Abstract
Considerable insight into phosphoinositide-regulated cytoplasmic functions has been gained by identifying phosphoinositide-effector proteins. Phosphoinositide-regulated nuclear functions however are fewer and less clear. To address this, we established a proteomic method based on neomycin extraction of intact nuclei to enrich for nuclear phosphoinositide-effector proteins. We identified 168 proteins harboring phosphoinositide-binding domains. Although the vast majority of these contained lysine/arginine-rich patches with the following motif, K/R-(Xn = 3–7)-K-X-K/R-K/R, we also identified a smaller subset of known phosphoinositide-binding proteins containing pleckstrin homology or plant homeodomain modules. Proteins with no prior history of phosphoinositide interaction were identified, some of which have functional roles in RNA splicing and processing and chromatin assembly. The remaining proteins represent potentially other novel nuclear phosphoinositide-effector proteins and as such strengthen our appreciation of phosphoinositide-regulated nuclear functions. DNA topology was exemplar among these: Biochemical assays validated our proteomic data supporting a direct interaction between phosphatidylinositol 4,5-bisphosphate and DNA Topoisomerase IIα. In addition, a subset of neomycin extracted proteins were further validated as phosphatidyl 4,5-bisphosphate-interacting proteins by quantitative lipid pull downs. In summary, data sets such as this serve as a resource for a global view of phosphoinositide-regulated nuclear functions.
Phosphoinositides (PIs)1 are lipid second messengers unique among phospholipids: Their inositol head group is rapidly phosphorylated by specific lipid kinases yielding seven distinct biologically relevant phosphatidylinositol derivatives. The coordinated activities of the PI-specific kinases and phosphatases generate PI profiles, which contribute to downstream signaling events regulating a variety of cellular processes such as proliferation, cell survival, migration, and vesicular trafficking (1–4). Impairment of PI metabolism is associated with cancer as well as neurological and immunological disorders (5–7). PIs are not only substrates for the generation of second messengers but are also second messengers themselves. They have emerged as sensors for specific PI-binding domains present in a diverse array of proteins: PH (pleckstrin homology), epsin N-terminal homology, FYVE (Fab1, YOTB, Vac1, EEA1), Phox homology, PHD (plant homeodomain), PDZ domains as well as unstructured lysine/arginine-rich patches. These domains display a range of heterogeneity in terms of their specificity for the different PIs (8–10) and recruit target, domain-containing, effector proteins in a temporal and spatial manner to sites of PI synthesis at many cytoplasmic locations (11).
PIs (notably phosphatidylinositol(3)phosphate (PtdIns(3)P), PtdIns(4)P, PtdIns(5)P, PtdIns(4,5)P2, PtdIns(3,4)P2 and PtdIns(3,4,5)P3) have also been identified within the confines of the nucleus, together with the enzymes responsible for their metabolism (12–18). They are regulated independently of the cytoplasmic PI pool (19, 20) and have been localized to defined nuclear regions (21–25). Emerging data point to significant roles for nuclear PIs. Increases in nuclear PtdIns(5)P mass levels result in the nuclear localization of the transcription factor inhibitor of growth protein 2 via an interaction between its PHD domain and the lipid (26, 27). Nuclear PtdIns(4,5)P2 binds to and regulates the activity of the poly(A) polymerase Star-PAP (nuclear speckle targeted PIPKIα regulated-poly(A) polymerase), an enzyme that also binds directly to the enzyme responsible for the synthesis of PtdIns(4,5)P2, namely, the Type I PtdIns(4)P 5-kinase (28). The chromatin remodeling protein BRG1 binds to PtdIns(4,5)P2 (29, 30), whereas other data link nuclear PIs to cell cycle progression (31, 32), apoptosis via an interaction between nucleophosmin and PtdIns(3,4,5)P3 (33), and pre-mRNA processing via interaction of nuclear speckle pools of PtdIns(4,5)P2 (22–25). Nuclear speckles are enriched in small nuclear ribonucleoproteins (snRNPs) and splicing factors (34) and constituent proteins that have been identified as PtdIns(4,5)P2 effector proteins, such as Syntenin-2 (35). These authors suggest that this interaction sequesters PtdIns(4,5)P2 to these nuclear structures. Furthermore the mRNA export factor Aly binds to both PtdIns(4,5)P2 and PtdIns(3,4,5)P3, an interaction essential for its localization to nuclear speckles (36).
Together these data point to a diverse set of nuclear activities regulated, in part, by the presence of PIs within this organelle. The global significance of nuclear PI-protein interactions however is still poorly understood, due largely to few known nuclear effector proteins. However in cases where this has been investigated in detail, it appears that such interactions have profound physiological effects (27–29). The identification of other nuclear PI effectors is therefore likely to shed more light on these and importantly other nuclear PI functions. To address this we have developed a proteomic approach to enrich for and identify potential nuclear PI-binding proteins by nano liquid chromatography (LC)-tandem MS (MS/MS). We have used the aminoglycoside neomycin to pull out this subset of proteins from isolated, intact nuclei and combined this approach with quantitative MS to determine specificity of the extraction procedure. Lipid affinity matrices were used to validate, biochemically, our proteomics data on target proteins and bioinformatics clustering analysis of the identified proteins allowed us to speculate the functional significance of nuclear PIs on a proteomic scale. Using this principle, we have identified 349 nuclear proteins, 48% of which harbor PI binding domains. Clustering analysis of these proteins revealed overrepresented functions related to RNA splicing, chromatin assembly, and DNA topological change. Furthermore, we have validated this method first by identifying a subset of proteins displaced by neomycin as PtdIns(4,5)P2 interacting proteins using quantitative lipid pull downs and second by biochemically characterizing the interaction of PtdIns(4,5)P2 with DNA topoisomerase IIα (Topo IIα), an enzyme with hitherto indirect links with nuclear PI metabolism. This proteomics approach provides compelling evidence suggesting that nuclear PIs interact with a wide range of nuclear proteins regulating numerous nuclear functions.
EXPERIMENTAL PROCEDURES
Reagents
Plasmids
The plasmid encoding GST (glutathione S-transferase)-tagged plekstrin homology domain of phospholipase Cδ1 (GST-PLCδ1-PH) (in pGEX-4T) was obtained from Dr AZ Gray (University of Dundee, UK) and GST-tagged PH domain of general receptor for phosphoinositides-1 (GST-GRP1-PH) (in pGEX-4T3) was obtained from Dr J Hastie (MRC, University of Dundee, UK). EGFP-RGS-6xHis-TopoIIα (in pEGFP-C3) was from Dr WT Beck (University of Illinois, Chicago, USA) (37). GST-TopoIIα-C-terminal domain (GST-TopoIIα-CTD) (aa 1171–1531, in pGEX-6P-1) was obtained from Dr. Susan PC Cole (Queen's University at Kingston, Ontario, Canada) (38).
Antibodies
commercial antibodies used were anti-Topo IIα (Ab1, #NA14, Calbiochem, La Jolla, CA), anti-GST (sc-138, Santa Cruz, Santa Cruz, CA), anti-nucleophosmin (32–5200, Zymed Laboratories Inc.), and HRP-coupled donkey anti-mouse and anti-rabbit secondary antibodies (Jackson ImmunoResearch, West Grove, PA). Anti-NF45 and anti-hnRNP D0B (peptide 3) were kind gifts from Dr PN Kao (Stanford University Medical Center) and Dr M Tolnay (Food and Drug Administration, Rockville, MD) respectively.
Lipid preparation
Phosphoinositide lipids used for surface plasmon resonance (SPR) analysis were purchased from Cell Signals Inc. l-α-d-myo-phosphatidylinositol (PtdIns), l-α-d-myo-phosphatidylinositol 3-monophosphate (PtdIns3P), l-α-d-myo-phosphatidylinositol 5-monophosphate (PtdIns5P), l-α-d-myo-phosphatidylinositol 3,5-bisphosphate (PtdIns(3,5)P2), and l-α-d-myo-phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) were stored as dry aliquots of 0.5 nmoles at −70 °C. For SPR analysis, lipid aliquots were equilibrated to room temperature, before being rehydrated in 100 μl 10 mm Tris pH 7.4 for 1 h. The solution was sonicated at high frequency for 5 min (cycles of 15 s ON and 7.5 s OFF) using a Bioruptor UCD200 (Diagenode, Denville, NJ). Finally, 200 μl 50 mm HCl were added and lipids were loaded onto an activated gas-liquid chromatography sensor chip at 30 μl/min for 300 s. Phosphoinositide lipids used in competition in either lipid pull downs or DNA decatenation assays were short chain PIs of eight carbons from Echelon Biosciences Inc. (Salt Lake City, UT).
Protein Purification
Expression and Purification of GST-Tagged Proteins Used in Lipid Pull Downs
Purification of GST-PLCδ1-PH and GST-GRP1-PH was as described (39) but with some modifications. GST-PLCδ-PH and GST-GRP1-PH constructs were transformed into Escherichia coli strain BL21 RIL DE3 and bacterial cultures were grown at 37 °C and induced with 0.5 mm isopropyl-β-d-thiogalactopyranoside (IPTG) for 3 h at 37 °C. Bacterial pellets were resuspended in 50 mm Tris pH 7.5, 1 mm EDTA, 0.27 mm sucrose, 1% Triton X-100, 1 mm Na3VO4 and 2 mm NaF, including 1 mm dithiotreitol and complete protease inhibitor mixture from Roche added fresh, lysed by one round of freeze-thawing and sonication and debris were removed by centrifugation. GST-tagged proteins were purified with glutathione-agarose 4B beads from an overnight pull down, and analyzed by SDS-PAGE and Coomassie staining for purity.
Expression and Purification of GST-Tagged Proteins Used in SPR Analyses
A glycerol stock of E. coli DH5α cells transformed with the corresponding GST-protein construct was used to inoculate 10 ml LB supplemented with 100 μg/ml ampicillin. The culture was grown overnight at 37 °C with shaking. Cells were diluted 10-fold and grown for another 2 h at 37 °C and induced with 100 μm isopropyl-β-d-thiogalactopyranoside at 25 °C overnight. Cells were collected by centrifugation at 4000 rpm for 15 min and lysed into 2 ml ice-cold lysis buffer (PBS, 1% Triton X-100, and 10 mm dithiotreitol). The lysate was frozen in liquid nitrogen, thawed at 37 °C and sonicated using a Bioruptor UCD200 (Diagenode). The lysate was cleared by centrifugation at 4 °C and the supernatant was mixed with 50 μl packed glutathione Sepharose 4B beads (GE Healthcare), previously equilibrated in lysis buffer. The mixture was rotated at 4 °C for 1.5 h, the beads were collected by centrifugation and washed three times with lysis buffer followed by two washes with wash buffer (50 mm Tris pH 8.0 and 300 mm NaCl). Proteins were eluted with 240 μl elution buffer (wash buffer supplemented with 10 mm reduced glutathione and 10 mm dithiotreitol). Aliquots were snap frozen in liquid nitrogen and kept at −70 °C.
Purification of His-Topo IIα
COS-1 cells were transfected with EGFP-RGS-6xHis-TopoIIα plasmid and lysed with buffer A (50 mm NaH2PO4 pH 8, 300 mm NaCl, 0.5% Tween 20, including 20 mm imidazole and protease inhibitors) with one freeze-thaw cycle. Cell extracts were incubated with Ni-NTA agarose beads for 16 h at 4 °C on a rotator. Sedimented beads were washed five times with buffer B (same as buffer A but with 0.1% Tween 20) and Topo IIα was eluted with buffer B including 250 mm imidazole). Topo IIα was identified by MS and detected by Western immunoblotting.
Cell Preparation
Cell Culture and SILAC (Stable Isotope Labeling with Amino Acids in Cell Culture) Labeling
Jurkat cells were grown in RPMI 1640 and 10% FCS and MEL cells in Dulbecco modified Eagle medium (DMEM) and 10% FCS in 5% CO2 at 37 °C. For isotopic metabolic labeling, MEL cells were transferred to DMEM (minus lysine and arginine) supplemented with 10% dialyzed fetal bovine serum with either 12C l-arginine and 12C l-lysine (light medium) or 13C l-arginine and 13C l-lysine (heavy medium) and subcultured for at least 10 population doublings that ensured complete incorporation of isotopic amino acids.
Nuclear Fractionation and Neomycin Extraction
Intact nuclei were prepared as described previously (32, 40) in the absence of Triton X-100. An equivalent number of nuclei (106 nuclei per 100 μl) were washed twice with retention buffer (20 mm Tris pH7.5, 70 mm NaCl, 20 mm KCl, 5 mm MgCl2, and 3 mm CaCl2) (41) and incubated in the same buffer in the absence (control) or presence of 5 mm neomycin (trisulfate salt, Sigma) for 30 min at room temperature. Nuclei were sedimented by centrifugation at 13,000 rpm for 5 min and supernatants were collected and used in Western analyses or in quantitative proteomic analyses. For quantitative proteomic analyses (Fig. 2), nuclei obtained from 12C-labeled cells were incubated with neomycin whereas nuclei obtained from 13C-labeled cells were incubated with retention buffer only (2.5 × 108 cells were used per condition).
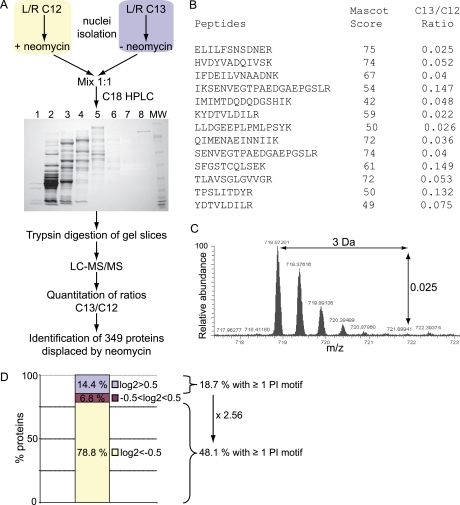
Fig. 2.
Workflow for the proteomic screening of nuclear proteins displaced by neomycin. A, Cell populations were metabolically labeled using SILAC to allow discrimination based on different peptide masses. The 13C K/R-labeled nuclei were incubated with control nuclear retention buffer, whereas the 12C K/R-labeled nuclei were incubated with 5 mm neomycin and proteins displaced out of nuclei were collected. Proteins from those parallel incubations were combined with equal concentration, fractionated on a macroporous C18 RP column, separated on one-dimensional SDS-PAGE, sliced and digested with trypsin. Peptides were analyzed by LC-MS/MS and isotope ratios for each peptide were quantified using MSQuant. See Table S1 for the identified proteins. B, Peptides identified and quantified for Topo IIα listing their respective Mascot score and 13C/12C ratio. C, Example for the first peptide (ELILFSNSDNER), doubly charged, which is about 40 times more abundant in the light form than in the heavy form. D, Protein classification according to their Log2 13C/12C ratios: proteins with Log2 > 0.5 (displaced by buffer, 64 proteins), Log2 < −0.5 (displaced by neomycin, 349 proteins) and proteins with −0.5 < Log2 < 0.5 (equally displaced by buffer and neomycin, 30 proteins).
Western immunoblotting: Proteins were resolved on 4%–12% NuPAGE Novex Bis-Tris gels with MOPS running buffer (Invitrogen) and transferred to nitrocellulose membranes. Membranes were blocked with 5% nonfat milk, incubated with primary antibodies overnight at 4 °C and with secondary antibodies conjugated to HRP for 1 h at room temperature. Revelation was performed by chemiluminescence using the SuperSignal West Pico Chemiluminescent Substrate (Pierce) and scanned with a LAS-3000 imaging system (Fuji).
Multi fractionation of proteins: Nuclear supernatants were dried down by vacuum centrifugation and resuspended in 200 μl of 6 m urea and 1% acetic acid. Proteins were separated by high performance liquid chromatography (HPLC) on a macroporous C18 reverse phase column (Agilent Technologies) with a 30 min multistep gradient elution of buffer A (2% acetonitrile, 0.1% trifluoroacetic acid) to buffer B (95% acetonitrile, 0.08% trifluoroacetic acid) at 300 μl/min. Eight fractions were collected, dried down, and resuspended in SDS sample buffer before separation by SDS-PAGE.
Protein-PI Interaction Analyses
Lipid pull downs: Purified-recombinant GST-tagged proteins (2 μg for GST-PLCδ1-PH and GST-GRP1-PH) or EGFP-His-Topo IIα full length (5 μg) were diluted in 400 μl of binding buffer (20 mm HEPES pH 7.5, 200 mm NaCl, 5 mm EDTA, 0.1% Nonidet P-40, 5 mm β-glycerophosphate, 1 mm orthovanadate and 2 mm NaF) with 20 μl PI-conjugated agarose beads or control agarose beads (Echelon Biosciences Inc) for 1 h at 4 °C. Sedimented beads were washed twice with the same buffer and proteins were eluted and resolved by SDS-PAGE. For competition studies with free lipids, short chain C8 PtdIns(4,5)P2 was incubated with recombinant proteins for 15 min on ice prior to the addition of the lipid beads. For lipid pull down assays of control and neomycin supernatants, three rounds of dialysis in binding buffer using Slide-A-Lyser® Mini dialysis units (Pierce) was carried out. For the quantitative lipid pull downs, dialyzed neomycin extracts (750 μg) obtained from either 12C-labeled cells or 13C-labeled cells were incubated with control-conjugated beads and PtdIns(4,5)P2-conjugated beads respectively.
Surface Plasmon Resonance (SPR)
A ProteOn XPR36 (BioRad) was used for SPR analysis. To prepare hydrophobic SPR sensor chips, undecylamine (C11H25N) was covalently coupled to a GLC sensor chip (BioRad) using a three-step amine coupling procedure in PBS running buffer at 25 °C. After cleaning the sensor surface with two horizontal and two vertical injections of 0.5% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid (CHAPS) at 100 μl/min for 36 s, a freshly mixed solution of 50 mm sulfo-NHS and 200 mm EDC was injected at 30 μl/min for 360 s. Undecylamine (Sigma Aldrich) was prepared for immobilization by making up a 1% solution in dimethyl sulfoxide, which was then diluted 20-fold in 10 mm sodium acetate pH 5.0 and injected at 30 μl/min for 400 s in the second step of the immobilization procedure. One molar ethanolamine hydrochloride pH 8.5 injected at 30 μl/min for 360 s was used to deactivate excess activated carboxylic groups and to remove electrostatically bound undecylamine.
After the sensor surface was preconditioned with two injections of 50 mm NaOH at 100 μl/min for 36 s, phosphoinositide lipids were injected onto the activated GLC sensor chip surface at 30 μl/min for 300 s at 25 °C. If necessary this step was repeated until the desired lipid loading level was obtained. The ligand sensor surface was postconditioned with one injection of 1 m NaCl and two injections of 50 mm NaOH at 100 μl/min for 18 s.
The running buffer used for analyte injections was PBS. Recombinant GST-tagged proteins were diluted in PBS. All analyte injections were performed at a flow rate of 50 μl/min and 25 °C sensor chip temperature. Association of proteins to phosphoinositide lipids was measured for 140 s, whereas dissociation was measured for 280 s. To avoid protein degradation, the autosampler temperature was maintained at 4 °C for the duration of analyte analysis. Following each analyte injection, the sensor chip surface was regenerated with one pulse of 1 m NaCl and two pulses of 50 mm NaOH at 100 μl/min for 18 s.
Measurements of protein binding to phosphoinositides were referenced by subtracting the binding response generated in the reference PtdIns channel employing ProteOn manager software (BioRad).
Topo IIα Activity Assay
DNA Decatenation Assay
Purified Topo IIα (TopoGEN, Inc, Port Orange, FL) (1 U per reaction) was incubated on ice in the absence (−) or presence of the indicated PIs (all C8 from Echelon) for 15 min prior to the addition of 200 ng of kinetoplast DNA (kDNA, TopoGEN), followed by the incubation of the samples for 30 min at 37 °C, according to the manufacturer's instructions (TopoGEN). The samples were resolved by agarose electrophoresis. Unprocessed kDNA remains in the wells, whereas decatenated kDNA migrates through the agarose gel.
Proteomics Sample Preparation
Protein Digestion
Proteins were separated by SDS-PAGE and the coomassie stained gel lanes were cut into 16–20 sections. Each gel section was cut into 2 mm cubes and washed sequentially with water, 100% acetonitrile, 100 mm ammonium bicarbonate and acetonitrile. Proteins were then reduced with 10 mm dithiotreitol in 100 mm ammonium bicarbonate for 45 min at 56 °C and then alkylated with 50 mm iodoacetamide in 100 mm ammonium bicarbonate for 45 min at room temperature. The gel pieces were then washed with 100 mm ammonium bicarbonate and finally with acetonitrile prior to drying under vacuum. The gel pieces were rehydrated in 6 μg/ml trypsin (Promega, Madison, WI) in 100 mm ammonium bicarbonate and incubated at 37 °C for 16 h.
Peptide separation, mass spectrometry and database analysis: Digested peptide mixtures were subjected to LC-MS/MS performed using QToF (Ultima Global, Waters, Denver, CO) or LTQ-Orbitrap XL (ThermoScientific) mass spectrometers.
The QTof was operated in positive ion mode and equipped with a Z-spray nano-ESI source and coupled to an LC Packings Ultimate 3000 nano-HPLC system using a ten port, zero dead volume valve (Valco vici) enabling fast sample loading onto a precolumn cartridge (LC Packings C18 PepMap 100 5 μm 100 Å 300 μmID × 5 mm). An isocratic flow rate of 30 μl/min of solvent A was delivered by the loading pump. The composition of the solvents A and B was 0.1% (v/v) formic acid in 5% (v/v) acetonitrile or 90% (v/v) acetonitrile respectively. After washing of the pre-column, the ten-port valve was switched allowing delivery of a biphasic acetonitrile gradient at 300 nl/min onto the analytical column (Reprosil-Pur 3.5 μm C18 resin Dr.Maisch Gmbh, Germany packed in a in a 15 cm × 75 μm ID fused silica capillary) by back flushing the precolumn. The gradient composition was 5%B to 80%B over 35 min followed by 10 min at 100%B. The QToF was operated in Data Directed Aquisition mode using a 1s MS survey scan. Collision-induced dissociation spectrum acquisition was allowed for up to a total of 2 s on each precursor ion or stopped when the signal intensity fell below ten counts per second respectively before a new MS to MS/MS cycle was started. Fused silica capillaries with 20 μm inner diameter served as spray emitters. Precursors were excluded from MS/MS experiments for 1 min and singly charged ions were excluded as precursors for MS/MS. Raw data were processed using MassLynx 4.0 ProteinLynx and the MS/MS data exported in Micromass pkl format.
The LTQ-Orbitrap XL was coupled to a Ultimate 3000 nano-HPLC system (Dionex, Sunnyvale, CA) fitted with an Acclaim PepMap 100 column (C18, 3 μm, 100 Å) (Dionex, Sunnyvale, CA) with a capillary of 12 cm bed length. A flow rate of 300 nl/min was employed with a solvent gradient of 7% B to 35% B in 77 min and subsequently to 50% B in 10 min. Solvent A was 0.1% formic acid, whereas aqueous 90% acetonitrile in 0.1% formic acid was used as solvent B. The mass spectrometer was operated in the data-dependent mode to automatically switch between Orbitrap-MS and LTQ-MS/MS acquisition. Survey full scan MS spectra (from m/z 300 to 2000) were acquired in the Orbitrap with resolution r = 60,000 at m/z 400 (after accumulation to a target of 1,000,000 charges in the LTQ). The method used allowed sequential isolation of the most intense ions, up to six, depending on signal intensity, for fragmentation on the linear ion trap using collision-induced dissociation at a target value of 100,000 charges. For accurate mass measurements the lock mass option was enabled in MS mode and the polydimethylcyclosiloxane ions generated in the electrospray process from ambient air were used for internal recalibration during the analysis. Target ions already selected for MS/MS were dynamically excluded for 60 s. General mass spectrometry conditions were: electrospray voltage, 1.5 kV; no sheath and auxiliary gas flow. Ion selection threshold was 500 counts for MS/MS, and an activation Q-value of 0.25 and activation time of 30 ms were also applied for MS/MS.
Raw files were processed to generate mascot generic files using the program DTASupercharge v.1.29 and merged with MGM combiner to obtain one mgf file per gel lane.
Computational Analyses
Database Search
The resulting pkl files (International Protein Index) (Q-ToF) and mgf files (LTQ-orbitrap) were searched against the IPI mouse database v3.46 (55272 entries) using the Mascot search engine version 2.1.04. Possible structure modifications allowed were carbamidomethyl cysteine as a fixed modification. Oxidized methionine, deamidation of Asn and Gln, 13C(6) Lys and Arg were searched as variable modifications. Searches were done with tryptic specificity allowing up to 1 miscleavage and a tolerance on mass measurement of 75 ppm in MS mode and 0.4 Da for MS/MS ions (Q-ToF), and 7 ppm in MS mode and 0.4 Da for MS/MS ions (LTQ-orbitrap). Only peptide identifications with a p value <0.05 and with ion scores over 42 (as determined by Mascot), and protein identifications based on no less than two peptides were used further. The false discovery rate was 0.082% using a reversed database under the same conditions.
Protein Quantitation
Protein quantitation was achieved using the MSQuant software v1.4.2a23 (http://msquant.sourceforge.net/), using raw data files together with the corresponding HTML result files generated with Mascot. Peptide ratios were calculated for each arginine- and/or lysine-containing peptide as the peak intensity of 13C labeled arginine/lysine divided by the peak intensity of nonlabeled 12C arginine/lysine (13C/12C) for each single-scan mass spectrum. Peptide ratios obtained for each protein were averaged and the standard deviation determined. Only proteins identified with LOG2 ratios <- 0.5 (Fig. 2) and LOG2 ratios > 0.5 (Fig. 4) were kept. The results were exported to Excel for further analysis. Statistical analyses were performed with StatQuant (42) for the quantitative lipid pull down.
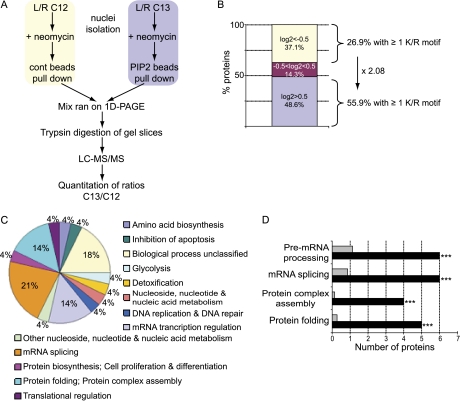
Fig. 4.
Quantitative and functional analysis of PtdIns(4,5)P2 pulled down proteins from neomycin extracts. A, Workflow of the quantitative pull-down strategy. 13C K/R-labeled and 12C K/R-labeled nuclei were incubated with 5 mm neomycin. Displaced proteins were pulled down at equal concentration with control beads (12C) or PtdIns(4,5)P2-conjugated beads (13C). Proteins in mixed eluates were resolved by SDS-PAGE, Coomassie stained and trypsin digested. Peptides were analyzed by LC-MS/MS and 13C/12C ratios were quantified using MSQuant and statistics were determined with StatQuant (42). Seventy proteins with at least two peptides and with significant (p < 0.05) Log2 13C/12 C ratios were retained for further analysis. Log2 > 0.5 represent proteins specifically pulled down by PtdIns(4,5)P2-beads (listed in Table I). B, Protein classification according to their Log2 values: proteins with Log2 < −0.5 (binding to control beads, 26 proteins), Log2 > 0.5 (binding to PtdIns(4,5)P2, 34 proteins) and proteins with −0.5 < Log2 < 0.5 (equal interaction with either control or PI beads, 10 proteins). C, Functional classification of PtdIns(4,5)P2-interacting proteins according to their biological process. D, Overrepresented biological processes for the PtdIns(4,5)P2-interacting proteins compared with all proteins annotated to the nucleus compartment (p values: *** p < 0.001).
Bioinformatic Analyses
Searches for K/R motifs: the IPI accession numbers were extracted and submitted to the Sequence Retrieval System (SRS) (http://srs.ebi.ac.uk/) and the corresponding fasta format sequences were retrieved. The fasta formatted sequences file was then parsed by an in-house script (using Python 2.5.2 (3)) to find occurrences of the following pattern K/R-X(n3–7)-K-X-K/R-K/R.
For the biological networks analyses, the identified IPI entries were first reassigned to gene entries (EntrezGene ID) annotated in the EnsEMBL database using Biomart (www.ensembl.org) (43). Gene Ontology (GO) identifiers were assigned to each IPI/gene entry and overrepresentation of GO categories were statistically analyzed using Cytoscape 2.6.1 and its plug-in BiNGO 2.0 (Biological Networks Gene Ontology tool) (44) by comparing the GO annotations obtained in our dataset to those of the mouse genome restricted to entries annotated to the nucleus compartment (GO :0005634). The representation for each GO category was calculated as the ratio between the cluster (neomycin dataset) frequency and the reference dataset (murine nucleome) frequency, the frequency being the percentage of gene entries in a particular GO term category compared with the respective total number of entries. These analyses were performed for both Biological processes and Molecular functions. The significance of overrepresented GO categories was assessed with a hypergeometric test and the Benjamini and Hochberg false discovery rate correction. A corrected p value < 0.05 was considered significant and only significantly overrepresented GO categories are presented (Fig. 3).
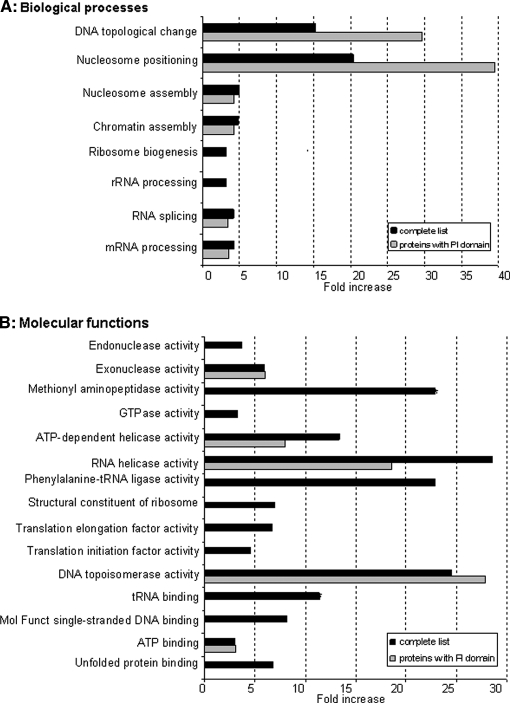
Fig. 3.
GO functional analysis for overrepresented biological processes and molecular functions. Statistically significant overrepresented GO categories for both biological processes (A) and molecular functions (B), as a result of the comparison of the neomycin-extracted protein dataset to the murine nucleome reference data set. Each GO category is represented as the ratio between % neomycin (= % of annotated proteins of the whole neomycin set) and % nucleome (= % of annotated proteins of the whole nucleome) and are visualized as black bars (complete list). The gray bars represent the statistically significant overrepresented GO categories when the same analysis was done but with only proteins containing K/R motifs, PH and PHD domains as the test dataset (proteins with PI domain). See Supplemental Figs. S2 and S3 for a visualization as hierarchy networks.
The functional classification of PtdIns(4,5)P2 interacting proteins and the over-representation of their biological processes presented in Fig. 2 were performed using PANTHER classification system/gene expression analysis (http://www.pantherdb.org/tools/genexAnalysis.jsp) (45, 46).
RESULTS
Extraction of Potential Nuclear PI-Binding Proteins with Neomycin
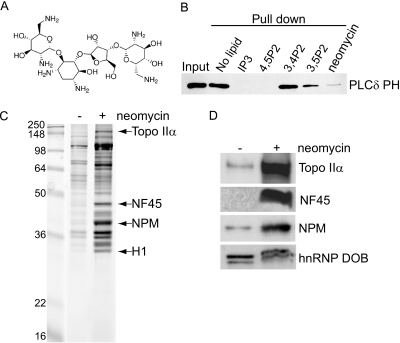
Neomycin (Fig. 1A) is known to bind to PIs with high affinity by way of electrostatic interactions between the basic amino groups of the compound and the negatively charged phosphate groups of PIs (47–50). Proteins that bind to PIs do so via similar interactions involving basic residues found in their PI-binding domains (9, 51). We speculated therefore that neomycin should compete for PI binding with PI-binding proteins, displacing them from their site of interaction. To establish if neomycin can displace PI bound proteins, immobilized PtdIns(4,5)P2 affinity beads were incubated with GST-tagged PH domain of PLC δ1 (Fig. 1B). In the absence of any competing lipids GST-PLCδ1-PH bound efficiently to PtdIns(4,5)P2 affinity beads. However, in the presence of free PtdIns(4,5)P2 and the water soluble head group, Ins(1,4,5)P3, this interaction was completely abolished. PtdIns(3,4)P2 was unable to compete for binding whereas only partial binding was observed in the presence of PtdIns(3,5)P2 demonstrating specificity for PtdIns(4,5)P2. Importantly, in the presence of neomycin the binding of GST-PLCδ1-PH domain was significantly reduced. These data demonstrate that neomycin can compete with characterized PI binding domains for binding to their target lipids. We extended these findings to isolated intact nuclei preparations. Supernatants recovered from nuclei incubated in the absence or presence of neomycin showed marked differences in their protein composition, as assessed by Coomassie blue staining (Fig. 1C). Protein bands were visible in control (no neomycin) supernatants, presumably as a result of nonspecific extraction or leakage. In contrast many more protein bands were visible from supernatants obtained following neomycin incubation. These differences in protein extraction were the same even when nuclei were prepared in the presence of 0.1% Triton X-100 (Supplemental Fig. S1), suggesting that the majority of these proteins are not extractable by detergent and derive from within the nuclear matrix. MS was used to identify prominent proteins specific for the neomycin extract (Topo IIα, NF45, nucleophosmin and histone H1; Fig. 1C), which were validated by Western immunoblot analyses (Fig. 1D). In contrast nonspecific proteins, such as the heterogeneous nuclear ribonucleoprotein (hnRNP) DOB, were equally present in supernatants obtained from nuclei incubation with or without neomycin.
Fig. 1.
Neomycin extraction of nuclear proteins. A, Molecular structure of neomycin, a polybasic molecule with 6 amino groups. B, 2 μg GST-PLCδ PH was incubated with PtdIns(4,5)P2-conjugated beads in the absence or presence of 20 μm free lipids or 5 mm neomycin. Eluates were analyzed by Western blotting using an anti-GST antibody. C, Isolated Jurkat nuclei (107) were washed and incubated in the absence (−) and presence (+) of 5 mm neomycin, releasing several specific proteins into the supernatant. These proteins were separated by SDS-PAGE and a few proteins, released by neomycin, were identified by nanoLC MSMS as Topo IIα, NF45, nucleophosmin (NPM) and Histone H1 (H1). See also Supplemental Fig. S1. D, Control (−) and neomycin (+) supernatants were analyzed by Western blotting for Topo IIα, NPM, NF45 and hnRNP DOB as a loading control.
Identification of Neomycin-Displaced Nuclear Proteins
To establish a substantial inventory of proteins that are specifically displaced from intact nuclei by neomycin, we have combined a SILAC-based quantitative proteomic approach with our extraction procedure. The workflow is outlined in Fig. 2A. An equal number of isolated nuclei from 12C and 13C labeled cells were extracted with neomycin and buffer respectively. The supernatants were combined at equal protein concentration and the protein complexity of the sample reduced by an initial round of fractionation using macroporous C18 reverse phase LC. Resulting fractions were further separated by one-dimensional-PAGE. Using this approach we were able to provide a more in depth analysis of the protein content of these samples. Each of the gel lanes were sliced into 16–22 fractions and the proteins within each gel section digested by trypsin. The extracted peptides were analyzed by nano LC-MS/MS and quantified with MSQuant. All proteins with decreased 13C/12C ratios were considered to be specific for the neomycin extract and 349 proteins were found to be specifically displaced by neomycin (Supplemental Table S1). The average for the ratios of all 349 proteins was 0.16 ± 0.15, demonstrating a clear validation for the neomycin extraction of these proteins. An example is shown for Topo IIα, which was identified by 13 peptides with a mean ratio of 0.065 ± 0.046 (Fig. 2B). The raw data used in MSQuant for one of these peptides is shown in Fig. 2C.
We predicted that some of the neomycin specific proteins would be candidate PI-binding proteins, either via direct or indirect binding, and a bioinformatics approach was used to search for the presence of known PI binding domains. Seven proteins were found to harbor PHD or PH domains (dynamin-2, Bruton's tyrosine kinase (BTK), PHD finger protein 8 (Phf8), the E3 ubiquitin ligase UHRF1, Fbxl11 (or 133 kDa protein), Src kinase associated phosphoprotein 2 (Skap2) and Rho/Rac guanine exchange factor 2 (ArhGEF2)), whereas 165 proteins, contained at least one K/R-rich motif (K/R- (Xn = 3–7)-K-X-K/R-K/R), known to bind PtdIns(4,5)P2 in several effector proteins (52) (Supplemental Table S1). Overall, 48% of all proteins displaced by neomycin displayed a K/R motif. In comparison, 18.7% of the proteins displaced by buffer alone contained such motifs (Fig. 2D). In addition, 30.2% of all known proteins that are annotated to the nucleus contain K/R motifs. The use of neomycin achieved therefore a 1.6-fold enrichment of proteins containing a K/R motif.
Functional Characterization of Neomycin-Displaced Proteins
To have some functional insight of our data, each of the 349 neomycin specific proteins was annotated by a Gene Ontology (GO) database for biological processes and molecular functions. The over-representation of each GO category was determined using Cytoscape and its Plug-in BiNGO, in the context of GO categories annotated to the nuclear compartment. Categories that were significantly over-represented statistically are shown in Figs. 3A and 3B (p < 0.05). These categories are also visualized as interaction networks and appear in downstream nodes in Supplemental Figs. S2 and S3. The GO categories, DNA topological change (represented by DNA topoisomerases) and nucleosome positioning (represented by histones) are particularly over-represented in the neomycin dataset (Fig. 3A). Several categories in molecular functions were also over-represented (Fig. 3B) and may represent functions attributed to PI effector proteins. We performed the same analyses, but on the subset of proteins containing PI-binding motifs, which resulted in the discovery of several common categories with the whole list (shown as gray bars in Figs. 3A and 3B and also visualized as blue circles in GO hierarchy networks in Supplemental Figs. S2 and S3). The proteins present in these categories are listed in Supplemental Tables S2 and S3. mRNA processing and especially RNA splicing, as well as chromatin assembly, were found to be significantly overrepresented, consistent with other data (24, 28, 53). The GO category, DNA topological change, was highlighted in our analysis and to our knowledge proteins listed in this category have not so far been reported to bind to PIs.
Identification of PtdIns(4,5)P2-Interacting Proteins from Neomycin Extracts
Our previous data suggests that within the neomycin extracted proteins exits a subset of proteins that bind directly to PIs, in particular PtdIns(4,5)P2. Quantitative lipid pull-down experiments were run to identify specific PtdIns(4,5)P2-interacting proteins from neomycin supernatants (Fig. 4A). Neomycin supernatants obtained from either 12C and 13C-labeled cells were incubated with control beads (beads conjugated to the fatty acid moieties only) or PtdIns(4,5)P2-conjugated beads respectively. Quantitative and statistical analyses revealed the identification of a total of 70 proteins, of which 28 proteins, annotated to the nucleus compartment, were specifically pulled down by PtdIns(4,5)P2 beads. These proteins are listed in Table I. Nineteen of these harbor a K/R motif that represented a twofold enrichment of PtdIns(4,5)P2 interacting proteins containing such motifs compared with noninteracting proteins (Fig. 4B). Functional classification of these proteins pointed to roles in mRNA transcription regulation, mRNA splicing and protein folding (Fig. 4C), the last two categories being over-represented (Fig. 2D) in this dataset.
Table I. PtdIns(4,5)P2 interacting proteins dataset. Proteins pulled down by PtdIns(4,5)P2, identified with at least 2 peptides, with p < 0.05 after analysis with StatQuant, and with C13/C12 log2 ratios >0.5, are indicated in this table.
| IPI ID | Gene name | Uniprot | Ratio | Log ratio | (log) STDEV | Number peptides | p value | % protein coverage | Description | K/R motif sequence | K/R motif position | GO cell compartment | GO biological process | Note |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IPI00399961 | Api5 | O35841 | 1.507 | 0.580 | 0.206 | 6 | 9.84E-04 | 12 | Putative uncharacterized protein | KGGTKEKR | 36–43 | Nucleus, cytoplasm | Inhibition of apoptosis | |
| IPI00319992 | Hspa5 | P20029 | 1.630 | 0.657 | 0.397 | 8 | 0.00225 | 16 | 78 kDa glucose-regulated protein precursor | RREVEKAKR RKDNRAVQKLRR | 290–298, 280–291 | Nucleus, cytoplasm | Protein folding; Protein complex assembly; Stress response | |
| IPI00120691 | Ddx21 | Q9JIK5 | 1.626 | 0.673 | 0.298 | 11 | 2.05E-05 | 18 | Nucleolar RNA helicase 2 | KPSEKKTRK | 24–33 | Nucleus | Nucleoside, nucleotide and nucleic acid metabolism | |
| IPI00225961 | Phgdh | Q61753 | 1.704 | 0.762 | 0.166 | 4 | 0.00275 | 9 | d-3-phosphoglycerate dehydrogenase | No pattern | Unannotated | Amino acid biosynthesis | ||
| IPI00153212 | Ccdc137 | Q8R0K4 | 1.721 | 0.782 | 0.087 | 2 | 0.04999 | 9 | Coiled-coil domain-containing protein 137 | KKTLSNKKRKK | 78–88 | Unannotated | Biological process unclassified | K/R motif within coil-coil domain1 |
| IPI00121311 | Csda | Q61478 | 1.987 | 0.873 | 0.638 | 6 | 0.02036 | 40 | DbpA murine homologue | No pattern | Nucleus, cytoplasm | mRNA transcription regulation | ||
| IPI00222509 | Rcc2 | Q8BK67 | 1.869 | 0.897 | 0.143 | 3 | 0.00838 | 5 | Regulator of chromosome condensation 2 | No pattern | Nucleus, cytoplasm | Biological process unclassified | ||
| IPI00331092 | Rps4x | P62702 | 1.929 | 0.927 | 0.279 | 5 | 0.00176 | 22 | 40S ribosomal protein S4, X isoform | KYKLCKVRK | 120–128 | Cytoplasm | ||
| IPI00154084 | Myef2 | Q8C854 | 1.928 | 0.933 | 0.248 | 3 | 0.02267 | 5 | Isoform 2 of Myelin expression factor 2 | No pattern | Nucleus | mRNA splicing | ||
| IPI00133801 | Sarnp | Q9D1J3 | 1.955 | 0.947 | 0.284 | 4 | 0.00687 | 23 | SAP domain-containing ribonucleoprotein | RKSEDDEKLKK | 165–176 | Nucleus, cytoplasm | mRNA transcription regulation | |
| IPI00136883 | Ptbp1 | Q3T984 | 1.964 | 0.952 | 0.303 | 3 | 0.0321 | 6 | Polypyrimidine tract binding protein 1 | KGFKFFQKDRK | 481–491 | Nucleus | mRNA splicing | |
| IPI00406118 | Syncrip | Q7TMK9 | 2.033 | 1.022 | 0.074 | 4 | 0.0001 | 11 | Isoform 2 of Heterogeneous nuclear ribonucleoprotein Q | KPPDQKRKERK | 406–417 | Nucleus, cytoplasm | mRNA splicing | |
| IPI00407130 | Pkm2 | P52480 | 2.040 | 1.024 | 0.137 | 3 | 0.00589 | 6 | Isoform M2 of Pyruvate kinase isozymes M1/M2 | No pattern | Nucleus, cytoplasm | Glycolysis | ||
| IPI00307837 | Eef1a1 | P10126 | 2.072 | 1.048 | 0.100 | 4 | 0.00024 | 9 | Elongation factor 1-alpha 1 | No pattern | Nucleus, cytoplasm | Translational regulation | ||
| IPI00126148 | Mafk | Q61827 | 2.077 | 1.054 | 0.083 | 2 | 0.03524 | 24 | Transcription factor MafK | KPNKALKVKK KEEVTRLKQRR | 6–15, 46–56 | Nucleus | mRNA transcription regulation; Cell proliferation and differentiation | K/R motif partly in DNA binding domain aa 51–76 |
| IPI00410836 | Fen1 | P39749 | 2.177 | 1.122 | 0.013 | 2 | 0.00513 | 5 | Flap endonuclease 1 | KAKTGGAGKFRR | 366–378 | Nucleus, cytoplasm | DNA replication; DNA repair | |
| IPI00116283 | Cct3 | P80318 | 2.217 | 1.125 | 0.319 | 3 | 0.02571 | T-complex protein 1 subunit gamma | No pattern | Nucleus, cytoplasm | Protein folding; Protein complex assembly | |||
| IPI00117877 | Mafg | O54790 | 2.221 | 1.140 | 0.215 | 3 | 0.01163 | 17 | Transcription factor MafG | KGNKALKVKR KEEIIQLKQRR | 6–15, 46–56 | Nucleus | mRNA transcription regulation; Cell proliferation and differentiation | K/R motif partly in DNA binding domain aa 53–83 |
| IPI00420261 | Hmgb1 | P63158 | 2.287 | 1.176 | 0.280 | 3 | 0.01839 | 20 | High mobility group protein B1 | KKGVVKAEKSKKKK | 172–185 | Nucleus, cytoplasm | Biological process unclassified | |
| IPI00117288 | Hnrnpab | Q99020 | 2.272 | 1.182 | 0.089 | 6 | 5.2E-07 | 21 | Heterogeneous nuclear ribonucleoprotein A/B | No pattern | Nucleus, cytoplasm | mRNA splicing | ||
| IPI00119305 | Pa2g4 | P50580 | 2.326 | 1.218 | 0.016 | 2 | 0.00602 | 4 | Proliferation-associated protein 2G4 | RKTQKKKKKK | 363–373 | Nucleus, cytoplasm | Protein biosynthesis; Cell proliferation and differentiation | K/R motif within region of interaction with RNA aa 361–375 |
| IPI00116281 | Cct6a | P80317 | 2.357 | 1.237 | 0.007 | 2 | 0.00247 | 3 | T-complex protein 1 subunit zeta | No pattern | Nucleus, cytoplasm | Protein folding; Protein complex assembly | ||
| IPI00469268 | Cct8 | P42932 | 2.378 | 1.248 | 0.081 | 3 | 0.00139 | 5 | T-complex protein 1 subunit theta | No pattern | Nucleus, cytoplasm | Protein folding; Protein complex assembly | ||
| IPI00230086 | Hnrnpd | Q60668 | 2.44 | 1.286 | 0.075 | 2 | 0.0262 | 9 | Isoform 2 of Heterogeneous nuclear ribonucleoprotein D0 | No pattern | Nucleus, cytoplasm | mRNA splicing | ||
| IPI00135686 | Ppib | P24369 | 2.450 | 1.291 | 0.097 | 3 | 0.00186 | 18 | Peptidylprolyl isomerase B | No pattern | Cytoplasm | |||
| IPI00462291 | Hmgb2 | P30681 | 2.599 | 1.372 | 0.160 | 3 | 0.00448 | 27 | High mobility group protein B2 | KGDKKGKK | 82–89 | Nucleus, cytoplasm | Biological process unclassified | |
| IPI00129430 | Sfpq | Q8VIJ6 | 2.612 | 1.383 | 0.110 | 2 | 0.03592 | 2 | Splicing factor, proline- and glutamine-rich | No pattern | Nucleus, cytoplasm | mRNA splicing | ||
| IPI00458583 | Hnrnpu | Q8VEK3 | 2.676 | 1.401 | 0.251 | 8 | 9.77E-07 | 10 | Heterogeneous nuclear ribonucleoprotein U | KEELKKRR | 17–24 | Nucleus, cytoplasm | Other nucleoside, nucleotide and nucleic acid metabolism | K/R motif within SAP domain aa 8–42 |
| IPI00318841 | Eef1g | Q9D8N0 | 2.840 | 1.506 | 0.047 | 2 | 0.01396 | 6 | Elongation factor 1-gamma | No pattern | Cytoplasm | |||
| IPI00555069 | Pgk1 | P09411 | 2.862 | 1.513 | 0.150 | 2 | 0.04441 | 8 | Phosphoglycerate kinase 1 | KLDVKGKR | 11–18 | Cytoplasm | ||
| IPI00118875 | Eef1d | P57776 | 2.890 | 1.531 | 0.011 | 3 | 1.58E-05 | 6 | Eukaryotic translation elongation factor 1 delta isoform a | RQYAEKKAKK | 180–189 | Cytoplasm | ||
| IPI00230108 | Pdia3 | P27773 | 3.579 | 1.836 | 0.116 | 8 | 7.1E-10 | 19 | Protein disulfide-isomerase A3 precursor | KKMTSGKIKK | 225–234 | Cytoplasm | ||
| IPI00555023 | Gstp1 | P19157 | 3.643 | 1.864 | 0.082 | 2 | 0.01972 | 12 | Glutathione S-transferase P 1 | No pattern | Unannotated | Detoxification | ||
| IPI00453827 | Tbc1d24 | Q3UUG6 | 13.07 | 3.703 | 0.168 | 2 | 0.02037 | 6 | Isoform 2 of TBC1 domain family member 24 | RQKGITVKQKR | 312–322 | Unannotated | Biological process unclassified |
Topo IIα Associates with PtdIns(4,5)P2
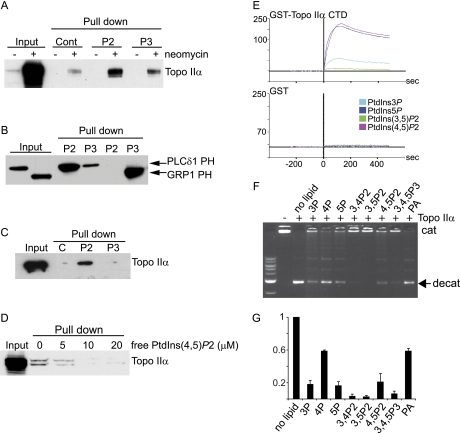
Although previous studies indicated a potential link between the PI pathway and the regulation of Topo IIα activity (54), no data exists to suggest that an interaction between Topo IIα with PIs exists. To demonstrate an interaction between Topo IIα and PIs, neomycin supernatants were dialyzed (removing neomycin) and incubated with control beads or beads conjugated with either PtdIns(4,5)P2 or PtdIns(3,4,5)P3 (Fig. 5A). Proteins bound to lipid beads were eluted and resolved by SDS-PAGE. A positive band for Topo IIα by Western blotting showed the interaction with PtdIns(4.5)P2-conjugated beads, whereas binding to PtdIns(3,4,5)P3-conjugated beads was marginally greater than control beads (Fig 5A). These data demonstrated specificity for binding to PtdIns(4,5)P2. As a control, specific interactions with PtdIns(4.5)P2 or PtdIns(3,4,5)P3 beads were demonstrated by lipid pull-down assays of known proteins harboring PH domains interacting specifically with either PtdIns(4,5)P2 or PtdIns(3,4,5)P3, i.e. PLCδ-PH and GRP1-PH respectively (Fig 5B). The same analysis was performed with purified Topo IIα. In this case, Topo IIα specifically bound to PtdIns(4,5)P2 whereas binding to PtdIns(3,4,5)P3 was at control level (Fig 5C). We also investigated the ability of free PIs to compete with PtdIns(4,5)P2-beads for binding to purified Topo IIα (Fig. 5D). PtdIns(4,5)P2 competed for binding of Topo IIα to PtdIns(4,5)P2 beads in a dose dependent manner.
Fig. 5.
Topo IIα interacts with PtdIns(4,5)P2. A, Control (−) and neomycin (+) dialyzed supernatants were incubated with control beads (Cont, only DiC6), PtdIns(4,5)P2 (P2)- or PtdIns(3,4,5)P3 (P3)-conjugated beads (pull down). Five percent input and eluates were analyzed by Western blotting using an anti- Topo IIα antibody. B, GST-PLCδ PH and GST-GRP1-PH were incubated with PtdIns(4,5)P2 (P2)- or PtdIns(3,4,5)P3(P3)-conjugated beads (pull down). Inputs and eluates were analyzed by Western blotting using an anti-GST antibody. C, EGFP-His-Topo IIα was incubated with control beads or PtdIns(4,5)P2 (P2)- and PtdIns(3,4,5)P3 (P3)-conjugated beads (pull down) (D) or with PtdIns(4,5)P2-conjugated beads in the presence or absence of free lipids. Inputs and eluates were analyzed by Western blotting using an anti- Topo IIα antibody. E, Analysis of binding of GST-TopoIIα-CTD (1 μm) and GST (1 μm), using SPR to PtdIns(3)P (light blue), PtdIns(5)P (dark blue), PtdIns(3,5)P2 (green) and PtdIns(4,5)P2 (pink). See also Supplemental Fig. S4. F, Topo IIα activity determined in the absence (−) or presence of free PIs (10 μm) and apparent in the form of decatenated kDNA (indicated with a filled arrow). G, Quantification of Topo IIα activity from 3 independent experiments.
Examination of the Topo IIα protein sequence highlighted the presence of 7 K/R motifs, all located in the regulatory C-terminal domain (CTD). Sequence alignment of these motifs are shown with other known PtdIns(4,5)P2 binding proteins with the same K/R motif, such as gelsolin (55), cofilin (56), villin (57), and myristoylated alanine-rich protein kinase C substrate (58) (Supplemental Table S4). To further determine if Topo IIα could interact with PIs, GST fused Topo IIα-CTD protein was purified and used for analysis by SPR (Fig. 5E). GST-proteins with known PI binding affinities were used to validate the assay: The PH domain from PLCδ1 interacted with PtdIns(4,5)P2, the 2xFYVE domain interacted with PtdIns(3)P and the PHD finger from ING2 interacted with PtdIns(5)P (Supplemental Fig. S4). GST alone did not interact with any of the lipids (Fig 5E). GST-Topo IIα-CTD interacted with PtdIns(4,5)P2 and PtdIns(5)P (Fig 5E) as well as with PtdIns(3,4,5)P3 (data not shown). We attempted to demonstrate binding of Topo IIα to liposomes consisting of PIs and phosphatidylcholine in a ratio of 1 to 100. Although we were able to demonstrate interaction of liposomes with the previously characterized PI binding proteins (PH-PLCδ1, 2xFYVE and PH-GRP1), no interaction was seen with the Topo IIα CTD domain or the 2xING2 PHD finger. Why the CTD and PHD finger should only recognize PIs when presented on their own is not clear but may be because of several factors: the properties of the lipids or vesicles used in the assay or the use of isolated domain instead of the full length protein. In addition, it may also be related to the fact that nuclear proteins may not be suited to interacting with PIs in the context of a membrane as this is unlikely to be the physico-chemical nature of PIs in the nucleus.
The ability of PIs to modulate Topo IIα activity in vitro was assessed by kDNA decatenation assays and visualized by agarose electrophoresis (Figs. 5F and 5G). In the absence of the enzyme, the catenated kDNA was restricted from entering the gel (lane 1), whereas the decatenated kDNA resulted in the presence of two processed DNA bands in the presence of Topo IIα (lane 2), as expected. The addition of PtdIns(3)P, PtdIns(5)P, PtdIns(3,4)P2, PtdIns(3,5)P2, PtdIns(4,5)P2, and PtdIns(3,4,5)P3 dramatically reduced the enzyme activity whereas the inhibition was less pronounced with PtdIns(4)P and phosphatidic acid.
DISCUSSION
Emerging data point to significant roles for PIs within the context of mammalian nuclei (24, 26–29) and identifying effector proteins for these lipids represents an important first step in deciphering PI-regulated nuclear functions. Currently the most successful methods used to identify effector proteins include the direct affinity purification of PI-binding proteins using lipid-conjugated beads. A prerequisite for the successful application of affinity-bead based enrichment protocols is generating a suitable starting protein extract, i.e. one that has reduced protein complexity whereas maintaining an enriched pool of target proteins (PI-binding proteins in this case). However, affinity-purifying PI effector proteins directly from total nuclear lysates is, at least from a proteomic point of view, fraught with issues of sample complexity and protein dynamic range. The overwhelming presence of abundant polybasic proteins present in the total nuclear extract would exclude or mask the binding of other less abundant bona-fide PI binding proteins by nonspecifically binding to the PI conjugated beads. Furthermore, because the physico-chemical nature of the protein-PI interaction within the nucleus is unclear, we cannot rule out that endogenous PIs remain bound to their effector proteins, thereby competing with the bead-conjugated PI during lipid pull downs. To address these issues, we used neomycin extraction as a means of both reducing sample complexity and enriching for a pool of potential PI-binding proteins devoid of bound PI. Several prior observations suggested that incubating isolated nuclei in the presence of excess neomycin would produce an extract that is both relatively less complex than a total nuclear lysate and importantly enriched in PI-binding proteins whose PI-binding domain(s) is or are devoid of any endogenous nuclear derived PI. First, neomycin binds PIs with high affinity and immobilized neomycin has been extensively used to purify PIs from tissue and cellular lipid extracts (47, 49, 50). Second, neomycin can compete with PtdIns(4,5)P2 specific antibodies for nuclear PtdIns(4,5)P2 binding sites in intact cell nuclei (24). Third, fluorescently labeled neomycin can bind to PtdIns(4,5)P2 and be used to visualize the lipid in model membrane preparations (59), plasma membrane and nuclear structures (60). Our nano LC-MS/MS data identified prominent proteins, with apparent specificity for the neomycin extract, such as histone H1 and nucleophosmin, two proteins reported to bind PIs (33, 53), substantiating our working hypothesis. Gel-based comparative analysis however is an inefficient means of determining specificity and we combined our neomycin extraction with SILAC labeling of cells (61) to provide greater sensitivity, throughput and depth of analytical coverage (62). SILAC is a powerful technique, not only for quantitative proteomic analyses, but also as a tool for comparative experiments assessing specificity. SILAC analysis allowed the identification of 349 proteins that were specific for the neomycin extract by our criteria, dramatically increasing the repertoire of neomycin extracted proteins from our initial Coomassie blue gel-staining analysis.
The neomycin extracted proteins included those with authentic PI-binding domains such as dynamin-2 (63) and the PtdIns(3,4,5)P3 binding protein BTK (63). Using either the SMART or Pfam software we also identified proteins that, to our knowledge, have yet to be characterized as PI-binding proteins but which nevertheless have PHD or PH domains within their sequence: Phf8, UHRF1, Fbxl11, Skap2 and ArhGEF2. By far, the majority of the identified proteins possessed at least one K/R motif. Histone H1 for example was identified and has been shown to bind PtdIns(4,5)P2 at the C-terminal domain that contains four K/R-rich motifs (53). All four Histone H1 variants (a,b,c and d) were identified in neomycin extracts. K/R motifs are of particular interest here, especially as they were first described as PtdIns(4,5)P2 binding domains for the actin-binding protein gelsolin (55) and were subsequently extended to include other cytoskeletal binding proteins, such as villin and cofilin (56–57). How these cytoskeletal proteins bind PIs is still unclear. It is not known if these interactions occur either as a component of a membrane structure or via a pool of cytosolic PtdIns(4,5)P2 distinct from any membrane structures. This has some similarity within the largely membrane free nuclear matrix and it is intriguing to speculate that a similar mechanism of interaction exists for both nuclear-PI and cytoskeletal-PI interactions via these K/R motifs, within the context of a membrane-free environment. The K/R motifs identified also share some similarities to nuclear localization sequences (NLS): the bipartite NLS sequence of KR-(X9–29)-KKK and K-K/R-X-K/R for monopartite NLS (65). We cannot rule out the possibility that some of these motifs may function as a NLS and have no relevance to nuclear PI metabolism. There are however examples of proteins, such as SAP30L and SAP30, which contain a NLS that has subsequently been shown to function as a binding motif for monophosphoinositides in the context of DNA binding regulation (64), thereby setting a precedent for a multifunctional role for these motifs. Finally the absence of a recognized PI-binding motif does not exclude these candidate proteins from being PI effectors. Nucleophosmin, for example, binds to PtdIns(3,4,5)P3 via several lysine residues within its C terminus in the absence of any characterized PI-binding domain (33). We identified nucleophosmin in our list highlighting the use of neomycin extraction as a non-biased approach for extracting nuclear PI effector proteins.
Cluster analysis of the neomycin specific proteins highlighted several biological processes represented by proteins in these extracts: DNA topology, nucleosome positioning/assembly and DNA packaging, chromatin assembly/disassembly, mRNA processing/RNA splicing and proteins involved in ribosomal related functions (Supplemental Table S2). A growing body of data links nuclear PI metabolism to RNA splicing and mRNA processing events via their colocalization to proteins that define nuclear subdomains called nuclear speckles (34). These domains consist of interchromatin granule clusters, are largely devoid of DNA and have been suggested to be sites for the assembly/storage/modification of predominantly splicing factors that can be released to sites of active transcription. These structures contain snRNPs, mRNA-splicing factors, and a hyperphosphorylated form of RNA polymerase II (66). PIPKs and their product PtdIns(4,5)P2 reorganize identically with speckles, both spatially and temporally (23, 24), suggesting direct interaction of PIPKs with speckle component(s). Several speckle associated proteins involved in RNA splicing were also identified in our neomycin extracts; U2 snRNP A', U4/U6 snRNP Prp4, SF3A1, U2AF65, DDX5/p68, SPF27, hnRNPQ and Bat1/UAP56. U2 snRNP A' and U4/U6 snRNP Prp4 proteins bind U2, U4 and U6 snRNAs. Significantly, U2, U4, and U6 snRNAs were identified in protein complexes immunoprecipitated using an anti-PtdIns(4,5)P2 antibody (24). Speckles are dynamic structures that are variable in size and shape and recent proteomic data suggests that the protein components of these structures are equally dynamic and include proteins resident in other structures such as spliceosomes: transcription factors, 3′-end RNA processing factors and other structural proteins (67). From this point of view several proteins in our data stand out. First, CPSF6 (Cleavage and polyadenylation specificity factor subunit 6 or CPSF 68 kDa) is a subunit of the 3′ pre-mRNA processing machinery, which also includes CPSF-73, CPSF-100 and the canonical poly(A) polymerase (PAP) (68). Both CPSF-73 and CPSF-100 interact directly with the noncanonical Star-PAP, a protein that is not only regulated by PtdIns(4,5)P2 (28) but also binds PIPKIα, the kinase responsible for the synthesis of the lipid. Second, we identified the RNA helicase Bat1/UAP56 that is known to recruit mRNAs to Aly, a nuclear speckle protein implicated in mRNA export. Aly binds to PIs and this interaction localizes Aly to nuclear speckles (36). Although the physiological role(s) for the presence of PtdIns(4,5)P2 in nuclear speckles is unclear, data suggest that they may contribute to the regulation of diverse activities of proteins involved in the processing of RNA: splicing, stabilization by poly(A)adenylation and finally export to the cytoplasm.
Proteins that control the level of DNA supercoiling and therefore DNA topology were enriched in the neomycin extracts. DNA topoisomerases I and IIα/β catalyze the passage of individual DNA strands (Topo I) or double strands (Topo II) through one another and contribute to the regulation of replication, transcription, recombination and chromosome segregation at mitosis (69, 70). Of specific interest here was the identification of Topo IIα. This enzyme, in vitro, is a direct target for protein kinase C α, β, and γ isoforms with the serine-29 suggested to be phosphorylated during G2/M phase of the cell cycle, thereby increasing enzyme activity (54). There is a large body of evidence that points to these diacylglycerol (DAG)-dependent protein kinase C isoforms as regulators of cell cycle progression (71–73) drawing upon a pool of DAG derived from the hydrolysis of nuclear PtdIns(4,5)P2 (74). Topo IIα is also the direct target for the cancer therapeutic agent, etoposide. In addition to inhibiting the enzyme activity, etoposide has been shown to induce changes in nuclear PI profiles. Treatment of HT1080 cells with etoposide leads to an increase in the nuclear mass levels of PtdIns(5)P (27) and subsequent translocation of the transcription factor ING2 to the nucleus (26). Etoposide treatment of HeLa cells also leads to the nuclear translocation of a Type I PtdIns(4,5)P2 4-phosphatase (75), the enzymatic product of which is PtdIns(5)P. This may contribute to the etoposide induced increases in PtdIns(5)P levels, in addition perhaps to the inhibition of a nuclear PtdIns(5)P 4-kinase activity (27). Protein sequence analysis of Topo IIα revealed seven K/R motifs located in the regulatory C-terminal domain, of which five are well conserved between the human, mouse and rat proteins. The location of the K/R motifs in the regulatory domain and our SPR data suggest that PtdIns(4,5)P2 may regulate the activity of Topo IIα either directly or as a component of large multisubunit protein complexes. Our biochemical data support a Topo IIα/PtdIns(4,5)P2 interaction in a variety of formats: in (i) lipid pull down experiments using purified Topo IIα, (ii) SPR analysis using the CTD of Topo IIα and (iii) in vitro enzyme activity assays suggesting that PtdIns(4,5)P2 interacts with and inhibits Topo IIα activity. This later effect was however also seen in response to other PIs, suggesting a lack of clear specificity for one PI with the observed inhibition. It is not clear how this reflects the situation in vivo. Because we know little about the the physico-chemical nature of the nuclear environment in which these lipids reside, the heterogeneity in binding we observe may reflect a complex level of regulation involving the dynamic regulation of more than one pool of nuclear PIs.
We have stated above that it is unlikely that the entire range of proteins extracted by neomycin represent PI-binding proteins. Proteins that bind to other negatively charged sites (DNA and RNA) via basic, lysine and arginine rich motifs are also likely to be displaced by neomycin. Nevertheless an assumption is that at least a sub-set of proteins extracted represent candidate PI effectors. Neomycin extracted proteins therefore represent an ideal preparation from which to affinity-purify PI-effector proteins using specific lipid conjugated matrices, avoiding the issues of sample complexity and dynamic range addressed previously. Probing SILAC labeled neomycin extracts with PtdIns(4,5)P2 conjugated beads resulted in the identification of 28 novel, nuclear PtdIns(4,5)P2-interacting proteins. More than half of these proteins were characterized by the presence of K/R motifs and more importantly, the proportion of proteins presenting such motifs was twofold increased in PtdIns(4,5)P2-interacting proteins compared with noninteracting proteins, which may suggest a mode of interaction for PI binding proteins localized in the nucleus.
In summary, we have shown that neomycin displaced over 300 proteins from isolated nuclei, and almost half of these harbor potential PI binding domains. These PI binding domains were mostly represented by unstructured positively charged K/R motifs and these motifs may represent predominant interaction sites for nuclear proteins with PI. Some of these proteins have been implicated in PI binding but many proteins with no previous history of PI binding were also identified. Furthermore, cluster analysis for biological functions highlighted several categories such as mRNA processing and splicing, DNA topological changes and chromatin assembly. To this end our proteomic methods have also enabled us to identify nuclear PtdIns(4,5)P2 interacting proteins that indeed provide an important resource on which to base future investigations. This work not only represents a global approach to identifying nuclear PI-effector proteins, it also represents a global approach to identify the array of nuclear functions dependent on nuclear PI turnover.
Acknowledgments
We thank W Beck (University of Illinois, USA), AZ Gray (University of Dundee, UK), PN Kao (Stanford University Medical Center, USA), SPC Cole (Queen's University at Kingston, Canada) and M Tolnay (FDA, USA) for kind gifts of reagents, P Puntervoll (Bergen Center for Computational Science, University of Bergen, Norway) for his help in the initial phase of the K/R pattern search and B Thiede (University of Oslo, Norway) for the MS analysis of lipid pull down.
Footnotes
* This research was supported by funding from the Functional Genomics Programme (FUGE) of the Research Council of Norway.
 This article contains supplemental Figs. 1–4 and Tables 1–4.
This article contains supplemental Figs. 1–4 and Tables 1–4.
1 The abbreviations used are:
- PI
- phosphoinositides
- NLS
- nuclear localization sequence
- PH
- pleckstrin homology
- PHD
- plant homeodomain
- PLC
- phospholipase C
- PtdIns(3)P
- phosphatidylinositol 3-monophosphate
- PtdIns(4)P
- phosphatidylinositol 4-monophosphate
- PtdIns(5)P
- phosphatidylinositol 5-monophosphate
- PtdIns(3,4)P2
- phosphatidylinositol 3,4-bisphosphate
- PtdIns(3,5)P2
- phosphatidylinositol 3,5-bisphosphate
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PtdIns(3,4,5)P3
- phosphatidylinositol 3,4,5-trisphosphate
- SILAC
- stable isotope labelling with amino acids in cell culture
- SPR
- surface plasmon resonance
- Topo IIα
- DNA Topoisomerase IIα.
REFERENCES
- 1. Janmey P. A., Lindberg U. (2004) Cytoskeletal regulation: rich in lipids. Nat. Rev. Mol. Cell Biol. 5, 658–666 [DOI] [PubMed] [Google Scholar]
- 2. Toker A. (2002) Phosphoinositides and signal transduction. Cell Mol Life Sci 59, 761–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Di Paolo G., De Camilli P. (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 4. Poccia D., Larijani B. (2009) Phosphatidylinositol metabolism and membrane fusion. Biochem. J. 418, 233–246 [DOI] [PubMed] [Google Scholar]
- 5. Pendaries C., Tronchère H., Plantavid M., Payrastre B. (2003) Phosphoinositide signaling disorders in human diseases. FEBS Lett. 546, 25–31 [DOI] [PubMed] [Google Scholar]
- 6. Skwarek L. C., Boulianne G. L. (2009) Great expectations for PIP: phosphoinositides as regulators of signaling during development and disease. Dev Cell 16, 12–20 [DOI] [PubMed] [Google Scholar]
- 7. Vicinanza M., D'Angelo G., Di Campli A., De Matteis M. A. (2008) Function and dysfunction of the PI system in membrane trafficking. EMBO J. 27, 2457–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lemmon M. A. (2008) Membrane recognition by phospholipid-binding domains. Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 9. McLaughlin S., Murray D. (2005) Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438, 605–611 [DOI] [PubMed] [Google Scholar]
- 10. Zimmermann P. (2006) The prevalence and significance of PDZ domain-phosphoinositide interactions. Biochim Biophys Acta 1761, 947–956 [DOI] [PubMed] [Google Scholar]
- 11. Lindmo K., Stenmark H. (2006) Regulation of membrane traffic by phosphoinositide 3-kinases. J. Cell Sci. 119, 605–614 [DOI] [PubMed] [Google Scholar]
- 12. Irvine R. F. (2003) Nuclear lipid signalling. Nat. Rev. Mol. Cell Biol. 4, 349–360 [DOI] [PubMed] [Google Scholar]
- 13. Hammond G., Thomas C. L., Schiavo G. (2004) Nuclear phosphoinositides and their functions. Curr Top Microbiol Immunol 282, 177–206 [DOI] [PubMed] [Google Scholar]
- 14. Jones D. R., Divecha N. (2004) Linking lipids to chromatin. Curr. Opin. Genet. Dev. 14, 196–202 [DOI] [PubMed] [Google Scholar]
- 15. Gonzales M. L., Anderson R. A. (2006) Nuclear phosphoinositide kinases and inositol phospholipids. J. Cell. Biochem. 97, 252–260 [DOI] [PubMed] [Google Scholar]
- 16. Cocco L., Martelli A. M., Fiume R., Faenza I., Billi A. M., Manzoli F. A. (2006) Signal transduction within the nucleus: revisiting phosphoinositide inositide-specific phospholipase Cbeta1. Adv Enzyme Regul 46, 2–11 [DOI] [PubMed] [Google Scholar]
- 17. Ye K., Ahn J. Y. (2008) Nuclear phosphoinositide signaling. Front Biosci 13, 540–548 [DOI] [PubMed] [Google Scholar]
- 18. Planchon S. M., Waite K. A., Eng C. (2008) The nuclear affairs of PTEN. J. Cell Sci. 121, 249–253 [DOI] [PubMed] [Google Scholar]
- 19. Cocco L., Martelli A. M., Gilmour R. S., Ognibene A., Manzoli F. A., Irvine R. F. (1989) Changes in nuclear inositol phospholipids induced in intact cells by insulin-like growth factor I. Biochem. Biophys. Res. Commun. 159, 720–725 [DOI] [PubMed] [Google Scholar]
- 20. Divecha N., Banfić H., Irvine R. F. (1991) The polyphosphoinositide cycle exists in the nuclei of Swiss 3T3 cells under the control of a receptor (for IGF-I) in the plasma membrane, and stimulation of the cycle increases nuclear diacylglycerol and apparently induces translocation of protein kinase C to the nucleus. EMBO J. 10, 3207–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Payrastre B., Nievers M., Boonstra J., Breton M., Verkleij A. J., Van Bergen en Henegouwen P. M. (1992) A differential location of phosphoinositide kinases, diacylglycerol kinase, and phospholipase C in the nuclear matrix. J. Biol. Chem. 267, 5078–5084 [PubMed] [Google Scholar]
- 22. Mazzotti G., Zini N., Rizzi E., Rizzoli R., Galanzi A., Ognibene A., Santi S., Matteucci A., Martelli A. M., Maraldi N. M. (1995) Immunocytochemical detection of phosphatidylinositol 4,5-bisphosphate localization sites within the nucleus. J Histochem Cytochem 43, 181–191 [DOI] [PubMed] [Google Scholar]
- 23. Boronenkov I. V., Loijens J. C., Umeda M., Anderson R. A. (1998) Phosphoinositide signaling pathways in nuclei are associated with nuclear speckles containing pre-mRNA processing factors. Mol. Biol. Cell 9, 3547–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Osborne S. L., Thomas C. L., Gschmeissner S., Schiavo G. (2001) Nuclear PtdIns(4,5)P2 assembles in a mitotically regulated particle involved in pre-mRNA splicing. J. Cell Sci. 114, 2501–2511 [DOI] [PubMed] [Google Scholar]
- 25. Watt S. A., Kular G., Fleming I. N., Downes C. P., Lucocq J. M. (2002) Subcellular localization of phosphatidylinositol 4,5-bisphosphate using the pleckstrin homology domain of phospholipase C delta1. Biochem. J. 363, 657–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gozani O., Karuman P., Jones D. R., Ivanov D., Cha J., Lugovskoy A. A., Baird C. L., Zhu H., Field S. J., Lessnick S. L., Villasenor J., Mehrotra B., Chen J., Rao V. R., Brugge J. S., Ferguson C. G., Payrastre B., Myszka D. G., Cantley L. C., Wagner G., Divecha N., Prestwich G. D., Yuan J. (2003) The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 114, 99–111 [DOI] [PubMed] [Google Scholar]
- 27. Jones D. R., Bultsma Y., Keune W. J., Halstead J. R., Elouarrat D., Mohammed S., Heck A. J., D'Santos C. S., Divecha N. (2006) Nuclear PtdIns5P as a transducer of stress signaling: an in vivo role for PIP4Kbeta. Mol Cell 23, 685–695 [DOI] [PubMed] [Google Scholar]
- 28. Mellman D. L., Gonzales M. L., Song C., Barlow C. A., Wang P., Kendziorski C., Anderson R. A. (2008) A PtdIns4,5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature 451, 1013–1017 [DOI] [PubMed] [Google Scholar]
- 29. Zhao K., Wang W., Rando O. J., Xue Y., Swiderek K., Kuo A., Crabtree G. R. (1998) Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95, 625–636 [DOI] [PubMed] [Google Scholar]
- 30. Rando O. J., Zhao K., Janmey P., Crabtree G. R. (2002) Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc. Natl. Acad. Sci. U.S.A. 99, 2824–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martelli A. M., Gilmour R. S., Neri L. M., Manzoli L., Corps A. N., Cocco L. (1991) Mitogen-stimulated events in nuclei of Swiss 3T3 cells. Evidence for a direct link between changes of inositol lipids, protein kinase C requirement and the onset of DNA synthesis. FEBS Lett. 283, 243–246 [DOI] [PubMed] [Google Scholar]
- 32. Clarke J. H., Letcher A. J., D'Santos C.S., Halstead J. R., Irvine R. F., Divecha N. (2001) Inositol lipids are regulated during cell cycle progression in the nuclei of murine erythroleukaemia cells. Biochem. J. 357, 905–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ahn J. Y., Liu X., Cheng D., Peng J., Chan P. K., Wade P. A., Ye K. (2005) Nucleophosmin/B23, a nuclear PI(3,4,5)P(3) receptor, mediates the antiapoptotic actions of NGF by inhibiting CAD. Mol Cell 18, 435–445 [DOI] [PubMed] [Google Scholar]
- 34. Lamond A. I., Spector D. L. (2003) Nuclear speckles: a model for nuclear organelles. Nat. Rev. Mol. Cell Biol. 4, 605–612 [DOI] [PubMed] [Google Scholar]
- 35. Mortier E., Wuytens G., Leenaerts I., Hannes F., Heung M. Y., Degeest G., David G., Zimmermann P. (2005) Nuclear speckles and nucleoli targeting by PIP2-PDZ domain interactions. EMBO J. 24, 2556–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Okada M., Jang S. W., Ye K. (2008) Akt phosphorylation and nuclear phosphoinositide association mediate mRNA export and cell proliferation activities by ALY. Proc. Natl. Acad. Sci. U.S.A. 105, 8649–8654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mo Y. Y., Beck W. T. (1999) Association of human DNA topoisomerase IIalpha with mitotic chromosomes in mammalian cells is independent of its catalytic activity. Exp. Cell Res. 252, 50–62 [DOI] [PubMed] [Google Scholar]
- 38. Mirski S. E., Sparks K. E., Friedrich B., Köhler M., Mo Y. Y., Beck W. T., Cole S. P. (2007) Topoisomerase II binds importin alpha isoforms and exportin/CRM1 but does not shuttle between the nucleus and cytoplasm in proliferating cells. Exp. Cell Res. 313, 627–637 [DOI] [PubMed] [Google Scholar]
- 39. Dowler S., Currie R. A., Campbell D. G., Deak M., Kular G., Downes C. P., Alessi D. R. (2000) Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 351, 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vann L. R., Wooding F. B., Irvine R. F., Divecha N. (1997) Metabolism and possible compartmentalization of inositol lipids in isolated rat-liver nuclei. Biochem. J. 327 (Pt 2), 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reyes J. C., Muchardt C., Yaniv M. (1997) Components of the human SWI/SNF complex are enriched in active chromatin and are associated with the nuclear matrix. J. Cell Biol. 137, 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. van Breukelen B., van den Toorn H. W., Drugan M. M., Heck A. J. (2009) StatQuant: a post-quantification analysis toolbox for improving quantitative mass spectrometry. Bioinformatics 25, 1472–1473 [DOI] [PubMed] [Google Scholar]
- 43. Flicek P., Aken B. L., Beal K., Ballester B., Caccamo M., Chen Y., Clarke L., Coates G., Cunningham F., Cutts T., Down T., Dyer S. C., Eyre T., Fitzgerald S., Fernandez-Banet J., Gräf S., Haider S., Hammond M., Holland R., Howe K. L., Howe K., Johnson N., Jenkinson A., Kähäri A., Keefe D., Kokocinski F., Kulesha E., Lawson D., Longden I., Megy K., Meidl P., Overduin B., Parker A., Pritchard B., Prlic A., Rice S., Rios D., Schuster M., Sealy I., Slater G., Smedley D., Spudich G., Trevanion S., Vilella A. J., Vogel J., White S., Wood M., Birney E., Cox T., Curwen V., Durbin R., Fernandez-Suarez X. M., Herrero J., Hubbard T. J., Kasprzyk A., Proctor G., Smith J., Ureta-Vidal A., Searle S. (2008) Ensembl 2008. Nucleic Acids Res. 36, D707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maere S., Heymans K., Kuiper M. (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 [DOI] [PubMed] [Google Scholar]
- 45. Thomas P. D., Campbell M. J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A. (2003) PANTHER: a library of protein families and subfamilies indexed by function. Genome Res 13, 2129–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thomas P. D., Kejariwal A., Guo N., Mi H., Campbell M. J., Muruganujan A., Lazareva-Ulitsky B. (2006) Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 34, W645–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schacht J. (1976) Inhibition by neomycin of polyphosphoinositide turnover in subcellular fractions of guinea-pig cerebral cortex in vitro. J Neurochem 27, 1119–1124 [DOI] [PubMed] [Google Scholar]
- 48. Gabev E., Kasianowicz J., Abbott T., McLaughlin S. (1989) Binding of neomycin to phosphatidylinositol 4,5-bisphosphate (PIP2). Biochim Biophys Acta 979, 105–112 [DOI] [PubMed] [Google Scholar]
- 49. Schacht J. (1978) Purification of polyphosphoinositides by chromatography on immobilized neomycin. J. Lipid Res. 19, 1063–1067 [PubMed] [Google Scholar]
- 50. Schacht J. (1979) Isolation of an aminoglycoside receptor from guinea pig inner ear tissues and kidney. Arch Otorhinolaryngol 224, 129–134 [DOI] [PubMed] [Google Scholar]
- 51. Rosenhouse-Dantsker A., Logothetis D. E. (2007) Molecular characteristics of phosphoinositide binding. Pflugers Arch 455, 45–53 [DOI] [PubMed] [Google Scholar]
- 52. Martin T. F. (1998) Phosphoinositide lipids as signaling molecules: common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu. Rev. Cell Dev. Biol. 14, 231–264 [DOI] [PubMed] [Google Scholar]
- 53. Yu H., Fukami K., Watanabe Y., Ozaki C., Takenawa T. (1998) Phosphatidylinositol 4,5-bisphosphate reverses the inhibition of RNA transcription caused by histone H1. Eur J Biochem. 251, 281–287 [DOI] [PubMed] [Google Scholar]
- 54. Wells N. J., Fry A. M., Guano F., Norbury C., Hickson I. D. (1995) Cell cycle phase-specific phosphorylation of human topoisomerase II alpha. Evidence of a role for protein kinase C. J. Biol. Chem. 270, 28357–28363 [DOI] [PubMed] [Google Scholar]
- 55. Yu F. X., Sun H. Q., Janmey P. A., Yin H. L. (1992) Identification of a polyphosphoinositide-binding sequence in an actin monomer-binding domain of gelsolin. J. Biol. Chem. 267, 14616–14621 [PubMed] [Google Scholar]
- 56. Kusano K., Abe H., Obinata T. (1999) Detection of a sequence involved in actin-binding and phosphoinositide-binding in the N-terminal side of cofilin. Mol Cell Biochem. 190, 133–141 [PubMed] [Google Scholar]
- 57. Kumar N., Zhao P., Tomar A., Galea C. A., Khurana S. (2004) Association of villin with phosphatidylinositol 4,5-bisphosphate regulates the actin cytoskeleton. J. Biol. Chem. 279, 3096–3110 [DOI] [PubMed] [Google Scholar]
- 58. Wang J., Arbuzova A., Hangyás-Mihályné G., McLaughlin S. (2001) The effector domain of myristoylated alanine-rich C kinase substrate binds strongly to phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 276, 5012–5019 [DOI] [PubMed] [Google Scholar]
- 59. Arbuzova A., Martushova K., Hangyás-Mihályné G., Morris A. J., Ozaki S., Prestwich G. D., McLaughlin S. (2000) Fluorescently labeled neomycin as a probe of phosphatidylinositol-4, 5-bisphosphate in membranes. Biochim Biophys Acta 1464, 35–48 [DOI] [PubMed] [Google Scholar]
- 60. Holz R. W., Hlubek M. D., Sorensen S. D., Fisher S. K., Balla T., Ozaki S., Prestwich G. D., Stuenkel E. L., Bittner M. A. (2000) A pleckstrin homology domain specific for phosphatidylinositol 4, 5-bisphosphate (PtdIns-4,5-P2) and fused to green fluorescent protein identifies plasma membrane PtdIns-4,5-P2 as being important in exocytosis. J. Biol. Chem. 275, 17878–17885 [DOI] [PubMed] [Google Scholar]
- 61. Harsha H. C., Molina H., Pandey A. (2008) Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nat Protoc 3, 505–516 [DOI] [PubMed] [Google Scholar]
- 62. Ong S. E., Foster L. J., Mann M. (2003) Mass spectrometric-based approaches in quantitative proteomics. Methods 29, 124–130 [DOI] [PubMed] [Google Scholar]
- 63. Salim K., Bottomley M. J., Querfurth E., Zvelebil M. J., Gout I., Scaife R., Margolis R. L., Gigg R., Smith C. I., Driscoll P. C., Waterfield M. D., Panayotou G. (1996) Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton's tyrosine kinase. EMBO J. 15, 6241–6250 [PMC free article] [PubMed] [Google Scholar]
- 64. Viiri K. M., Janis J., Siggers T., Heinonen T. Y., Valjakka J., Bulyk M. L., Maki M., Lohi O. (2009) DNA-binding and -bending activities of SAP30L and SAP30 are mediated by a zinc-dependent module and monophosphoinositides. Mol. Cell. Biol. 29, 342–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McLane L. M., Corbett A. H. (2009) Nuclear localization signals and human disease. IUBMB Life 61, 697–706 [DOI] [PubMed] [Google Scholar]
- 66. Mortillaro M. J., Blencowe B. J., Wei X., Nakayasu H., Du L., Warren S. L., Sharp P. A., Berezney R. (1996) A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc. Natl. Acad. Sci. U.S.A. 93, 8253–8257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Saitoh N., Spahr C. S., Patterson S. D., Bubulya P., Neuwald A. F., Spector D. L. (2004) Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell 15, 3876–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mandel C. R., Bai Y., Tong L. (2008) Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci 65, 1099–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Champoux J. J. (2001) DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 [DOI] [PubMed] [Google Scholar]
- 70. Wang J. C. (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat. Rev. Mol. Cell Biol. 3, 430–440 [DOI] [PubMed] [Google Scholar]
- 71. Goss V. L., Hocevar B. A., Thompson L. J., Stratton C. A., Burns D. J., Fields A. P. (1994) Identification of nuclear beta II protein kinase C as a mitotic lamin kinase. J. Biol. Chem. 269, 19074–19080 [PubMed] [Google Scholar]
- 72. Thompson L. J., Fields A. P. (1996) betaII protein kinase C is required for the G2/M phase transition of cell cycle. J. Biol. Chem. 271, 15045–15053 [DOI] [PubMed] [Google Scholar]
- 73. Sun B., Murray N. R., Fields A. P. (1997) A role for nuclear phosphatidylinositol-specific phospholipase C in the G2/M phase transition. J. Biol. Chem. 272, 26313–26317 [DOI] [PubMed] [Google Scholar]
- 74. D'Santos C. S., Clarke J. H., Irvine R. F., Divecha N. (1999) Nuclei contain two differentially regulated pools of diacylglycerol. Curr. Biol. 9, 437–440 [DOI] [PubMed] [Google Scholar]
- 75. Zou J., Marjanovic J., Kisseleva M. V., Wilson M., Majerus P. W. (2007) Type I phosphatidylinositol-4,5-bisphosphate 4-phosphatase regulates stress-induced apoptosis. Proc. Natl. Acad. Sci. U.S.A. 104, 16834–16839 [DOI] [PMC free article] [PubMed] [Google Scholar]