Abstract
The ability to decipher the dynamic protein component of any system is determined by the inherent limitations of the technologies used, the complexity of the sample, and the existence of an annotated genome. In the absence of an annotated genome, large-scale proteomic investigations can be technically difficult. Yet the functional and biological species differences across animal models can lead to selection of partially or nonannotated organisms over those with an annotated genome. The outweighing of biology over technology leads us to investigate the degree to which a parallel approach can facilitate proteome coverage in the absence of complete genome annotation. When studying species without complete genome annotation, a particular challenge is how to ensure high proteome coverage while meeting the bioinformatic stringencies of high-throughput proteomics. A protein inventory of Oryctolagus cuniculus mitochondria was created by overlapping “protein-centric” and “peptide-centric” one-dimensional and two-dimensional liquid chromatography strategies; with additional partitioning into membrane-enriched and soluble fractions. With the use of these five parallel approaches, 2934 unique peptides were identified, corresponding to 558 nonredundant protein groups. 230 of these proteins (41%) were identified by only a single technical approach, confirming the need for parallel techniques to improve annotation. To determine the extent of coverage, a side-by-side comparison with human and mouse cardiomyocyte mitochondrial studies was performed. A nonredundant list of 995 discrete proteins was compiled, of which 244 (25%) were common across species. The current investigation identified 142 unique protein groups, the majority of which were detected here by only one technical approach, in particular peptide- and protein-centric two-dimensional liquid chromatography. Although no single approach achieved more than 40% coverage, the combination of three approaches (protein- and peptide-centric two-dimensional liquid chromatography and subfractionation) contributed 96% of all identifications. Parallel techniques ensured minimal false discovery, and reduced single peptide-based identifications while maximizing sequence coverage in the absence of the annotated rabbit proteome.
The ability to detect the dynamic protein component of any system is determined by the inherent limitations of the technologies used, the complexity of the sample, and the existence of an annotated genome. Although there are numerous annotated species, species variations can on occasion, make it impossible to compromise. With the underlying biological question an important factor in experimental design, a significant limitation to high-throughput proteomics is the inherent difficulties associated with the selection of a non- or partially sequenced and/or annotated species. Species differences result in cellular and molecular heterogeneity, even though broad functional homogeneity is retained. The heart for example, although functioning as a muscular pump, shows vast species heterogeneity, including major distinctions in the rate of contracture between rodents (600 beats/minute; mouse) and larger mammals (60 beats/minute; dog) (1). The species discrepancy in heart rate has been shown to be unrelated to myofibrillar density (2). Instead there is a close relationship between heart rate and mitochondrial content, oxygen consumption and oxidative capacity (1–3). These species differences increase the possibility for differences in their respective cardiomyocyte proteomes.
Oryctolagus cuniculus (rabbit) is often selected for myocardial studies, along with other species lacking deep genome coverage, including dog (3081 UniProtKB identifiers), pig (7948 identifiers), and to a lesser extent, bovine (15,575 identifiers). With the rabbit whole genome shotgun not complete, there is incomplete annotation of the proteins associated. The challenges that this presents to proteomic studies are often outweighed by the physiology and pathophysiology of the species being more similar to human. Animal models are often used because the ability to obtain sufficient amounts of appropriate human tissue can be challenging. Human myocardium is generally scarce and rarely obtained in large quantities from nonpathophysiological states, potentially influencing protein localization and modification status, thereby altering the types of protein identified. The applicability to human tissue is the gold-standard measurement of any animal model. For cardiac studies, functional discrepancy between rodents and large mammals is a major consideration for species selection (4–8). Rabbit myocardium is a particularly suitable cardiac model, as it is the smallest mammalian species that accurately mimics several human physiological parameters including heart rate, electron transport chain coupling and mitochondrial density (2). For the purposes of the current study, we aimed to “reverse engineer” the rabbit cardiomyocyte mitochondrial proteome through the combined use of (i) multiple high-throughput proteomic techniques and (ii) sequence homology by mammalian phylogeny (9).
The mitochondria have been implicated in numerous disease models including diabetes (10), cardiovascular disease (as reviewed by (11), cancer (12), and neurological disorders (13). Although the overall role is generally uniform, mitochondria have evolved to facilitate diverse morphological and functional roles (14), meeting the specific needs of not only the individual tissue, but also the species (15–17). Previous mitochondrial proteomic studies have observed this tissue distinction at the protein level (18–20). Of the numerous protein inventories created of the mitochondria, where proteomics and GFP targeting are the most commonly used techniques, species with sequenced genomes are utilized.
Previous mitochondrial proteomic investigations have utilized a range of approaches, both gel-based and gel-free. The combination of one-dimensional gel electrophoresis (1-DE)1 and on-line peptide fractionation (GeLC-MS; gel-based) has been successfully applied to the study of the cardiomyocyte mitochondria from well-annotated species, Mus musculus (537 protein groups;(20) and Homo sapiens 542 protein groups;(21)) whereas a gel-free approach identified 406 protein groups in Homo sapiens (22). Mitochondrial proteomic profiles have also been generated across multiple organs obtained from mouse (591 protein groups from brain, heart, liver, and kidney; (23)) and rat (689 protein groups from muscle, heart, and liver; (18)) with the use of GeLC-MS. In investigations of mitochondria enriched from the liver, gel-free methods resolved 297 protein groups (mouse; (24)), with further partitioning of the inner mitochondrial membrane, identifying 348 and 182 protein groups from rat and mouse liver respectively (25, 26). The commonality across these large proteomic studies is the use of species with well annotated genomes. By comparison, mitochondrial proteomics performed in species with incomplete genome annotation, has to-date, been achieved using traditional two-dimensional electrophoresis (27, 28).
To help circumvent the need for annotated genomes, traditional two-dimensional gel electrophoresis (2-DE) methods can be utilized (8, 29, 30), whereby with adequate separation, single proteins can be resolved for analysis with mass spectrometry. The mitochondria can however, pose distinct challenges for such approaches, because of the bias toward basic proteins (31, 32) and the extensive inner and outer mitochondrial membranes (inner mitochondrial membrane and outer mitochondrial membrane respectively) (33). These membranes create functionally dissimilar compartments, with the mitochondrial matrix (internal to the inner mitochondrial membrane) and intermembrane space (between the inner mitochondrial membrane and outer mitochondrial membrane) containing additional proteins with essential functions. Previous studies have shown the impact of the intrinsic properties of mitochondrial proteins on 2-DE analysis, with only 77 unique inner mitochondrial membrane proteins identified by comparison with 342 identified using gel-free methodologies (32). Although 1-DE approaches are not subject to the same limitations, alone they are insufficient to observe low-abundance proteins. Gel-free methods are assumed to be more amenable to such investigations (as reviewed by (34, 35) but as mentioned previously, have the prerequisite of an annotated genome.
We hypothesize that depth and breadth of coverage of the partially annotated rabbit mitochondrion would be enhanced by using more than gel free protein or peptide separation method of the intact or subproteomes of the mitochondria that are ultimately dependent upon the ability of the selected technique to adequately solublize and partition (as reviewed by (36, 37)) the mitochondrial proteins. Furthermore, because of the lack of complete genome coverage of the rabbit, protein sequence homology from other mammals with phylogenetic relationships (9) must be used. We therefore also aim to determine the ability of these parallel methodologies for minimizing (i) false discovery rates; (ii) redundancy at numerous levels; and (iii) “one-hit wonders,” while (iv) maximizing sequence coverage; these being factors that influence the ultimate success of such a study.
EXPERIMENTAL PROCEDURES
All chemicals were purchased from Bio-Rad (Hercules, CA) and Sigma (St Louis, MO) unless stated otherwise.
Isolation and Enrichment of Intact Mitochondria
Mitochondria were isolated from frozen rabbit left ventricular myocardium (wet weight 3 g) as previously described (29, 31, 32). Briefly, myocardial samples were pulverized and then gently homogenized in eight volumes of ice-cold sucrose-mannitol buffer solution (220 mmol/L mannitol, 70 mmol/L sucrose, and 2 mmol/L HEPES, pH 7.4) in the presence of protease inhibitors (Sigma). Differential centrifugation sequentially partitioned, insoluble material and nuclear-associated structures (1100 × g); soluble and cytosolic proteins (20,000 × g), and myofilament-associated components (7000 × g), with the final pellet enriched for intact mitochondria (15,000 × g) (29, 31, 32). A fraction of the mitochondria pellet was set aside for further subfractionation (see below). The remaining mitochondrial pellet (or “intact mitochondria”) was resolublized in either 5% ASB-14 (protein-centric) or 6 mol/L Urea/2 mol/L thiourea (peptide-centric). Prior to protein-centric 2-DLC, the solublized mitochondria proteins required precipitation in ice-cold acetone to remove excess detergent. Following this, the protein pellet was solublized in LC-compatible buffer containing 2.5% n-octyl β-d-glucopyranoside (Sigma), prior to desalting and buffer exchange through SephadexTM G-25 Medium (PD-10 columns; Amersham Biosciences, Uppsala, Sweden) with PF2D chromatofocussing buffer (pH 8.5) (Beckman Coulter, Carlsbad, CA).
Subfractionation of Membrane-Enriched Proteins of the Mitochondria
Inner mitochondrial membrane were isolated according to previously reported protocols (38); and mitochondrial membranes were enriched by the modified sodium carbonate precipitation method as previously described (34, 39) using the Bio-Rad Membrane II Protein Extraction Kit (Bio-Rad, Hercules, CA). The membrane-enriched pellets from both preparations were resolublised in either 2.5% n-octyl β-d-glucopyranoside (protein-centric) or 6 mol/L Urea/2 mol/L thiourea (peptide-centric). To limit potential interference from lipids associated with the mitochondrial membranes, delipidation was performed by phase partitioning using the ProteoSolveLRS Kit (Pressure Biosciences Inc., West Bridgewater, MA. This step was only performed on the membrane-enriched preparations because of the relatively high ratio of lipid to protein suggested in the mitochondrial membranes (39). In addition to investigating the membrane-enriched fraction, the soluble fraction produced from the sodium carbonate precipitation method was also analyzed to observe loosely associated and soluble proteins. Soluble and associate proteins were concentrated and desalted using solid phase extraction (C-18 Sep-Pak cartridges, Waters Corp., Milford, MA) prior to analysis. Collectively, these fractionated samples will be regarded as “fractionated mitochondria.”
Protein-centric 2-DLC (PF2D)
Analysis of denatured, intact mitochondrial proteins was carried out on a PF2D (Beckman-Coulter, Fullerton, CA) as previously described (32, 40, 41). Briefly, proteins were separated in the first dimension through a chromatofocussing column (250 mm × 2.1 mm, Eprogen, Darien, IL), where proteins were fractionated through a gradient formed through the transition from 100% start buffer (6 mol/L urea, 25 mmol/L Bis-Tris, pH 8.5) to 100% eluent buffer (6 mol/L urea, 10% v/v Polybuffer 74 (GE Healthcare), pH 4 in 20% isopropanol) at a flow rate of 0.2 ml/min as previously described (40, 41). Following sample injection, a stable baseline was established at pH 8.5 for 20 min prior to the initiation of the pH gradient, following which the column was washed with 1 mol/L NaCl. Elution profiles were monitored at 280 nm. Fractions were collected every 5 mins while stable pH was detected (pH 8.5 and 4.0), and during pH gradient fractions were collected at 0.3 pH intervals. Fractions from the first dimension were sequentially injected onto the second dimension reverse phase-high performance liquid chromatography (RP-HPLC) column (33 mm × 4.6 mm, 1.5 mm nonporous ODS-IIIE C18 silica beads, Eprogen). Protein elution from RP-HPLC at 50 °C constant was monitored at 214 nm from injection (minute 0) through until 100% solvent B. Fractions were collected every 0.25 mins during 15 min linear gradient from 25% to 75% solvent B (solvent A: 0.1% trifluoroacetic acid in ddH2O; solvent B 0.08% trifluoroacetic acid in 100% acetonitrile) at a flow rate of 0.75 ml/min. Those chromatofocussing fractions collected during stable pH were pooled prior to injection onto RP-HPLC. In this case, fractions were collected every 0.25 mins during a 40 min linear gradient from 30%to 70% solvent B at a flow rate of 0.75 ml/min. All reverse-phase fractions were stored at −80 °C for further analysis.
Protein-centric 1-DLC
Both intact and fractionated mitochondria were separated by RP-HPLC (33 mm × 4.6 mm, 1.5 mm nonporous ODS-IIIE C18 silica beads, Eprogen). A 125-μg aliquot of solublized proteins were diluted in 0.1% trifluoroacetic acid /10% acetonitrile prior to loading onto RP-HPLC, kept constant at 50 °C. Fractions were collected every 0.4 mins during a 40 min linear gradient from 30% to 70% solvent B at a flow rate of 0.75 ml/min. All reverse phase fractions were stored at −80 °C for further analysis. To complement the linear separation of proteins, a stepwise (sawtooth) gradient from 10% to 100% B, was also implemented. Proteins were eluted from the RP-HPLC in packets, as determined by prolonged (5 mins) steps of solvent B. Fractions were collected every 1.25 mins from 10%, 20%, 30%, 40%, 50%, and 60% solvent B. Using electrospray ionization-tandem mass spectrometry (ESI-MS/MS), it was determined that proteins were only detectable in fractions collected at 10%–40% solvent B (data not shown). The four fractions collected during these steps were pooled prior to storage at −80 °C.
For RPLC ESI-MS/MS analysis, fractions generated from both 1-DLC and 2-DLC protein-centric approaches were dried using a SpeedVac concentrator (ThermoSavant). To re-solublize and neutralize each fraction, 25mmol/L ammonium bicarbonate was added prior to reduction and alkylation with 5 mmol/L tris(2-carboxyethyl)phosphine and 10mmol/L iodoacetamide. Proteins underwent proteolysis in sequencing grade modified porcine trypsin (1:25 ratio; Promega, Madison, WI) at 37 °C for 16 h.
Peptide-centric Approaches
Following urea/thiourea solublization of both intact and fractionated mitochondria, proteins (125 μg) were reduced and alkylated with 5 mmol/L tris(2-carboxyethyl)phosphine and 10mmol/L iodoacetamide. Proteins were initially digested in limiting amounts of Lys-C for 6 h at room temperature. Given the high concentration of urea, the mixture required dilution in five volumes of 50 mmol/L ammonium bicarbonate prior to proteolysis in sequencing grade modified porcine trypsin. To prevent chemical modifications, the digestion was performed at 25 °C for 24 h. Prior to peptide-centric 1-DLC and 2-DLC, peptides were desalted and concentrated with the use of C-18 SPE Sep-Pak cartridges (Waters Corp., Milford, MA), following which the samples were dried and stored at −80 °C. Peptides prepared for peptide-centric 1-DLC were loaded directly into on-line RP-HPLC ESI-MS/MS. For peptide-centric 2-DLC, the peptide mixture was fractionated by strong cation exchange chromatography (SCX) on a 1100 RP-HPLC system (Agilent, Santa Clara, CA) using a PolySulfoethyl A column (2.1 × 100 mm, 5 μm, 300 Å, PolyLC, Columbia, MD), by first dissolving the sample in 4 ml SCX loading buffer (25% v/v acetonitrile, 10 mmol/L KH2PO4 pH 2.8, adjusted with 1 N phosphoric acid). The sample was loaded onto the column and washed isocratically for 30 min at 250 μl/min. Peptides were eluted by a gradient of 0–350 mmol/L KCl (25% v/v acetonitrile, 10 mm KH2PO4 pH 2.8) over 40 min at a flow rate of 250 μl/min. The 214 nm absorbance was monitored and 15 SCX fractions were collected along the gradient. Each SCX fraction was dried down prior to the second dimension.
On-line RPLC ESI-MS/MS of Peptides
Protein identification by ESI-MS/MS analysis of peptides was performed using an LTQ ion trap MS (ThermoFisher, San Jose, CA) interfaced with a two-dimensional nanoLC system (Eksigent Technologies, Livermore, CA), or an LTQ Orbitrap (ThermoFisher) interfaced with a 1200 series nanoflow RPLC system (Agilent). Each sample (all preparations) was dissolved in 8 μl of 0.1% formic acid prior to analysis. Peptides were fractionated by RP-HPLC on a 75 μm × 100 mm C18 column with a 10 μm emitter using 0%–60% acetonitrile/0.5% formic acid gradient over 30 min or 60 min (peptide-centric 1-DLC alone) at 300 nl/min.
Identification of Unique Peptides and Proteins
Tandem mass spectra were extracted within Sorcerer (Sage-N, Milpitas, CA) using the ReAdW program. Charge state deconvolution and deisotoping were performed. All tandem mass spectrometry (MS/MS) samples were analyzed using Sequest (ThermoFinnigan, San Jose, CA; version v.27, rev. 11). Sequest was set up to search against the SWISS-PROT mammalian database (70894 entries; Protein Knowledgebase release 55.0) from UniProt knowledgebase (UniProtKB; SWISS-PROT + TrEMBL), assuming trypsin digestion, with a maximum of two missed cleavages. Reverse database searching was performed simultaneously to ensure false discovery rates (FDR) were minimal (FDR ≤ 3.7%). The rate was slightly relaxed, given the lack of the annotated rabbit genome. Sequest was searched with parent ion tolerance set to either 0.06 Da (Orbitrap) or 1.2 Da (LTQ; monoisotopic) and fragment tolerance set to 1.0 Da. Oxidation of methionine was specified in Sequest as a variable modification. Iodoacetamide derivative of cysteine was specified in Sequest as a fixed modification. In total 763,475 scanning events took place across all preparations and including all technical replicates. Scaffold (version Scaffold_2_02_00, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. All Sequest-derived peptide identifications were analyzed manually (44,098 spectra) if they exceeded deltaCn scores greater than 0.10 and XCorr scores greater than 2.1, 3.5, and 3.5 for doubly, triply, and quadruply charged peptides respectively. Those that did not meet these criteria were excluded immediately (719,377 scanning events). For inclusion, spectra were required to fulfill numerous requirements as outlined previously (42). Briefly, these requirements included (i) assignment of the majority of ions detected, including the most intense peaks, to parent or daughter ions and their associated peaks (arising from loss of water or amine); (ii) assignment of large ions (not related to the parent ion) to Pro/Asp; (iii) at least five isotopically resolved ions in sequential order, from both b- and y-ion series, matching theoretical peptide fragments and; (iv) maximum of one peak resolving with sufficient s/n ratios, that could not be assigned to parent and/or daughter ions and associated peaks. A total of 6521 spectra failed to meet these criteria. For those spectra that met requirements (iii) and (iv) but failed to meet (i) and (ii), de novo sequencing was attempted (3947 spectra), as previously described (8). Briefly, amino acid sequences were deduced by the mass differences between y- or b-ion “ladder,” following which, peptide sequences were then used to search the UniProtKB (SwissProt and TrEMBL) database using the program BLASTP “short nearly exact matches” (43). For inclusion of these de novo sequenced peptides, requirements (i) and (ii) needed to be met. Those that did not meet these criteria were excluded (10,459 spectra). In the end, 33,639 spectra met all the requirements for inclusion. No singly charged peptides were examined, given that ESI imparts a single charge, in addition to the charged C-terminus created as a result of tryptic digestion. Protein identifications were accepted if they contained at least one identified peptide (Supplemental Table 1). As previously described, single identifying peptides (or one-hit wonders) were excluded if they were: (i) not present in both technical replicates; (ii) only sequenced and matched once within an individual dataset (44). For those peptides with only one identifying peptide (Supplemental Fig. 1), that met the criteria for inclusion, verification was performed by manual interpretation of the MS/MS spectra. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Removal of possible sources of redundancy was achieved at three levels; naming redundancy, sequence redundancy, and species redundancy, by external validation of every sequenced peptide with the use of the BLASTP program, searching the UniProtKB. Within each experiment, numerous individual peptides were sequenced on multiple occasions (Supplemental Table 2). The number of MS/MS replicates for each peptide ranged from 2 to 35. As well, identification of a specific protein isoform was included if a peptide or an amino acid sequence unique to that particular isoform was observed. To enable cross-species comparisons with previously published human (21, 22) and mouse (20) cardiomyocyte preparations, all proteins detected were mapped to Mouse UniProtKB (Taxonomy 10090; 59,533 identifiers) identifiers. To limit the species effects, all proteins detected were mapped also to Human UniProtKB (Taxonomy 9606; 95,621 identifiers) identifiers. When a protein could not be mapped with 99% homology, the BLASTP “short nearly exact matches” program was used to identify the protein with the closest homology. If the homology was less than 90%, then it was included as a separate entry.
RESULTS AND DISCUSSION
The aim of the current study was to use proteomic technologies to improve coverage of the partially annotated rabbit mitochondrial proteome through the use of five parallel methodological approaches. Current coverage of the Oryctolagus cuniculus (rabbit) protein database represents <3% of the human sequence (2501 rabbit versus 95,621 human identifiers in UniProtKB database). Traditionally, this would limit the ability to undertake high-throughput proteomics, however with the use of multidimensional fractionation to ensure adequate partitioning of the complex sample and sequence homology across mammalian groups with phylogenetic relationships to rabbits (including rodentia and primates), we have attempted to circumvent this. To determine the success of the current study to achieve this aim, we performed a cross-experimental comparison with previous cardiomyocyte mitochondrial studies, undertaken in species with deep genome coverage.
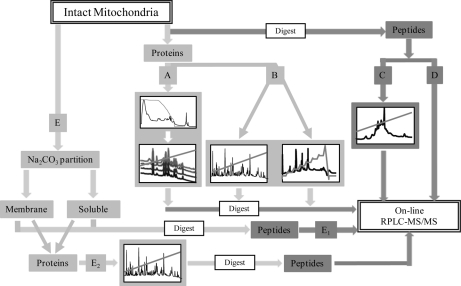
The current study used between 1 (peptide 1-DLC) and 3 dimensions (subfractionation) of separation (Fig. 1) including: (i) peptide-centric 1-DLC; (ii) peptide-centric 2-DLC; (iii) protein-centric 1-DLC; (iv) protein-centric 2-DLC and; (v) subfractionated mitochondria (Table I) to partition mitochondrial proteins and peptides prior to mass spectrometry. When taken together, these five approaches enabled identification of 558 nonredundant proteins and 2934 unique peptides, in the absence of a fully annotated genome (Table I). The overall coverage of the cardiomyocyte mitochondrial proteome was improved by this parallel approach, where the total number of nonredundant peptides was nearly two fold greater than the number of peptides identified by any single technique (subfractionation; 1520 peptides). This was also reflected in the total number of nonredundant proteins, which was more than 1.5-fold greater than any single approach (peptide-centric 2-DLC; 379 proteins). Of the 558 nonredundant proteins that were identified, there was a bimodal distribution across the pH spectrum (Supplemental Fig. 2a), with a skew toward the low molecular mass range, (10–20 kDa; 21%; Supplemental Fig. 2b). Each method also showed a partiality for a specific biochemical property, for example protein-centric 2-DLC identified a significant proportion of proteins with high pI (>9.5), and peptide-centric 2-DLC displayed a clear propensity for neutral proteins (pH 7) (Supplemental Fig. 3). We found that the multifaceted approach, one that utilizes multiple separation methods and the intrinsic biological features of mitochondria, was required to create a protein inventory that is equivalent to those from annotated species, as no single method would provide sufficient depth of coverage.
Fig. 1.
Schematic representation of multidimensional fractionation of mitochondria. Mitochondria were subjected to five orthogonal approaches. Following initial preparation of intact mitochondria, a small subset was further fractionated on the basis of topographical location (E). For protein-centric analysis (light gray), proteins were solublized and subject to either 2-DLC (A; HPCF and RP-HPLC) or 1-DLC (B; RP-HPLC alone). 1-DLC was performed using both linear and sawtooth RP gradients. Each resulting fraction was then digest and subject to RPLC-MS/MS. Peptide-centric analysis (dark gray) was performed on mitochondrial tryptic peptides. Samples were fractionated by 2-DLC (C; SCX and on-line RPLC-MS/MS) and 1-DLC (D; on-line RPLC-MS/MS alone). Subfractionated mitochondria (E) whereby membrane-associated and soluble partitions were created, were separated by either peptide-centric 1-DLC (E1) or protein-centric 1-DLC (E2). Protein-centric 2-DLC of intact mitochondria (A) and protein-centric 1-DLC of fractionated mitochondria (E2) are the strategies that resulted in three dimensions of fractionation.
Table I. Total number of nonredundant peptides and proteins from each technique. The total number of peptides and proteins is calculated on unique peptides and proteins across all techniques. Mitochondrial subfractionation consists of sodium carbonate enriched membrane proteins, inner mitochondrial membrane-enriched proteins, soluble and integral proteins. NOTE: Protein names have been clustered to remove redundancy.
| Preparation | Peptides | Proteins |
|---|---|---|
| Peptide-centric 1DLC | 1173 | 239 |
| Peptide-centric 2DLC | 1507 | 379 |
| Protein-centric 1DLC | 792 | 215 |
| Protein-centric 2DLC | 776 | 246 |
| Mitochondrial Subfractionation | 1520 | 306 |
| Total non-redundant | 2934 | 558 |
To ascertain the ability of the current study to sufficiently cover the mitochondrial proteome, in the absence of the complete rabbit genome annotation (2501 identifiers, SWISS-PROT + TrEMBL), it was necessary to compare it with large-scale investigations of human and mouse cardiomyocyte mitochondria, both of which have fully annotated genomes (59,533 mouse identifiers; 95,621 human identifiers; SWISS-PROT + TrEMBL). Although there is mitochondrial heterogeneity between species and tissues at the level of protein expression, there is sufficient homology in protein sequences across mammals to enable this comparison. This homology was deemed to be of sufficient value as to circumvent the use of de novo sequencing algorithms, which can be problematic when analyzing ion trap data (45).
Proteomic Comparison of Partially and Fully Annotated Genomes
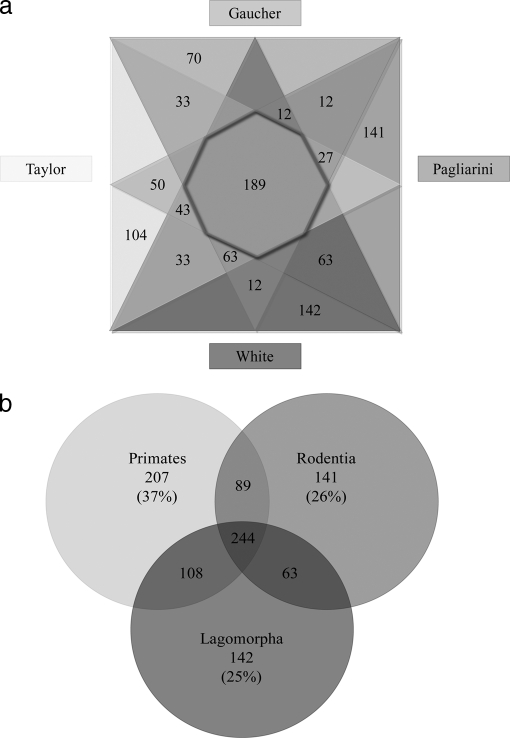
The ability to perform a cross-experimental comparison is dependent upon factors ranging from sample selection through to proteomic experimental conditions (including sample preparation, separation, and identification techniques). The rabbit cardiomyocyte mitochondrion that was used in this study was compared with protein inventories of human (21, 22) and mouse (20) cardiomyocyte preparations. Of the 995 nonredundant proteins identified, only 19% (189 proteins) were observed by all (Fig. 2a). By comparison with the proteins that were observed across the five orthogonal strategies used in the current study (108 proteins), over 90% (99 proteins) were also observed in mouse and human cardiomyocyte mitochondria studies (20–22). This may indicate that observation of proteins across multiple technologies improves the likelihood of true mitochondrial localization and may suggest that these proteins are highly abundant within the mitochondria, independent of species and tissue differences.
Fig. 2.
Direct comparison of mitochondrial protein identifications across four investigations. To illustrate the overlap and unique identifications from the current study and those undertaken in species with complete genome annotation we produced a four-way Venn diagram, comparing the individual studies (a) and a 3-way Venn diagram to illustrate the species to species variation (b). Fig. 2a shows the current study; and those undertaken by Gaucher et al. (22); Pagliarini et al. (20) and Taylor et al. (21). A total of 188 proteins were identified with across all investigations (as indicated by the intersection of the four triangles). The current study indentified 142 proteins, the study by Pagliarini et al. showed 141 unique proteins, whereas Taylor et al. and Gaucher et al. observed 104 and 70 unique proteins respectively. Overlapping segments of the triangles, represent the proteins shared between studies. Fig. 2b shows the comparison between primates (21, 22), rodents (20); and Lagomorpha. In this species comparison, 244 protein groups (25%) were identified across all investigations. Each species identified a unique subset of protein groups, which were compared with the total number of proteins identified within that species to give the percentages presented.
Perhaps a more valid comparison is one that investigates, not the results of these individual proteomic studies, but the proteins observed at the level of the organisms. Rabbits (Lagomorpha) have been traditionally grouped with rodentia (including rats and mice) under the superorder of Glires on the basis of morphological features (as reviewed by (46). At the protein level, studies have suggested Lagomorpha are more closely related to primates than rodents, based on sequence homology across 88 proteins (9). By comparing the mitochondrial proteins identified, rather than the sequences comprising them, mouse (20), rabbit and human (21, 22) proteome studies, each identified a similar number of proteins (535, 556, and 555 proteins respectively). As can be observed in Fig. 2b, this proteomic comparison may suggest more shared features between primates (human) and lagomorpha (rabbit), concurring with the results of Graur et al. The current study failed to detect 89 proteins that were identified in human and mouse mitochondrial studies, including 16 subunits of the 28S and 39S ribosome. Although it is difficult to determine the reason for this absence, if we compare the two primate studies, where one utilized gel-based (21) and the other, a gel-free (22) approach, we can observe the same absence of 28S ribosome subunits in the Gaucher et al. investigation, as the current study. We therefore hypothesize that this is a result of an experimental discrepancy between gel-based and gel-free methods, as we were otherwise able to detect low-abundance proteins, including other proteins contained in the 28S and 39S ribosomes.
The current study contributed 142 unique proteins, a similar proportion to that of both mouse (Pagliarini et al., 141 proteins) and human (Taylor et al., 104 proteins; Gaucher et al., 70 proteins), suggesting a level of divergence between experiments or species. Comparison with MitoMiner (v1.1; Supplemental Fig. 4), a compendium of 31 mitochondrial inventories across diverse tissues and species (47), showed that of the 142 unique proteins, 69% (99 proteins) had been suggested to associate with the mitochondria, confirming the sequence homology/expression heterogeneity across mammals. This group included proteins involved in mitochondrial fission (mitochondrial fission factor; MFF HUMAN) and fusion (mitofusin-2; MFN2 HUMAN), which are vital to the regulation of mitochondrial morphology and distribution (as reviewed by (11); and multiple components of the 39S mitochondrial ribosomal complex (RM16, RM42, RM45, RM50, and RM51 subunits). The vast majority of these unique proteins (108 proteins) were identified by a single orthogonal approach, including the low abundance proteins of the 39S ribosomal complex. This suggests that to ensure depth of coverage, the parallel approach is essential. Overall, both side-by-side comparisons (study comparison and species comparison) showed that even in the absence of complete genome annotation, a high-throughput proteomics study is possible, detecting a comparable number of proteins as those undertaken in species with annotated genomes. This was only possible however, with the (i) use of 5 different separation strategies and (ii) common amino acid sequence similarities across mammalian species. Combining the results from peptide- and protein-centric 2-DLC and subfractionated approaches, 96% of protein groups and all but 372 peptides (13%) were identified.
To decipher the strengths of each approach, we performed a five-way comparison (Supplemental Fig. 5). By comparison with the total number of proteins identified by each separation strategy (Table I), peptide-centric 2-DLC provided the highest proportion of unique protein groups (28%; 106 protein groups), followed by protein-centric 2-DLC (23%; 56 protein groups), subfractionation (14%; 44 protein groups), protein-centric 1-DLC (10%; 21 proteins), and peptide-centric 1-DLC (1%; 3 protein groups). The majority of the 230 proteins that were detected by a single method (Table II) were identified by multiple observations of a single peptide (with caveats outlined in Methods and discussed below). As mentioned previously, numerous components of the 39S mitochondrial ribosomal complex and transporter inner membrane complex were only observed by one strategy. With the exception of peptide-centric 1-DLC, all strategies contributed at least two unique subunits of the 39S complex and at least one unique transporter inner membrane subunit (Table II). Given the relatively low abundance of the proteins contained in these complexes, these results may suggest that proteins identified by a single approach represent those expressed at low levels. It is therefore important not to overlook these proteins as they specifically enhance coverage of the rabbit cardiomyocyte mitochondrial proteome.
Table II. Proteins uniquely observed using a single method. Numerous proteins were only observed in a single method. Proteins are sorted according to the method by which they were observed. Mitochondrial subfractionation consists of sodium carbonate enriched membrane proteins, inner mitochondrial membrane-enriched proteins, soluble and integral proteins. NOTE: Protein names have been clustered to remove redundancy.
| UniProtKB Accession number | Protein name | pI | Molecular Mass (Da) |
|---|---|---|---|
| Peptide-centric 1-DLC | |||
| DHSD_BOVIN | Succinate dehydrogenase complex subunit D, mitochondrial | 7.75 | 10996.82 |
| HS71A_BOVIN | Heat shock 70 kDa protein 1A | 5.68 | 70258.51 |
| MTCH1_HUMAN | Mitochondrial carrier homolog 1 | 9.4 | 41544.26 |
| Peptide-centric 2-DLC | |||
| ABCB8_HUMAN | ATP-binding cassette sub-family B member 8, mitochondrial | 9.21 | 79988.84 |
| ABLM1_HUMAN | Actin-binding LIM protein 1 | 8.91 | 87687.53 |
| ACAD8_HUMAN | Isobutyryl-CoA dehydrogenase, mitochondrial | 6.78 | 42681.86 |
| ACOT9_HUMAN | Acyl-coenzyme A thioesterase 9 | 7.79 | 47694 |
| ACOX1_CAVPO | Acyl-coenzyme A oxidase 1, peroxisomal | 8.81 | 74389.55 |
| ACSM5_HUMAN | Acyl-coenzyme A synthetase ACSM5, mitochondrial | 7.99 | 61718.45 |
| ADCY5_RABIT | Adenylate cyclase type 5 | 7.16 | 139623.6 |
| AL1A1_HUMAN | Retinal dehydrogenase 1 | 6.29 | 54730.65 |
| ANX10_MOUSE | Annexin A10 | 5.4 | 37300.66 |
| ANXA4_MOUSE | Annexin A4 | 5.43 | 35858.67 |
| AT1B1_RABIT | Sodium/potassium-transporting ATPase subunit β-1 | 8.61 | 34940.09 |
| ATP5S_BOVIN | ATP synthase subunit s, mitochondrial | 7.18 | 20324.55 |
| BAG3_HUMAN | BAG family molecular chaperone regulator 3 | 6.46 | 61594.69 |
| BUB3_BOVIN | Mitotic checkpoint protein BUB3 | 6.37 | 36954.52 |
| CA2D1_RABIT | Voltage-dependent calcium channel subunit α-2/Δ-1 precursor | 5.16 | 122178.14 |
| CAC1A_HUMAN | Voltage-dependent P/Q-type calcium channel subunit α-1A | 9 | 282364.63 |
| CALX_HUMAN | Calnexin precursor | 4.47 | 65395.57 |
| CAP1_RAT | Adenylyl cyclase-associated protein 1 (CAP 1) | 7.3 | 51457.7 |
| CHCH2_MOUSE | Coiled-coil-helix-coiled-coil-helix domain-containing protein 2, mitochondrial | 5.53 | 12375.92 |
| CIA30_MOUSE | Complex I intermediate-associated protein 30, mitochondrial | 6.74 | 35047.49 |
| COQ6_HUMAN | Ubiquinone biosynthesis monooxygenase COQ6 | 6.81 | 50869.93 |
| COX3_RABIT | Cytochrome c oxidase subunit 3 | 6.4 | 29730.37 |
| CQ10A_HUMAN | Protein COQ10 A, mitochondrial | 9.53 | 26117.41 |
| CTNB1_BOVIN | Catenin β-1 | 5.53 | 85510.61 |
| CXA1_RABIT | Connexin-43 | 8.96 | 42902.25 |
| D3D2_MOUSE | 3,2-trans-enoyl-CoA isomerase, mitochondrial | 7.77 | 29110.72 |
| DHB4_HUMAN | Peroxisomal multifunctional enzyme type 2 | 8.96 | 79555.16 |
| EFTS_BOVIN | Elongation factor Ts, mitochondrial | 6.26 | 30739.12 |
| EHD1_HUMAN | EH domain-containing protein 1 | 6.35 | 60626.86 |
| FA36A_MOUSE | Protein FAM36A | 9.21 | 13163.23 |
| FBN1_PIG | Fibrillin-1 | 4.83 | 309698.33 |
| G6PI_RABIT | Glucose-6-phosphate isomerase | 7.18 | 62615.54 |
| GBB1_BOVIN | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit β-1 | 5.6 | 37245.77 |
| GCSH_HUMAN | Glycine cleavage system H protein, mitochondrial | 4.36 | 13813.36 |
| GHC2_HUMAN | Mitochondrial glutamate carrier 2 | 9.39 | 33848.76 |
| GHITM_HUMAN | Growth hormone-inducible transmembrane protein | 9.95 | 37205.19 |
| GPDM_MOUSE | Glycerol-3-phosphate dehydrogenase, mitochondrial | 5.82 | 76551.52 |
| GRP78_BOVIN | 78 kDa glucose-regulated protein precursor | 5.01 | 70464.55 |
| GSTK1_HUMAN | Glutathione S-transferase κ1 | 8.53 | 25365.66 |
| INMT_RABIT | Indolethylamine N-methyltransferase | 5.16 | 28955.03 |
| ITB1_BOVIN | Integrin beta-1 precursor | 5.29 | 85868.18 |
| K6PF_RABIT | 6-phosphofructokinase, muscle type | 8.48 | 85072.2 |
| KBL_BOVIN | 2-amino-3-ketobutyrate coenzyme A ligase, mitochondrial | 6.36 | 42820.04 |
| KINH_HUMAN | Kinesin-1 heavy chain | 6.12 | 109684.86 |
| LAMA2_HUMAN | Laminin subunit α-2 precursor | 6.01 | 341915.98 |
| LAMB1_HUMAN | Laminin subunit β-1 precursor | 4.81 | 195734.5 |
| LAMC1_HUMAN | Laminin subunit gamma-1 precursor | 4.94 | 174281.89 |
| LPPRC_HUMAN | Leucine-rich PPR motif-containing protein, mitochondrial | 5.53 | 151839.79 |
| LUM_RABIT | Lumican | 5.93 | 37001.3 |
| MAON_HUMAN | NADP-dependent malic enzyme, mitochondrial | 8.16 | 67068.44 |
| MCAT_RAT | Mitochondrial carnitine/acylcarnitine carrier protein | 9.55 | 33153.99 |
| MCCB_MOUSE | Methylcrotonoyl-CoA carboxylase β chain, mitochondrial | 8.2 | 61379.05 |
| MDHC_MOUSE | Malate dehydrogenase, cytoplasmic | 6.16 | 36379.97 |
| MOES_BOVIN | Moesin | 5.91 | 67843.99 |
| MUTA_BOVIN | Methylmalonyl-CoA mutase, mitochondrial | 6.02 | 79441.22 |
| NALD2_MOUSE | N-acetylated-alpha-linked acidic dipeptidase 2 (EC 3.4.17.21) | 8.45 | 82801.05 |
| NDRG2_MOUSE | Protein NDRG2 | 5.23 | 40789.21 |
| NEUL_RABIT | Neurolysin, mitochondrial | 5.41 | 76618.5 |
| NIPS1_HUMAN | Protein NipSnap1 | 9.35 | 33309.98 |
| NU3M_RABIT | NADH-ubiquinone oxidoreductase chain 3 | 4.5 | 13067.82 |
| OAT_MOUSE | Ornithine aminotransferase, mitochondrial | 5.73 | 45790.52 |
| OPA3_HUMAN | Optic atrophy 3 protein | 9.07 | 19996.23 |
| PCCA_MOUSE | Propionyl-CoA carboxylase α chain, mitochondrial | 6.13 | 75158.18 |
| PCKGM_MOUSE | Phosphoenolpyruvate carboxykinase [GTP], mitochondrial | 6.24 | 66808.43 |
| PDIP2_HUMAN | Polymerase Δ-interacting protein 2 | 8.80 | 42033.28 |
| PDK1_HUMAN | Pyruvate dehydrogenase [lipoamide] kinase isozyme 1, mitochondrial | 7.68 | 46391.82 |
| PGBM_HUMAN | Perlecan | 6.03 | 466599.18 |
| PLAP_HUMAN | Phospholipase A-2-activating protein | 5.96 | 87157.07 |
| PPIA_BOVIN | Peptidyl-prolyl cis-trans isomerase A | 8.37 | 17738.16 |
| PTH2_HUMAN | Peptidyl-tRNA hydrolase 2, mitochondrial | 9.33 | 12679.91 |
| PYC_BOVIN | Pyruvate carboxylase, mitochondrial | 6.16 | 127368.67 |
| PYGM_MOUSE | Glycogen phosphorylase, muscle form | 6.65 | 97155.11 |
| RHOC_BOVIN | Rho-related GTP-binding protein RhoC precursor | 6.2 | 21682.93 |
| RL14_BOVIN | 60S ribosomal protein L14 | 10.79 | 23304.77 |
| RL28_RAT | 60S ribosomal protein L28 | 12.02 | 15717.42 |
| RLA0_BOVIN | 60S acidic ribosomal protein P0 | 5.72 | 34370.62 |
| RM13_HUMAN | 39S ribosomal protein L13, mitochondrial | 9.18 | 20691.96 |
| RM18_HUMAN | 39S ribosomal protein L18, mitochondrial | 9.63 | 20576.54 |
| RM50_BOVIN | Mitochondrial 39S ribosomal protein L50 | 8.95 | 18066.87 |
| RRBP1_HUMAN | Ribosome-binding protein 1 | 8.69 | 152472.22 |
| RS10_BOVIN | 40S ribosomal protein S10 | 10.15 | 18897.77 |
| RS20_BOVIN | 40S ribosomal protein S20 | 9.95 | 13241.52 |
| RT18A_BOVIN | 28S ribosomal protein S18a, mitochondrial | 10.11 | 18772.83 |
| RT34_BOVIN | 28S ribosomal protein S33, mitochondrial | 10.16 | 25700.51 |
| RTN3_BOVIN | Reticulon-3 | 8.35 | 27454.89 |
| RTN4_HUMAN | Reticulon-4 | 4.43 | 129931.42 |
| RYR2_RABIT | Ryanodine receptor 2 | 5.85 | 565380.92 |
| SCO1_BOVIN | SCO1 protein homolog, mitochondrial | 5.91 | 26499.31 |
| SEP11_BOVIN | Septin-11 | 6.38 | 48860.62 |
| SFXN1_BOVIN | Sideroflexin-1 | 9.42 | 35558.36 |
| SIRT3_MOUSE | NAD-dependent deacetylase sirtuin-3 | 5.81 | 28822.35 |
| SRBS1_HUMAN | Sorbin and SH3 domain-containing protein 1 | 6.4 | 142496.81 |
| SSPN_HUMAN | Sarcospan | 8.12 | 26618.48 |
| SYIM_HUMAN | Isoleucyl-tRNA synthetase, mitochondrial | 6.14 | 108737.58 |
| SYMM_HUMAN | Methionyl-tRNA synthetase, mitochondrial | 7.86 | 63267.02 |
| TAU_BOVIN | Microtubule-associated protein tau | 6.32 | 76243.42 |
| TBB2A_HUMAN | Tubulin beta-2A chain | 4.78 | 49906.97 |
| TBB5_HUMAN | Tubulin beta chain | 4.78 | 49670.82 |
| TFAM_PIG | Transcription factor A, mitochondrial | 9.52 | 24197.83 |
| THTR_BOVIN | Thiosulfate sulfurtransferase | 6.78 | 33164.63 |
| TIM23_HUMAN | Mitochondrial import inner membrane translocase subunit TIM23 | 8.81 | 21943.15 |
| TIM8A_RAT | Mitochondrial import inner membrane translocase subunit TIM8A | 5.11 | 11042.45 |
| TMM65_HUMAN | Transmembrane protein 65 | 9.1 | 25542.92 |
| UGPA_BOVIN | UTP-glucose-1-phosphate uridylyltransferase | 7.68 | 56772.04 |
| VPP1_BOVIN | Vacuolar proton translocating ATPase 116 kDa subunit a isoform 1 | 6.19 | 96301.69 |
| ZADH2_MOUSE | Zinc-binding alcohol dehydrogenase domain-containing protein 2 | 7.01 | 40528.85 |
| Protein-centric 1-DLC | |||
| ANXA1_RABIT | Annexin A1 | 6.28 | 38735.27 |
| B2MG_RABIT | β-2-microglobulin | 7.06 | 11654.13 |
| CN159_MOUSE | UPF0317 protein C14orf159 homolog | 6.15 | 63466.16 |
| COQ5_HUMAN | Ubiquinone biosynthesis methyltransferase COQ5, mitochondrial | 5.67 | 31803.21 |
| COXAM_HUMAN | COX assembly mitochondrial protein homolog (Cmc1p) | 8.89 | 12489.62 |
| ELMD1_BOVIN | ELMO domain-containing protein 1 | 8.88 | 38036.4 |
| GA2L2_HUMAN | GAS2-like protein 2 | 9.3 | 96519.85 |
| HNRPK_BOVIN | Heterogeneous nuclear ribonucleoprotein K | 5.14 | 51019.22 |
| HS105_BOVIN | Heat shock protein 105 kDa | 5.24 | 96725.87 |
| IPYR2_HUMAN | Inorganic pyrophosphatase 2, mitochondrial | 5.97 | 34707.39 |
| LDB3_HUMAN | LIM domain-binding protein 3 | 8.47 | 77134.87 |
| NQO1_PONAB | NAD(P)H dehydrogenase [quinone] 1 | 8.72 | 30939.71 |
| PIMT_HUMAN | Protein-l-isoaspartate(d-aspartate) O-methyltransferase | 6.78 | 24519.21 |
| PPIB_BOVIN | Peptidyl-prolyl cis-trans isomerase B precursor | 9.13 | 20200.15 |
| RM12_HUMAN | 39S ribosomal protein L12, mitochondrial | 5.37 | 16394.18 |
| RM16_RAT | 39S ribosomal protein L16, mitochondrial | 10.17 | 24807 |
| RM44_PONAB | 39S ribosomal protein L44, mitochondrial | 7.01 | 34429.51 |
| RM52_HUMAN | 39S ribosomal protein L52, mitochondrial | 10.08 | 11250.78 |
| SARDH_MOUSE | Sarcosine dehydrogenase, mitochondrial | 6.28 | 101682.28 |
| THEM2_HUMAN | Thioesterase superfamily member 2 | 9.23 | 14960.48 |
| TIM10_BOVIN | Mitochondrial import inner membrane translocase subunit TIM10 | 5.89 | 10332.95 |
| Protein-centric 2-DLC | |||
| A1CF_HUMAN | APOBEC1 complementation factor | 8.6 | 65202.48 |
| ABCB7_MOUSE | ATP-binding cassette subfamily B member 7, mitochondrial | 9.36 | 82594.95 |
| ADRO_HUMAN | NADPH:adrenodoxin oxidoreductase, mitochondrial | 7.65 | 49995.38 |
| ADT3_HUMAN | ADP/ATP translocase 3 | 9.76 | 32735.07 |
| ALDOC_RAT | Fructose-bisphosphate aldolase | 6.79 | 39152.59 |
| BOLA3_HUMAN | BolA-like protein 3 | 9.66 | 12114.17 |
| CATA_HUMAN | Catalase | 6.95 | 59624.98 |
| CHCH7_MOUSE | Coiled-coil-helix-coiled-coil-helix domain-containing protein 7 | 8.81 | 10101.52 |
| CSRP1_MOUSE | Cysteine and glycine-rich protein 1 | 8.92 | 20452.2 |
| DC1I2_BOVIN | Cytoplasmic dynein 1 intermediate chain 2 | 5.2 | 68377.26 |
| DNJA3_MOUSE | DnaJ homolog subfamily A member 3, mitochondrial | 9.34 | 52443.46 |
| DNJC4_HUMAN | DnaJ homolog subfamily C member 4 | 10.56 | 27593.36 |
| EF1A1_BOVIN | Elongation factor 1-α1 | 9.1 | 50140.86 |
| FABPH_RAT | Fatty acid-binding protein, heart | 5.92 | 14643.52 |
| GPX4_HUMAN | Phospholipid hydroperoxide glutathione peroxidase, mitochondrial | 8.69 | 22024.65 |
| GRPE1_HUMAN | GrpE protein homolog 1, mitochondrial | 6.03 | 21336.46 |
| H31_BOVIN | Histone H3.1 | 11.13 | 15272.89 |
| HBB_HUMAN | Hemoglobin subunit β | 6.81 | 15867.22 |
| HSC20_HUMAN | Cochaperone protein HscB, mitochondrial | 5.35 | 19435.12 |
| HXK3_HUMAN | Hexokinase-3 | 5.23 | 99025.36 |
| IF3M_BOVIN | Translation initiation factor IF-3, mitochondrial | 9.69 | 20190.16 |
| INO1_HUMAN | Inositol-3-phosphate synthase | 5.52 | 61067.81 |
| ISCU_HUMAN | Iron-sulfur cluster assembly enzyme ISCU, mitochondrial | 8.83 | 14385.66 |
| KCNC3_RAT | Potassium voltage-gated channel subfamily C member 3 | 6.62 | 94393.4 |
| MFF_HUMAN | Mitochondrial fission factor | 9.01 | 38464.55 |
| MTO1_MOUSE | Protein MTO1 homolog, mitochondrial | 8.52 | 71389.11 |
| NUCG_HUMAN | Endonuclease G, mitochondrial | 8.92 | 27717.43 |
| PLCB1_BOVIN | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta-1 | 5.86 | 138715.05 |
| PYRG1_HUMAN | CTP synthase 1 | 6.02 | 66690.36 |
| RHG20_HUMAN | Rho GTPase-activating protein 20 | 8.26 | 132607.97 |
| RL23_BOVIN | 60S ribosomal protein L23 | 10.51 | 14865.44 |
| RL23A_BOVIN | 60S ribosomal protein L23a | 10.44 | 17695.06 |
| RM17_MOUSE | 39S ribosomal protein L17, mitochondrial | 9.1 | 13927.02 |
| RM49_HUMAN | 39S ribosomal protein L49, mitochondrial | 9.47 | 19198.05 |
| RM51_BOVIN | 39S ribosomal protein L51, mitochondrial | 11.23 | 11762.89 |
| RRFM_HUMAN | Ribosome-recycling factor, mitochondrial | 9.54 | 22855.51 |
| RS13_PIG | 40S ribosomal protein S13 | 10.58 | 12034.24 |
| RS14_HUMAN | 40S ribosomal protein S14 | 10.08 | 16141.51 |
| RS18_BOVIN | 40S ribosomal protein S18 | 10.99 | 17587.48 |
| RS19_BOVIN | 40S ribosomal protein S19 | 10.31 | 15929.31 |
| RS25_BOVIN | 40S ribosomal protein S25 | 10.12 | 13742.13 |
| RS3A_BOVIN | 40S ribosomal protein S3a | 9.75 | 29813.72 |
| RT06_BOVIN | 28S ribosomal protein S6, mitochondrial | 9.49 | 14016.33 |
| RT18C_HUMAN | 28S ribosomal protein S18c, mitochondrial | 9.63 | 15849.72 |
| RT63_HUMAN | Mitochondrial ribosomal protein 63 | 11.45 | 12266.28 |
| S10A8_HUMAN | Protein S100-A8 | 6.51 | 10834.51 |
| S10AA_PIG | Protein S100-A10 | 6.35 | 10943.74 |
| S10AB_HUMAN | Protein S100-A11 | 6.56 | 11740.44 |
| SDHF2_HUMAN | Succinate dehydrogenase assembly factor 2, mitochondrial | 5.79 | 16567.82 |
| TAGL_HUMAN | Transgelin | 8.88 | 22479.71 |
| TAGL2_HUMAN | Transgelin-2 | 8.45 | 22260.25 |
| TENS1_HUMAN | Tensin-1 | 7.55 | 185701.35 |
| TIM44_MOUSE | Mitochondrial import inner membrane translocase subunit TIM44 | 8.48 | 51176.43 |
| TM109_RABIT | Transmembrane protein 109 | 9.96 | 22812.67 |
| TPPP3_MOUSE | Tubulin polymerization-promoting protein family member 3 | 9.18 | 18965.48 |
| UD11_MOUSE | UDP-glucuronosyltransferase 1-1 precursor | 8.87 | 56874.45 |
| Mitochondrial Subfractionation | |||
| ABHDA_HUMAN | Abhydrolase domain-containing protein 10, mitochondrial | 6.29 | 28203.62 |
| ACBD5_HUMAN | Acyl-CoA-binding domain-containing protein 5 | 5.19 | 60091.84 |
| ACD10_HUMAN | Acyl-CoA dehydrogenase family member 10 | 8.33 | 118834.15 |
| AKT1_BOVIN | RAC-α serine/threonine-protein kinase | 5.64 | 55748.42 |
| APOOL_BOVIN | Apolipoprotein O-like precursor | 9.41 | 26470.4 |
| AT5G1_PIG | ATP synthase lipid-binding protein, mitochondrial | 9.14 | 24668.72 |
| ATD3B_HUMAN | ATPase family AAA domain-containing protein 3B | 9.3 | 72572.92 |
| AUHM_HUMAN | Methylglutaconyl-CoA hydratase, mitochondrial | 9.15 | 29196.1 |
| BASI_RABIT | Basigin precursor | 5.76 | 27095.29 |
| CALL5_HUMAN | Calmodulin-like protein 5 | 4.34 | 15761.32 |
| CC90B_HUMAN | Coiled-coil domain-containing protein 90B, mitochondrial | 5.36 | 24455.92 |
| CCD72_BOVIN | Coiled-coil domain-containing protein 72 | 10 | 7066.3 |
| CCHL_HUMAN | Cytochrome c-type heme lyase | 6.25 | 30601.58 |
| CI046_HUMAN | Transmembrane protein C9orf46 | 9.58 | 17201.32 |
| CX6A1_RABIT | Cytochrome c oxidase subunit 6A1, mitochondrial | 6.41 | 9618.8 |
| CYB5_RABIT | Cytochrome b5 | 5.16 | 15218.05 |
| EFGM_HUMAN | Elongation factor G 1, mitochondrial | 6.58 | 83471.49 |
| FIS1_BOVIN | Mitochondrial fission 1 protein | 9.04 | 16937.7 |
| GBG12_HUMAN | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit γ-12 | 9.14 | 7535.55 |
| LETM1_RAT | LETM1 and EF-hand domain-containing protein 1, mitochondrial | 5.38 | 70763.58 |
| MPPA_BOVIN | Mitochondrial-processing peptidase subunit α | 6 | 54441.56 |
| MYO1A_RAT | Myosin-Ia | 9.39 | 97210.81 |
| NB5R1_HUMAN | NADH-cytochrome b5 reductase 1 | 9.41 | 34094.84 |
| NDUB6_BOVIN | NADH dehydrogenase [ubiquinone] 1β subcomplex subunit 6 | 9.62 | 15392.89 |
| NDUB7_BOVIN | NADH dehydrogenase [ubiquinone] 1β subcomplex subunit 7 | 8.38 | 16266.57 |
| NU6M_RABIT | NADH-ubiquinone oxidoreductase chain 6 | 3.94 | 18728.3 |
| OCAD1_BOVIN | OCIA domain-containing protein 1 | 6.67 | 27829.16 |
| RAB21_BOVIN | Ras-related protein Rab-21 | 8.16 | 23783.99 |
| RAB7A_BOVIN | Ras-related protein Rab-7a | 6.39 | 23543.84 |
| RB11A_BOVIN | Ras-related protein Rab-11A | 6.14 | 23983.03 |
| RL13_HUMAN | 60S ribosomal protein L13 | 11.65 | 24130.28 |
| RM14_MOUSE | 39S ribosomal protein L14, mitochondrial | 10.8 | 12728.89 |
| RM40_HUMAN | 39S ribosomal protein L40, mitochondrial | 9.35 | 19254.22 |
| SMC1B_HUMAN | Structural maintenance of chromosomes protein 1B | 7.69 | 143907.84 |
| SPTN4_HUMAN | Spectrin β chain, brain 3 | 5.72 | 288985.33 |
| SURF1_HUMAN | Surfeit locus protein 1 | 9.64 | 33331.41 |
| TFB2M_MOUSE | Dimethyladenosine transferase 2, mitochondrial | 8.37 | 41187.46 |
| THIOM_MOUSE | Thioredoxin, mitochondrial | 4.88 | 11835.58 |
| TIM14_BOVIN | Mitochondrial import inner membrane translocase subunit TIM14 | 10.1 | 12367.43 |
| TIM16_HUMAN | Mitochondrial import inner membrane translocase subunit TIM16 | 9.69 | 13824.73 |
| TIM50_BOVIN | Mitochondrial import inner membrane translocase subunit TIM50 | 6.61 | 35594.54 |
| TPP1_HUMAN | Tripeptidyl-peptidase 1 precursor | 5.74 | 39790.38 |
| TRI25_MOUSE | Tripartite motif-containing protein 25 | 8.62 | 71772.3 |
| YMEL1_HUMAN | ATP-dependent metalloprotease YME1L1 | 8.86 | 86455.28 |
At the peptide level, once again peptide-centric 2D-LC resulted in the highest proportion of unique peptides observed (35%; 535 peptides), followed by protein-centric 2D-LC (30%; 234 peptides) and subfractionation (27%; 412 peptides). Protein-centric 1-DLC (20%; 163 peptides) and peptide-centric 1-DLC (15%; 181 peptides) contributed the fewest unique peptides overall. When taken together, these results tend to indicate, that peptide-centric 1D-LC is not a feasible approach as a stand-alone technique, which is unsurprising given that all potential mitochondrial proteins are separated across 1 dimension alone. The strengths of peptide-centric 1D-LC lie in its ability to provide validation for the remaining preparations when proteins are identified by a single peptide or one-hit wonders and by enhancing protein sequence coverage. With large proteomic studies becoming increasingly popular, the need to ensure accurate representation of the data and by proxy, the proteome of interest, is increasingly imperative (48), as such one-hit wonders are traditionally not included in such investigations. We found it necessary to include such proteins in the current study however.
Bioinformatic Challenges: Protein Sequence Coverage and “One-Hit Wonders”
Traditionally, protein matches where less than two discrete peptides can be successfully identified are removed from large proteomic studies. There are obvious caveats to this arising in relation to post-translational modifications, where a single peptide within a protein may exist in a modified form, or alternatively in the case of incomplete genome annotation. In the current study, when considering the approaches individually, 68% of the total number of protein groups identified (380 out of 556) were identified by a single peptide or one-hit wonder. By combining the five approaches however, 143 of these one-hit wonders were identified with more peptides, contributed by each of the approaches. For example, ATP synthase e (ATP5I_PIG), was identified by a single peptide following peptide-centric 2-DLC, however both protein-centric 1- and 2-DLC contributed an additional unique peptide, with the subfractionated preparation contributing an additional two peptides. In the end, we identified ATP synthase e using three discrete peptides (48% sequence coverage) through the combination of these four approaches. As discussed above, in the current study, where the rabbit genome was unavailable because of incomplete annotation, to ensure that the minimum number of proteins were identified by one-hit wonders, it was important to combine the results from the multistage approach, rather than selecting one or two methods alone.
Not only did this multistage approach ensure proteins were identified by more than one peptide on several occasions, but it also facilitated improved sequence coverage. We found that 113 proteins showed improved sequence coverage by using the combination of methodologies. This included NAD(P) transhydrogenase (NNTM_BOVIN), which following separation by protein-centric 1-DLC was identified by four unique peptides (5% sequence coverage). This increased to 39 discrete peptides (36% sequence coverage) with the addition of the remaining four methodologies. The increased sequence coverage could be observed by investigating the overlap of the five orthogonal strategies at both the protein and peptide level (Supplemental Fig. 5). There were only 159 peptides that were observed by all 5 orthogonal methods, a relatively small contribution (5%) by comparison with the total number of unique peptides (see Table I). These peptides represent the most abundant proteins detected in the current study, with multiple peptides identified from ATP synthase-α and -β. At the protein level however, the 5 strategies combined to identify more proteins (108 protein groups) than any of the individual approaches (peptide-centric 2-DLC; 106 unique protein groups). Of the 108 proteins that were observed across all preparations, 98 (91%) were identified with more than one unique peptide contributed by any single strategy (Supplemental Table 3). These unique peptides increased protein sequence coverage by up to 40% (e.g. NDUA8_HUMAN). This suggests that even though there was redundancy across the five strategies at the protein level, the combination of approaches shows limited redundancy at the peptide level, therefore contributing toward the increased sequence coverage. In the current study, where high-throughput proteomics was performed in the absence of an annotated genome, the increased sequence coverage achieved makes this a valuable contribution to the overall protein identification confidence.
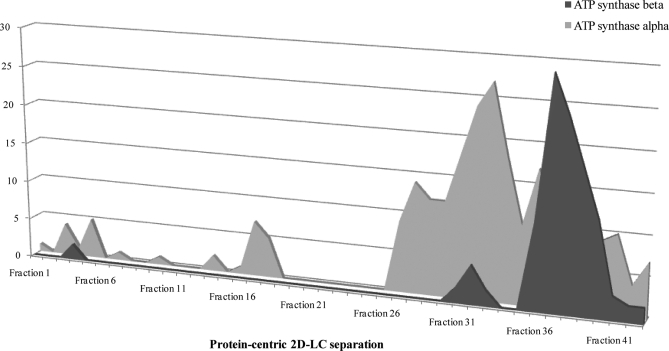
One consideration of the current study is the increased “observation” redundancy within protein-centric approaches. By maintaining intact proteins, information pertaining to discrete populations of protein variants is possible. At the same time however, protein-centric approaches lead to over six observations (based on tandem-MS spectra) per peptide, whereas peptide-centric approaches resulted in a significantly lower number (3.4 observations/peptide). This is most likely because of distinct populations of proteins resolving independently because of a biochemical characteristic including the presence of a post-translational modification. As previously described (32), discrete populations of proteins can be observed with altered retention times in protein-centric approaches, indicating changes in biochemical properties (Fig. 3) (49). Therefore the partitioning of proteins into numerous fractions by protein-centric methods (1-DLC in particular) may lead to the reduced number of unique peptides observed because of the increased observations of the same peptide.
Fig. 3.
Subpopulations of mitochondrial proteins resolved by protein-centric approaches. To determine the ability of protein-centric approaches to identify protein subpopulations, the number of peptides (y axis) attributable to ATP synthase-α and -β were plotted against the protein-centric fractions (x axis) they were observed in.
Bioinformatic Challenges: False Discovery Rates
Large proteomic studies can be computationally validated with the use of FDR whereby the generation of reverse databases facilitates prediction of the likelihood of false positives. In the current study, the FDR was relaxed slightly given the absence of genome annotation and subsequent inclusion of additional species to ensure maximal coverage. The caveat with such an approach was the introduction of sequence redundancy. This resulted in a true versus false one-hit wonder paradox, whereby the nature of searching against multiple species with high sequence homology, but not sequence identity resulted in false one-hit wonders. Although the majority of peptides were correctly assigned to the species with the closest homology, peptides with amino acid substitutions were assigned to a divergent species. This is best exemplified by trifunctional enzyme subunit α (ECHA MOUSE), where the majority of the 22 peptides sequenced in the course of the current investigation, contained three peptides that were homologous to different species (441ADMVIEAVFEDLSLK455 from ECHA HUMAN; 327FGELAMTK334 from ECHA PIG and; 520DTTASAVAVGLK530 from ECHA RAT). Analysis of these sequences reveals low homology in response to single amino acid substitutions (Fig. 4). The potential for falsely elevated FDR is possible in the absence of an annotated genome and therefore must be considered. In the current study, we found that the proportion of false one-hit wonders was higher when protein-centric 2-DLC approaches (59% miss-assigned) were used.
Fig. 4.

Species variations within trifunctional enzyme subunit α. In the current study we observed an increased number of one-hit wonders where a single peptide was used to identify a given protein. This included false one-hit wonders, where species variations resulted in single peptides attributed to divergent species (e.g. trifunctional enzyme subunit α). The majority of the 22 peptides sequenced were attributed to the sequenced mouse protein (ECHA_MOUSE). However, 3 peptides showed species specific variations. The peptide spanning residues 327–334 was identified as ECHA_PIG, showing sequence divergence at residues 331 (Ala) and 332 (Met). A similar divergence was observed for the peptides spanning residues 441–455 (ECHA_HUMAN; 451Asp; 453Ser; 454Leu) and 520–530 (ECHA_RAT; 521Thr; 522Thr; 530Lys). Species variations resulted in each peptide being attributed to one species uniquely.
CONCLUSIONS
The current study has applied one-, two-, and three-dimensions of separation to improve coverage of the partially-annotated rabbit cardiomyocyte mitochondrial genome and found that (i) for adequate coverage and bioinformatic confidence in the absence of an annotated genome, all five strategies combined are required; (ii) 25% of mitochondrial proteins are common to rabbit, mouse, and human studies of cardiomyocte mitochondria; and (iii) the converse was also true, with 25% of proteins uniquely identified within each species. In the current study, we found that bioinformatic confidence was improved by (i) the significantly reduced population of proteins identified by a single peptide (one-hit wonder) by combining the five approaches; (ii) increased sequence coverage through the reduced peptide redundancy across the five strategies; (iii) “observation” redundancy, a factor of discrete protein variants resolving individually with protein-centric approaches (at the cost of unique peptides and proteins) and; (iv) identifying false one-hit wonders, caused by poor sequence conservation with fully annotated genomes, the removal of which resulted in reduced FDR and enhanced sequence coverage. Thus, a combination of high dimensional separation for protein and peptides in parallel with further subfractionation is a successful approach to provide proteome coverage for rabbit, achieving a similar number of proteins identified as those from species with annotated genomes, while meeting the bioinformatic requirements of large-scale proteomic studies.
Footnotes
* This research was supported in part by funding from the National Health and Medical Research Foundation (NHMRC) of Australia (MYW is a CJ Martin Postdoctoral Research Fellow 464905). J.V.E. would like to thank the following funding sources: The National Heart Lung and Blood Institute Proteomic Initiative (contract N0-HV-28180), NIH (contracts P01HL081427 and P01HL077180) and the Donald P. Amos Family Foundation.
 This article contains supplemental Figs 1–5 and Tables 1–3.
This article contains supplemental Figs 1–5 and Tables 1–3.
1 The abbreviations used are:
- 1-DE
- one-dimensional gel electrophoresis
- 1-DLC
- one-dimensional liquid chromatography
- 2-DE
- two-dimensional gel electrophoresis
- 2-DLC
- two-dimensional liquid chromatography
- FDR
- false discovery rate
- ESI-MS/MS
- electrospray ionization-tandem mass spectrometry
- Mr
- molecular mass
- MS/MS
- tandem mass spectrometry
- PTM
- post translational modifications
- RP-HPLC
- reverse phase-HPLC
- SCX
- strong cation exchange.
REFERENCES
- 1. Schaper J., Meiser E., Stämmler G. (1985) Ultrastructural morphometric analysis of myocardium from dogs, rats, hamsters, mice, and from human hearts. Circulation Research 56, 377–391 [DOI] [PubMed] [Google Scholar]
- 2. Barth E., Stämmler G., Speiser B., Schaper J. (1992) Ultrastructural quantitation of mitochondria and myofilaments in cardiac muscle from 10 different animal species including man. J. Mol. Cellular Cardiol. 24, 669–681 [DOI] [PubMed] [Google Scholar]
- 3. Adair T. H., Gay W. J., Montani J. P. (1990) Growth regulation of the vascular system: evidence for a metabolic hypothesis. Am. J. Physiol. 259, R393–404 [DOI] [PubMed] [Google Scholar]
- 4. Hamilton N., Ianuzzo C. D. (1991) Contractile and calcium regulating capacities of myocardia of different sized mammals scale with resting heart rate. Mol. Cell. Biochem. 106, 133–141 [DOI] [PubMed] [Google Scholar]
- 5. Rouslin W. (1987) The mitochondrial adenosine 5′-triphosphatase in slow and fast heart rate hearts. Am. J. Physiol. 252, H622–627 [DOI] [PubMed] [Google Scholar]
- 6. Rouslin W. (1988) Factors affecting the loss of mitochondrial function during zero-flow ischemia (autolysis) in slow and fast heart-rate hearts. J. Mol. Cell. Cardiol. 20, 999–1007 [DOI] [PubMed] [Google Scholar]
- 7. Rouslin W., Broge C. W. (1989) Regulation of mitochondrial matrix pH and adenosine 5′-triphosphatase activity during ischemia in slow heart-rate hearts. Role of Pi/H+ symport. J. Biol. Chem. 264, 15224–15229 [PubMed] [Google Scholar]
- 8. White M. Y., Cordwell S. J., McCarron H. C., Prasan A. M., Craft G., Hambly B. D., Jeremy R. W. (2005) Proteomics of ischemia/reperfusion injury in rabbit myocardium reveals alterations to proteins of essential functional systems. Proteomics 5, 1395–1410 [DOI] [PubMed] [Google Scholar]
- 9. Graur D., Duret L., Gouy M. (1996) Phylogenetic position of the order Lagomorpha (rabbits, hares and allies). Nature 379, 333–335 [DOI] [PubMed] [Google Scholar]
- 10. Lowell B. B., Shulman G. I. (2005) Mitochondrial dysfunction and type 2 diabetes. Science 307, 384–387 [DOI] [PubMed] [Google Scholar]
- 11. White M. Y., Edwards A. V., Cordwell S. J., Van Eyk J. E. (2008) Mitochondria: A mirror into cellular dysfunction in heart disease. Proteomics 2, 845–861 [DOI] [PubMed] [Google Scholar]
- 12. Wallace D. C. (2005) A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Ann. Rev. Gen. 39, 359–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen H., Chan D. C. (2004) Mitochondrial dynamics in mammals. Current Topics Develop. Biol. 59, 119–144 [DOI] [PubMed] [Google Scholar]
- 14. Collins T. J., Berridge M. J., Lipp P., Bootman M. D. (2002) Mitochondria are morphologically and functionally heterogeneous within cells. EMBO J. 21, 1616–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hamilton N., Ashton M. L., Ianuzzo C. D. (1991) Cell size of mammalian myocardia is not related to physiological demand. Experientia 47, 1070–1072 [DOI] [PubMed] [Google Scholar]
- 16. Porter R. K., Brand M. D. (1993) Body mass dependence of H+ leak in mitochondria and its relevance to metabolic rate. Nature 362, 628–630 [DOI] [PubMed] [Google Scholar]
- 17. Sohal R. S., Svensson I., Sohal B. H., Brunk U. T. (1989) Superoxide anion radical production in different animal species. Mech. Ageing Develop. 49, 129–135 [DOI] [PubMed] [Google Scholar]
- 18. Forner F., Foster L. J., Campanaro S., Valle G., Mann M. (2006) Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol. Cell. Proteomics 5, 608–619 [DOI] [PubMed] [Google Scholar]
- 19. Johnson D. T., Harris R. A., French S., Blair P. V., You J., Bemis K. G., Wang M., Balaban R. S. (2007) Tissue heterogeneity of the mammalian mitochondrial proteome. Am. J. Physiol. 292, C689–697 [DOI] [PubMed] [Google Scholar]
- 20. Pagliarini D. J., Calvo S. E., Chang B., Sheth S. A., Vafai S. B., Ong S. E., Walford G. A., Sugiana C., Boneh A., Chen W. K., Hill D. E., Vidal M., Evans J. G., Thorburn D. R., Carr S. A., Mootha V. K. (2008) A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Taylor S. W., Fahy E., Zhang B., Glenn G. M., Warnock D. E., Wiley S., Murphy A. N., Gaucher S. P., Capaldi R. A., Gibson B. W., Ghosh S. S. (2003) Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 21, 281–286 [DOI] [PubMed] [Google Scholar]
- 22. Gaucher S. P., Taylor S. W., Fahy E., Zhang B., Warnock D. E., Ghosh S. S., Gibson B. W. (2004) Expanded coverage of the human heart mitochondrial proteome using multidimensional liquid chromatography coupled with tandem mass spectrometry. J. Proteome Res. 3, 495–505 [DOI] [PubMed] [Google Scholar]
- 23. Mootha V. K., Bunkenborg J., Olsen J. V., Hjerrild M., Wisniewski J. R., Stahl E., Bolouri M. S., Ray H. N., Sihag S., Kamal M., Patterson N., Lander E. S., Mann M. (2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115, 629–640 [DOI] [PubMed] [Google Scholar]
- 24. Foster L. J., de Hoog C. L., Zhang Y., Zhang Y., Xie X., Mootha V. K., Mann M. (2006) A mammalian organelle map by protein correlation profiling. Cell 125, 187–199 [DOI] [PubMed] [Google Scholar]
- 25. Da Cruz S., Xenarios I., Langridge J., Vilbois F., Parone P. A., Martinou J. C. (2003) Proteomic analysis of the mouse liver mitochondrial inner membrane. J. Biol. Chem. 278, 41566–41571 [DOI] [PubMed] [Google Scholar]
- 26. McDonald T. G., Van Eyk J. E. (2003) Mitochondrial proteomics Undercover in the lipid bilayer. Basic Res. Cardiol. 98, 219–227 [DOI] [PubMed] [Google Scholar]
- 27. Agnetti G., Kaludercic N., Kane L. A., Elliott S. T., Guo Y., Chakir K., Samantapudi D., Paolocci N., Tomaselli G. F., Kass D. A., Van Eyk J. E. (2010) Modulation of mitochondrial proteome and improved mitochondrial function by biventricular pacing of dyssynchronous failing hearts. Circulation 3, 78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim N., Lee Y., Kim H., Joo H., Youm J. B., Park W. S., Warda M., Cuong D. V., Han J. (2006) Potential biomarkers for ischemic heart damage identified in mitochondrial proteins by comparative proteomics. Proteomics 6, 1237–1249 [DOI] [PubMed] [Google Scholar]
- 29. Arrell D. K., Elliott S. T., Kane L. A., Guo Y., Ko Y. H., Pedersen P. L., Robinson J., Murata M., Murphy A. M., Marbán E., Van Eyk J. E. (2006) Proteomic analysis of pharmacological preconditioning: novel protein targets converge to mitochondrial metabolism pathways. Circulation Res. 99, 706–714 [DOI] [PubMed] [Google Scholar]
- 30. White M. Y., Tchen A. S., McCarron H. C., Hambly B. D., Jeremy R. W., Cordwell S. J. (2006) Proteomics of ischemia and reperfusion injuries in rabbit myocardium with and without intervention by an oxygen-free radical scavenger. Proteomics 6, 6221–6233 [DOI] [PubMed] [Google Scholar]
- 31. Kane L. A., Yung C. K., Agnetti G., Neverova I., Van Eyk J. E. (2006) Optimization of paper bridge loading for 2-DE analysis in the basic pH region: Application to the mitochondrial subproteome. Proteomics 6, 5683–5687 [DOI] [PubMed] [Google Scholar]
- 32. McDonald T., Sheng S., Stanley B., Chen D., Ko Y., Cole R. N., Pedersen P., Van Eyk J. E. (2006) Expanding the subproteome of the inner mitochondria using protein separation technologies: one- and two-dimensional liquid chromatography and two-dimensional gel electrophoresis. Mol. Cell. Proteomics 5, 2392–2411 [DOI] [PubMed] [Google Scholar]
- 33. Scheffler N. K., Miller S. W., Carroll A. K., Anderson C., Davis R. E., Ghosh S. S., Gibson B. W. (2001) Two-dimensional electrophoresis and mass spectrometric identification of mitochondrial proteins from an SH-SY5Y neuroblastoma cell line. Mitochondrion 1, 161–179 [DOI] [PubMed] [Google Scholar]
- 34. Cordwell S. J. (2006) Technologies for bacterial surface proteomics. Current Opinions Microbiol. 9, 320–329 [DOI] [PubMed] [Google Scholar]
- 35. Wu C. C., Yates J. R., 3rd (2003) The application of mass spectrometry to membrane proteomics. Nat. Biotechnol. 21, 262–267 [DOI] [PubMed] [Google Scholar]
- 36. Cordwell S. J. (2008) Sequential extraction of proteins by chemical reagents. Methods in Mol. Biol. 424, 139–146 [DOI] [PubMed] [Google Scholar]
- 37. Santoni V., Molloy M., Rabilloud T. (2000) Membrane proteins and proteomics: un amour impossible? Electrophoresis 21, 1054–1070 [DOI] [PubMed] [Google Scholar]
- 38. Pedersen P. L., Greenawalt J. W., Reynafarje B., Hullihen J., Decker G. L., Soper J. W., Bustamente E. (1978) Methods in cell biology, Volume 20, pp. 411–481, Academic Press Inc., New York, NY: [DOI] [PubMed] [Google Scholar]
- 39. Fujiki Y., Fowler S., Shio H., Hubbard A. L., Lazarow P. B. (1982) Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes: comparison with endoplasmic reticulum and mitochondrial membranes. J. Cell Biol. 93, 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barré O., Solioz M. (2006) Improved protocol for chromatofocusing on the ProteomeLab PF2D. Proteomics 6, 5096–5098 [DOI] [PubMed] [Google Scholar]
- 41. Sheng S., Chen D., Van Eyk J. E. (2006) Multidimensional liquid chromatography separation of intact proteins by chromatographic focusing and reversed phase of the human serum proteome: optimization and protein database. Mol. Cell. Proteomics 5, 26–34 [DOI] [PubMed] [Google Scholar]
- 42. Chen Y., Kwon S. W., Kim S. C., Zhao Y. (2005) Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J. Proteome Res. 4, 998–1005 [DOI] [PubMed] [Google Scholar]
- 43. Altschul S. F., Lipman D. J. (1990) Protein database searches for multiple alignments. Proc. Natl Acad. Sci. U. S. A. 87, 5509–5513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cordwell S. J., Len A. C., Touma R. G., Scott N. E., Falconer L., Jones D., Connolly A., Crossett B., Djordjevic S. P. (2008) Identification of membrane-associated proteins from Campylobacter jejuni strains using complementary proteomics technologies. Proteomics 8, 122–139 [DOI] [PubMed] [Google Scholar]
- 45. Pevtsov S., Fedulova I., Mirzaei H., Buck C., Zhang X. (2006) Performance evaluation of existing de novo sequencing algorithms. J. Proteome Res. 5, 3018–3028 [DOI] [PubMed] [Google Scholar]
- 46. Novacek M. J. (1992) Mammalian phylogeny: shaking the tree. Nature 356, 121–125 [DOI] [PubMed] [Google Scholar]
- 47. Smith A. C., Robinson A. J. (2009) MitoMiner, an integrated database for the storage and analysis of mitochondrial proteomics data. Mol. Cell. Proteomics 8, 1324–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Carr S., Aebersold R., Baldwin M., Burlingame A., Clauser K., Nesvizhskii A. (2004) The need for guidelines in publication of peptide and protein identification data: Working Group on Publication Guidelines for Peptide and Protein Identification Data. Mol. Cell. Proteomics 3, 531–533 [DOI] [PubMed] [Google Scholar]
- 49. Nesvizhskii A. I., Aebersold R. (2005) Interpretation of shotgun proteomic data: the protein inference problem. Mol. Cell. Proteomics. 4, 1419–1440 [DOI] [PubMed] [Google Scholar]