Abstract
Pine wilt disease (PWD) is one of the most devastating forest diseases in Asia and Europe. The pine wood nematode, Bursaphelenchus xylophilus, has been identified as the pathogen underlying PWD, although the pathology is not completely understood. At present, diagnosis and confirmation of PWD are time consuming tasks that require nematode extraction and microscopic examination. To develop a more efficient detection method for B. xylophilus, we first generated monoclonal antibodies (MAbs) specific to B. xylophilus. Among 2304 hybridoma fusions screened, a hybridoma clone named 3-2A7-2H5 recognized a single protein from B. xylophilus specifically, but not those from other closely related nematodes. We finally selected the MAb clone 3-2A7-2H5-D9-F10 (D9-F10) for further studies. To identify the antigenic target of MAb-D9-F10, we analyzed proteins in spots, fractions, or bands isolated from SDS-PAGE, two-dimensional electrophoresis, anion exchange chromatography, and immunoprecipitation via nano liquid chromatography electrospray ionization quadrupole ion trap mass spectrometry (nano-LC-ESI-Q-IT-MS). Peptides of galactose-binding lectin-1 of B. xylophilus (Bx-LEC-1) were commonly detected in several proteomic analyses, demonstrating that this LEC-1 is the antigenic target of MAb-D9-F10. The localization of MAb-D9-F10 immunoreactivities at the area of the median bulb and esophageal glands suggested that the Bx-LEC-1 may be involved in food perception and digestion. The Bx-LEC-1 has two nonidentical galactose-binding lectin domains important for carbohydrate binding. The affinity of the Bx-LEC-1 to d-(+)-raffinose and N-acetyllactosamine were much higher than that to l-(+)-rhamnose. Based on this combination of evidences, MAb-D9-F10 is the first identified molecular biomarker specific to the Bx-LEC-1.
The pinewood nematode, Bursaphelenchus xylophilus (Steiner and Buhrer) Nickle (1), is a serious pathogen of the forest, particularly of Pinus species, causing pine wilt disease (PWD)1. Even though PWD was first reported in Japan in 1905 (2), the relationship between B. xylophilus and PWD was not elucidated until 1971 (3). Currently, PWD has spread to Far East Asian countries including Korea (4), China (5), and Taiwan (6) as well as to Portugal in the European Union (7). The distribution of B. xylophilus is currently extensive, explaining the seriousness of PWD.
Distribution and development of PWD within pine trees have been described (8–10). The Japanese pine sawyer beetle, Monochamus alternatus, is the known vector for B. xylophilus (9, 11). Once B. xylophilus infects pine trees via wounds made by sawyer beetles, it kills epithelial cells in resin canals, resulting in the death of neighboring parenchymal cells. Following B. xylophilus infection, the hydraulic conductivity of pine trees is rapidly reduced because colonized B. xylophilus inhibit the conduction of water through the xylem (8, 12). Thus, B. xylophilus-infected trees wilt or die in autumn, and M. alternatus uses them as oviposition grounds where the host beetles complete their life cycle. B. xylophilus moves to the tracheal system of emerging adult beetles in dead pine trees and can be distributed to new pines in the forest. The repetition of these mutualistic life cycles between B. xylophilus and M. alternatus makes it possible for B. xylophilus to become widely distributed in the forest.
Although the route of transmission of B. xylophilus is understood and expressed sequence tags of B. xylophilus have been reported (13, 14), it is still challenging to diagnose and confirm B. xylophilus infection in pine trees. In practice, it is very difficult to discriminate pathogenic B. xylophilus from closely related nonpathogenic pine tree-resident nematodes. For example, B. mucronatus, very closely related to B. xylophilus, is widely distributed in Asia and Europe, and coexists with B. xylophilus in the forest. B. mucronatus is morphologically similar to B. xylophilus, and the only morphological difference is found in the mucro on the tail of B. mucronatus female. Interspecies forms have also been found, making it even more difficult to distinguish between these two closely related species (15). To distinguish B. xylophilus from B. mucronatus, several DNA-based methods have been developed, such as genomic DNA hybridization (16), PCR-restriction fragment length polymorphism (17–19) and species-specific PCR (ssPCR) (20). More recently, a real-time PCR protocol using the 5S rRNA as a probe was developed (21). Although these DNA- and real-time reverse transcription-PCR-based methods differentiate B. xylophilus from B. mucronatus, there are several barriers to the application of these methods to practical on-site diagnosis: prolonged time in detection and analysis, requirement for equipments such as a PCR machine and electrophoresis apparatus, and high probability of false positive signals from PCR-based technologies. Therefore, these DNA- and real-time reverse transcription PCR-based methods do not meet the key requirements of diagnosis such as promptness, convenience, and accuracy. For specificity and speed in the detection of pathogenic B. xylophilus, antibodies specifically recognizing B. xylophilus proteins may be attractive and advantageous candidates as compared with currently available diagnostic methods.
In this study, to generate and isolate hybridoma clones that secrete antibodies specific to B. xylophilus and not other closely related Bursaphelenchus species, we began by injecting mice with whole proteins of B. xylophilus. One monoclonal antibody (MAb) clone, 3-2A7-2H5-D9-F10 (D9-F10), discriminated B. xylophilus from other Bursaphelenchus species as well as from other organisms such as human, mouse, and Drosophila. We applied several biochemical approaches followed by proteomic analysis to identify the target protein and found the antigenic target to be galactose-binding lectin-1 (LEC-1), which was the first identified LEC of B. xylophilus. These results suggest that the MAb-D9-F10 clone is the first B. xylophilus-specific molecular biomarker and can be used to detect pathogenic B. xylophilus.
EXPERIMENTAL PROCEDURES
Nematodes
Bursaphelenchus xylophilus initially isolated from infected pinewoods and other Bursaphelenchus spp. including B. mucronatus, B. thailandae and B. doui were grown on a lawn of Botrytis cinerea cultured on potato-dextrose-agar media at 25 °C and separated according to the same methods as in previous reports (22, 23).
Generation, Screening, and Isotyping of Monoclonal Antibodies Specific to Bursaphelenchus xylophilus
Hybridomas producing B. xylophilus-specific monoclonal antibodies (MAbs) were generated as previously described (24). Specific MAb screenings were performed with ELISA, dot blot screenings and Western blot analysis. Briefly, each well of the 96-well Maxisorp plate (Thermo Fisher Scientific, Rochester, NY) was coated with 0.1 μg of B. xylophilus proteins in coating buffer (0.1 m NaHCO3, pH 8.6). Each well was blocked with 1% bovine serum albumin (BSA; Amnesco, Solon, OH) in phosphate buffered saline (PBS) (pH 7.4) and then incubated with a primary antibody (1:1000 dilution) in blocking solution. The plate was then incubated with horse radish peroxidase-conjugated anti-mouse IgG heavy chain (1:2000; Thermo Fisher Scientific) in blocking solution. Then, 1-Step™ Ultra TMB-ELISA solution (Thermo Fisher Scientific) was added to develop colors, and absorbance at 450 nm was monitored.
For dot blot screenings, nematodes were directly boiled with SDS sample buffer without bromphenol blue. Protein concentration was quantified with Bradford's method (Bio-Rad, Hercules, CA) (25). For this procedure, 100 ng of proteins was applied to each well on the nitrocellulose membrane (GE Healthcare, Piscataway, NJ) in dot blotters (Bio-Rad). The membrane was blotted with supernatants of primary hybridoma clones and horse radish peroxidase-conjugated goat anti-mouse IgG heavy chain (Thermo Fisher Scientific). The membrane signal was detected with Supersignal West Pico (Thermo Fisher Scientific).
For Western blot analysis, 10 μg of total proteins for each sample was separated with 10% tris-glycine polyacrylamide SDS gel and then transferred to the nitrocellulose membrane (GE Healthcare). The membrane was blocked with 3% nonfat dried milk in PBST. The procedures for antibody treatment and detection were as described above. The immunoglobulin (Ig) isotype and subclass of the selected clone were determined using the ImmunoPure Monoclonal Antibody Isotyping Kit (Thermo Fisher Scientific). The plate wells were coated with antibody working solution (2 μg/ml goat anti-mouse Ig(G + A + M) in 0.1 m sodium bicarbonate, pH 9.5). Following blocking with 0.5% BSA in TBS, culture supernatants were added into each well. Subclass-specific anti-mouse IgGs were added, and alkaline phosphatase-conjugated goat anti-rabbit IgG was used as a secondary antibody. The substrate, para-nitrophenylphosphate solution, was added, and absorbance at 405 nm was monitored.
Blotting of Bursaphelenchus spp., Human, Mouse, and Drosophila melanogaster Proteins with MAb-D9-F10
Proteins from Bursaphelenchus spp., D. melanogaster, nerve cords, and brains from mice, and a human cervix carcinoma cell line were ground in PBS with a proteinase inhibitor mixture (Roche) and then boiled in SDS sample buffer for 5 min. The procedures for SDS-PAGE, protein transfer and Western blot were as described above. Two identical gels were generated and processed for MAb-D9-F10 immunoblotting or Coomassie brilliant blue staining. The gel was incubated in staining solution (0.25% Coomassie brilliant R-250, 5% methanol, and 7.5% acetic acid) for at least 4 h and decolorized with destaining solution (25% methanol and 7.5% acetic acid).
Two-dimensional Electrophoresis (2-DE) of B. xylophilus Proteins
Two-DE was performed with ZOOM® immobilized pH gradient runner (Invitrogen). The sample was mixed with ampholyte (nonlinear pH 3–10) for rehydration, and then isoelectric focusing was performed with the following steps: 175 V for 15 min, 175–2,000 V ramp for 45 min and 2,000 V for 40 min. The IEF strip was incubated with NuPAGE® reducing buffer (Invitrogen) for 15 min on a rotary shaker and treated with alkylating solution [125 mm iodoacetamide (Armresco, Solon, OH), 1X NuPAGE LDS Sample Buffer (Invitrogen)] for 15 min on a rotary shaker. The strip was immediately applied to SDS-PAGE. Two identical 2-DE gels were generated and processed for MAb-D9-F10 immunoblotting or Coomassie brilliant blue staining as described above.
Anion Exchange Chromatography and Positive Fraction Identification
Whole proteins of B. xylophilus were extracted in buffer A (10 mm Tris, pH 7.4) with repetitive freeze-thawing and sonication on ice for 15 s in five rounds, each with 50% power (Branson, Danbury, CT). Whole proteins were filtered with an Ultrafree-MC centrifugal filter device (Millipore, Billerica, MA), and the supernatant was immediately applied to a Resource Q column (GE Healthcare), at a flow rate of 0.2 ml/min at 4 °C. Proteins were eluted with an increasing linear gradient of NaCl from 0 to 1.0 m in buffer A, and each fraction (0.5 ml) was collected by an FPLC AKTA Purifier (GE Healthcare). Each fraction was probed with MAb-D9-F10 through Western blot analysis.
MAb-D9-F10 Immunoprecipitation (IP)
Whole protein extracts of B. xylophilus and positive fractions from anion exchange chromatography were mixed with 10 μl of MAb-D9-F10 and rolled overnight at 4 °C. Agarose-protein A+G beads (Calbiochem Co., Darmstadt, Germany) were blocked with 1% BSA (Armresco) in radioimmunoprecipitation assay buffer [150 mm NaCl (Sigma), 1% IGEPAL CA-630 (Sigma), 0.5% Na-oxycholate (Sigma), 0.1% SDS (Sigma), 50 mm Tris, pH 8.0 (Sigma), 1× protease inhibitor mixture (Roche)] for 2 h at 4 °C. The mixtures of B. xylophilus protein extracts and MAb-D9-F10 were added to agarose-protein A+G beads and rolled for 2 h. Following washing with radioimmunoprecipitation assay buffer, samples were boiled for 5 min, electrophoresed, and blotted with MAb-D9-F10 or stained with Coomassie brilliant blue staining solution as described above.
In-Gel Digestion
Stained protein spots from one-dimensional electrophoresis (DE) or 2-DE were excised from gels and washed with 10 mm ammonium bicarbonate and 50% acetonitrile (Sigma). The sliced gels were digested in a buffer containing 50 mm ammonium bicarbonate, 5 mm calcium chloride, and 12.5 ng/ml trypsin (Promega, Madison, WI) at 37 °C for 12–16 h. The digested peptides were recovered by extraction twice with 50 mm ammonium bicarbonate and 100% acetonitrile. The resulting peptide extracts were pooled, lyophilized in a vacuum centrifuge and stored at 4 °C for subsequent amino acid sequencing.
Protein Identification by Nano-liquid Chromatography (LC)-tandem MS (MS/MS) and Data Analysis
All MS/MS experiments for peptide identification in this study were performed using a nano LC/MS system consisting of a Surveyor HPLC system (Thermo Scientific, Waltham, MA) and electrospray ionization (ESI)-quadrupole ion trap (IT) MS (LCQ Deca XP-Plus, Thermo Scientific, Waltham, MA) equipped with a nano-ESI source. For each experiment, 10 μl of sample was loaded by the auto sampler onto a C18 trap column (I.D 300 μm, length 5 mm, particle size 5 μm; LC Packings, Amsterdam, Netherlands) for desalting and concentration at a flow rate of 20 μl/min. The trapped peptides were then back-flushed and separated on a homemade microcapillary column (150 mm in length) packed with C18 resin (particle size 5 μm) in 75-μm silica tubing (8-μm id orifice). The mobile phases, A and B, were composed of 0% and 80% of acetonitrile containing 0.02% formic acid and 0.5% acetic acid, respectively. The gradient began at 5% of the mobile phase B for 15 min and was ramped to 20% for 3 min, 50% for 32 min, 60% for 5 min, 80% for 5 min, 100% for 2 min, and finally held at 100% for 8 min. The column was equilibrated with 5% acetonitrile for 10 min before the next run. MS and MS/MS spectra were obtained at a heated capillary temperature of 220 °C, an ESI voltage of 2.5 kV, and a collision energy setting of 35%. Data-dependent peak selection of the three most abundant MS ions from MS was used. Dynamic exclusion was enabled with a repeat count of two, a repeat duration of 0.5 min, and 3 min exclusion duration. Mass spectrometer scan functions and HPLC solvent gradients were controlled using the Xcalibur data system (Therom Finnigan, USA). MS/MS mass peak lists were analyzed for b and y ions using SEQUEST (version 3.3.1, Thermo Electron Corporation, USA) software. SEQUEST was used to match MS/MS spectra to peptides in an unpublished translated contig database (with 11,862 entries) and EST (with 26,224 entries) of B. xylophilus (http://PinewoodNematode.Net/EST). Searches for peptides were first performed with the following parameters: a mass tolerance of 2.0 Da on the parent ion and 1.0 Da on the MS/MS, one missed cleavage per peptide was allowed, and modifications of proteins were not taken into account. The validity of peptide/spectrum matches was hence assessed using the SEQUEST defined parameters, the cross-correlation score (XCorr), and the normalized difference in cross-correlation scores. Matched peptide sequences had to pass the following filters for provisional identification: 1) the uniqueness scores of matches normalized difference in cross-correlation scores) were at least 0.1 and 2) minimum cross-correlation scores of 1.9, 2.2, and 3.75 for charge states +1, +2, and +3, respectively. SEQUEST automatically saves search results. An SRF file including merging of proteins, filter and sort settings, ratios and protein area/height values was used to select and sort peptide/spectrum matches passing this set of criteria. Proteins were considered detected if they were identified by more than two peptides per spot. In the case of a protein that was identified with only one peptide, a BLAST search was performed, and some peptide sequences were found to be shared with a multiprotein family. When significant matches were found in the translated contig database, we performed a similarity search with the contig sequence to obtain information on related proteins. For the similarity search, we used Blastp from NCBI and installed the contig database into the BLAST database.
Immunohistochemistry
Cleaned B. xylophilus were put on ice for 2 min and fixed with 4% paraformaldehyde (Sigma) in phosphate buffer. The worms were frozen with liquid nitrogen, defrosted with warm water and shaken at 4 °C overnight. The worms were treated with β-mercaptoethanol solution (5% β-mercaptoethanol, 1% Triton X-100, 120 mm Tris, pH 7.0) for 2 h. They were then treated with collagenase solution [100 mm Tris base, 1 mm CaCl2 dihydrate, 0.1% Triton X-100, collagenase (1000 unit/ml)] for 2 h. They were blocked in solution [5% normal goat serum in antibody buffer (0.5% Triton X-100, 1 mm M EDTA, pH 8.0), 0.1% BSA, 0.05% sodium azide in PBS] overnight at 4 °C and incubated with primary antibody (1:5–1:100 dilution) in blocking solution overnight at 4 °C. The worms were incubated with goat anti-mouse alexa-568 (1:400 dilution; Invitrogen) for 1 h at RT. After washed with antibody buffer, the worms were mounted with VECTASHIELD® mounting medium (Vector Laboratories, Burlingame, CA). A Zeiss LSM510 confocal laser scanning microscope (Zeiss, Oberkochen, Germany) was used to take immunofluorescence images. Identical settings were used to acquire digital images from B. xylophilus and B. mucronatus. Digital images were processed using the Zeiss image browser (Zeiss) and Adobe Photoshop (Adobe Systems Inc., San Jose, CA).
Multiple Alignments and Phylogenetic Analysis of the Bx-LEC-1
To identify conserved domains in the galectin-1 of B. xylophilus (Bx-LEC-1) as well as evolutionary relationships to other related proteins, the predicted amino acid sequence of Bx-LEC-1 was searched using NCBI's Entrez database. Amino acid sequences of eight evolutionarily conserved LECs were extracted from NCBI's Entrez database and multiply aligned by CLC DNA Workbench 6 (CLC bio, Katrinebjerg, Denmark).
Two highly conserved galactose-binding domains (GBDs) in the Bx-LEC-1 were searched independently using NCBI's conserved domain database. By reverse position-specific BLAST, two GBDs were assigned into cd00070. The phylogenetic sequence trees of cd00070, including two GBDs in the Bx-LEC-1, were generated, examined and modified using CDTree 3.1 (26).
Prediction of Three-dimensional Structures of Galactose-Binding Lectin Domains
Three-dimensional homology models of GBDs in the Bx-LEC-1 were generated using the Swiss Model workspace (http://swissmodel.expasy.org/workspace) and MATRAS: Protein 3-D Structure Comparison (http://biunit.aist-nara.ac.jp/matras). Several three-dimensional structures of GBDs were automatically generated. Predicted three-dimensional structures of GBDs were evaluated using the Qualitative Model Energy Analysis (QMEAN) server (http;//swissmodel.expasy.org/qmean). Estimated per-residue errors of experimental three-dimensional structures and homology modeled three-dimensional structures of GBDs were also considered. The homology model that received the highest QMEAN score with the smallest estimated per-residue errors was chosen to represent the three-dimensional structure of each GBD domain. Visualization and modification of models and structures was done with Swiss PDB viewer 4.0.1. (Swiss Institute of Bioinformatics).
Cloning, Expression and Purification of Bx-LEC-1
To amplify the candidate gene, Bx-LEC-1, two specific primers, LEC-1-F [5′-GCATGCATGACTGAGGAAAAGAAAAC-3′, the sequence includes a SphI site (underlined) and an initiation codon (bold)] and LEC-1-R [5′-AAGCTTTTAATGGATCTGGATGCC-3′, the sequence includes stop codon (bold) and a HindIII site (underlined)] were designed. The EST clone T3–2163, containing the full-length of the Bx-LEC-1 cDNA, was used as a PCR template. All PCRs were performed using Pfx Taq polymerase (Invitrogen). The resulting PCR product was cloned into the pGEMT-easy vector (Promega), and both strands were sequenced. The cloned gene was subcloned into the pQE30 vector (Qiagen Inc., Valencia, CA) and polyhistidine (His) was fused with the N-terminal region of the Bx-LEC-1 in the resulting expression vector, which was transformed into Escherichia coli M15 strain (Qiagen Inc.). The Bx-LEC-1 protein expression was primarily induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) (Final concentration, 1 mm; Sigma). Cells were harvested every hour to check the amount of expressed proteins. Then, 10 μg of total proteins was loaded into 10% tris-glycine SDS-polyacrylamide gel. The procedures for SDS-PAGE, protein transfer, and immunoblotting with MAb-D9-F10 were as described above. The bacteria induced by IPTG for 6 h were harvested by centrifugation at 2000 × g for 10 min. Cell pellets were resuspended with NP10 buffer (50 mm NaH2PO4, 300 mm NaCl, 10 mm imidazole (pH 8.0)) and disrupted using a Vibra cellTM (Sonics and Materials, Inc., Newtown, CT). Following centrifugation at 10,000 × g for 20 min at 4 °C, the supernatant was applied to a size exclusion column, Superdex 75 HR 10/30 (GE Healthcare) with FPLC AKTA Purifier (GE Healthcare). The fractions containing the Bx-LEC-1 protein were subsequently applied to Ni-NTA superflow column (Qiagen). After washing the column with NP20 buffer (50 mm NaH2PO4, 300 mm NaCl, 20 mm imidazole (pH 8.0)), the His-Bx-LEC-1 was eluted with NP250 buffer (50 mm NaH2PO4, 300 mm NaCl, 250 mm imidazole (pH 8.0)) in FPLC AKTA Purifier (GE Healthcare). The purified His-Bx-LEC-1 was concentrated by filtration with a microcon centrifugal filter device (Millipore) with PBS. Protein concentration was determined using a Bio-Rad Protein Assay dye reagent and BSA as a standard.
Estimation of Kinetic Constants Based on Surface Plasmon Resonance
The dissociation constants (KDs) between the Bx-LEC-1 and sugars were measured using the His-Bx-LEC-1 immobilized sensor in a BIAcore 2000 (GE Healthcare, Upsaala, Sweden). The purified His-Bx-LEC-1 was immobilized on the CM5 sensor surface at pH4.5, following the manufacturer's instructions. The amount of the His-Bx-LEC-1 immobilized by amine coupling was ∼14,000 response unit. Various carbohydrates including d-(+)-raffinose (O-α-d-Galactopyranosyl [1→6] α-d-glucopyranosyl β-d-fructofuranoside, Sigma), N-acetyllactosamine (Lac-NAc; Sigma) and l-(+)-rhamnose (Sigma), in HBS-EP (0.01 m HEPES-NaOH (pH 6.8), 0.15 m NaCl, 3 mm EDTA, 0.005% (v/v) polysorbate 20) buffer were introduced onto the surface at a flow rate of 20 μl/min. BSA was used as a reference protein. The interaction between the immobilized Bx-LEC-1 and various carbohydrates was monitored for 2 min at 25 °C by subtracting the signal obtained from the reference surface. The KDs were calculated using BIA evaluation 3.1 software (GE Healthcare).
RESULTS
A B. xylophilus Specific Monoclonal Antibody, 3-2A7-2H5-D9-F10
To develop biomarkers for distinguishing the pinewood nematode B. xylophilus from other closely related pine tree-resident nematodes, we first generated monoclonal antibodies (MAbs) specific to B. xylophilus by injecting mice with whole protein extracts of B. xylophilus. We generated and screened 2304 fusion cells using ELISA and dot blot screening (data not shown). Three hundred seventy clones secreted antibodies that recognized B. xylophilus extracts. We further characterized them by performing dot blot analysis using B. mucronatus extracts as the negative control. We found that one MAb clone named 3-2A7-2H5 specifically recognized B. xylophilus extract, and not B. mucronatus. To isolate monoclonal hybridoma cells, we performed end point dilution using MAb-3-2A7-2H5 and then carried out dot blot screening and immunoblotting. Among several clones tested, MAb-3-2A7-2H5-D9-F10 (MAb-D9-F10) showed the highest specificity and the strongest reactivity to B. xylophilus (Supplemental Fig. S1), and was therefore selected for further studies. The immunoglobulin (Ig) isotype and subclass of MAb-D9-F10 were determined to be IgG κ-light.
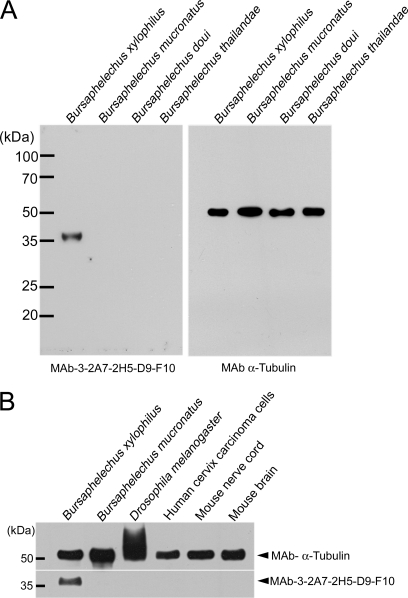
In order to further confirm the specificities of MAb-D9-F10, we performed Western blot analysis using protein extracts of Bursaphelenchus species, Drosophila melanogaster, nerve cords and brains from mice, and a human cervix carcinoma cell line (Fig. 1). MAb-D9-F10 specifically recognized a protein expressed in B. xylophilus with no cross-reactivity with other closely related nematodes, even though a similar amount of α-tubulin was detected in other samples (Fig. 1A). In addition, MAb-D9-F10 did not show any cross-reactivity with other proteins originating from Drosophila, mice, or human cells (Fig. 1B). The molecular weight of the target protein for MAb-D9-F10 was estimated to be ∼35 kDa. After total proteins from B. xylophilus were separated by SDS-PAGE, a band bearing the MAb-D9-F10 positive signals was excised and analyzed by nano-LC-ESI-Q-IT-MS. Twenty seven proteins showing credible cross-correlation scores were identified, including subunits of ATP synthases, ribosomal proteins and elongation factors, enzymes, house-keeping proteins, and galactose-binding lectin-1 (LEC-1) (Table I, Supplemental Excel File S1).
Fig. 1.
Monoclonal antibody 3-2A7-2H5-D9-F10 (MAb-D9-F10) recognized only Bursaphelenchus xylophilus. A, Even though similar amounts of proteins from Bursaphelenchus xylophilus, B. mucronatus, B. doui and B. thailandae were loaded, MAb-D9-F10 recognized only one band from B. xylophilus. B, To assess the specificity of MAb-D9-F10, various samples including B. xylophilus, B. mucronatus, Drosophila melanogaster, human cervix carcinoma cells, mouse nerve cord, and mouse brain were tested with MAb-D9-F10. MAb-D9-F10 recognized only one band in B. xylophilus, even though similar or greater amounts of other proteins were loaded. For loading controls, α-tubulin was used.
Table I. Summary of biochemical and proteomic identification results.
| Biochemical/Proteomic analysis | GeneBank accession number | Name | Sequence coverage % | Unique peptide detected | Predicted M.W. (kDa) |
|---|---|---|---|---|---|
| 1 DE/Nano-LC-ESI Q-IT-MS | Gu130151 | Tubulin α chain | 6.49 | 2 | 60.57 |
| Gu130146 | ATP synthase α chain | 4.06 | 1 | 58.97 | |
| Gu130131 | ATP synthase β chain | 6.38 | 2 | 57.16 | |
| Gu103155 | Eukaryotic translation elongation factor 1A | 6.91 | 3 | 50.01 | |
| Gu130144 | Elongation factor 1 γ | 2.24 | 1 | 45.56 | |
| Gu130154 | Prolyl carboxy Peptidase like | 5.49 | 1 | 45.31 | |
| Gu130149 | Hydroxyacyl dehydrogenase | <3.67 | 1 | >44.76 | |
| Gu130153 | Cathepsin B-like cysteine proteinase | 3.21 | 1 | 44.70 | |
| Gu130147 | OV9m | 8.59 | 42.42 | ||
| Gu130148 | Actin-4 | 38.83 | 2 | 41.81 | |
| Gu130152 | Aldolase | 6.93 | 10 | 39.29 | |
| Gu130145 | 60S ribosomal protein L7a | 9.77 | 1 | 35.32 | |
| Gu130143 | Malate DeHydrogenase | 28.57 | 3 | 35.15 | |
| Gu130150 | ATP Synthase B homolog | 4.25 | 6 | 34.59 | |
| Gu130161 | Electron-transfer-flavoprotein | <10.19 | 1 | >33.95 | |
| Gu130142 | 60S ribosomal protein L5 | 4.34 | 2 | 33.80 | |
| Gu130141 | Tropomyosin | 11.97 | 1 | 33.14 | |
| Gu130139 | ATP synthase γ chain | 14.43 | 3 | 32.51 | |
| Gu130138 | galactose-binding lectin-1 (LEC-1) | 13.31 | 2 | 31.87 | |
| Gu130137 | AnNEXin | 7.42 | 3 | 31.75 | |
| Gu130136 | 40S ribosomal protein S3 | 5.15 | 30.43 | ||
| Gu130140 | 60S ribosomal protein L8 | 6.96 | 1 | 30.10 | |
| Gu130135 | 40S ribosomal protein S2 | 8.57 | 1 | 29.41 | |
| Gu130130 | 40S ribosomal protein S3a | 8.98 | 2 | 28.99 | |
| Gu130134 | 60S ribosomal protein L7 | 6.02 | 1 | 28.57 | |
| Gu130133 | C. elegans protein Y43F8B.1b | 4.84 | 2 | 28.32 | |
| Gu130132 | Elongation factor 1 β | 5.14 | 1 | 23.66 | |
| 2 DE/Nano-LC-ESI Q-IT-MS | GU130159 | HSP90 | <3.13 | 1 | >51.84 |
| GU130160 | DEAD/DEAH box helicase family | <5.81 | 1 | >35.51 | |
| GU130138 | LEC-1 | 6.47 | 1 | 31.87 | |
| GU130158 | 14–3-3 protein | 5.18 | 1 | 28.53 | |
| GU130157 | Small HSP21 homologue | 59.76 | 7 | 18.32 | |
| GU130156 | Histone H4 gene | 21.36 | 2 | 11.37 | |
| IP/1 DE/Nano-LC-ESI Q-ITMS | GU130148 | Actin | 12.77 | 2 | 41.81 |
| GU130138 | LEC-1 | 6.12 | 1 | 31.87 | |
| Anion Exchange Chromatography/IP/1 DE/Nano-LC-ESI Q-IT-MS | GU130138 | LEC-1 | 8.30 | 1 | 31.87 |
| E. coli-expressed LEC-1/1DE/Nano-LC-ESI Q-IT-MS | GU130138 | LEC-1 | 86.81 | 11 | 31.87 |
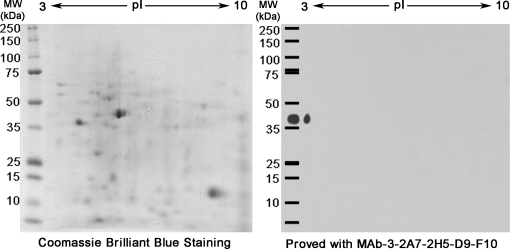
Two-dimensional Electrophoresis and Protein Identification
To collect biochemical information on the antigenic target of MAb-D9-F10, two identical gels were generated by performing two-dimensional electrophoresis (2-DE) simultaneously. One gel was stained with Coomassie brilliant blue, and the other was processed for MAb-D9-F10 immuno-blotting analysis. There was only one positive spot in the 2-DE immuno-blot, indicating that MAb-D9-F10 specifically recognized an antigenic protein. The antigenic protein of MAb-D9-F10 was highly acidic, and its pI was very close to 3.0 (Fig. 2). Based on the location of the positive spot of MAb-D9-F10 in Western blot analysis, the corresponding spot from the stained 2-DE gel was excised and analyzed with nano-LC-ESI-Q-IT-MS. Six proteins exhibiting high cross-correlation scores were identified as putative target proteins of MAb-D9-F10, including enzymes, heat-shock proteins, a histone subunit, the LEC-1, and a highly conserved eukaryotic protein (Table I, Supplemental Excel File S2).
Fig. 2.
A single spot was detected by MAb-D9-F10 when total proteins of B. xylophilus were separated by two-dimensional electrophoresis. Two identical two-dimensional electrophoresis (2-DE) gels were generated using total proteins from B. xylophilus. One 2-DE gel was processed with Coomassie Brilliant Blue staining to reveal protein spots. Another gel was processed with immunoblotting analysis using MAb-D9-F10. Only one spot very close to pI 3 was detected with MAb-D9-F10. As a reference, total proteins from B. xylophilus were loaded together with protein molecular weight markers.
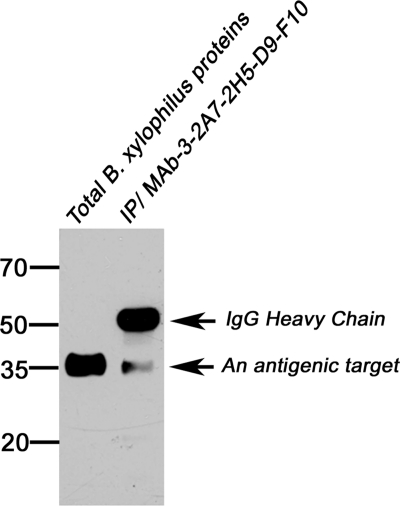
Purification of the Antigenic Target by MAb-D9-F10 Immunoprecipitation and Protein Identification
Immunoprecipitation is another way to purify target proteins for antibodies (27). The total proteins of B. xylophilus were mixed with MAb-D9-F10 to precipitate the antigenic target protein as its binding partner. When immunoprecipitates were separated by SDS-PAGE and blotted with MAb-D9-F10, two bands were detected (Fig. 3). One band contained Ig heavy chains, and the other was the antigenic target. The antigenic target band in an identical gel stained with Coomassie brilliant blue was excised and directly analyzed with nano-LC-ESI-Q-IT-MS. Only two proteins, LEC-1 and Actin, were identified, and they were also found in 1-DE or 2-DE/nano-LC-ESI-Q-IT-MS (Table I, Supplemental Excel File S1, S2, and S3).
Fig. 3.
Antigenic target proteins were precipitated together with MAb-D9-F10. To identify the antigenic target of MAb-D9-F10, total proteins of B. xylophilus and proteins precipitated with MAb-D9-F10 were separated and immunoblotted with MAb-D9-F10. One single band corresponding to the antigenic target was detected.
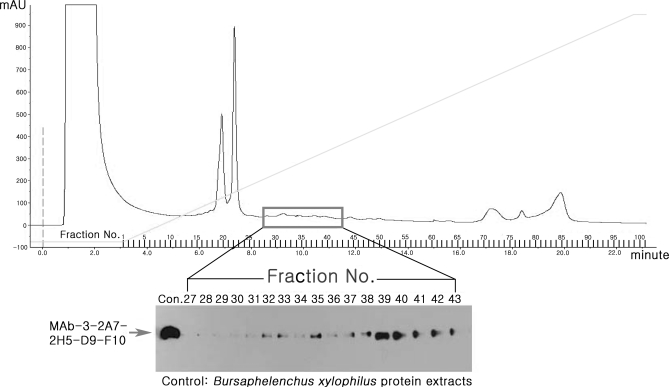
Because the antigenic target is a very acidic protein, the total proteins were fractionated by anion exchange chromatography, and each fraction was immunoblotted with MAb-D9-F10. MAb-D9-F10 specific antigens were present in fraction numbers 27 to 42 (Fig. 4). Fraction nos. 39 to 41 were selected, pooled, and used for IP with MAb-D9-F10. The MAb-D9-F10 corresponding band was excised and analyzed with nano-LC-ESI-Q-IT-MS. Only one protein, LEC-1, was identified as the target protein; this protein was found in all identification analyses (Table I, Excel File S4). Fragmentation ion spectra of two peptides with parent ion molecular masses 1147.67214 and 1530.69316 Da, were identified as LEC-1 peptides, whose sequences were identified as K162KLVIYGTPEKK173 and R202NALQANEWGNEERE217 with y ion and b ion fragmentation data, respectively (Supplemental Fig. S2). These results also suggested that LEC-1 is the antigenic target of MAb-D9-F10.
Fig. 4.
Separation and isolation of fractions containing antigenic targets for MAb-D9-F10 clone. To further characterize and isolate antigenic targets, B. xylophilus protein extracts were fractionated by anion exchange chromatography. In total, 101 fractions were recovered and tested by performing Western blot analysis with MAb-D9-F10. Fraction numbers 27 to 43 contained an antigenic target for MAb-D9-F10. The arrow indicates the MAb-D9-F10-specific bands. The line indicates the increasing linear gradient of NaCl. The y axis indicates relative protein concentration.
The Bx-LEC-1 Expressed in E. coli was Recognized by MAb-D9-F10
To verify that the antigenic target of MAb-D9-F10 is the Bx-LEC-1, we attempted to express the Bx-LEC-1 proteins using an E. coli expression system. The full-length cDNA of the Bx-LEC-1 was obtained from an EST clone of B. xylophilus, T3–2163. The predicted amino acid sequence showed the highest similarity to LECs from nematodes. The cloned Bx-LEC-1 was expressed in the E. coli M15 strain, and its expression was induced by spiking with IPTG. Expression of the Bx-LEC-1 was first confirmed with Coomassie blue staining. The expression amount gradually increased for 6 h following IPTG induction (Supplemental Fig. S3A). In addition, the Bx-LEC-1 expressed in E. coli was recognized by MAb-D9-F10 (Supplemental Fig. S3B). The intensity of the MAb-D9-F10-specific band gradually increased. The Bx-LEC-1 peptides from the IPTG-inducing band showed high cross-correlation scores covered more than 80% of the full length of the Bx-LEC-1 by nano-LC-ESI-Q-IT-MS (Supplemental Excel file S5). These results confirmed that the expressed Bx-LEC-1 is the antigenic target of MAb-D9-F10.
Localization of Bx-LEC-1 in the B. xylophilus
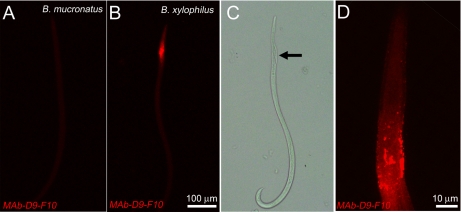
The localization of the Bx-LEC-1 was examined by MAb-D9-F10 immunohistochemistry. No MAb-D9-F10 positive immunoreactivities were observed when B. mucronatus was processed for MAb-D9-F10 immunocytochemistry (Fig. 5A), suggesting the specificity of MAb-D9-F10 for the Bx-LEC-1. Strong MAb-D9-F10 immunoreactivities were observed from the anterior body of B. xylophilus (Fig. 5B). The comparison of this confocal image (Fig. 5B) with a Nomarski differential interference image of the same B. xylophilus (Fig. 5C) as well as high-magnification observation of MAb-D9-F10 immunoreactivities (Fig. 5D) suggested that MAb-D9-F10 immunoreactivity was present at the neighboring areas of the median bulb, which also contains esophageal glands. The large cells of the esophageal glands were intensely labeled with MAb-D9-F10 (Fig. 5D). These localization results suggested that the Bx-LEC-1 might be involved in food perception and digestion.
Fig. 5.
Localization of the LEC-1 in B. xylophilus. A, No MAb-D9-F10-positive immunoreactivities were observed in B. mucronatus. B, MAb-D9-F10-positive immunoreactivities were found in neighboring areas of the median bulb. C, The Nomarski differential interference contrast image of the same nematode at the same magnification revealed the median bulb (arrow) and the esophageal gland. D, The MAb-D9-F10 immunoreactivities at the anterior body of B. xylophilus, magnified.
Evolutionarily Conserved Galactose-binding Lectin Domains of the Bx-LEC-1
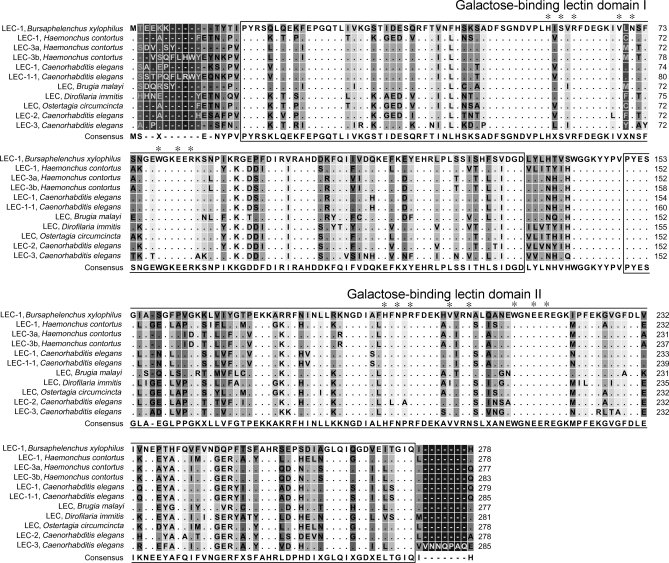
Recent studies have suggested that understanding evolutionarily conserved domains and motifs in proteins is the first step toward elucidating protein functions in silico (26). The predicted amino acid sequence of the Bx-LEC-1 was blasted with NCBI's Entrez database and conserved domain database to identify evolutionarily conserved domains and related proteins. The sequence identity and similarity of the Bx-LEC-1 to LEC-1 and LEC-3 from Caenorhabditis elegans, Caenorhabditis briggsae and Haemonchus contortus were very high: over 80% and 90%, respectively. Two nonidentical galactose-binding lectin domains (GBDs) linked with a short peptide sequence were identified in the Bx-LEC-1. The sequence identity and similarity between the two GBDs in the Bx-LEC-1 were 30% and 51%, respectively. In addition, eight amino acids shown to be important for the binding of sugars were completely conserved in the GBDs of all LECs (Fig. 6).
Fig. 6.
Multiple alignment of deduced amino acid sequences of the Bx-LEC-1 and other evolutionarily conserved LECs. Two nonidentical GBDs connected by a short linker peptide sequence were boxed. Dots represent amino acids identical to those of the Bx-LEC-1. Background grayscale indicates the degree of amino acid residue conservation among 11 GaLECtins. White indicates 100% conservation, whereas black represents 0%. Asterisks (*) identify the eight highly conserved residues (H, S/N, R, V, N, W, E, and R) that have been shown to play key roles in the binding of sugars.
Each nonidentical GBD of the Bx-LEC-1 was independently compared with the GBDs of the galectins of various organisms, including mammals, amphibians, invertebrates, and plants, using NCBI's CDD library (Supplemental Fig. S4). The N-terminal GBD (GBD1) of the Bx-LEC-1 was clustered tightly with the N-terminal GBD of LEC-3 of H. contortus, LEC in Ostertagia circumcincta, and LEC-3 in C. elegans. The C-terminal GBD (GBD2) of the Bx-LEC-1 largely formed a cluster with GBDs from C. elegans, H. contortus, Globodera rosteochinensis and Onchocera volvulus (Supplemental Fig. S4). These results showed that each GBD of the Bx-LEC-1 is closely associated with each GBD of other tandem-repeat type of nematode LECs, indicating that these GBDs are generated tandemly in the ancestral genes as shown in C. elegans (28). However, the chimera type of LECs such as LEC-7, -8, -9 and -10 in C. elegans were quite distal, forming the independent phylogeny (Supplemental Fig. S4). When the GBDs of vertebrate galectins independently clustered in our phylogenetic analysis were considered, N-terminal GBDs of mammalian galectin-4, -9, -14 and -12 formed the phylogeny adjacent to C-terminal GBDs of galectin-4, -6, -7, and -9 (Supplemental Fig. S4). Those GBDs were not associated in the phylogenetic analysis of vertebrate galectins reported by Houzelstein et al. (2004). Those differences may be explained by differences in the methods used to generate the phylogenetic trees and species of galectins compared. Nevertheless, GBDs from nematodes, mammals, amphibians, insects, and plants formed an independent phylogeny (Supplemental Fig. S4).
Two GBDs of the Bx-LEC-1 Exhibited a β-Sandwich Fold
To address the functions of the two GBDs in the Bx-LEC-1, protein structure homology modeling was performed using two web-based homology modeling servers, Swiss-Model Workspace and MATRAS: Protein 3-D Comparison. The QMEAN results revealed that five models failed to pass quality estimation (data not shown) whereas 15 homology models produced credible QMEAN scores (Supplemental Table S1). QMEAN scores represent a global score of the whole model, reflecting predicted model reliability ranging from 0 to 1 (30). The QMEAN score of a GBD1 model based on 2D6P A (2D6P A-GBD1) was the greatest among 10 homology models for GBD1 and greater than the QMEAN score of its template, 2D6P A (Supplemental Table S1). The QMEAN score of a model for GBD2 based on 2DYC (2DYC-GBD2) was the greatest among five homology models, but slightly smaller than the QMEAN score of its template, 2DYC (Supplemental Table S1). The estimated per-residue errors of two experimental three-dimensional structures, 2D6P A and 2DYC, and other modeled three-dimensional structures of GBDs were also considered (Supplemental Fig. S5). The estimated per-residue errors of 2D6P A-GBD1 and 2DYC-GBD2 were similar to those of their templates.
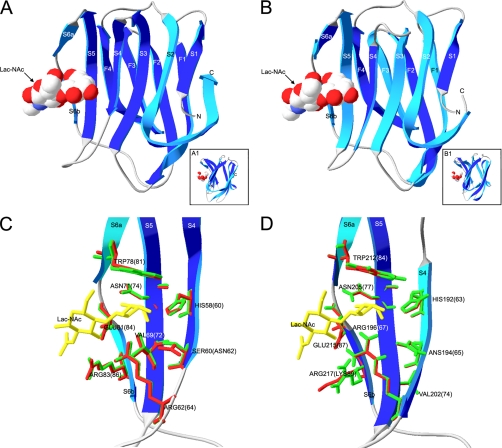
The predicted 3-D protein structures of GBD1 and GBD2 exhibited a β-sandwich fold comprising two antiparallel β-sheets, one four-stranded (F1–F4) and the other six-stranded (S1–S6b) (Figs. 7A and 7B). Insets show the side views of each domain, clearly displaying β-sandwich forms, the positions of β-galactosides and their binding pockets in the GBDs (Fig. 7A1 and 7B1). The eight amino acid residues identified to be evolutionarily conserved and important for binding to carbohydrates were distributed in four β sheets (S4, S5, S6a, and S6b) in GBD 1 and GBD 2. The side chains of amino acid residues of modeled GBDs, colored green, and those of templates, colored red, were superimposed to show how the positions of those residues were conserved (Figs. 7C and 7D). Except for two amino acid residues, Asn-62 in the N-terminal GBD of mouse galectin-9 and Lys-89 in the N-terminal GBD of mouse galectin-4, which were replaced by Ser 60 in GBD1 of the Bx-LEC-1 (C) and Arg-217 in GBD2 of the Bx-LEC-1 (D), respectively, the other residues were completely conserved and overlapped. These predicted three-dimensional protein structures of GBD1, GBD2, and their ligands suggested that the Bx-LEC-1 may be able to bind to small β-galactosides.
Fig. 7.
Three-dimensional homology models of two galactose-binding lectin domains in the Bx-LEC-1. Ribbon models of the N- and C- terminal galactose-binding lectin domains (GBD1, A; GBD2, B) are predicted together with space-filled models of N-acetyllactosamine (Lac-NAc). The four-stranded (F1–F4) and six-stranded (S1–S6) β-sheets are indicated by the letter-number code. The face and back of β-sheet structures are colored blue and aqua, respectively. Insets show side views of GBD1 (A1) and GBD2 (A2), clearly showing the anti-parallel β sheets of a β-sandwich fold. Amino acid residues forming the Lac-NAc binding sites of GBD1 (C) and GBD2 (D) are shown by rod model (green color) and labeled. Red colors are rod models of the amino acid residues of mouse galectin-9 (C) and mouse galectin-4 (D). The first residue number always belongs to the Bx-LEC-1, and the numbers and amino acid residues in parentheses belong to mouse galectin-9 (C) and mouse galectin-4 (D). Rod models of Lac-NAc are yellow.
Carbohydrate Binding Activity of the Bx-LEC-1
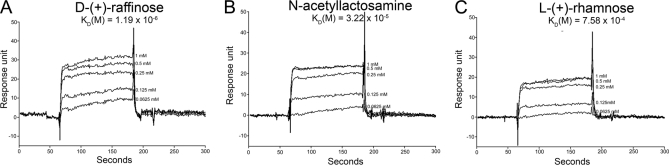
In order to investigate the carbohydrate binding ability of the Bx-LEC-1, the dissociation constants (KDs) of the binding between Bx-LEC-1 and three carbohydrates were measured on the basis of the Bx-LEC-1 immobilized assay system using a BIAcore 2000 instrument. Fig. 8 depicted the sensograms and KDs of each binding experiment. The KDs of between the Bx-LEC-1 and d-(+)-raffinose and N-acetyllactosamine was ∼637.0, and ∼23.5 times, respectively, lower than that of between the Bx-LEC-1 and l-(+)-rhamnose (Fig. 8). These results suggested that the Bx-LEC-1 has strong binding ability to small β-galactosides because l-(+)-rhamnose is a naturally occurring deoxy sugar without a galactose moiety whereas others had a galactose moiety.
Fig. 8.
Sensograms and KDs of tested carbohydrates for the immobilized Bx-LEC-1. Various concentrations of d-(+)-raffinose (A), N-acetyllactosamine (B), and l-(+)-rhamnose (C) solutions were introduced on the Bx-LEC-1-immobilized surface for 120 s at a flow rate 20 μl/min. The response unit was indicated as subtraction of the reference protein obtained on the immobilized surface from the values obtained on the Bx-LEC-1 immobilized surface.
DISCUSSION
The major aim of this study was to develop monoclonal antibodies (MAbs) specific to pathogenic Bursaphelenchus xylophilus as molecular biomarkers, and to characterize their antigenic targets with biochemical and proteomic analyses. To date, the most efficient way to prevent and control PWD is the complete removal of all pine trees near B. xylophilus-infected trees, resulting in severe environmental disruption and economic losses (31). If MAbs specific only to B. xylophilus and not to other closely related pine tree-resident nematodes such as B. muscronatus, B. thailandae, and B. doui are available, they would be used as molecular markers to detect pathogenic B. xylophilus. Thus, our most significant accomplishment in this study was the establishment of MAb-D9-F10 as specifically discriminating B. xylophilus from closely related pine tree-resident nematodes, as well as other organisms that might contribute to false positive signals (Fig. 1). Our data confirmed the specificity of MAb-D9-F10 and supported its possible use as a molecular bio-marker for B. xylophilus.
The specificity of MAb-D9-F10 attracted our scientific interest because identifying its antigenic target would increase our understanding of the biological and pathogenic features of B. xylophilus. Thus, we employed several biochemical and proteomic methods to isolate and identify the antigenic target. We identified 27 proteins including LEC-1, subunits of ATP synthases, ribosomal proteins, and elongation factors from a MAb-D9-F10-specific band isolated from 1-DE of B. xylophilus protein extracts (Table I). To identify candidates for the antigenic target, the proteins were separated by 2-DE, and six proteins were identified from the MAb-D9-F10 specific spot (Fig. 2, Table I). The molecular weights of two proteins, Bx-LEC-1 and 14-3-3 protein, were similar to the predicted molecular weights of antigenic targets, around 35 kDa. The fact that 14-3-3 protein was not detected in the 1-DE analysis suggested that the antigenic target protein of MAb-D9-F10 might be the Bx-LEC-1. MAb-D9-F10 IP using whole B. xylophilus protein extracts coprecipitated with the Bx-LEC-1 and Actin. The other MAb-D9-F10 IP using anion exchange chromatography fractions containing antigenic targets for MAb-D9-F10 only contained the Bx-LEC-1 (Table I). Further confirmation of the Bx-LEC-1 as the antigenic target was accomplished by expressing the full-length Bx-LEC-1 in an E. coli expression system and probing the E. coli-expressed Bx-LEC-1 with MAb-D9-F10 (Supplemental Fig. S3). Taken together, several lines of biochemical and proteomic analysis results suggested that the antigenic target of MAb-D9-F10 is the Bx-LEC-1.
The galectins modulate distinct cellular process such as inflammation, cell adhesion, cell proliferation, and cell death, and belong to a family of animal lectins that share evolutionarily conserved carbohydrate recognition domains of around 130 amino acids (32, 33). The galectins are localized in both the cytoplasm and extracellular regions, and they bind to small β-galactosides. According to their molecular domain organization, the galectins in mammals are divided into three subfamilies as proto-, chimera-, and tandem-repeat types. Proto- and chimera-type galectins have a single galactose-binding lectin domain (GBD; also known as carbohydrate-binding domain) with a short N-terminal sequence and an extended C-terminal tail containing several repeats of a Pro-Tyr-Gly rich motif, respectively. Tandem-repeat-type galectins have two nonidentical GBDs connected by a short linker peptide sequence (33, 34) and also known as bi-GBD galectins (29). Even though LECs having characteristics of both chimera- and tandem-repeat type galectins are found in C. elegans (28), the molecular domain structures based on the deduced amino acid residues of the Bx-LEC-1 suggested that it belongs to the tandem-repeat type of LEC subfamily, because two nonidentical GBDs with a short linker peptide sequence were predicted (Fig. 6) and the chimera type of LECs including LEC-7, -8, -9, and -10 in C. elegans were clustered into the independent phylogeny (Supplemental Fig. S4). In addition, eight amino acid residues that have been shown to play pivotal roles in binding to carbohydrates were completely conserved in the Bx-LEC-1. Indeed, in this study we have shown that Bx-LEC-1 had strong binding affinity to small β-galactosides such as d-(+)-rhaffinose and Lac-NAc (Fig. 8). The affinity of the Bx-LEC-1 for small β-galactosides can be explained by its primary and tertiary structural similarity to tandem-repeat type of mammalian galectins and nematode LECs and phylogenetic analysis (Figs. 6 and 7, and Supplemental Fig. S4).
Three-dimensional homology modeling of important protein domains is of great interest for understanding molecular and cellular features and functions of domains when no experimental three-dimensional structures are available (35–37). Because more than 70 experimental three-dimensional structures of the GBDs of galectins were currently available (a search result from Research Collaboratory for Structural Bioinformatics Protein Data bank; http://www.rcsb.org), we obtained three-dimensional structure predictions for GBD1 and GBD2 of the Bx-LEC-1 by performing structure modeling. Because two modeled three-dimensional structures of each GBD showed greater or similar QMEAN scores and similar estimated per-residue errors to those of two experimental three-dimensional structures, and because the sequence identity (34 ∼38%) and similarity (57%) between templates and modeled GBDs were relatively high, the relevant physiological three-dimensional structures of GBDs might be very close to those depicted in Figs. 7A and 7B. The three-dimensional structures of GBD1 and GBD2 exhibited a β-sandwich fold, as previously reported for GBDs in other galectins (38–40). The similar precise positions of eight amino acid residues in GBD1 and GBD2 of the Bx-LEC-1 were revealed by superimposing modeled structures and their templates (Fig. 7C and 7D). By examining interactions between several different carbohydrates and the N-terminal GBD in mouse galectin-9 at atomic levels, Nagae et al. (38) showed that certain amino acid residues play central roles in forming hydrogen bonds with carbohydrates containing β-galactoside. The β-galactoside moieties in carbohydrates were deeply buried in the binding sites formed by β strands S4–S6, and their positions were aligned by two residues with planar side chains (His-60 and Trp-81). His-60 and Asn-62 formed hydrogen bonds with the β-galactoside moiety, whereas Arg-64, Glu-84, and Arg-86 interacted with the glucose moiety of the lactose. In addition, Arg-64 and Glu-84 formed hydrogen bonds with the N-acetylglucosamine (GlcNAc) moiety in N-acetyllactosamine (Lac-NAc), whereas the guanidino head group of Arg-86 made a van der Waals contact with the methyl group (38). Interestingly, the amino acid residues in the N-terminal GBD of mouse galectin-9 were conserved in GBD1 and GBD2 of the Bx-LEC-1, except for Asn-62. Asn-62 in the N-terminal GBD of mouse galectin-9 was replaced by Ser-60 in the N-terminal GBD1 of the Bx-LEC-1 (Fig. 6 and Fig. 7C). Because a Ser residue has a short side chain compared with Asn, replacement of Asn with Ser may alter hydrogen bond formation between GBD1 and β-galactosides. Indeed, studies of the galactose binding activity of LEC-like genes in C. elegans showed that GBD1 and GBD2 of C. elegans LEC like-1 had different sugar binding activities, and GBDs of other LECs that lack the eight conserved amino acid residues showed poor binding activities (28).
Based on the results from the confocal microscopic observations and a Nomarski differential interference contrast image (Fig. 5), the localization pattern of MAb-D9-F10 immunoreactivities in the areas of the median bulb and esophageal glands of B. xylophilus was elucidated. The localization pattern suggested that the Bx-LEC-1 might be involved in feeding and digestion processes, because the median bulb is a valve allowing food to move from the stylet to the intestine, and the large cells of the esophageal glands are thought to release substances that help with digestion and feeding (41–43). It was also reported that the LEC-1 in C. elegans is localized at the terminal bulb of the pharynx, which functions as a valve controlling food movement from the mouth to the intestine (44). The localization patterns of the Bx-LEC-1 suggested that it may be involved in detecting the type and quality of foods, and in regulating secretion of substances from the esophageal glands.
In conclusion, we developed MAb-D9-F10 as a molecular biomarker specific to pathogenic B. xylophilus. Several lines of evidences presented in this study supported the conclusion that the antigenic target of MAb-D9-F10 was Bx-LEC-1. Phylogenetic sequence analysis, multiple alignments, immuno-localization patterns, strong affinity to small β-galactosides and three-dimensional homology models of the GBDs in the Bx-LEC-1 suggested that the Bx-LEC-1 may be able to bind to carbohydrates with different specificity and may be involved in the feeding and digestion of nematodes. In addition, the approaches used in this study could be applied to other researches aimed at elucidating the identity and characteristics of unknown specific proteins or markers from the pathogens or organisms whose full genomic sequences are not available.
Footnotes
* This study was carried out with the support of Forest Science and Technology Projects (Project No. S110708L0503706C) provided by Korea Forest Service and a grant (KRF-2006-521-E00103) from National Research Foundation of Korea, Republic of Korea.
 This article contains supplemental Figs. 1–5 and Table 1.
This article contains supplemental Figs. 1–5 and Table 1.
1 The abbreviations used are:
- PWD
- pine wilt disease
- MAbs
- monoclonal antibodies
- nano-LC-ESI-Q-IT-MS
- nano liquid chromatography electrospray ionization quadrupole ion trap mass spectrometry
- LEC-1
- galactose-binding lectin-1
- BSA
- bovine serum albumin
- DE
- dimensional electrophoresis
- Bx-LEC-1
- galectin-1 from B. xylophilus
- IPTG
- isopropyl-β-D-thiogalactopyranoside
- CDD
- conserved domain database
- GBD
- galactose-binding lectin domain
- Ig
- immunoglobulin
- IP
- immuno-precipitation
- QMEAN
- qualitative model energy analysis
- Lac-NAc
- N-acetyllactosamine.
REFERENCES
- 1. Steiner G., Buhrer E. M. (1934) Aphelenchoides xylophilus, n. sp., a nematode associated with blue-stain and other fungi in timber. J. Agric. Res. 48, 949–951 [Google Scholar]
- 2. Yano S. (1913) Investigation on pine death in Nagasaki prefecture. Sanrin-Kouhou 4, 1–14 [Google Scholar]
- 3. Kiyohara T., Tokushige Y. (1971) Inoculation experiments of a nematode, Bursaphelenchus sp., onto pine trees. J. Jpn. For. Soc. 53, 210–218 [Google Scholar]
- 4. Enda N. (1989) Current status of pine wilt disease in Korea. Forest Pests 38, 148–152 [Google Scholar]
- 5. Yang B., Wang Q. (1989) Distribution of the pine wood nematode in China and susceptibility of some Chinese and exotic pine to the nematode. Can. J. For. Res. 19, 1527–1530 [Google Scholar]
- 6. Enda N. (1988) Current status of pine wilt disease in Taiwan. Forest Pests 37, 161–166 [Google Scholar]
- 7. Mota M. M., Braasch H., Bravo M. A., Penas A. C., Burgermeister W., Metge K., Sousa E. (1999) First report of Bursaphelenchus xylophilus in Portugal and in Europe. Nematology 1, 727–734 [Google Scholar]
- 8. Fukuda K. (1997) Physiological process of the symptom development and resistance mechanism in pine wilt disease. J. For. Res. 2, 171–181 [Google Scholar]
- 9. Linit M. J. (1988) Nematode—vector relationships in the pine wilt disease system. J. Nematol. 20, 227–235 [PMC free article] [PubMed] [Google Scholar]
- 10. Mamiya Y. (1983) Pathology of the pine wilt disease caused by Bursaphelenchus xylophilus. Annu. Rev. Phytopathol. 21, 201–220 [DOI] [PubMed] [Google Scholar]
- 11. Mamiya Y. (1988) History of pine wilt disease in Japan. J. Nematol. 20, 219–226 [PMC free article] [PubMed] [Google Scholar]
- 12. Mamiya Y. (1976) Pine wilting disease caused by the pine wood nematode, Bursaphelenchus lignnicolus in Japan. Jpn. Agric. Res. Quart. 10, 206–211 [Google Scholar]
- 13. Kikuchi T., Aikawa T., Kosaka H., Pritchard L., Ogura N., Jones J. T. (2007) Expressed sequence tag (EST) analysis of the pine wood nematode Bursaphelenchus xylophilus and B. mucronatus. Mol. Biochem. Parasitol. 155, 9–17 [DOI] [PubMed] [Google Scholar]
- 14. Kang J. S., Lee H., Moon I. S., Lee Y., Koh Y. H., Je Y. H., Lim K. J., Lee S. H. (2009) Construction and characterization of subtractive stage-specific expressed sequence tag (EST) libraries of the pinewood nematode Bursaphelenchus xylophilus. Genomics. 94, 70–77 [DOI] [PubMed] [Google Scholar]
- 15. Webster J. M., Anderson R. V., Baillie D. L., Beckenbach K., Curran J., Rutherford T. A. (1990) DNA probes for differentiating isolates of the pinewood nematode species complex. Rev. Nématol. 13, 255–263 [Google Scholar]
- 16. Tares S., Lemontey J. M., Deguiran G., Abad P. (1994) Use of species-specific satellite DNA from Bursaphelenchus xylophilus as a diagnostic probe. Phytopathology. 84, 294–298 [Google Scholar]
- 17. Hoyer U., Burgermeister W., Braasch H. (1998) Identification of Bursaphelenchus species (Nematoda, Aphelenchoideae) on the basis of amplified ribosomal DNA (ITS-RFLP). Nachr. Dtsch. Pflanzenschutzd. 50, 273–277 [Google Scholar]
- 18. Iwahori H., Tsuda K., Kanzaki N., Izui K., Futai K. (1998) PCR-RFLP and sequencing analysis of ribosomal DNA of Bursaphelenchus nematodes related to pine wilt disease. Fund. Appl. Nematol. 21, 655–666 [Google Scholar]
- 19. Iwahori H., Kanzaki N., Futai K. (2000) A simple, polymerase chain reaction-restriction fragment length polymorphism-aided diagnosis method for pine wilt disease. For. Pathol. 30, 157–164 [Google Scholar]
- 20. Kang J. S., Choi K. S., Shin S. C., Moon I. S., Lee S. G., Lee S. H. (2004) Development of an efficient PCR-based diagnosis protocol for the identification of the pinewood nematode, Bursaphelenchus xylophilus (Nematoda : Aphelenchoididae). Nematology 6, 279–285 [Google Scholar]
- 21. Kang J. S., Moon I. S., Lee S. G., Shin S. C., Lee S. H. (2009) Rapid and accurate prediction of the frequencies of Bursaphelenchus xylophilus and B. mucronatus in mixed nematode samples using real-time species-specific PCR. Nematology 11, 289–299 [Google Scholar]
- 22. Chawla M. L., Prasad S. K. (1975) Techniques in nematology. II. Comparative efficiency of sampling tools and nematode extraction methods. Indian J. Nematol. 4, 115–123 [Google Scholar]
- 23. Viglierchio D. R., Schmitt R. V. (1983) On the methodology of nematode extraction from field samples: baermann funnel modifications. J. Nematol. 15, 438–444 [PMC free article] [PubMed] [Google Scholar]
- 24. Kascsak R. J., Rubenstein R., Merz P. A., Tonna-DeMasi M., Fersko R., Carp R. I., Wisniewski H. M., Diringer H. (1987) Mouse polyclonal and monoclonal antibody to scrapie-associated fibril proteins. J. Virol. 61, 3688–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bradford M. M. (1976) A Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 26. Marchler-Bauer A., Anderson J. B., Derbyshire M. K., DeWeese-Scott C., Gonzales N. R., Gwadz M., Hao L., He S., Hurwitz D. I., Jackson J. D., Ke Z., Krylov D., Lanczycki C. J., Liebert C. A., Liu C., Lu F., Lu S., Marchler G. H., Mullokandov M., Song J. S., Thanki N., Yamashita R. A., Yin J. J., Zhang D., Bryant S. H. (2007) CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 35, D237–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Springer T. A. (1996) Current Protocols in Immunology, pp. 8.3.1–8.3.11 John Wiley and Sons, NY [Google Scholar]
- 28. Nemoto-Sasaki Y., Hayama K., Ohya H., Arata Y., Kaneko M. K., Saitou N., Hirabayashi J., Kasai K. -I. (2008) Caenorhabditis elegans galectins LEC-1–LEC-11: Structural features and sugar-binding properties. Biochim. Biophys. Acta. 1780, 1131–1142 [DOI] [PubMed] [Google Scholar]
- 29. Houzelstein D., Gonçalves I. R., Fadden A. J., Sidhu S. S., Cooper D. N., Drickamer K., Leffler H., Poirier F. (2004) Phylogenetic analysis of the vertebrate galectin family. Mol. Biol. Evol. 21, 1177–1187 [DOI] [PubMed] [Google Scholar]
- 30. Benkert P., Künzli M., Schwede T. (2009) QMEAN server for protein model quality estimation. Nucleic Acids Res. 37, W510–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dwinell L. D. (1997) The pinewood nematode: Regulation and mitigation. Annu. Rev. Phytopathol. 35, 153–166 [DOI] [PubMed] [Google Scholar]
- 32. Delacour D., Koch A., Jacob R. (2009) The role of galectins in protein trafficking. Traffic. 10, 1405–1413 [DOI] [PubMed] [Google Scholar]
- 33. Vasta G. R. (2009) Roles of galectins in infection. Nat. Rev. Microbiol. 7, 424–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hirabayashi J., Kasai K. (1993) The family of metazoan metal-independent beta-galactoside-binding lectins: structure, function and molecular evolution. Glycobiology. 3, 297–304 [DOI] [PubMed] [Google Scholar]
- 35. Al-Lazikani B., Jung J., Xiang Z., Honig B. (2001) Protein structure prediction. Curr. Opin. Chem. Biol. 5, 51–56 [DOI] [PubMed] [Google Scholar]
- 36. Kopp J., Bordoli L., Battey J. N., Kiefer F., Schwede T. (2007) Assessment of CASP7 predictions for template-based modeling targets. Proteins 69, 38–56 [DOI] [PubMed] [Google Scholar]
- 37. Tramontano A., Morea V. (2003) Assessment of Homology-Based Predictions in CASP5. Proteins 53, 352–368 [DOI] [PubMed] [Google Scholar]
- 38. Nagae M., Nishi N., Murata T., Usui T., Nakamura T., Wakatsuki S., Kato R. (2006) Crystal structure of the galectin-9 N-terminal carbohydrate recognition domain from Mus musculus reveals the basic mechanism of carbohydrate recognition. J. Biol. Chem. 281, 35884–35893 [DOI] [PubMed] [Google Scholar]
- 39. Wälti M. A., Thore S., Aebi M., Künzler M. (2008) Crystal structure of the putative carbohydrate recognition domain of human galectin-related protein. Proteins. 72, 804–808 [DOI] [PubMed] [Google Scholar]
- 40. Seetharaman J., Kanigsberg A., Slaaby R., Leffler H., Barondes S. H., Rini J. M. (1998) X-ray crystal structure of the human galectin-3 carbohydrate recognition domain at 2.1-Å resolution. J. Biol. Chem. 273, 13047–13052 [DOI] [PubMed] [Google Scholar]
- 41. Davis E. L., Hussey R. S., Baum T. J., Bakker J., Schots A., Rosso M. N., Abad P. (2000) Nematode parasitism genes. Annu. Rev. Phytopathol. 38, 365–396 [DOI] [PubMed] [Google Scholar]
- 42. Rosso M. N., Dubrana M. P., Cimbolini N., Jaubert S., Abad P. (2005) Application of RNA interference to root-knot nematode genes encoding esophageal gland proteins. Mol. Plant-Microbe Interact. 18, 615–620 [DOI] [PubMed] [Google Scholar]
- 43. Lambert K. N., Allen K. D., Sussex I. M. (1999) Cloning and characterization of an esophageal-gland-specific chorismate mutase from the phytoparasitic nematode Meloidogyne javanica. Mol. Plant-Microbe Interact. 12, 328–336 [DOI] [PubMed] [Google Scholar]
- 44. Arata Y., Akimoto Y., Hirabayashi J., Kasai K., Hirano H. (1996) An immunohistochemical study of the 32-kDa galectin (beta-galactoside-binding lectin) in the nematode Caenorhabditis elegans. Histochem. J. 28, 201–207 [DOI] [PubMed] [Google Scholar]