Abstract
Significant progress in instrumentation and sample preparation approaches have recently expanded the potential of MALDI imaging mass spectrometry to the analysis of phospholipids and other endogenous metabolites naturally occurring in tissue specimens. Here we explore some of the requirements necessary for the successful analysis and imaging of phospholipids from thin tissue sections of various dimensions by MALDI time-of-flight mass spectrometry. We address methodology issues relative to the imaging of whole-body sections such as those cut from model laboratory animals, sections of intermediate dimensions typically prepared from individual organs, as well as the requirements for imaging areas of interests from these sections at a cellular scale spatial resolution. We also review existing limitations of MALDI imaging MS technology relative to compound identification. Finally, we conclude with a perspective on important issues relative to data exploitation and management that need to be solved to maximize biological understanding of the tissue specimen investigated.
Since its introduction in the late 90s (1), MALDI imaging mass spectrometry (MS) technology has witnessed a phenomenal expansion. Initially introduced for the mapping of intact proteins from fresh frozen tissue sections (2), imaging MS is now routinely applied to a wide range of different compounds including peptides, proteins, lipids, metabolites, and xenobiotics (3–7). Numerous compound-specific sample preparation protocols and analytical strategies have been developed. These include tissue sectioning and handling (8–14), automated matrix deposition approaches and data acquisition strategies (15–21), and the emergence of in situ tissue chemistries (22–25). Originally performed on sections cut from fresh frozen tissue specimens, methodologies incorporating an in situ enzymatic digestion step prior to matrix application have been optimized to access the proteome locked in formalin-fixed paraffin-embedded tissue biopsies (25–29). The possibility to use tissues preserved using non-cross-linking approaches has also been demonstrated (30–32). These methodologies are of high importance for the study of numerous diseases because they potentially allow the retrospective analysis for biomarker validation and discovery of the millions of tissue biopsies currently stored worldwide in tissue banks and repositories.
In the past decade, instrumentation for imaging MS has also greatly evolved. Whereas the first MS images were collected with time-of-flight instruments (TOF) capable of repetition rates of a few hertz, modern systems are today capable of acquiring data in the kilohertz range and above with improved sensitivity, mass resolving power, and accuracy, significantly reducing acquisition time and improving image quality (33, 34). Beyond time-of-flight analyzers, other MALDI-based instruments have been used such as ion traps (35–37), Qq TOF instruments (38–40), and trap-TOF (16, 41). Ion mobility technology has also been used in conjunction with imaging MS (42–44). More recently, MALDI FT/ICR and Orbitrap mass spectrometers have been demonstrated to be extremely valuable instruments for the performance of imaging MS at very high mass resolving power (45–47). These non-TOF-based systems have proven to be extremely powerful for the imaging of lower molecular weight compounds such as lipids, drugs, and metabolites. Home-built instrumentation and analytical approaches to probe tissues at higher spatial resolution (1–10 μm) have also been described (48–50). In parallel to instrumentation developments, automated data acquisition, image visualization, and processing software packages have now also been developed by most manufacturers.
To date, a wide range of biological systems have been studied using imaging MS as a primary methodology. Of strong interest are the organization and identification of the molecular composition of diseased tissues in direct correlation with the underlying histology and how it differs from healthy tissues. Such an approach has been used for the study of cancers (51–54), neurologic disorders (55–57), and other diseases (58, 59). The clinical potential of the imaging MS technology is enormous (7, 60, 61). Results give insights into the onset and progression of diseases, identify novel sets of disease-specific markers, and can provide a molecular confirmation of diagnosis as well as aide in outcome prediction (62–64). Imaging MS has also been extensively used to study the development, functioning, and aging of different organs such as the kidney, prostate, epididymis, and eye lens (65–70). Beyond the study of isolated tissues or organs, whole-body sections from several model animals such as leeches, mice, and rats have been investigated (71–74). For these analyses, specialized instrumentation and protocols are necessary for tissue sectioning and handling (72, 73). Whole-body imaging MS opens the door to the study of the localization and accumulation of administered pharmaceuticals and their known metabolites at the level of entire organisms as well as the monitoring of their efficacy or toxicity as a function of time or dose (72, 73, 75, 76).
There is considerable interest in determining the identification and localization of small biomolecules such as lipids in tissues because they are involved in many essential biological functions including cell signaling, energy storage, and membrane structure and function. Defects in lipid metabolism play a role in many diseases such as muscular dystrophy and cardiovascular disease. Phospholipids in tissues have been intensively studied by several groups (37, 40, 77–83). In this respect, for optimal recovery of signal, several variables such as the choice of matrix for both imaging and fragmentation, solvent system, and instrument polarity have been investigated (20, 84). Particularly, the use of lithium cation adducts to facilitate phospholipid identification by tandem MS directly from tissue has also been reported (85). Of significant interest is the recent emergence of two new solvent-free matrix deposition approaches that perform exceptionally well for phospholipid imaging analyses. The first approach, described by Hankin et al. (86), consists in depositing the matrix on the sections through a sublimation process. The described sublimation system consists of sublimation glassware, a heated sand or oil bath (100–200 °C), and a primary vacuum pump (∼5 × 10−2 torr). Within a few minutes of initiating the sublimation process, an exceptionally homogeneous film of matrix forms on the section. The thickness of the matrix may be controlled by regulating pressure, temperature, and sublimation time. The second approach, described by Puolitaival et al.(87), uses a fine mesh sieve (≤20 μm) to filter finely ground matrix on the tissue sections. Agitation of the sieve results in passage of the matrix through the mesh and the deposition of a fairly homogeneous layer of submicrometer matrix crystals of the surface of the sections. The matrix density on the sections is controlled by direct observation using a standard light microscope. This matrix deposition approach was also found to be ideal to image certain drug compounds (88, 89). Both strategies allow very rapid production of homogeneous matrix coatings on tissue sections with a fairly inexpensive setup. Signal recovery was found to be comparable with those obtained by conventional spray deposition. With the appropriate size sublimation device or sieve, larger sections with dimensions of several centimeters such as those cut from mouse or rat whole bodies can also be rapidly and homogeneously coated.
Here we present several examples of MALDI imaging MS of phospholipids from tissue sections using TOF mass spectrometers over a wide range of dimensions from whole-body sections (several centimeters), to individual organs (several millimeters), down to high spatial resolution imaging of selected tissue areas (hundreds of micrometers) at 10-μm lateral resolution and below. For all of these dimension ranges, technological considerations and practical aspects are discussed. In light of the imaging MS results, we also address issues faced for compound identification by tandem MS analysis performed directly on the sections. Finally, we discuss under “Perspective” our vision of the future of the field as well as the technological improvements and analytical tools that need to be improved upon and developed.
MATERIALS AND METHODS
Chemicals
Matrix, 2,5-dihydroxybenzoic acid (DHB),1 was purchased from Sigma-Aldrich and recrystallized in house.
Tissue Samples and Sectioning
Mouse brains were purchased frozen from Pel-Freez Biologicals (Rogers, AR). One-day-old whole ICR mouse pups and one female ICR adult were a gift from H. Scott Baldwin (Vanderbilt University). All mice were sacrificed according to Vanderbilt University Institutional Animal Care and Use Committee-approved protocols. After sacrifice, (a) the pups were immediately flash frozen by slowly plunging them into liquid nitrogen to avoid shattering, and (b) the heart was dissected from the adult mouse and immediately flash frozen by immersion in liquid nitrogen.
For sectioning, all tissue samples were immobilized with embedding medium, cut on a Leica CM3050 cryostat (Leica Microsystems GmbH, Wetzlar, Germany), and thaw-mounted on a gold-coated MALDI target plate (mouse brain) or on indium-tin oxide-coated glass slides (Delta Technologies, Stillwater, MN) (mouse pup and heart). Brain sections were cut at a thickness of 10 μm, whereas the mouse pup sections and adult heart were cut at a thickness of 12 μm. All sections were allowed to dry in a desiccator prior to matrix application. To assess the histological accuracy of the imaging MS data, serial sections were also collected and stained with hematoxylin and eosin (H&E) according to standard protocols.
Matrix Application
Matrix application was performed according to published protocols (86, 87). Imaging MS data applications were performed on the same day as sectioning.
Sublimation
Matrix deposition was performed in a sublimation apparatus (Chemglass Life Science, Vineland, NJ) (supplemental Fig. 1). The gold-coated MALDI plates or glass slides were mounted on the bottom of the condenser using thermally conductive double sided tape (3M, St. Paul, MN). The target plate was kept cold (≥0 °C) by adding ice slush to the reservoir. Approximately 300 mg of in-house recrystallized 2,5-DHB matrix was deposited in the bottom of the condenser sleeve. Matrix was distributed evenly to ensure an even and homogenous matrix coating on the section. The sublimation apparatus was connected to a primary vacuum pump (E2M12, Edwards, Tewksbury, MA) via a vacuum connection valve. A cold trap was installed between the primary pump and the sublimation apparatus to capture sublimated matrix particles carried by the vacuum flow. This significantly helps in maintaining a clean and properly functioning vacuum gauge. The condenser apparatus and the bottom flask were sealed via a Viton O-ring. The primary pump was started, and a ∼5 × 10−2-torr vacuum was established in the apparatus. The system was allowed to equilibrate under vacuum for a period of 2 min. After stabilization of vacuum was assured, sublimation was initiated by placing the bottom of the sublimation apparatus in a sand bath heated to ∼120 °C. 2,5-DHB sublimation was allowed to progress for ∼4 min. This resulted in a thin homogeneous 5–10-μm-thick coating of matrix on the sections. The process was halted by removing the sublimation apparatus from the sand bath and closing the vacuum valve. The sublimation apparatus was allowed to slowly re-equilibrate to atmospheric pressure before removal of the sample plate.
Dry Coating
Dry coating was performed as described previously (87) (supplemental Fig. 2). Briefly, a slide with a thaw-mounted tissue section was attached by double sided tape to the receiving pan of an automated shaker. Approximately 1 g of finely ground 2,5-DHB was added to a 20-μm mesh U. S. A. standard stainless steel test sieve (Hogentogler and Co., Columbia, MD) with five glass milling balls. The sieve with 2,5-DHB was attached to the receiving pan, covered, and placed onto an automated shaker (Analysette 3 Spartan, Fritsch, Idar-Oberstein, Germany). The amplitude of the milling balls was manually adjusted to a height of 1 inch, and sieving was performed for a total of 20 min when a uniform coating was observed across the tissue.
MALDI Imaging Mass Spectrometry
Low Spatial Resolution Imaging
Low spatial resolution imaging MS (100–200 μm) was performed on a MALDI-TOF/TOF Ultraflex III mass spectrometer equipped with SmartBeam technology operating at 200 Hz (Bruker Daltonics, Billerica, MA). MALDI MS data acquisition was performed in reflectron geometry under optimized delayed extraction conditions (focused at m/z ∼800) with a source accelerating voltage of ±20 kV. With optimal source settings, a mass resolving power of M/ΔM ∼20,000 was achieved in the studied mass range for phospholipids between m/z 600 and 1000. External mass calibration was performed using 100 nl of a mixture of standard peptides deposited onto the sample plate after matrix deposition. With both positive and negative polarities, this mixture provides nine calibration points in the mass range between 428 and 1460 Da and allows mass accuracy for phospholipids to be typically better than 40 ppm across a whole-body image. Imaging MS data acquisition and image reconstruction and visualization were performed with FlexImaging 2.1 software.
High Spatial Resolution Imaging
High spatial resolution imaging MS (25–5 μm) was performed on a previously characterized home-built MALDI-TOF system (48). This system was recently upgraded with a higher performance N2 laser (Marathon 103-PD, LTB Lasertechnik Berlin, Berlin, Germany). Briefly, the low divergence of the laser beam, the coaxial laser illumination ion source geometry, and the control of the position by a Piezo stage of the sample plate on the time-of-flight axis achieve on-target surface irradiation of less than 5 μm in diameter. For all experiments, precise adjustment of the laser energy and number of laser shots per spot was predetermined with respect to desired spatial resolution. MALDI MS data acquisition was performed in linear geometry under optimized delayed extraction conditions (focused at m/z ∼800) with a source accelerating voltage of 15 kV. With optimal source settings, a mass resolving power of M/ΔM ∼2000 was achieved in the studied mass range for phospholipids between m/z 600 and 1000. Mass calibration was performed with the peptide mixture mentioned above. Imaging MS data were acquired with MMSIT software and visualized in Biomap (http://maldi-msi.org/).
Approaches to Glycerophospholipid Imaging Analysis
Whole-body Imaging MS
The primary goal of whole-body imaging by MS is to rapidly obtain compound localization throughout the body with sufficient spatial resolution to clearly delineate the different organs present. Several aspects must be considered when imaging whole-body sections from model animals by MALDI MS. Of primary concern are tissue sectioning, overall matrix deposition, and data acquisition time. For smaller whole-body specimens (≤3 cm in length), standard specimen cryostats can be used as described under “Materials and Methods.” For sectioning larger animals such as adult mice and rats, whole-body cryostats are used. Because of the size of the resulting sections, specialized handling protocols have been adapted (73). To minimize overall processing time, the time taken by the matrix deposition step needs to be taken into account. Most reports of whole-body imaging MS examples shown in the literature involve a very quick matrix deposition step such as manual spray coating (40). In this regard, for whole-body imaging of phospholipids, both matrix sublimation and dry coating methods are ideal approaches. Based on the dimension of the sections to be analyzed, the choice of spatial resolution will significantly affect the total time for data acquisition. A higher pixel density may produce a better looking image but in the end may not give more information. Another significant aspect to consider is the stability of the sample with time, especially in the high vacuum environment of MALDI time-of-flight instruments.
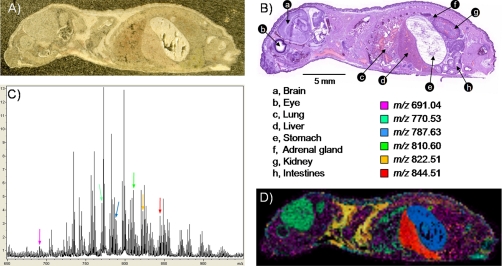
The example shown here relates to the imaging of phospholipids from a 1-day-old mouse pup section. The section is ∼28 × 8 mm2 in dimension. Matrix deposition was performed using the dry matrix coating approach. The coating procedure including setup, agitation, and cleaning took less than 30 min. After dry coating, a relatively homogeneous coating was observed throughout the sections with an 80–90% coating coverage. The percentage of coverage was observed to be tissue- and organ-dependent (Fig. 1A). The stability over time of the 2,5-DHB dry coating under high vacuum conditions was found to be excellent with no significant signal alteration within a 24-h period. Imaging MS data were acquired with a spatial resolution of 200 μm, representing 5444 pixels. Imaging MS data were acquired by summing the signals from 300 consecutive laser shots per pixel over the course of 5.5 h. Fig. 1B displays a photomicrograph of a serial H&E-stained section where numerous organs can be observed including the eye, brain, lung, liver, stomach, kidney, adrenal gland, and intestines. Fig. 1C presents the sum spectrum resulting from imaging MS data acquisition, displaying a typical phospholipid profile with intense signals from m/z 650 to 900 (87). Fig. 1D presents an overlay of six different ion images localizing in some of the different organs or tissues found throughout the section. The signal at m/z 691.04 was overall found ubiquitously in muscles throughout the section; m/z 770.53 was predominantly found in the eye socket, brain, and lungs; m/z 783.63 was exclusively observed in the stomach; m/z 810.60 was found to be very intense in the adrenal gland; m/z 822.51 was essentially found in the fatty tissue around the collarbone; and m/z 844.51 was observed to be very intense in the liver. Note that no normalization or postacquisition processing was performed. For whole-body imaging MS data, variations observed in total ion current (TIC) across the different organs and tissues encountered during the course of analysis may generate misleading ion images. Some tissues of the sections such as bones produce very little signal in the phospholipid mass range. Examples of some of the potential adverse effects of TIC normalization are addressed below for the mouse brain (see supplemental Fig. 4). For some of the observed signals in Figs. 1 and 2 and supplemental Figs. 3 and 4, tentative identifications based on molecular weight may be found elsewhere (20, 40, 80).
Fig. 1.
Imaging MS of phospholipids in positive ionization mode from a whole-body 1-day-old mouse pup section. A, photomicrograph of the section after 2,5-DHB matrix deposition by dry coating. B, photomicrograph of a H&E-stained serial section. Numerous organs constituent of the animal are observed. C, sum spectrum after imaging MS data acquisition with a lateral resolution of 200 μm. D, composite ion image made from the overlay of six different lipid ion distributions with observed abundances in various tissues or organs.
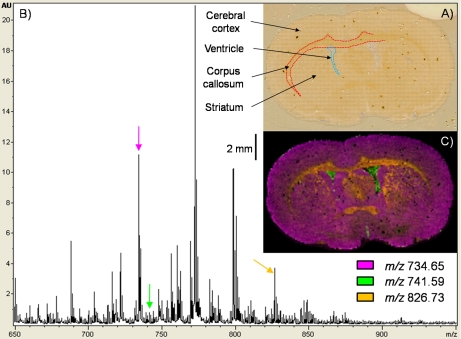
Fig. 2.
Imaging MS of phospholipids in positive ionization mode from a coronal mouse brain section. A, photomicrograph of the section after 2,5-DHB matrix sublimation and imaging MS data acquisition with a lateral resolution of 100 μm. The outlines of the major brain substructures are clearly visible. B, phospholipid sum spectrum obtained after imaging MS data acquisition. C, overlay of three different phospholipid ion images expressed in different regions of the section.
Intermediate Dimension Imaging MS
MALDI imaging MS of intermediate dimension sections (∼1 cm in length) is often the starting point for most users. Tissue specimens are for the most part easily handled and cut. For the imaging of phospholipids, no organic rinsing protocol is applied as these have been shown to extensively remove the lipid component of sections, although in some cases, brief washes in aqueous solution have been found to be useful (90). After a brief dehydration step in a desiccator, the sections are ready for matrix deposition. Several matrix coating strategies may be considered, ranging from manual or automated spray, droplet array printing, sublimation, or dry deposition. In all cases, coating can be performed within a few hours. Imaging MS of midsize sections is typically performed with resolutions between 250 and 50 μm. The spatial resolution required will greatly affect the overall data acquisition time. Acquisition time may again become critical when considering the stability of the sample, especially in the high vacuum environments of time-of-flight instruments. This is especially true for matrix deposited by sublimation. In this case, the thin layer of matrix coating the section may progressively “back-sublimate” and with time create gradient artifacts as imaging acquisition progresses. From our experience, for sublimated 2,5-DHB with a thickness of ∼7 μm, such effects start to be significant for time periods exceeding 24 h.
Fig. 2 and supplemental Fig. 3 present imaging MS results serially acquired from a coronal mouse brain section in positive and negative ionization mode, respectively. Imaging MS data were acquired with a spatial resolution of 100 μm. For imaging MS in the negative mode, to avoid ablation pattern overlaps, the grid array was aligned with an offset of 50 μm in both the x and y dimensions with respect to the array defined for positive data acquisition. In both cases, to maximize the information, the number of shots per pixel was kept constant at 400. Per pixel, the resulting ablated area was ∼40 μm in diameter. The oval-shaped brain section analyzed had dimensions of ∼13 × 8 mm2; when imaging at a spatial resolution of 100 μm, this represents ∼10,850 pixels. Fig. 2A presents a photomicrograph of the section after data acquisition where the matrix ablation pattern is clearly visible. The total acquisition time (positive and negative polarities) was ∼22 h. Fig. 2B presents the sum spectrum after acquisition in the positive mode. A typical phospholipid profile in the 650–900 m/z range was observed (86). Fig. 2C presents an overlay of three phospholipid images expressed in different regions of the section. The signal at m/z 734.65 was predominantly found in the cerebral cortex and striatum, m/z 741.59 was exclusively found in the ventricles, and m/z 826.73 was found in part of the striatum and the corpus callosum. Again, signal intensity normalization was not performed. Although normalization by TIC may improve the image contrast for some phospholipid signals, we have observed that even when restricted to the phospholipid m/z range TIC normalization can significantly distort localization. Some effects of TIC normalization are given in supplemental Fig. 4. Whereas in some cases normalization seems to have improved the quality of the image (m/z 660.07 and 776.16), in other cases, obvious variations within the images were observed such as edge effects (m/z 746.66 and 820.66), oversaturation (m/z 772.67), loss of information (m/z 746.66 and 797.67), and of greater concern “fabrication” of information (m/z 820.66). The necessity and effects of imaging MS data set normalization are worthy of additional study, particularly for the relatively narrow phospholipid mass range. Normalization using TIC is not the only strategy that needs to be thoroughly explored (91).
Supplemental Fig. 3 presents the imaging MS results obtained in negative ionization. Although 2,5-DHB is not the optimal matrix for negative phospholipid analysis by MALDI MS, some signals are nevertheless observed. An advantage to using 2,5-DHB is that it is fairly stable under high vacuum conditions for time periods of several hours, allowing collection of large images in both positive and negative modes. Supplemental Fig. 3A presents the sum spectrum obtained after imaging MS data acquisition. Again, signals in the phospholipid range (m/z 650–1400) are observed but with overall less intensity than in the positive ionization mode. Supplemental Fig. 3C presents an overlay of three phospholipid images (computed without normalization) expressed in different regions of the section. The signal at m/z 883.85 was predominantly found in the outer layers of the cerebral cortex and in the striatum, m/z 888.90 was found in the inner layers of the cerebral cortex and was very abundant in the corpus callosum, and m/z 1236.07 was exclusively found in the ventricles.
High Resolution Imaging MS
High resolution imaging is necessary for analysis of small anatomical features (a few hundreds of micrometers in dimension) within a tissue section with a lateral resolution down to the dimensions of individual cells (≤10 μm). The ability to analyze tissue sections at high resolution is primarily dependent upon laser spot size for laser microprobe systems and application of an ultrafine homogenous coating without inducing analyte lateral migration. To date, only a handful of MALDI instruments capable of acquiring imaging MS data at cellular length scales (≤10 μm) have been reported (48–50). Current commercially available instruments are limited to spot sizes of at best 15–20 μm. However, limitations on laser spot size may be compensated for using an oversampling approach (92). In this technique, the raster width is narrower than the laser spot diameter. Because this causes the laser spot to overlap several pixels, full depletion of the matrix is necessary to produce sufficient signal for image calculation before movement to the next adjacent pixel. With oversampling, resolutions may be reasonably reduced to 4 times smaller than the laser spot size. High resolution imaging utilizes the same tissue sectioning procedures as intermediate imaging. However, because the area of interest has finer details than gross anatomy of an organ, matrix coatings are limited to those that produce very homogenous coatings of submicrometer crystals (<1 μm) such as obtained by sublimation (86).
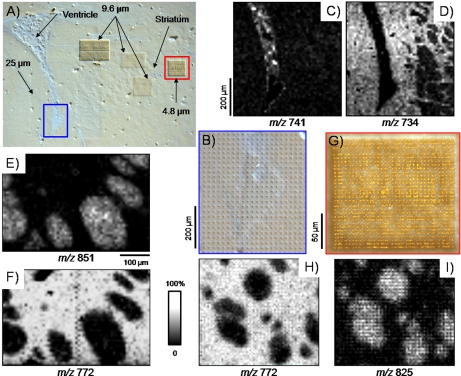
Examples of high resolution imaging MS are given in Fig. 3. Several serial measurements were performed in positive ion mode on selected areas of a coronal mouse brain section coated with 2,5-DHB by sublimation (Fig. 3A). In this case, matrix sublimation was used as the coating technique with respect to the dry coating approach because it gives near complete tissue coverage. Data were acquired using a prototype linear MALDI-TOF mass spectrometer capable of focusing the laser probe on target to better than 5 μm in diameter (supplemental Fig. 5A) (48). The ability to manipulate the stage in the z direction allowed for precise control of laser spot focus onto the sample target. This system is also capable of operating under delayed extraction conditions, and unit mass separation on phospholipid signals can be achieved (supplemental Fig. 5B). Measurements were performed with a lateral resolution of 25 μm over the tip of the ventricle and at 9.6 and 4.8 μm within the striatum. Best results were obtained for laser energy-number of shots combinations for which matrix ablation went through the sample down to the target plate (see Fig. 3, B and G). Ion images obtained from the imaging of the tip of the ventricle are presented in Fig. 3, C and D; the signal at m/z 741 was found to be specific to the ventricle, whereas the signal at m/z 734 was found to be uniquely expressed in the surrounding tissues. With an imaging resolution of 25 μm, some pattern is already visible within the striatum (right side of images). This patterning was further explored by imaging over the striatum with lateral resolutions of 9.6 and 4.8 μm. At these resolutions, some signals were clearly found to be expressed within the nerve bundles of the striatum (m/z 825 and 851; Fig. 3, I and E), whereas others were uniquely detected within the surrounding tissues (m/z 772; Fig. 3, F and H). A closer observation of the ablation pattern obtained after imaging at 4.8-μm spatial resolution (Fig. 3G) shows regular rows and columns of drilled holes, indicating that no overlap between adjacent pixels occurred. It is also interesting to note that for the brain, when imaging sections at spatial resolutions of 5–10 μm, no obvious phospholipid signal delocalization was observed. This indicates that no delocalization is induced during the freezing, sectioning, mounting, and matrix application steps.
Fig. 3.
High resolution imaging MS of phospholipids in positive ionization mode from a coronal mouse brain section. A, photomicrograph of the section after 2,5-DHB matrix deposition by sublimation and imaging MS data acquisition. Measurements were performed with a lateral resolution of 25 μm over the tip of the ventricle and at 9.6 and 4.8 μm within the striatum. B, magnified view of the tip of the ventricle showing the matrix ablation pattern after data acquisition. C and D, phospholipid ion images acquired with a lateral resolution of 25 μm over the tip of the ventricle. E and F, phospholipid ion images acquired with a lateral resolution of 9.6 μm over the striatum. G, magnified view of the ablation pattern after imaging MS of an area of the striatum with a lateral resolution of 4.8 μm. H and I, phospholipid ion images acquired with a lateral resolution of 4.8 μm over an area of the striatum.
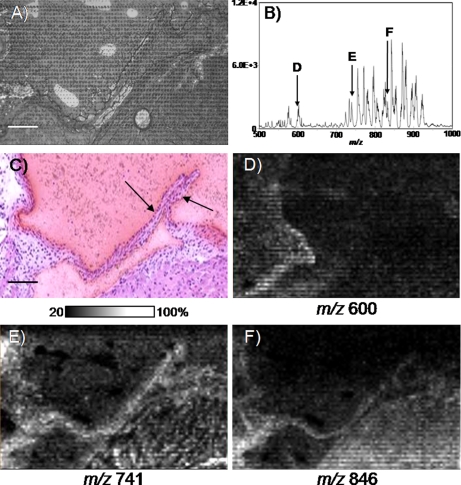
Fig. 4 presents the analysis of an adult mouse aortic valve acquired in the positive ion mode at a 9.6-μm lateral resolution. The size of one valve cusp is 500-μm length × 15-μm width. The adult mouse aortic valve consists of an outer layer of endothelial cells with an internal single cell layer of vascular interstitial cells. The valve cusp is essentially three cell layers in thickness so imaging the valve requires exceptional sensitivity as well as high spatial resolution techniques in instrumentation and matrix application. For imaging the aortic valve, a 12-μm cryosection was collected from the heart and coated with 2,5-DHB by sublimation. An area of 540 × 1100 μm encompassing both leaflets, representing 4697 pixels, was imaged over a period of 6.5 h. Laser ablation of sublimated matrix resulted in a circular spot size with an internal diameter of 2.93 ± 0.34 μm impinging into the tissue and an outer diameter of 6.48 ± 0.41 μm (Fig. 4A). These spots did not overlap between pixels, indicating achievement of a cellular level of analysis. Fig. 4B presents the sum spectrum of the image with many intense peaks occurring from m/z 550 to 600 and from m/z 700 to 925, matching a typical phospholipid signature. These data were not normalized to TIC. Fig. 4C presents an H&E staining of a neighboring section. Fig. 4D maps the intensity of the ion signal at m/z 600 occurring in the hinge region of the aortic valve. Ion intensity maps from m/z 741 (Fig. 4E) and m/z 846 (Fig. 4F) are good examples of ion signal occurring in both the hinge region and the cups of the valve. These data pose an exciting application toward investigations involving small anatomical features. For instance, numerous mouse models exist for the study of cardiovascular disorders and defects (93). Further improvements in resolving power, mass accuracy, and sensitivity to such instruments will greatly increase the utility for phospholipid imaging MS analyses.
Fig. 4.
High resolution imaging MS of phospholipids in positive ionization mode from an adult mouse aortic valve. A, photomicrograph of the section after 2,5-DHB matrix deposition by sublimation and imaging MS data acquisition with a lateral resolution of 9.6 μm. Scale bar, 100 μm. B, phospholipid sum spectrum obtained after imaging MS data acquisition. C, photomicrograph of a matching H&E-stained serial section. The aortic valve cusps are clearly visible (arrows). D–F, ion images from phospholipid signals, some of which were observed expressed in the aortic valve (E and F).
PERSPECTIVE
Interest in imaging lipids from biological tissue sections by MALDI MS is growing, and the recent advances in matrix deposition strategies and instrumentation have significantly contributed to developments in the field. Imaging lipids with MALDI-TOF instruments provides distribution maps of at least the most abundant species, bringing a wealth of new molecular information. However, a typical MALDI lipid profile contains a wide range of molecular species with the presence of numerous species within a nominal isobar. Resolving images from these will require alternative imaging strategies that incorporate high mass-resolving MS instrumentation such as MALDI FT/ICR or Orbitraps in conjunction with high spatial resolution capabilities. Ion mobility mass spectrometry offers the potential for broadband resolution of molecular isomers without the need for tandem MS. Already the technique has demonstrated an extra dimension of separation for imaging MS and is foreseen to significantly help in separating and mapping isobars.
An alternative approach can be used to image isobaric phospholipid species (and other potential compounds) within a nominal isobar by imaging in the MS/MS mode (35, 47). In this case, imaging MS is performed by selecting a parent ion window of typically 1 amu and fragmenting its content. An image of the parent compound is then acquired by monitoring the intensity of one or several of its specific fragment ions. This approach is widely used to image xenobiotics such as pharmaceutical compounds and their known metabolites, monitoring for each compound a specific previously characterized parent ion → fragment ion transition. In the case of phospholipids, by fragmenting the molecular content of a nominal isobar, several of the compounds present within that isobar can be individually imaged by monitoring the intensity of the different fragment ions produced (35, 47). This is only possible after characterization of the phospholipid-specific fragmentation transitions.
Identification of phospholipids from tissues by MALDI tandem MS is relatively straightforward at least for some of the most intense signals. In positive ionization, MALDI tandem MS can be enhanced by doping the matrix with lithium chloride, which can be manually or automatically spotted onto tissues (85). This approach can be ideal for both whole-body and intermediate imaging because laser spot sizes in the tens of micrometers produce an intense phospholipid ion yield. Identifying phospholipids observed from high resolution imaging can be difficult if the signal is coming from a small area and sample is limited, especially after the imaging process. Alternate cryosections are recommended for identification purposes. Furthermore, with decreasing laser spot size, the overall ion yield drops and may hinder the generation of quality MS/MS data.
There are several instruments that are useful for on-tissue fragmentation of lipids, but all have limitations in precursor selection for nominally isobaric phospholipids. The MALDI-TOF/TOF and MALDI ion trap instruments produce suitable fragmentation spectra from tissue with a typical precursor selection window of ±1 amu in the m/z 700–900 phospholipid region. In this case, only the most abundant phospholipid signal within that mass selection window is usually successfully identified versus lesser intense isobaric species. Narrowing the selection window typically results in significant loss in parent ion signal intensity and lower fragment ion yields, limiting identification potential. For these low abundance isobaric species, the use of high mass accuracy instrumentation may allow classification of some of these lipid species. Adding an extra dimension of separation using an ion mobility cell may also allow sufficient separation and isolation of low abundance isobars for identification by tandem MS.
Although imaging lipids can bring important insight into some extremely complex biological processes, there is a more general question that needs to be addressed. That is, how do we make optimum use of the wealth of information contained within an imaging MS experiment? Strict comparison with the underlying histology may highlight subsets of signals that are differentially expressed. Even after identification of a majority of the compounds involved, we must ask how that information is correlated to the physiological state of the tissue. How does it complement imaging information obtained from proteins? How do we integrate all of that information with previous findings related to the same biological system or disease? New bioinformatics tools need to be developed to rapidly interrogate imaging MS data and, upon compound identification, interface the results with other sets of data using a systems biology approach.
Since the introduction of MALDI imaging MS over a decade ago, tremendous advances in technological development have propelled the discipline to new heights. We foresee that in the next decade MALDI imaging MS will become a primary tool in biological and biomedical research, enabling a better understanding of molecular mechanisms over a wide range of biological systems and facilitating a vast array of new applications.
Acknowledgments
We thank Kenneth E. Schriver, Ph.D. (Department of Physics and Astronomy, Vanderbilt University) for help with the high resolution imaging experiments.
Footnotes
* This work was supported, in whole or in part, by National Institutes of Health Grant 5R01 GM 058008-11 and NIH/NHLBI 5RL1 HL092551–02.
 This article contains supplemental Figs. 1–5.
This article contains supplemental Figs. 1–5.
1 The abbreviations used are:
- DHB
- dihydroxybenzoic acid
- TIC
- total ion current.
REFERENCES
- 1. Caprioli R. M., Farmer T. B., Gile J. (1997) Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 69, 4751–4760 [DOI] [PubMed] [Google Scholar]
- 2. Stoeckli M., Chaurand P., Hallahan D. E., Caprioli R. M. (2001) Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat. Med. 7, 493–496 [DOI] [PubMed] [Google Scholar]
- 3. Cornett D. S., Reyzer M. L., Chaurand P., Caprioli R. M. (2007) MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat. Methods 4, 828–833 [DOI] [PubMed] [Google Scholar]
- 4. McDonnell L. A., Heeren R. M. (2007) Imaging mass spectrometry. Mass Spectrom. Rev. 26, 606–643 [DOI] [PubMed] [Google Scholar]
- 5. Seeley E. H., Caprioli R. M. (2008) Molecular imaging of proteins in tissues by mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 105, 18126–18131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Amstalden van Hove E. R., Smith D. F., Heeren R. M. (2010) A concise review of mass spectrometry imaging. J. Chromatogr. A 1217, 3946–3954 [DOI] [PubMed] [Google Scholar]
- 7. Chughtai K., Heeren R. M. (2010) Mass spectrometric imaging for biomedical tissue analysis. Chem. Rev. 110, 3237–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kruse R., Sweedler J. V. (2003) Spatial profiling invertebrate ganglia using MALDI MS. J. Am. Soc. Mass Spectrom. 14, 752–759 [DOI] [PubMed] [Google Scholar]
- 9. Schwartz S. A., Reyzer M. L., Caprioli R. M. (2003) Direct tissue analysis using matrix-assisted laser desorption/ionization mass spectrometry: practical aspects of sample preparation. J. Mass Spectrom. 38, 699–708 [DOI] [PubMed] [Google Scholar]
- 10. Chaurand P., Norris J. L., Cornett D. S., Mobley J. A., Caprioli R. M. (2006) New developments in profiling and imaging of proteins from tissue sections by MALDI mass spectrometry. J. Proteome Res. 5, 2889–2900 [DOI] [PubMed] [Google Scholar]
- 11. Lemaire R., Wisztorski M., Desmons A., Tabet J. C., Day R., Salzet M., Fournier I. (2006) MALDI-MS direct tissue analysis of proteins: improving signal sensitivity using organic treatments. Anal. Chem. 78, 7145–7153 [DOI] [PubMed] [Google Scholar]
- 12. Seeley E. H., Oppenheimer S. R., Mi D., Chaurand P., Caprioli R. M. (2008) Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. J. Am. Soc. Mass Spectrom. 19, 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kaleta° B. K., van der Wiel I. M., Stauber J., Güzel C., Kros J. M., Luider T. M., Heeren R. M. (2009) Sample preparation issues for tissue imaging by imaging MS. Proteomics 9, 2622–2633 [DOI] [PubMed] [Google Scholar]
- 14. Végvári A., Fehniger T. E., Gustavsson L., Nilsson A., Andrén P. E., Kenne K., Nilsson J., Laurell T., Marko-Varga G. (2010) Essential tactics of tissue preparation and matrix nano-spotting for successful compound imaging mass spectrometry. J. Proteomics 73, 1270–1278 [DOI] [PubMed] [Google Scholar]
- 15. Aerni H. R., Cornett D. S., Caprioli R. M. (2006) Automated acoustic matrix deposition for MALDI sample preparation. Anal. Chem. 78, 827–834 [DOI] [PubMed] [Google Scholar]
- 16. Shimma S., Furuta M., Ichimura K., Yoshida Y., Setou M. (2006) Direct MS/MS analysis in mammalian tissue sections using MALDI-QIT-TOFMS and chemical inkjet technology. Surf. Interface Anal. 38, 1712–1714 [Google Scholar]
- 17. Sugiura Y., Shimma S., Setou M. (2006) Two-step matrix application technique to improve ionization efficiency for matrix-assisted laser desorption/ionization in imaging mass spectrometry. Anal. Chem. 78, 8227–8235 [DOI] [PubMed] [Google Scholar]
- 18. Baluya D. L., Garrett T. J., Yost R. A. (2007) Automated MALDI matrix deposition method with inkjet printing for imaging mass spectrometry. Anal. Chem. 79, 6862–6867 [DOI] [PubMed] [Google Scholar]
- 19. Franck J., Arafah K., Barnes A., Wisztorski M., Salzet M., Fournier I. (2009) Improving tissue preparation for matrix-assisted laser desorption ionization mass spectrometry imaging. Part 1: using microspotting. Anal. Chem. 81, 8193–8202 [DOI] [PubMed] [Google Scholar]
- 20. Meriaux C., Franck J., Wisztorski M., Salzet M., Fournier I. (2010) Liquid ionic matrixes for MALDI mass spectrometry imaging of lipids. J. Proteomics 73, 1204–1218 [DOI] [PubMed] [Google Scholar]
- 21. McDonnell L. A., van Remoortere A., van Zeijl R. J., Dalebout H., Bladergroen M. R., Deelder A. M. (2010) Automated imaging MS: toward high throughput imaging mass spectrometry. J. Proteomics 73, 1279–1282 [DOI] [PubMed] [Google Scholar]
- 22. Groseclose M. R., Andersson M., Hardesty W. M., Caprioli R. M. (2007) Identification of proteins directly from tissue: in situ tryptic digestions coupled with imaging mass spectrometry. J. Mass Spectrom. 42, 254–262 [DOI] [PubMed] [Google Scholar]
- 23. Setou M., Hayasaka T., Shimma S., Sugiura Y., Matsumoto M. (2008) Protein denaturation improves enzymatic digestion efficiency for direct tissue analysis using mass spectrometry. Appl. Surf. Sci. 255, 1555–1559 [Google Scholar]
- 24. Franck J., El Ayed M., Wisztorski M., Salzet M., Fournier I. (2009) On-tissue N-terminal peptide derivatizations for enhancing protein identification in MALDI mass spectrometric imaging strategies. Anal. Chem. 81, 8305–8317 [DOI] [PubMed] [Google Scholar]
- 25. Morita Y., Ikegami K., Goto-Inoue N., Hayasaka T., Zaima N., Tanaka H., Uehara T., Setoguchi T., Sakaguchi T., Igarashi H., Sugimura H., Setou M., Konno H. (2010) Imaging mass spectrometry of gastric carcinoma in formalin-fixed paraffin-embedded tissue microarray. Cancer Sci. 101, 267–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Groseclose M. R., Massion P. P., Chaurand P., Caprioli R. M. (2008) High-throughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics 8, 3715–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lemaire R., Desmons A., Tabet J. C., Day R., Salzet M., Fournier I. (2007) Direct analysis and MALDI imaging of formalin-fixed, paraffin-embedded tissue sections. J. Proteome Res. 6, 1295–1305 [DOI] [PubMed] [Google Scholar]
- 28. Stauber J., Lemaire R., Franck J., Bonnel D., Croix D., Day R., Wisztorski M., Fournier I., Salzet M. (2008) MALDI Imaging of formalin-fixed paraffin-embedded tissues: application to model animals of Parkinson disease for biomarker hunting. J. Proteome Res. 7, 969–978 [DOI] [PubMed] [Google Scholar]
- 29. Djidja M. C., Francese S., Loadman P. M., Sutton C. W., Scriven P., Claude E., Snel M. F., Franck J., Salzet M., Clench M. R. (2009) Detergent addition to tryptic digests and ion mobility separation prior to MS/MS improves peptide yield and protein identification for in situ proteomic investigation of frozen and formalin-fixed paraffin-embedded adenocarcinoma tissue sections. Proteomics 9, 2750–2763 [DOI] [PubMed] [Google Scholar]
- 30. Chaurand P., Latham J. C., Lane K. B., Mobley J. A., Polosukhin V. V., Wirth P. S., Nanney L. B., Caprioli R. M. (2008) Imaging mass spectrometry of intact proteins from alcohol-preserved tissue specimens: bypassing formalin fixation. J. Proteome Res. 7, 3543–3555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mangé A., Chaurand P., Perrochia H., Roger P., Caprioli R. M., Solassol J. (2009) Liquid chromatography-tandem and MALDI imaging mass spectrometry analyses of RCL2/CS100-fixed, paraffin-embedded tissues: proteomics evaluation of an alternate fixative for biomarker discovery. J. Proteome Res. 8, 5619–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grey A. C., Gelasco A. K., Section J., Moreno-Rodriguez R. A., Krug E. L., Schey K. L. (2010) Molecular morphology of the chick heart visualized by MALDI imaging mass spectrometry. Anat. Rec. 293, 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moskovets E., Preisler J., Chen H. S., Rejtar T., Andreev V., Karger B. L. (2006) High-throughput axial MALDI-TOF MS using a 2-kHz repetition rate laser. Anal. Chem. 78, 912–919 [DOI] [PubMed] [Google Scholar]
- 34. Vestal M. L. (2009) Modern MALDI time-of-flight mass spectrometry. J. Mass Spectrom. 44, 303–317 [DOI] [PubMed] [Google Scholar]
- 35. Garrett T. J., Prieto-Conaway M. C., Kovtoun V., Bui H., Izgarian N., Stafford G., Yost R. A. (2007) Imaging of small molecules in tissue sections with a new intermediate-pressure MALDI linear ion trap mass spectrometer. Int. J. Mass Spectrom. 260, 166–176 [Google Scholar]
- 36. Drexler D. M., Garrett T. J., Cantone J. L., Diters R. W., Mitroka J. G., Prieto Conaway M. C., Adams S. P., Yost R. A., Sanders M. (2007) Utility of imaging mass spectrometry (IMS) by matrix-assisted laser desorption ionization (MALDI) on an ion trap mass spectrometer in the analysis of drugs and metabolites in biological tissues. J. Pharmacol. Toxicol. Methods 55, 279–288 [DOI] [PubMed] [Google Scholar]
- 37. Garrett T. J., Yost R. A. (2006) Analysis of intact tissue by intermediate-pressure MALDI on a linear ion trap mass spectrometer. Anal. Chem. 78, 2465–2469 [DOI] [PubMed] [Google Scholar]
- 38. Hsieh Y., Chen J., Korfmacher W. A. (2007) Mapping pharmaceuticals in tissues using MALDI imaging mass spectrometry. J. Pharmacol. Toxicol. Methods 55, 193–200 [DOI] [PubMed] [Google Scholar]
- 39. Reyzer M. L., Hsieh Y., Ng K., Korfmacher W. A., Caprioli R. M. (2003) Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 38, 1081–1092 [DOI] [PubMed] [Google Scholar]
- 40. Hayasaka T., Goto-Inoue N., Zaima N., Kimura Y., Setou M. (2009) Organ-specific distributions of lysophosphatidylcholine and triacylglycerol in mouse embryo. Lipids 44, 837–848 [DOI] [PubMed] [Google Scholar]
- 41. Goto-Inoue N., Hayasaka T., Sugiura Y., Taki T., Li Y. T., Matsumoto M., Setou M. (2008) High-sensitivity analysis of glycosphingolipids by matrix-assisted laser desorption/ionization quadrupole ion trap time-of-flight imaging mass spectrometry on transfer membranes. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 870, 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McLean J. A., Ridenour W. B., Caprioli R. M. (2007) Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J. Mass Spectrom. 42, 1099–1105 [DOI] [PubMed] [Google Scholar]
- 43. Jackson S. N., Ugarov M., Egan T., Post J. D., Langlais D., Albert Schultz J., Woods A. S. (2007) MALDI-ion mobility-TOFMS imaging of lipids in rat brain tissue. J. Mass Spectrom. 42, 1093–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stauber J., MacAleese L., Franck J., Claude E., Snel M., Kaletas B. K., Wiel I. M., Wisztorski M., Fournier I., Heeren R. M. (2010) On-tissue protein identification and imaging by MALDI-ion mobility mass spectrometry. J. Am. Soc. Mass Spectrom. 21, 338–347 [DOI] [PubMed] [Google Scholar]
- 45. Cornett D. S., Frappier S. L., Caprioli R. M. (2008) MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Anal. Chem. 80, 5648–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taban I. M., Altelaar A. F.., van der Burgt Y. E., McDonnell L. A., Heeren R. M., Fuchser J., Baykut G. (2007) Imaging of peptides in the rat brain using MALDI-FTICR mass spectrometry. J. Am. Soc. Mass Spectrom. 18, 145–151 [DOI] [PubMed] [Google Scholar]
- 47. Landgraf R. R., Prieto Conaway M. C., Garrett T. J., Stacpoole P. W., Yost R. A. (2009) Imaging of lipids in spinal cord using intermediate pressure matrix-assisted laser desorption-linear ion trap/orbitrap MS. Anal. Chem. 81, 8488–8495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chaurand P., Schriver K. E., Caprioli R. M. (2007) Instrument design and characterization for high resolution MALDI-MS imaging of tissue sections. J. Mass Spectrom. 42, 476–489 [DOI] [PubMed] [Google Scholar]
- 49. Luxembourg S. L., Mize T. H., McDonnell L. A., Heeren R. M. A. (2004) High-spatial resolution mass spectrometric imaging of peptide and protein distributions on a surface. Anal. Chem. 76, 5339–5344 [DOI] [PubMed] [Google Scholar]
- 50. Spengler B., Hubert M. (2002) Scanning microprobe matrix-assisted laser desorption ionization (SMALDI) mass spectrometry: instrumentation for sub-micrometer resolved LDI and MALDI surface analysis. J. Am. Soc. Mass Spectrom. 13, 735–748 [DOI] [PubMed] [Google Scholar]
- 51. Lemaire R., Menguellet S. A., Stauber J., Marchaudon V., Lucot J. P., Collinet P., Farine M. O., Vinatier D., Day R., Ducoroy P., Salzet M., Fournier I. (2007) Specific MALDI imaging and profiling for biomarker hunting and validation: fragment of the 11S proteasome activator complex, reg alpha fragment, is a new potential ovary cancer biomarker. J. Proteome Res. 6, 4127–4134 [DOI] [PubMed] [Google Scholar]
- 52. Schwartz S. A., Weil R. J., Thompson R. C., Shyr Y., Moore J. H., Toms S. A., Johnson M. D., Caprioli R. M. (2005) Proteomic-based prognosis of brain tumor patients using direct-tissue matrix-assisted laser desorption ionization mass spectrometry. Cancer Res. 65, 7674–7681 [DOI] [PubMed] [Google Scholar]
- 53. Yanagisawa K., Shyr Y., Xu B. J., Massion P. P., Larsen P. H., White B. C., Roberts J. R., Edgerton M., Gonzalez A., Nadaf S., Moore J. H., Caprioli R. M., Carbone D. P. (2003) Proteomic patterns of tumour subsets in non-small-cell lung cancer. Lancet 362, 433–439 [DOI] [PubMed] [Google Scholar]
- 54. Cazares L. H., Troyer D., Mendrinos S., Lance R. A., Nyalwidhe J. O., Beydoun H. A., Clements M. A., Drake R. R., Semmes O. J. (2009) Imaging mass spectrometry of a specific fragment of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase kinase 2 discriminates cancer from uninvolved prostate tissue. Clin. Cancer Res. 15, 5541–5551 [DOI] [PubMed] [Google Scholar]
- 55. Ceuppens R., Dumont D., Van Brussel L., Van de Plas B., Daniels R., Noben J. P., Verhaert P., Van der Gucht E., Robben J., Clerens S., Arckens L. (2007) Direct profiling of myelinated and demyelinated regions in mouse brain by imaging mass spectrometry. Int. J. Mass Spectrom. 260, 185–194 [Google Scholar]
- 56. Pierson J., Norris J. L., Aerni H. R., Svenningsson P., Caprioli R. M., Andrén P. E. (2004) Molecular profiling of experimental Parkinson's disease: direct analysis of peptides and proteins on brain tissue sections by MALDI mass spectrometry. J. Proteome Res. 3, 289–295 [DOI] [PubMed] [Google Scholar]
- 57. Stoeckli M., Staab D., Staufenbiel M., Wiederhold K. H., Signor L. (2002) Molecular imaging of amyloid beta peptides in mouse brain sections using mass spectrometry. Anal. Biochem. 311, 33–39 [DOI] [PubMed] [Google Scholar]
- 58. Meistermann H., Norris J. L., Aerni H. R., Cornett D. S., Friedlein A., Erskine A. R., Augustin A., De Vera Mudry M. C., Ruepp S., Suter L., Langen H., Caprioli R. M., Ducret A. (2006) Biomarker discovery by imaging mass spectrometry: transthyretin is a biomarker for gentamicin-induced nephrotoxicity in rat. Mol. Cell. Proteomics 5, 1876–1886 [DOI] [PubMed] [Google Scholar]
- 59. Minerva L., Clerens S., Baggerman G., Arckens L. (2008) Direct profiling and identification of peptide expression differences in the pancreas of control and ob/ob mice by imaging mass spectrometry. Proteomics 8, 3763–3774 [DOI] [PubMed] [Google Scholar]
- 60. Caprioli R. M. (2008) Perspectives on imaging mass spectrometry in biology and medicine. Proteomics 8, 3679–3680 [DOI] [PubMed] [Google Scholar]
- 61. Caprioli R. M. (2005) Deciphering protein molecular signatures in cancer tissues to aid in diagnosis, prognosis, and therapy. Cancer Res. 65, 10642–10645 [DOI] [PubMed] [Google Scholar]
- 62. Chaurand P., Sanders M. E., Jensen R. A., Caprioli R. M. (2004) Proteomics in diagnostic pathology: profiling and imaging proteins directly in tissue sections. Am. J. Pathol. 165, 1057–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chaurand P., Schwartz S. A., Caprioli R. M. (2004) Assessing protein patterns in disease using imaging mass spectrometry. J. Proteome Res. 3, 245–252 [DOI] [PubMed] [Google Scholar]
- 64. Chaurand P., Schwartz S. A., Reyzer M. L., Caprioli R. M. (2005) Imaging mass spectrometry: principles and potentials. Toxicol. Pathol. 33, 92–101 [DOI] [PubMed] [Google Scholar]
- 65. Chaurand P., Cornett D. S., Caprioli R. M. (2006) Molecular imaging of thin mammalian tissue sections by mass spectrometry. Curr. Opin. Biotechnol. 17, 431–436 [DOI] [PubMed] [Google Scholar]
- 66. Chaurand P., Rahman M. A., Hunt T., Mobley J. A., Gu G., Latham J. C., Caprioli R. M., Kasper S. (2008) Monitoring mouse prostate development by profiling and imaging mass spectrometry. Mol. Cell. Proteomics 7, 411–423 [DOI] [PubMed] [Google Scholar]
- 67. Chaurand P., Fouchécourt S., DaGue B. B., Xu B. J., Reyzer M. L., Orgebin-Crist M. C., Caprioli R. M. (2003) Profiling and imaging proteins in the mouse epididymis by imaging mass spectrometry. Proteomics 3, 2221–2239 [DOI] [PubMed] [Google Scholar]
- 68. Han J., Schey K. L. (2006) MALDI tissue imaging of ocular lens {alpha}-crystallin. Invest. Ophthalmol. Vis. Sci. 47, 2990–2996 [DOI] [PubMed] [Google Scholar]
- 69. Herring K. D., Oppenheimer S. R., Caprioli R. M. (2007) Direct tissue analysis by matrix-assisted laser desorption ionization mass spectrometry: application to kidney biology. Semin. Nephrol. 27, 597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grey A. C., Chaurand P., Caprioli R. M., Schey K. L. (2009) MALDI imaging mass spectrometry of integral membrane proteins from ocular lens and retinal tissue. J. Proteome Res. 8, 3278–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wisztorski M., Croix D., Macagno E., Fournier I., Salzet M. (2008) Molecular MALDI imaging: an emerging technology for neuroscience studies. Dev. Neurobiol. 68, 845–858 [DOI] [PubMed] [Google Scholar]
- 72. Stoeckli M., Staab D., Schweitzer A., Gardiner J., Seebach D. (2007) Imaging of a beta-peptide distribution in whole-body mice sections by MALDI mass spectrometry. J. Am. Soc. Mass Spectrom. 18, 1921–1924 [DOI] [PubMed] [Google Scholar]
- 73. Khatib-Shahidi S., Andersson M., Herman J. L., Gillespie T. A., Caprioli R. M. (2006) Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Anal. Chem. 78, 6448–6456 [DOI] [PubMed] [Google Scholar]
- 74. Trim P. J., Henson C. M., Avery J. L., McEwen A., Snel M. F., Claude E., Marshall P. S., West A., Princivalle A. P., Clench M. R. (2008) Matrix-assisted laser desorption/ionization-ion mobility separation-mass spectrometry imaging of vinblastine in whole body tissue sections. Anal. Chem. 80, 8628–8634 [DOI] [PubMed] [Google Scholar]
- 75. Reyzer M. L., Caldwell R. L., Dugger T. C., Forbes J. T., Ritter C. A., Guix M., Arteaga C. L., Caprioli R. M. (2004) Early changes in protein expression detected by mass spectrometry predict tumor response to molecular therapeutics. Cancer Res. 64, 9093–9100 [DOI] [PubMed] [Google Scholar]
- 76. Sugiura Y., Setou M. (2010) Imaging mass spectrometry for visualization of drug and endogenous metabolite distribution: toward in situ pharmacometabolomes. J. Neuroimmune Pharmacol. 5, 31–43 [DOI] [PubMed] [Google Scholar]
- 77. Burnum K. E., Cornett D. S., Puolitaival S. M., Milne S. B., Myers D. S., Tranguch S., Brown H. A., Dey S. K., Caprioli R. M. (2009) Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J. Lipid Res. 50, 2290–2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jackson S. N., Wang H. Y., Woods A. S. (2005) Direct profiling of lipid distribution in brain tissue using MALDI-TOFMS. Anal. Chem. 77, 4523–4527 [DOI] [PubMed] [Google Scholar]
- 79. Rujoi M., Estrada R., Yappert M. C. (2004) In situ MALDI-TOF MS regional analysis of neutral phospholipids in lens tissue. Anal. Chem. 76, 1657–1663 [DOI] [PubMed] [Google Scholar]
- 80. Sugiura Y., Setou M. (2009) Selective imaging of positively charged polar and nonpolar lipids by optimizing matrix solution composition. Rapid Commun. Mass Spectrom. 23, 3269–3278 [DOI] [PubMed] [Google Scholar]
- 81. Chen R., Hui L., Sturm R. M., Li L. (2009) Three dimensional mapping of neuropeptides and lipids in crustacean brain by mass spectral imaging. J. Am. Soc. Mass Spectrom. 20, 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Koizumi S., Yamamoto S., Hayasaka T., Konishi Y., Yamaguchi-Okada M., Goto-Inoue N., Sugiura Y., Setou M., Namba H. (2010) Imaging mass spectrometry revealed the production of lyso-phosphatidylcholine in the injured ischemic rat brain. Neuroscience 168, 219–225 [DOI] [PubMed] [Google Scholar]
- 83. Kim Y., Shanta S. R., Zhou L. H., Kim K. P. (2010) Mass spectrometry based cellular phosphoinositides profiling and phospholipid analysis: a brief review. Exp. Mol. Med. 42, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Astigarraga E., Barreda-Gómez G., Lombardero L., Fresnedo O., Castaño F., Giralt M. T., Ochoa B., Rodríguez-Puertas R., Fernández J. A. (2008) Profiling and imaging of lipids on brain and liver tissue by matrix-assisted laser desorption/ionization mass spectrometry using 2-mercaptobenzothiazole as a matrix. Anal. Chem. 80, 9105–9114 [DOI] [PubMed] [Google Scholar]
- 85. Jackson S. N., Wang H. Y., Woods A. S. (2005) In situ structural characterization of phosphatidylcholines in brain tissue using MALDI-MS/MS. J. Am. Soc. Mass Spectrom. 16, 2052–2056 [DOI] [PubMed] [Google Scholar]
- 86. Hankin J. A., Barkley R. M., Murphy R. C. (2007) Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 18, 1646–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Puolitaival S. M., Burnum K. E., Cornett D. S., Caprioli R. M. (2008) Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J. Am. Soc. Mass Spectrom. 19, 882–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Goodwin R. J., Macintyre L., Watson D. G., Scullion S. P., Pitt A. R. (2010) A solvent-free matrix application method for matrix-assisted laser desorption/ionization imaging of small molecules. Rapid Commun. Mass Spectrom. 24, 1682–1686 [DOI] [PubMed] [Google Scholar]
- 89. Goodwin R. J., Scullion P., Macintyre L., Watson D. G., Pitt A. R. (2010) Use of a solvent-free dry matrix coating for quantitative matrix-assisted laser desorption ionization imaging of 4-bromophenyl-1,4-diazabicyclo(3.2.2)nonane-4-carboxylate in rat brain and quantitative analysis of the drug from laser microdissected tissue regions. Anal. Chem. 82, 3868–3873 [DOI] [PubMed] [Google Scholar]
- 90. Jones E. A., Lockyer N. P., Vickerman J. C. (2008) Depth profiling brain tissue sections with a 40 keV C-60(+) primary ion beam. Anal. Chem. 80, 2125–2132 [DOI] [PubMed] [Google Scholar]
- 91. Deininger S. O., Becker M., Wolski E., Kaminski H., Paape R., Cornett D. S. (2010) Practical considerations on normalization in MALDI imaging, in Proceedings of the 58th ASMS Conference on Mass Spectrometry and Allied Topics, Salt Lake City, May 22–27, 2010, The American Society for Mass Spectrometry, Santa Fe, NM [Google Scholar]
- 92. Jurchen J. C., Rubakhin S. S., Sweedler J. V. (2005) MALDI-MS imaging of features smaller than the size of the laser beam. J. Am. Soc. Mass Spectrom. 16, 1654–1659 [DOI] [PubMed] [Google Scholar]
- 93. Russell J. C., Proctor S. D. (2006) Small animal models of cardiovascular disease: tools for the study of the roles of metabolic syndrome, dyslipidemia, and atherosclerosis. Cardiovasc. Pathol. 15, 318–330 [DOI] [PubMed] [Google Scholar]