide intensity ratios were considered as derived from efficient cleavage sites (see Results section).
Immunoblot Analysis
To inactivate endogenous caspases, and thus exclude caspase-mediated cleavage of tested substrates following granzyme B activation, all protein samples, except for a control sample and a sample incubated with 100 nm hGrB for 2 h (the latter samples demonstrate that the GrB-activity potential was unaltered when making use of alkylated protein lysates), were pretreated with iodoacetamide for 15 min in the dark at a final concentration of 5 mm (16, 17). Subsequently, all protein samples were prepared and treated with hGrB at different concentrations and for different time-points (see Results section). Following hGrB incubation, sample loading buffer was added and equal amounts of protein (50 μg/sample; protein concentrations were measured using the DC Protein Assay Kit from Bio-Rad (Munich, Germany)) were separated on 4%–12% or 12% polyacrylamide Criterion XT-gels in MOPS buffer (Bio-Rad) at 150 V. Subsequently, proteins were transferred onto a PVDF-membrane. Membranes were blocked for 30 min in 5% skimmed milk powder containing Tris-buffered saline and 0.1% Tween-20 (TBS-T) and probed for 1 h with primary antibody in 5% skimmed milk powder in TBS-T. Following three 10 min washes in TBS-T, membranes were incubated with species-specific HRP-conjugated antibody for 1 h in 5% skimmed milk powder containing TBS-T. Again following a threefold wash in TBS-T or TBS (last wash step), bands were visualized on ECL Hyperfilms (Amersham Biosciences, Buckinghamshire, UK) using an enhanced chemiluminiscence kit (PerkinElmer Life Sciences, Boston, MA). The caspase-3 antibody was from Calbiochem (AAP-113), Bid-antibody from R&D systems (AF860) (Minneapolis, MN), nucleolin antibody from Abcam (ab16940) (Cambridge, MA), Akap-8 and importin α-4 antibody from Abcam (ab32822 and ab6038). Recombinant glutathione S-transferase (GST)-tagged Bnip2 was from Abnova (H00000663-P01) (Taipei, Taiwan) and was assayed with a GST antibody from Abcam (ab6649). The degree of processing was determined by densitometry following Western blot analysis of various substrates that were processed with various concentrations of human GrB and following various time-points of incubation. Quantification was performed using the ChemiGenius imaging system (SynGene, Cambridge, UK) and signal intensity was quantified using GeneTools 3.07 image analysis software (SynGene).
Confirmation of hGrB-Specific Cleavage Events by Treating Synthesized Peptides with hGrB
The following peptides holding hGrB cleavage sites were synthesized in-house using the Fmoc-chemistry on an Applied Biosystems 433A Peptide Synthesizer: NH2. RLGRIEADSESQEDIIRY(NO2)R.OH (Bid), NH2.LPEDDSIEADILAITGY(NO2)R.OH (Bnip2), NH2.KETPEEVAADVLAEVIY(NO2)R.OH (Akap-8), NH2.VPHEDICEDSDIDGDY(NO2)R.OH (importin α-4), and NH2.FVKGLSEDTTEETLKY(NO2)R.OH (nucleolin). Substrate peptides were assayed for 2 h at 37 °C with various hGrB concentrations (10, 50, 200, and 500 nm) at a substrate concentration of 100 μm. The reactions were stopped by adding trifluoroacetic acid to a final concentration of 1%. Cleavage by hGrB was assessed by monitoring the UV absorbance signal (214 nm) of the precursor peptide and its fragments by reverse phase-HPLC separation. The extent of hydrolysis was determined by comparing the intensity of the product peak(s) with that of the unprocessed precursor.
Analysis of the Granzyme B Cleavage Efficiency of the Fluorogenic Peptide Ac-IEPD-AMC in a Buffer and in a Cell Lysate
Ac-IEPD-AMC was purchased from Bachem (Bubendorf, Switzerland) and a 20 mm stock solution was prepared in DMSO. Freeze-thaw lysates of human Jurkat T-cells were made in a buffer containing 100 mm NaCl, 50 mm Tris-HCl pH 7.9 and these lysates were S-alkylated and desalted as described above. Measurements were performed in black 96-well plates on a temperature-controlled Bio-Tek FLx800 fluorescence microplate reader (excitation at 360 nm, emission at 460 nm). A 40 μl aliquot of a 400 nm granzyme B solution (150 mm NaCl, 50 mm Tris-HCl, pH 7.9) was added in every well while the plate was kept on ice. In a different plate, Ac-IEPD-AMC was diluted to twice the indicated final concentration in buffer or in the cell lysate (the protein concentration in the cell lysate was 2.6 mg/ml). A 40 μl aliquot of the hence diluted Ac-IEPD-AMC was then added to the wells containing granzyme B. The plate was briefly mixed and placed in the microplate reader at 37 °C. Over a period of 50 min, fluorescence was measured every 78 s.
RESULTS
General Description of the Proteomics Setup
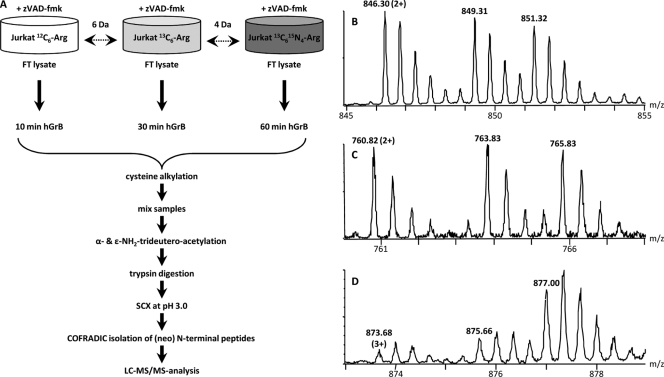
Human Jurkat cells were grown in three different SILAC media containing 12C6,13C6, or 13C615N4 l-arginine (Fig. 1A). Prior to freeze-thaw lysis, cells were treated with N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethyl ketone to prevent substrate processing by caspases that are known to get activated by granzyme B (18). These different cell lysates were incubated with hGrB for 10, 30 or 60 min respectively (three-plex setup, Fig. 1A) and following protease inactivation, equal sample amounts were mixed and N-terminal peptides were isolated by the N-terminal COFRADIC sorting procedure (7). Subsequent LC-MS/MS analysis resulted in the identification of 2666 MS/MS-spectra that were linked to 992 peptides. Of these, 295 peptides (29.7%) present in 261 proteins were trideutero-acetylated, did not start at protein position one or two and thus formed our primary pool of potential hGrB recognized cleavage sites and substrates. This list was cross-checked with our previously reported catalogue of GrB processing events (1) and in this way, 101 unique neo-N-termini in 86 proteins (i.e. 34.2% of all neo-N termini identified in this study) were assigned as resulting from hGrB-mediated processing (Supplemental Table 1).
Fig. 1.
Experimental setup to probe the extent of substrate proteolysis by hGrB. A, SILAC using 12C6, 13C6, and 13C615N4 l-arginine was used to label Jurkat cells and to then monitor concentration fluxes of (neo-)N-termini of zVAD-fmk pretreated Jurkat freeze-thaw (FT) cell lysates. 12C6, 13C6, and 13C615N4 l-arginine labeled lysates were in vitro incubated with 200 nm hGrB for respectively 10, 30, and 60 min. Following incubation and inactivation of hGrB by adding guanidinium hydrochloride, equal amounts of proteome preparations were mixed and N-terminal peptides were isolated by the N-terminal COFRADIC sorting procedure (7), the different key steps of which are indicated. B, A representative MS-spectrum showing the hGrB unaffected and thus unaltered N-terminus Ac-2MLGPEGGEGFVVKLR16 (Ac denotes an α-acetylated amino group) from the heterogeneous nuclear ribonucleoprotein F (hnRNPF). C, A similar MS-profile was observed for an hGrB generated neo-N-terminus (AcD3-692SGGNTSPGVTANGEAR707 (AcD3 denotes a trideutero-acetylated α-amino group)) from Rho/Rac guanine nucleotide exchange factor 2 (ARHG2), indicating an efficient cleavage event. D, Increasing ion intensities were observed for AcD3-484SGLITPGGFSSVPAGMETPELIELR508 from the splicing factor 3B subunit 2 (SF3B2) hinting to a less efficiently cleaved hGrB site.
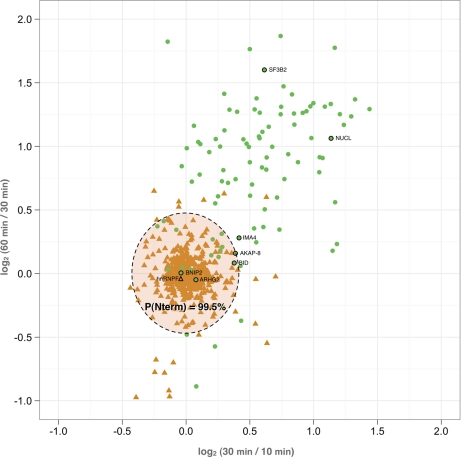
Our hypothesis was that efficient hGrB sites are cleaved to completeness at the shortest incubation time and thus the concentration of their corresponding neo-N-terminal peptides was not expected to increase upon prolonged hGrB incubation (Fig. 1C). Therefore, the MS pattern of these peptides was expected to resemble that of protein N-terminal peptides (Fig. 1B), whereas a less efficiently cleaved hGrB site should give rise to an increase of ion intensities in MS spectra at later time-points (Fig. 1D). Taking the intra-experimental ion signal variation of the protein N-termini into account, a category of efficiently cleaved hGrB sites was in this way identified within a 99.5% confidence interval. In fact, the averaged ratio values of 506 UniProt annotated protein N-terminal peptides (peptides with a protein start position of one or two) were calculated and used to delineate the most efficient hGrB cleavage sites (Fig. 2). The full list of hGrB-induced neo-N-termini, their ion intensity changes at the incubation times examined and their assignment as efficient or less efficient sites is given in Table I. Besides the well-characterized and highly efficient hGrB substrate BH3-interacting domain death agonist or Bid, the pro-apoptotic Bcl-2 family member Bnip2, and the protein kinase A anchor protein 8 (Akap-8) were found to hold efficiently cleaved hGrB sites (fold changes observed between 10 and 30 min, and 30 and 60 min were 0.97 and 1, and 1.31, and 1.21 respectively). Of note is that all these proteins were previously reported to play prominent roles in the process of apoptotic execution under physiological conditions (19, 21, 27).
Fig. 2.
Classification of the most efficiently cleaved hGrB sites. The distribution of 506 unique protein N-termini starting at position one or two (triangles) and 101 unique hGrB specific neo-N-termini (circles) of which the 13C6/12C6 and 13C615N4/13C6 ratio could be calculated, is plotted. The spreading of ratio values for the unaffected protein N-termini was used to set the area containing the most efficiently cleaved hGrB sites (p ≥ 99.5%). Neo-N-termini or database annotated N-termini corresponding to the MS-spectra depicted in Fig. 1B–D and Fig. 3A–E are indicated.
Table I. Categorization of 101 hGrB-generated neo-N-termini and their corresponding substrates. The UniProt database primary accession number, protein description, P10-P10' sequence and fold change of the neo-N-termini between 10 and 30 min and between 30 and 60 min are given. For each category, proteins are ranked alphabetically according to their protein description.
| SwissProt accession | Protein description | Human cleavage site P10-P10′ | Fold change 10–30 min | Fold change 30–60 min |
|---|---|---|---|---|
| Efficient hGrB sites | ||||
| P63220 | 40S ribosomal protein S21 | MQND(4)↓AGEFVDLYVP | 0.89 | 1.33 |
| O43823 | A-kinase anchor protein 8 | KETPEEVAAD(576)↓VLAEVITAAV | 1.31 | 1.12 |
| Q14562 | ATP-dependent RNA helicase DHX8 | DEDDVKVAVD(144)↓VLKELEALMP | 0.98 | 1.03 |
| Q12982 | BCL2/adenovirus E1B 19 kDa protein-interacting protein 2 | LPEDDSIEAD(28)↓ILAITGPEDQ | 0.97 | 1.00 |
| P55957 | BH3-interacting domain death agonist | HSRLGRIEAD(75)↓SESQEDIIRN | 1.30 | 1.06 |
| Q9NUL3 | Double-stranded RNA-binding protein Staufen homolog 2 | PKGILHLSPD(418)↓VYQEMEASRH | 1.03 | 1.02 |
| P29083 | General transcription factor IIE subunit 1 | MADPD(5)↓VLTEVPAALK | 1.21 | 1.24 |
| Q6PKG0 | La-related protein 1 | KDQDEQEELD(606)↓FLFDEEMEQM | 1.19 | 1.09 |
| P18615 | Negative elongation factor E | SDRLRELGPD(149)↓GEEAEGPGAG | 0.86 | 1.29 |
| P26599 | Polypyrimidine tract-binding protein 1 | MDGIVPD(7)↓IAVGTKRGSD | 1.22 | 1.16 |
| Q92733 | Proline-rich protein PRCC | LVPPQEIAPD(385)↓ASFIDDEAFK | 1.00 | 0.72 |
| Q92974 | Rho/Rac guanine nucleotide exchange factor 2 | REPALPLEPD(689)↓SGGNTSPGVT | 1.06 | 0.97 |
| O75533 | Splicing factor 3B subunit 1 | LDEAQGVGLD(34)↓STGYYDQEIY | 0.95 | 1.27 |
| Q9Y2W1 | Thyroid hormone receptor-associated protein 3 | REAQVNVRMD(574)↓SFDEDLARPS | 1.20 | 1.13 |
| Q9Y5J1 | U3 small nucleolar RNA-associated protein 18 homolog | RCLEELVFGD(98)↓VENDEDALLR | 1.22 | 0.84 |
| O43395 | U4/U6 small nuclear ribonucleoprotein Prp3 | KRELKEVFGD(97)↓DSEISKESSG | 0.91 | 1.02 |
| O95365 | Zinc finger and BTB domain-containing protein 7A | QILAADAGAD(139)↓AGQLDLVDQI | 1.15 | 1.10 |
| Q6NZY4 | Zinc finger CCHC domain-containing protein 8 | RTASGAVDED(513)↓ALTLEELEEQ | 1.16 | 1.20 |
| Less efficient hGrB sites | ||||
| P62280 | 40S ribosomal protein S11 | QNKKRVLLGE(27)↓TGKEKLPRYY | 1.45 | 2.45 |
| P62841 | 40S ribosomal protein S15 | KFTYRGVDLD(23)↓QLLDMSYEQL | 1.17 | 0.67 |
| RGVDLDQLLD(27)↓MSYEQLMQLY | 1.31 | 1.67 | ||
| LDQLLDMSYE(31)↓QLMQLYSARQ | 1.60 | 2.00 | ||
| P62244 | 40S ribosomal protein S15a | MVRMNVLAD(9)↓ALKSINNAEK | 2.25 | 1.47 |
| P61247 | 40S ribosomal protein S3a | GRVFEVSLAD(73)↓LQNDEVAFRK | 1.26 | 1.64 |
| P62701 | 40S ribosomal protein S4, X isoform | VAAPKHWMLD(21)↓KLTGVFAPRP | 1.17 | 2.08 |
| RFAVHRITPE(117)↓EAKYKLCKVR | 2.00 | 2.53 | ||
| P62081 | 40S ribosomal protein S7 | RSRTLTAVHD(127)↓AILEDLVFPS | 1.64 | 1.51 |
| P10809 | 60 kDa heat shock protein, mitochondrial precursor | RKPLVIIAED(277)↓VDGEALSTLV | 1.96 | 2.49 |
| P27635 | 60S ribosomal protein L10 | FPLCGHMVSD(55)↓EYEQLSSEAL | 1.46 | 1.19 |
| KWGFTKFNAD(179)↓EFEDMVAEKR | 1.08 | 1.71 | ||
| P26373 | 60S ribosomal protein L13 | RKPSAPKKGD(138)↓SSAEELKLAT | 1.51 | 1.78 |
| P84098 | 60S ribosomal protein L19 | GKKKVWLDPN(27)↓ETNEIANANS | 1.23 | 2.18 |
| P61254 | 60S ribosomal protein L26 | NVRSMPIRKD(52)↓DEVQVVRGHY | 1.41 | 3.40 |
| P39023 | 60S ribosomal protein L3 | VPVNQVFGQD(214)↓EMIDVIGVTK | 1.66 | 1.27 |
| P46777 | 60S ribosomal protein L5 | YEGQVEVTGD(128)↓EYNVESIDGQ | 1.41 | 1.99 |
| Q01813 | 6-phosphofructokinase type C | NMGGLAAGAD(601)↓AAYIFEEPFD | 1.78 | 2.65 |
| P17858 | 6-phosphofructokinase, liver type | PGTDFSLGSD(542)↓TAVNAAMESC | 2.45 | 2.35 |
| P08237 | 6-phosphofructokinase, muscle type | PGSDFSVGAD(543)↓TALNTICTTC | 1.55 | 1.76 |
| P60709 | Actin, cytoplasmic 1 | KEKLCYVALD(222)↓FEQEMATAAS | 1.22 | 8.26 |
| Q9UG63 | ATP-binding cassette sub-family F member 2 | ANGRETTEVD(54)↓LLTKELEDFE | 2.27 | 1.17 |
| Q92499 | ATP-dependent RNA helicase DDX1 | NCTISQVEPD(676)↓IKVPVDEFDG | 0.98 | 1.79 |
| P55210 | Caspase-7 precursor | TELDDGIQAD(198)↓SGPINDTDAN | 1.35 | 0.77 |
| Q99459 | Cell division cycle 5-like protein | PARPDPIDMD(145)↓EDELEMLSEA | 1.13 | 1.94 |
| Q14839 | Chromodomain-helicase-DNA-binding protein 4 | EDDDLDVESD(320)↓FDDASINSYS | 1.45 | 1.28 |
| O00273 | DNA fragmentation factor subunit alpha | MEVTGD(6)↓AGVPESGEIR | 1.92 | 2.42 |
| P49736 | DNA replication licensing factor MCM2 | RDYRAIPELD(88)↓AYEAEGLALD | 1.98 | 2.09 |
| P33992 | DNA replication licensing factor MCM5 | FDDPGIFYSD(13)↓SFGGDAQADE | 1.74 | 1.92 |
| P68104 | Elongation factor 1-alpha 1 | DGPKFLKSGD(398)↓AAIVDMVPGK | 1.47 | 2.60 |
| MVPGKPMCVE(413)↓SFSDYPPLGR | 1.32 | 2.41 | ||
| Q04637 | Eukaryotic translation initiation factor 4 gamma 1 | RLQGINCGPD(665)↓FTPSFANLGR | 1.19 | 1.52 |
| Q92945 | Far upstream element-binding protein 2 | RQIAAKIGGD(91)↓AATTVNNSTP | 1.08 | 2.03 |
| P85037 | Forkhead box protein K1 | SGGSSGVSGD(80)↓SAVAGAAPAL | 1.34 | 1.04 |
| P14314 | Glucosidase 2 subunit beta precursor | AEAQALLSGD(260)↓TQTDATSFYD | 1.22 | 1.76 |
| Q14527 | Helicase-like transcription factor | IPPDDFLTSD(52)↓EEVDSVLFGS | 1.71 | 1.67 |
| P61978 | Heterogeneous nuclear ribonucleoprotein K | GEFGKRPAED(26)↓MEEEQAFKRS | 1.42 | 1.53 |
| Q00839 | Heterogeneous nuclear ribonucleoprotein U | VAGFKKQMAD(547)↓TGKLNTLLQR | 1.67 | 3.65 |
| P0C0S5 | Histone H2A.Z | MAGGKAGKD(9)↓SGKAKTKAVS | 5.77 | 1.14 |
| O00629 | Importin subunit alpha-4 | NVPHEDICED(59)↓SDIDGDYRVQ | 1.33 | 1.21 |
| Q12906 | Interleukin enhancer-binding factor 3 | LFPDTPLALD(597)↓ANKKKRAPVP | 1.27 | 2.44 |
| Q32MZ4 | Leucine-rich repeat flightless-interacting protein 1 | AAEEVLADGD(675)↓TLDFEDDTVQ | 1.49 | 2.11 |
| O60711 | Leupaxin | MEELD(5)↓ALLEELERST | 1.70 | 2.77 |
| Q15046 | Lysyl-tRNA synthetase | HTTDNGVGPE(67)↓EESVDPNQYY | 1.49 | 2.40 |
| P56192 | Methionyl-tRNA synthetase, cytoplasmic | TPQQTKITQD(368)↓IFQQLLKRGF | 2.36 | 2.25 |
| P27816 | Microtubule-associated protein 4 | KKCSLPAEED(642)↓SVLEKLGERK | 1.57 | 2.23 |
| Q9BQG0 | Myb-binding protein 1A | LGDEAMMALD(790)↓QSLASLFAEQ | 1.49 | 2.09 |
| P67809 | Nuclease-sensitive element-binding protein 1 | QYSNPPVQGE(216)↓VMEGADNQGA | 1.55 | 1.88 |
| Q9NR30 | Nucleolar RNA helicase 2 | MPGKLRSD(8)↓AGLESDTAMK | 1.03 | 1.65 |
| P19338 | Nucleolin | KNLPYKVTQD(407)↓ELKEVFEDAA | 1.52 | 1.29 |
| AYIEFKTEAD(441)↓AEKTFEEKQG | 2.06 | 1.74 | ||
| TLFVKGLSED(582)↓TTEETLKESF | 2.20 | 2.09 | ||
| P55209 | Nucleosome assembly protein 1-like 1 | NKEQSELDQD(15)↓LDDVEEVEEE | 2.07 | 1.88 |
| O75475 | PC4 and SFRS1-interacting protein | KNMFLVGEGD(433)↓SVITQVLNKS | 1.37 | 2.08 |
| Q8WUA2 | Peptidyl-prolyl cis-trans isomerase-like 4 | QLDSGRIGAD(194)↓EEIDDFKGRS | 1.43 | 1.62 |
| O60828 | Polyglutamine-binding protein 1 | PEPEEEIIAE(31)↓DYDDDPVDYE | 1.41 | 1.83 |
| P26599 | Polypyrimidine tract-binding protein 1 | ALAASAAAVD(172)↓AGMAMAGQSP | 1.17 | 1.47 |
| Q96GQ7 | Probable ATP-dependent RNA helicase DDX27 | ADENILTKAD(187)↓TLKVKDRKKK | 1.06 | 0.54 |
| Q9UQ80 | Proliferation-associated protein 2G4 | RITSGPFEPD(341)↓LYKSEMEVQD | 2.19 | 2.52 |
| Q8N163 | Protein KIAA1967 | ETEPTEQAPD(469)↓ALEQAADTSR | 1.20 | 2.00 |
| O15355 | Protein phosphatase 1G | DNEEAALLHE(147)↓EATMTIEELL | 1.67 | 2.38 |
| MEGKEEPGSD(326)↓SGTTAVVALI | 2.24 | 3.42 | ||
| P06454 | Prothymosin alpha [Contains: Thymosin alpha-1] | MSDAAVD(7)↓TSSEITTKDL | 2.13 | 2.48 |
| P06400 | Retinoblastoma-associated protein | PPPEEDPEQD(36)↓SGPEDLPLVR | 1.84 | 1.83 |
| Q9H814 | RNA U small nuclear RNA export adapter protein | EQNQDAVATE(133)↓LGILGMEGTI | 1.80 | 2.39 |
| P98175 | RNA-binding protein 10 | DERRESATAD(691)↓AGYAILEKKG | 1.21 | 1.65 |
| O75533 | Splicing factor 3B subunit 1 | YFDKLLVDVD(483)↓ESTLSPEEQK | 1.53 | 1.42 |
| Q13435 | Splicing factor 3B subunit 2 | TKPKLTIHGD(566)↓LYYEGKEFET | 1.39 | 2.03 |
| DETGFITPAD(719)↓SGLITPGGFS | 1.53 | 3.04 | ||
| Q86YP4 | Transcriptional repressor p66-alpha | KSERRPPSPD(102)↓VIVLSDNEQP | 2.50 | 2.58 |
| Q71U36 | Tubulin alpha-1A chain | KHVPRAVFVD(69)↓LEPTVIDEVR | 0.90 | 3.54 |
| GTYRQLFHPE(90)↓QLITGKEDAA | 2.31 | 2.38 | ||
| LRFDGALNVD(251)↓LTEFQTNLVP | 1.68 | 2.48 | ||
| P07437 | Tubulin beta chain | IDPTGTYHGD(39)↓SDLQLDRISV | 2.71 | 2.45 |
| Q14694 | Ubiquitin carboxyl-terminal hydrolase 10 | EYQRIEFGVD(62)↓EVIEPSDTLP | 1.00 | 1.98 |
| Q9NZ63 | Uncharacterized protein C9orf78 | QMLSGIPEVD(177)↓LGIDAKIKNI | 1.88 | 2.40 |
| P53990 | Uncharacterized protein KIAA0174 | AEAPPGVETD(194)↓LIDVGFTDDV | 1.04 | 2.24 |
| P54725 | UV excision repair protein RAD23 homolog A | LADISDVEGE(300)↓VGAIGEEAPQ | 1.80 | 2.25 |
| P46459 | Vesicle-fusing ATPase | IKASTKVEVD(466)↓MEKAESLQVT | 2.10 | 1.88 |
| P08670 | Vimentin | QDSVDFSLAD(90)↓AINTEFKNTR | 1.23 | 2.66 |
| QIQEQHVQID(257)↓VDVSKPDLTA | 1.07 | 2.05 | ||
| QEQHVQIDVD(259)↓VSKPDLTAAL | 1.51 | 2.16 | ||
| Q9UHR6 | Zinc finger HIT domain-containing protein 2 | EELDNAPGSD(150)↓AAELELAPAR | 2.22 | 1.13 |
When searching for amino acid sequence biases between highly efficient and less efficiently cleaved sites, the inspection of P4 to P1 amino acid occurrences in cleavage sites categorized as efficient sites reveals a consensus recognition motif much alike the one deduced from positional scanning substrate combinatorial library data (22); IEPD (data not shown). In our study, an increased relative occurrence of proline at the substrate P2 position was observed for highly efficiently cleaved sites: 6 out of 18 of these preferred sites (33%) and only 9 out of 83 (10.8%) of the less preferred sites have Pro at P2. In addition, considering the relative occurrences of isoleucine, leucine and valine, these amino acids occupy P4 in 14 out of 18 (77.78%) preferred sites and only 42 out of 83 (50.6%) less preferred sites. Of further note is the fact that alternative P1 residue cleavages such as glutamate, as described in (1), were never categorized to efficiently cleaved sites (11 of the 83 less efficient sites harbored a glutamate at P1). Taken together however, these subtle differences indicate that at least for hGrB, experimental analysis as opposed to in silico prediction remains a prerequisite to assign the efficiency of cleavage.
Biochemical Characterization of Human Granzyme B Processing Efficiency by Substrate Immunoblot Analysis
To confirm our proteomics data, the efficiency of substrate cleavage was determined by applying densitometry following protein immunoblot analysis for Bid, Bnip2, Akap-8, importin α-4, and nucleolin. Substrate processing was assessed with various concentrations of hGrB and at various times of incubation. Immunoblot analysis of caspase-3 and its derived proteolytic fragments confirmed the activity of hGrB on a natural substrate and showed iodoacetamide-mediated inhibition of downstream caspase activation by hGrB (Supplemental Fig. 1).
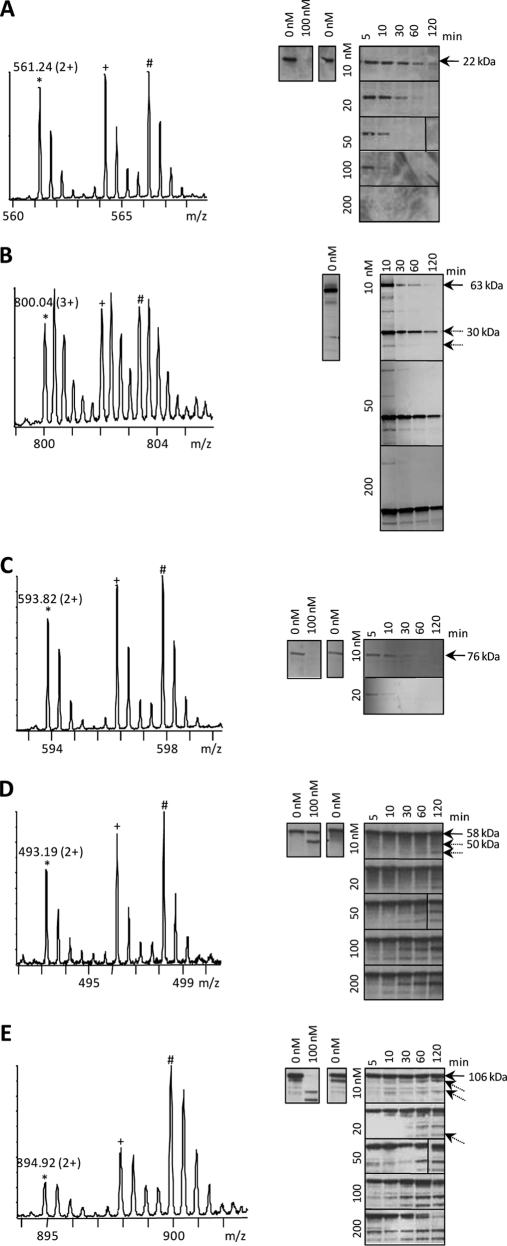
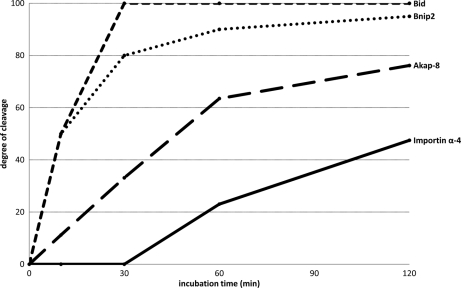
The MS-spectrum in Fig. 3A was assigned to the hGrB introduced neo-N-terminus 76SESQEDIIR84 of Bid, representing an early, signal-inducing cleavage event and an established key substrate in the hGrB mediated apoptotic pathway (23). In accordance to the proteomics data where no further increase in ion signal intensity at later time-points was observed, immunoblotting of Bid revealed complete precursor cleavage at the earliest time point assayed at 200 nm of hGrB incubation. A similar MS-pattern in addition to an identical P4-P1 cleavage site recognition motif (IEAD28) was observed for the neo-N-terminal peptide, 29ILAITGPEDQPGSLEVNGNKVR50, from Bnip2 (Fig. 3B). Immunoblotting of a 63 kDa N-terminal glutathione S-transferase (GST) tagged recombinant Bnip2 precursor protein showed the 30 kDa N-terminal fragment cleaved at IEAD28 as determined in the proteomics setup. Furthermore, densitometry confirmed Bnip2 as a highly efficient hGrB substrate as compared with Bid because the Bnip2 precursor was already cleaved for 95% following 2 h incubation with 10 nm of hGrB whereas Bid was cleaved for 76% under identical conditions (Fig. 4). The ion signals of 577VLAEVITAAVR587 of Akap-8 display a modest increase in function of time (Fig. 3C), and as such the observed MS-pattern still falls within the statistically delineated region of efficient hGrB cleavage sites (Fig. 2). As for Bid, no proteolytic Akap-8 fragments could be observed by immunoblot analysis, though efficient cleavage was confirmed by the complete absence of the precursor substrate as early as 30 min following the addition of 10 nm of hGrB (Fig. 3C and Fig. 4). Processing of importin α-4 was found to lie outside the region of efficient cleavage events (Fig. 2) and was indeed confirmed as being the result of a far less efficiently cleaved hGrB site (only 48% of cleavage was detected following 2 h of incubation with 10 nm hGrB (Fig. 4)) as deduced from the gradual increase of the 50 kDa protein fragment generated upon ICED59 cleavage at later time-points (Fig. 3D). Finally, the three different nucleolin neo-N-termini identified, displayed large differences in the MS-signal intensities upon prolonged hGrB incubation (here shown in Fig. 3E for the neo-N terminus 476TTEETLKESFDGSVR490). Although all three identified cleavage events were predicted to be quite inefficient, the combined action of hGrB on all cleavage sites still resulted in a significant reduction of the precursor protein when challenged with hGrB, nevertheless nucleolin was found to be a far less efficiently cleaved substrate as compared with Bid, Bnip2 and Akap-8. The presence of a cleavage fragment close to the precursor band on Western blot occluded straightforward densitometric analysis, implying that the degree of cleavage could not be determined.
Fig. 3.
Biochemical characterization of the categorized efficiency of cleavage of the hGrB substrates Bid, Bnip2, Akap-8 (all categorized as “efficiently cleaved”), importin α-4, and nucleolin (categorized as “less efficiently cleaved”). The peptide ion triplets shown in the MS-spectra represent hGrB-specific neo-N-terminal peptides identified in Jurkat freeze-thaw lysates treated with human recombinant GrB for 10 (12C6, *, m/z value and charge indicated), 30 (13C6, +) or 60 (13C615N4, #) minutes. The corresponding peptides were AcD3-76SESQEDIIR84 from Bid (A), AcD3-29ILAITGPEDQPGSLEVNGNKVR50 from Bnip2 (B), AcD3-577VLAEVITAAVR587 from Akap-8 (C), AcD3-60SDIDGDYR67 from the importin subunit alpha-4 (D), and AcD3-476TTEETLKESFDGSVR490 from nucleolin (E). GrB-mediated processing of the corresponding substrates was assayed by Western blotting using antibodies directed against these substrates (see also Methods section). In each panel, Western blots shown at the right side are from cell lysates that were pre-incubated with iodoacetamide and treated with different concentrations of hGrB for various incubation periods. When the molecular weights of protein fragments (indicated with striped arrows) agreed with the fragment sizes as determined by the degradomics screen, the molecular weight values are indicated. Western blots shown at the left side are from control cell lysates that were non pre-incubated with iodoacetamide.
Fig. 4.
Degrees of hGrB processing at various time points determined for Bid, Bnip2, Akap-8, and importin α-4. Percentages of substrate cleavage were determined by densitometry following Western blot analyses using 10 nm of hGrB for 10, 30, 60, and 120 min. MS/MS spectra of all categorized hGrB neo-N-termini are available in PDF format. The highest scoring spectra (n=101) and corresponding lower scoring spectra (n = 130) are available in the PDF-files: “MS-MS_highest_scoring_spectra” and “MS-MS_lower_scoring_spectra” respectively.
Validation of Cleavage Events using Peptide Mimetics
For some substrates the sizes of the generated proteolytic fragments observed on Western blot were not in accordance with the expected theoretical molecular weights based on the identified cleavage sites. Together with the presence of additional fragments for some of these substrates, this precluded unambiguous assignment and validation of hGrB cleavage efficiency as observed for single proteolytic events (e.g. Fig. 3E). Therefore, the degree of cleavage of peptide substrates holding the identified cleavage site was determined in a RP-HPLC assay. For the synthetic peptides holding the identified cleavage sites of Bid, Bnip-2, and Akap-8, complete cleavage was observed following incubation with 500 nm of hGrB for 2 h (Supplemental Figs. 2A–C), agreeing with our proteomics data and immunoblot analysis. The peptide holding the hGrB site of importin α-4 was here also found to be less efficiently processed (i.e. 59% processed at 500 nm of hGrB, Supplemental Fig. 2D), again agreeing with our proteomics and immunoblot data. For some other peptide substrates of which the proteomics analysis yielded fold-changes deviating significantly from those of protein N-termini, hGrB cleavage was not observed or only found to occur less than 5% even at the highest concentration of hGrB (examples are protein phosphatase 1G, nucleolin and microtubule-associated protein 4 (data not shown and Supplemental Fig. 2E)), verifying these as inefficiently cleaved hGrB sites. Overall, the degree of peptide cleavage followed the degree of protein processing and concomitantly validated the results from the proteomics screen.
DISCUSSION
In this study, a gel-free method, combining metabolic labeling with N-terminal COFRADIC was used to study quantitative aspects of substrate proteolysis by human granzyme B. Time-based concentration fluxes of previously assigned hGrB-specific neo-N-termini (1) were assayed against the distribution of database-annotated, hGrB unaffected protein N-termini. 101 hGrB-specific neo-N-termini from 86 human Jurkat proteins were categorized according to their cleavage efficiency. Our assay assigned the pro-apoptotic Bcl-2 family member Bid, which lies at a pivotal point in hGrB-induced apoptosis, as possessing one of hGrB's most efficient sites. This illustrates that our approach is capable of revealing a critical substrate cleavage event implicated in protease-mediated signaling. In this respect, the pro-apoptotic Bcl-2 family member Bnip2 and Akap-8, both of which are known to play prominent roles in apoptotic execution (20, 21), are here also identified as holding efficiently cleaved hGrB specific sites.
In a model of natural killer cytotoxic granule-mediated K-562 cell death, 14 hGrB-specific physiological cleavage sites were identified (1). Interestingly, in our current study six of these sites were identified and three—processing of Bid, Akap-8, and the zinc finger CCHC domain-containing protein 8—were categorized as efficient hGrB cleavage events. These results again hint to possible physiological more important cleavage sites. One could argue that similar kinetics studies could be applied to monitor substrate processing in such physiological more relevant cellular setups however, an increased variability of perforin-mediated delivery, in case of granzyme B, next to an overall reduced kinetics of proteolysis and possible decrease in the stability of generated proteolytic fragments in intact cells would make the assignment of critical cleavage events more troublesome in cellular systems.
Of note is that the observed efficiency for individual cleavage events cannot be directly extrapolated to the substrate level. By itself, the turnover of the precursor substrate reflects the total efficiency by which the substrate is cleaved at different sites as observed for nucleolin (Fig. 3E). On the other hand, the identification of one efficient cleavage event suffices to consider the substrate as an efficiently cleaved one. The opposite however, the identification of a rather inefficient cleavage event reflecting a poorly cleaved substrate, does not necessarily hold true.
Another shortcoming of our approach is exemplified by the identification of three neighboring hGrB cleavage sites in the 40S ribosomal protein S15 (VDLD24↓QLLD28↓MSYE32↓ QLMQLYSAR40, Supplemental Fig. 3). This is a situation in which neo-N-termini are identified that hold additional, nested hGrB cleavage sites. As a consequence the fold change observed in MS spectra for neo-N-terminal peptides holding such indirect cleavage sites might therefore lead to an erroneous categorization of such cleavages. In the same respect, although caspase-7 did not make it to the list of efficient substrates, Western blot analyses performed by Adrain et al. demonstrated that Bid and caspase-7 were cleaved by hGrB with equal efficiencies, and further revealed that caspase-7 was cleaved at least at two different sites (24). As such, caspase-7 probably would have made it on our list of efficiently cleaved substrates, but because of a secondary cleavage event, extrapolation of caspase-7 cleavage efficiency from the MS-spectrum resulting from the primary cleavage, is compromised. This observation further hints that substrate processing efficiencies of promiscuous proteases will be much more challenging to assay by our approach.
The apparent acceleration of cleavage at inefficient granzyme B recognition sites (Fig. 2), which does not follow the expected standard first order kinetics, was unanticipated. We reasoned that in complex mixtures such as the proteome assayed in this study, efficient substrates are initially consumed by granzyme B, after which granzyme B is “liberated” and starts to increasingly process inefficient sites. We tested our hypothesis by assaying granzyme B processing of the fluorogenic peptide Ac-IEPD-AMC, both in granzyme B buffer as well as in a Jurkat cell lysate using similar concentrations of granzyme B and proteome as used in our proteomics study. Ac-IEPD-AMC has a reported Km value of 370 μm ± 70 μm—note that we here measured a Km value of 461 μm ± 26 μm—and is thus an inefficient substrate, with low affinity for granzyme B (25). Overall, the absence of first-order cleavage of Ac-IEPD-AMC, deduced from the acceleration of cleavage observed at earlier time-points when the peptide was assayed in the cell lysate (Supplemental Fig. 4), corresponds with our observations in the proteomics setup (Fig. 2) and thus confirms that processing at inefficient sites in full proteomes is accelerated once efficient sites are processed.
Selected reaction monitoring (SRM) appears to be an attractive and alternative MS-based method to probe the extent of proteolysis. SRM allows quantification of peptides by targeting a predetermined set peptide-to-fragment transitions (26). When combined with spiked-in internal standard peptides, SRM allows for the specific (absolute) quantification of a modest to high number of peptides present in rather complex peptide mixtures. As for degradomics studies however, prior knowledge of cleavage events is mandatory and as such SRM may be more suited to study selected and already characterized proteolytic events in several cellular setups.
Overall, and because cell-free reconstruction of biochemical relevant proteolytic pathways have proven to be representative in a cellular context (24), we here show that our multiplexed degradomics screen by probing the efficiency of protease cleavage events might be an interesting asset to map the segment of critical protease substrates.
Acknowledgments
We would like to thank Niklaas Colaert for bioinformatics assistance.
Footnotes
* K.P. and K.H. are supported by Ph.D grants from the institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen). P.V.D. and F.I. are Postdoctoral Fellows of the Research Foundation - Flanders (FWO-Vlaanderen). We further acknowledge support of research grants from the Fund for Scientific Research - Flanders (Belgium) (project numbers G.0077.06 and G.0042.07), the Concerted Research Actions (project BOF07/GOA/012) from the Ghent University and the Inter University Attraction Poles (IUAP06).
 This article contains supplemental Figs. 1–4 and Table 1.
This article contains supplemental Figs. 1–4 and Table 1.
1 The abbreviations used are:
- (h)GrB
- (human) granzyme B
- (h)Bid
- (human) BH3-interacting domain death agonist
- AcD3
- trideutero-acetyl
- Akap-8
- A-kinase anchor protein 8
- Bnip2
- BCL2/adenovirus E1B 19 kDa protein-interacting protein 2
- COFRADIC
- combined fractional diagonal chromatography
- SILAC
- stable isotope labeling by amino acids in cell culture
- SRM
- selected reaction monitoring.
REFERENCES
- 1. Van Damme P., Maurer-Stroh S., Plasman K., Van Durme J., Colaert N., Timmerman E., De Bock P. J., Goethals M., Rousseau F., Schymkowitz J., Vandekerckhove J., Gevaert K. (2009) Analysis of protein processing by N-terminal proteomics reveals novel species-specific substrate determinants of granzyme B orthologs. Mol. Cell Proteomics 8, 258–272 [DOI] [PubMed] [Google Scholar]
- 2. Trapani J. A., Smyth M. J. (2002) Functional significance of the perforin/granzyme cell death pathway. Nat. Rev. Immunol. 2, 735–747 [DOI] [PubMed] [Google Scholar]
- 3. Romero V., Andrade F. (2008) Non-apoptotic functions of granzymes. Tissue Antigens 71, 409–416 [DOI] [PubMed] [Google Scholar]
- 4. Impens F., Colaert N., Helsens K., Plasman K., Van Damme P., Vandekerckhove J., Gevaert K. (2010) MS-driven protease substrate degradomics. Proteomics 10, 1284–1296 [DOI] [PubMed] [Google Scholar]
- 5. Enoksson M., Li J., Ivancic M. M., Timmer J. C., Wildfang E., Eroshkin A., Salvesen G. S., Tao W. A. (2007) Identification of Proteolytic Cleavage Sites by Quantitative Proteomics. J. Proteome Res. 6, 2850–2858 [DOI] [PubMed] [Google Scholar]
- 6. Timmer J. C., Salvesen G. S. (2007) Caspase substrates. Cell Death Differ. 14, 66–72 [DOI] [PubMed] [Google Scholar]
- 7. Staes A., Van Damme P., Helsens K., Demol H., Vandekerckhove J., Gevaert K. (2008) Improved recovery of proteome-informative, protein N-terminal peptides by combined fractional diagonal chromatography (COFRADIC). Proteomics 8, 1362–1370 [DOI] [PubMed] [Google Scholar]
- 8. Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G. R., Vandekerckhove J. (2003) Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol. 21, 566–569 [DOI] [PubMed] [Google Scholar]
- 9. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 10. Sun J., Bird C. H., Thia K. Y., Matthews A. Y., Trapani J. A., Bird P. I. (2004) Granzyme B encoded by the commonly occurring human RAH allele retains pro-apoptotic activity. J. Biol. Chem. 279, 16907–16911 [DOI] [PubMed] [Google Scholar]
- 11. Sun J., Bird C. H., Buzza M. S., McKee K. E., Whisstock J. C., Bird P. I. (1999) Expression and purification of recombinant human granzyme B from Pichia pastoris. Biochem. Biophys. Res. Commun. 261, 251–255 [DOI] [PubMed] [Google Scholar]
- 12. Ghesquière B., Van Damme J., Martens L., Vandekerckhove J., Gevaert K. (2006) Proteome-wide characterization of N-glycosylation events by diagonal chromatography. J. Proteome Res. 5, 2438–2447 [DOI] [PubMed] [Google Scholar]
- 13. Martens L., Vandekerckhove J., Gevaert K. (2005) DBToolkit: processing protein databases for peptide-centric proteomics. Bioinformatics 21, 3584–3585 [DOI] [PubMed] [Google Scholar]
- 14. Van Damme P., Martens L., Van Damme J., Hugelier K., Staes A., Vandekerckhove J., Gevaert K. (2005) Caspase-specific and nonspecific in vivo protein processing during Fas-induced apoptosis. Nat. Methods 2, 771–777 [DOI] [PubMed] [Google Scholar]
- 15. Huber P. J., Ronchetti E.M. (2009) Robust Statistics, 2nd Ed., Wiley, John and Sons [Google Scholar]
- 16. Demon D., Van Damme P., Vanden Berghe T., Deceuninck A., Van Durme J., Verspurten J., Helsens K., Impens F., Wejda M., Schymkowitz J., Rousseau F., Madder A., Vandekerckhove J., Declercq W., Gevaert K., Vandenabeele P. (2009) Proteome-wide substrate analysis indicates substrate exclusion as a mechanism to generate caspase-7 versus caspase-3 specificity. Mol. Cell Proteomics 8, 2700–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Casciola-Rosen L., Garcia-Calvo M., Bull H. G., Becker J. W., Hines T., Thornberry N. A., Rosen A. (2007) Mouse and human granzyme B have distinct tetrapeptide specificities and abilities to recruit the bid pathway. J. Biol. Chem. 282, 4545–4552 [DOI] [PubMed] [Google Scholar]
- 18. Andrade F., Roy S., Nicholson D., Thornberry N., Rosen A., Casciola-Rosen L. (1998) Granzyme B directly and efficiently cleaves several downstream caspase substrates: implications for CTL-induced apoptosis. Immunity 8, 451–460 [DOI] [PubMed] [Google Scholar]
- 19. Trapani J. A., Sutton V. R. (2003) Granzyme B: pro-apoptotic, antiviral and antitumor functions. Curr. Opin. Immunol. 15, 533–543 [DOI] [PubMed] [Google Scholar]
- 20. Valencia C. A., Cotten S. W., Liu R. (2007) Cleavage of BNIP-2 and BNIP-XL by caspases. Biochem. Biophys. Res. Commun. 364, 495–501 [DOI] [PubMed] [Google Scholar]
- 21. Kamada S., Kikkawa U., Tsujimoto Y., Hunter T. (2005) A-kinase-anchoring protein 95 functions as a potential carrier for the nuclear translocation of active caspase 3 through an enzyme-substrate-like association. Mol. Cell. Biol. 25, 9469–9477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thornberry N. A., Rano T. A., Peterson E. P., Rasper D. M., Timkey T., Garcia-Calvo M., Houtzager V. M., Nordstrom P. A., Roy S., Vaillancourt J. P., Chapman K. T., Nicholson D. W. (1997) A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 272, 17907–17911 [DOI] [PubMed] [Google Scholar]
- 23. Barry M., Heibein J. A., Pinkoski M. J., Lee S. F., Moyer R. W., Green D. R., Bleackley R. C. (2000) Granzyme B short-circuits the need for caspase 8 activity during granule-mediated cytotoxic T-lymphocyte killing by directly cleaving Bid. Mol. Cell. Biol. 20, 3781–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adrain C., Murphy B. M., Martin S. J. (2005) Molecular ordering of the caspase activation cascade initiated by the cytotoxic T lymphocyte/natural killer (CTL/NK) protease granzyme B. J. Biol. Chem. 280, 4663–4673 [DOI] [PubMed] [Google Scholar]
- 25. Ruggles S. W., Fletterick R. J., Craik C. S. (2004) Characterization of structural determinants of granzyme B reveals potent mediators of extended substrate specificity. J. Biol. Chem. 279, 30751–30759 [DOI] [PubMed] [Google Scholar]
- 26. Lange V., Picotti P., Domon B., Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Scott G. B., Bowles P. A., Wilson E. B., Meade J. L., Low B. C., Davison A., Blair G. E., Cook G. P. (2010) Identification of the BCL2/adenovirus E1B-19K protein-interacting protein 2 (BNIP-2) as a granzyme B target during human natural killer cell-mediated killing. Biochem. J. 431, 423–431 [DOI] [PubMed] [Google Scholar]