Abstract
CYP2A5 metabolizes xenobiotics and activates hepatocarcinogens, and induction occurs in response to hepatic damage and cellular stress. We evaluated whether ethanol can elevate CYP2A5 and whether CYP2E1 plays a role in the ethanol induction of CYP2A5. Wild-type (WT), CYP2E1 knockout (KO), and CYP2E1 knockin (KI) mice were fed ethanol for 3 weeks. Ethanol increased CYP2E1 and CYP2A5 protein and activity in WT mice but not in the KO mice. Induction of CYP2A5 (and CYP2E1) was restored in the KI mice. Ethanol induction of CYP2A5 occurred only after CYP2E1 was first induced. Immunohistochemical staining revealed that CYP2E1 and CYP2A5 colocalize to the same zones in the liver. Ethanol also elevated CYP2A5 mRNA levels in WT and KI mice but not in KO mice. Induction of CYP2A5 by cadmium was partially decreased in KO mice compared with WT or KI mice. Ethanol elevated CYP2A4 mRNA levels in all mice although the extent of induction was lowest in the KO mice. In summary, ethanol elevated mouse hepatic CYP2A5 levels, which may be of toxicological significance because CYP2A5 metabolizes nicotine and other drugs and activates hepatocarcinogens. Induction of CYP2A5 by ethanol is potentiated by the induction of CYP2E1. We speculate that ethanol induction of CYP2E1 followed by increases in reactive oxygen species and activation of Nrf2 are important steps in the mechanism by which ethanol induces CYP2A5. The possibility that induction of CYP2E1 is permissive for the induction of CYP2A5 may reflect a new contribution by CYP2E1 to the actions of ethanol.
Introduction
Mouse CYP2A5 and its human ortholog CYP2A6 metabolize several important xenobiotics including nicotine, coumarin, cotinine, testosterone, aflatoxin B1, and nitrosamines (Su and Ding, 2004). CYP2A5 is expressed in many tissues with high levels found in the respiratory tract, liver, and kidney. Increased expression of CYP2A5 occurs during viral, fulminant, or bacterial hepatitis, in certain tumors, and after treatment with a variety of hepatotoxins and heavy metals (Jounaïdi et al., 1994; Abu-Bakar et al., 2004; De-Oliveira et al., 2006; Lämsä et al., 2010). Induction of CYP2A5 has been suggested to occur in response to hepatocellular damage and to endoplasmic reticulum stress (Gilmore and Kirby, 2004). Depending on the inducer, the activation of hepatic CYP2A5 can occur via transcriptional and post-transcriptional mechanisms (Abu-Bakar et al., 2004, 2007; Gilmore and Kirby, 2004). Pyrazole, widely used as an inhibitor of alcohol dehydrogenase and of ethanol metabolism (Goldberg and Rydberg, 1969), induces CYP2A5 largely by a post-transcriptional mechanism involving stabilization of the CYP2A5 mRNA (Juvonen et al., 1985; Nichols and Kirby, 2008). Recent studies have indicated a role for cellular redox status and possible activation of stress-related transcription factors in activation of CYP2A5 (Gilmore and Kirby, 2004; Su and Ding, 2004; Abu-Bakar et al., 2007; Lämsä et al., 2010). For example, treatment of hepatocytes with menadione, a redox cycling agent, elevated CYP2A5 expression (Gilmore and Kirby, 2004). Pyrazole increases oxidative stress; pretreatment of hepatocytes with antioxidants such as N-acetylcysteine or vitamin E blunted pyrazole-mediated increases in CYP2A5 mRNA levels (Gilmore and Kirby, 2004). In a mouse hepatitis model, high CYP2A5 expression was found to colocalize with superoxide formation, suggesting that superoxide can up-regulate CYP2A5 (Sipowicz et al., 1997). A major advance was the finding that CYP2A5 constitutive expression and induction by heavy metals such as cadmium or lead or mercury were dependent on the redox-sensitive transcription factor Nrf2 in liver (Abu-Bakar et al., 2004, 2007; Lämsä et al., 2010). CYP2A5 was up-regulated by Nrf2 overexpression or deregulated by overexpression of Keap1, an inhibitor of Nrf2 translocation to the nucleus (Lämsä et al., 2010).
We previously found that chronic ethanol feeding to mice resulted in a 2-fold increase in Nrf2 levels in the liver (Gong and Cederbaum, 2006). Ethanol has long been known to elevate CYP2E1 levels (Lieber and DeCarli, 1972; Lieber, 1999); the 2-fold increase by ethanol of Nrf2 levels was associated with a 4-fold increase in CYP2E1 (Gong and Cederbaum, 2006). The potent inducer of CYP2A5, pyrazole, can also elevate CYP2E1 (Song et al., 1986; Yang et al., 1990; Winters and Cederbaum, 1992). Pyrazole treatment of mice or rats elevated Nrf2 levels approximately 3-fold in association with a similar increase in CYP2E1 (Gong and Cederbaum, 2006). CYP2E1 plays an important role in ethanol-induced oxidative stress (Lu and Cederbaum, 2008; Cederbaum et al., 2009). Chronic ethanol treatment can elevate other P450s besides CYP2E1, e.g., CYP2B (Johansson et al., 1988) or CYP3A (Niemelä et al., 1998). Hepatic levels of CYP2E1, CYP2A6, and CYP3A were elevated in patients with alcoholic and nonalcoholic liver diseases (Niemelä et al., 2000). One study in which ethanol was administered in the drinking water (10% ethanol) for 2 weeks reported that ethanol did not elevate CYP2A5 in DBA mice although pyrazole treatment did (Honkakoski et al., 1988). However, ethanol induction of CYP2E1 was modest (Honkakoski et al., 1988), and steatosis is generally not significantly produced by ethanol in drinking water models. In the current study, we evaluated whether chronic ethanol administration in a liquid diet model that produces fatty liver and prominent induction of CYP2E1 can induce CYP2A5. Such a possible elevation would be important in view of the toxicologically significant substrates that CYP2A5 metabolizes, e.g., nicotine, coumarin, aflatoxin B1, and nitrosamines. One goal of the current study was to determine whether chronic ethanol consumption can elevate CYP2A5. The other goal was to evaluate whether ethanol induction of CYP2E1 plays a role in the ethanol induction of CYP2A5.
Materials and Methods
Animals and Ethanol Treatment.
SV/129-background CYP2E1 knockout (Lee et al., 1996) and humanized CYP2E1 transgenic knockin mice (Cheung et al., 2005) were kindly provided by Dr. Frank J. Gonzalez (Laboratory of Metabolism, National Cancer Institute, Bethesda, MD). Colonies were established at Mount Sinai, and the female offspring of these mating pairs were used in this study. The female SV129 wild-type mice were purchased from Charles River Laboratories, Inc. (Wilmington, MA). Depending on the strain, female mice express much higher levels of CYP2A4 than male mice, whereas CYP2A5 is highly expressed in both male and female mice (Honkakoski and Negishi, 1997; Su and Ding, 2004). All mice were housed in temperature-controlled animal facilities with 12-h light/dark cycles and were permitted consumption of tap water and Purina standard chow ad libitum until being fed the liquid diets. The mice received humane care, and experiments were performed according to the criteria outlined in the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, 1996) and with approval of the Mount Sinai Animal Care and Use Committee.
All mice were initially fed the control liquid dextrose diet (Bio-Serv, Frenchtown, NJ) for 3 days to acclimate them to the liquid diet. Afterward, the mice were fed either the liquid ethanol diet (Bio-Serv) or the control liquid dextrose diet, as described by Lieber and DeCarli (1972), for 3 weeks. The content of ethanol was gradually increased every 3 to 4 days from 10% (1.77%, v/v) of total calories to 20% (3.54%, v/v), 25% (4.42%, v/v), 30% (5.31%, v/v), and finally 35% of total calories (6.2%, v/v). For experiments involving 1 or 2 weeks of feeding (time course), the mice were directly subjected to the diet containing ethanol as 35% of total calories. The control mice were pair-fed the control dextrose diet on an iso-energetic basis. The ethanol-fed mice had access to their rations ad libitum, and the conditions of wild-type, knockout, and humanized transgenic mice were comparable. The amount of food consumed by CYP2E1 knockout mice, the knockin mice, and the wild-type mice was approximately the same.
No mice died in any group after 3 weeks of feeding with the control or ethanol-containing diet. Before being sacrificed, the mice were fasted overnight and body weight was measured. Blood was collected, and serum was separated. As described by Lu et al. (2010), the ethanol feeding elevated serum transaminases, hepatic steatosis, and oxidant stress in the CYP2E1 KI mice to a much greater extent than in the CYP2E1 KO mice. Whole liver was removed, and liver weight was measured. Then the liver was rapidly excised into fragments and washed with cold saline, and 1 aliquot of tissue was placed in 10% formalin solution for paraffin blocking. Another aliquot of tissue was placed in RNAlater solution (Ambion, Austin, TX) for RNA extraction. The remaining aliquots were stored at −80°C for further assays. Liver homogenates were prepared in ice-cold 0.15 M KCl and stored at −80°C in aliquots.
Cadmium Induction of CYP2A5.
Cadmium chloride was injected once intraperitoneally at 3 mg/kg b.wt. to WT, CYP2E1 KO, and CYP2E1 KI mice. Before cadmium injection, some WT mice were given injections of saline or vitamin C (Vc) (125 mg/kg i.p., twice per day for 3 days; on day 3 cadmium was injected 30 min after the second Vc injection). Other WT mice were given injections of N-acetyl-cysteine (NAC) (150 mg/kg i.p., once per day for 3 days; on day 3, cadmium was injected 30 min after the NAC injection). Mice were killed 18 h after the cadmium injection.
Immunohistochemical Staining.
Immunohistochemical staining for CYP2E1 was performed by using anti-CYP2E1 antibody (a gift from Dr. Jerome Lasker, Hackensack Biomedical Research Institute, Hackensack, NJ) followed by an IHC Select HRP/DAB kit. Immunohistochemical staining for CYP2A5 was performed by using anti-CYP2A5 (a gift from Dr. Risto Juvonen, Department of Pharmacology and Toxicology, University of Kuopio, Kuopio, Finland) followed by a Broad Spectrum (AEC) Histostain-Plus kit (Invitrogen, Carlsbad, CA). No staining was observed in the absence of the primary antibody, either anti-CYP2E1 IgG or anti-CYP2A5 IgG.
Preparation of Hepatic Microsomes.
Hepatic microsomes were prepared by placing liver aliquots in 0.15 M KCl and homogenized in a Polytron homogenizer for 10 strokes. The homogenate was centrifuged at 9000g for 20 min, and then the resulting supernatant fraction was centrifuged further at 105,000g for 60 min. The resulting pellets (microsomes) were resuspended in 50 mM sodium phosphate buffer (pH 7.4). All procedures were performed under cold conditions.
CYP2E1 and CYP2A5 Activity.
CYP2E1 activity was measured by the rate of oxidation of 1 mM p-nitrophenol to p-nitrocatechol by 100 μg of microsomal protein for 15 min at 37°C (Lu and Cederbaum, 2006). CYP2A5 activity was measured by assessing coumarin 7-hydroxylase activity with 100 μM coumarin as substrate plus 100 μg of microsomal protein and incubation for 15 min at 37°C (Lu and Cederbaum, 2006, 2008).
Western Blotting.
Hepatic proteins in liver homogenates were separated by SDS-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. After 1 h of blocking with 2% fat-free milk, membranes were then incubated overnight with CYP2E1, CYP2A5, and β-actin (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) antibodies followed by a 1-h incubation with peroxidase secondary anti-rabbit, anti-chick, and anti-goat antibodies (Millipore Corporation, Billerica, MA), respectively. Chemiluminescence was detected by Image Reader LAS-4000 software (Fijifilm, Tokyo, Japan) after addition of Pierce EC Western Blotting Substrate (Thermo Fisher Scientific, Waltham, MA). β-Actin was used as a loading control. The bands of proteins were quantified with the Automated Digitizing System (ImageJ gel programs, version 1.34S; National Institutes of Health, Bethesda, MD).
Reverse Transcription and Real-Time Quantitative PCR Assay for the mRNA Levels of CYP2A5 and CYP2A4.
Total RNA from liver tissue was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. One microgram of RNA sample was reverse-transcribed to produce cDNA using random hexamer primers and ImProm-II reverse transcriptase (Promega, Madison, WI) according to the manufacturer's instructions. The primers were designed according to CYP2A5 (NM_007812.4) and CYP2A4 (NM_009997.2) mRNA sequences, which have 98% identity between them. The primers are located in exon 3 (forward) and exon 4 (reverse), respectively. To increase the specificity, the 3′-end base is different between CYP2A5 and CYP2A4 (shown in lower case in the CYP2A4 primer sequences). A real-time quantitative PCR assay for the expression of CYP2A5 with cDNA and LightCycler 480 SYBR Green I Master (Roche Diagnostics GmbH, Mannheim, Germany) was performed using the following primers: CYP2A5 (145 base pairs) forward 5′-TCGGAAGACGAACGGTGCTTTT-3′ (505–526) and reverse 5′-GCTTCCCAGCATCATTCGAAGC-3′ (649–628) and CYP2A4 (147 base pairs) forward 5′-CGGAAGACGAACGGTGCTTTc-3′ (507–527) and reverse 5′-GAgGCTTCCCAGCATCATTCtAAGa-3′ (653–629) in a LightCycler 480 system (Roche Diagnostics GmbH). The protocol for real-time PCR was activation of polymerase at 95°C for 10 min, then at 95°C for 10 s, at 55°C for 20 s, and at 72°C for 20 s for 50 cycles. All real-time PCR products were applied by melting curve analysis. The mRNA levels of CYP2A5 and CYP2A4 were normalized against the glyceraldehyde-3-phosphate dehydrogenase control levels using the comparative Ct (ΔΔCt) method (Livak and Schmittgen, 2001).
Statistics.
Results are expressed as means ± S.E.M. Statistical evaluation was performed by one-way analysis of variance followed by the Student-Newman-Keuls post hoc test. P < 0.05 was considered statistically significant. The number of experiments is indicated in the legends to figures.
Results
Levels of CYP2E1 and CYP2A5 in Wild-Type, CYP2E1 Knockout, and CYP2E1 Knockin Mice.
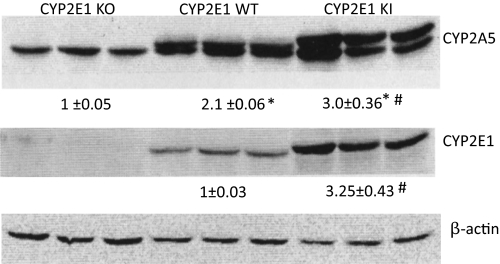
To evaluate biochemical and toxicological actions of CYP2E1, Gonzalez and colleagues (Lee et al., 1996; Cheung et al., 2005) have established CYP2E1 KO mice and CYP2E1 KI mice, in which the human CYP2E1 has been introduced into the CYP2E1 KO mice. The latter allows studies on the effects of human CYP2E1 in vivo in the absence of the mouse CYP2E1 (Cheung et al., 2005). Pyrazole, an inducer of CYP2E1, can also induce CYP2A5 (Juvonen et al., 1985; Nichols and Kirby, 2008). We initiated studies to evaluate whether CYP2E1 and CYP2A5 levels and induction are associated with each other. Figure 1 shows the levels of CYP2E1 in WT mice, CYP2E1 KO mice, and CYP2E1 KI mice. CYP2E1 was absent in the KO mice and present at high levels in the KI mice compared with those in the WT mice. The antibody used to detect CYP2A cannot distinguish between CYP2A4 and CYP2A5, both of which are present in female mice, and immunoblots detected two bands in livers of female wild-type control mice and CYP2E1 KI mice (Fig. 1). The upper band was barely detectable in livers from control male mice but was present in livers from female control mice (data not shown). The bottom band was present in high levels in livers of both male and female mice (data not shown). Because CYP2A5 is present in livers from male and female mice whereas CYP2A4 is very low or barely detectable in livers from male mice, the top band in the immunoblots for CYP2A is likely to be CYP2A4 and the bottom band is likely to be CYP2A5. Basal levels of CYP2A5 followed a pattern similar to that of the basal levels of CYP2E1: lowest in the CYP2E1 KO mice, intermediate in the WT mice, and highest in the CYP2E1 KI mice (Fig. 1). CYP2A5 was present in the absence of CYP2E1, but the highest levels of CYP2A5 expression occur when CYP2E1 is highly expressed. The top band in the immunoblots, presumably CYP2A4, was also present at the highest levels in the CYP2E1 KI mice and barely detectable in the CYP2E1 KO mice (Fig. 1).
Fig. 1.
Western blot analysis for the levels of CYP2A5 and CYP 2E1 in liver homogenates from untreated WT, CYP2E1 KO, and CYP2E1 KI mice. Results from three mice in each group are shown. Numbers under the blots refer to the CYP2A5 (lower bands)/β-actin or CYP2E1/β-actin ratio. *, P < 0.05 compared with the CYP2E1 KO group; #, P < 0.05 compared with the CYP2E1 WT group.
Induction of CYP2A5 by Chronic Ethanol Administration.
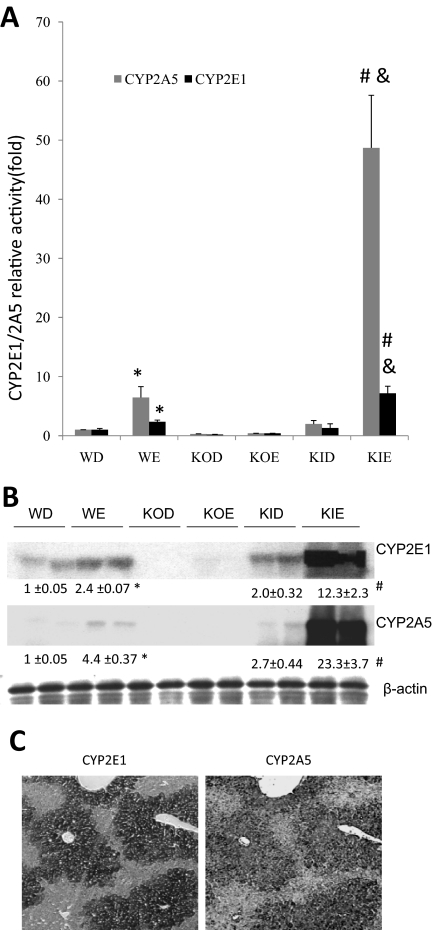
Because chronic ethanol feeding elevates CYP2E1, we evaluated whether ethanol also induces CYP2A5 and if it does, whether there was an association between induction of CYP2E1 and CYP2A5. Figure 2B shows that after 3 weeks of ethanol administration, levels of CYP2E1 were increased 2.4-fold by ethanol in WT mice compared with those in the dextrose-fed controls. Levels of CYP2A5 were also increased by ethanol in the WT mice (4.4-fold). However, ethanol did not significantly elevate CYP2A5 in the CYP2E1 KO mice (Fig. 2B). Oxidation of p-nitrophenol (PNP) and 7-hydroxylation of coumarin were assayed as reflections of CYP2E1 and CYP2A5 catalytic activities, respectively. The chronic ethanol treatment elevated PNP oxidation by approximately 3-fold in the WT mice: PNP oxidation was very low and was not elevated by ethanol in the CYP2E1 KO mice (Fig. 2A). Coumarin 7-hydroxylase activity was increased approximately 6-fold by ethanol in the WT mice, but no significant increase was observed in the CYP2E1 KO mice (Fig. 2A).
Fig. 2.
Chronic ethanol administration elevates CYP2A5 levels in SV/129 WT mice and CYP2E1 KI mice but not in CYP2E1 KO mice. WT, KO, and KI mice (n = 4 in each group) were fed the dextrose or ethanol diets for 3 weeks. A, microsomal oxidation of PNP or coumarin. B, Western blots to assay levels of CYP2E1 and CYP2A5 in liver homogenates. C, immunohistochemical staining of liver slices with anti-CYP2E1 or anti-CYP2A5 IgG to detect CYP2E1 or CYP2A5 in intact liver. *, P < 0.05 compared with WD group; #, P < 0.05 compared with KID group; &, P < 0.05 compared with WE group. WD, wild-type dextrose control; WE, wild-type mice fed ethanol; KOD, knockout mice fed dextrose; KOE, knockout mice fed ethanol; KID, knockin mice fed dextrose; KIE, knockin mice fed ethanol.
Ethanol Induction of CYP2A5 Is Restored in CYP2E1 KI Mice.
If the poor ability of ethanol to induce CYP2A5 in CYP2E1 KO mice is due to the absence of CYP2E1, restoring CYP2E1 should restore induction of CYP2A5 by ethanol. CYP2E1 KI mice were fed ethanol or dextrose for 3 weeks. Levels of CYP2E1 protein and activity were elevated in the CYP2E1 KI mice fed ethanol compared with those in the dextrose controls (Fig. 2, A and B) and were even higher than the levels and activity of CYP2E1 in the ethanol-fed WT mice. Levels of CYP2A5 and activity of CYP2A5 were elevated by ethanol in the CYP2E1 KI mice compared with those in the dextrose controls and, similar to CYP2E1, were even higher than the levels and activity of CYP2A5 in the ethanol-fed WT mice (Fig. 2, A and B). Thus, levels and activity of CYP2A5 after ethanol treatment mimic levels and activity of CYP2E1, both being low in CYP2E1 KO mice and both being high in CYP2E1 KI mice. Immunohistochemical staining showed that CYP2E1 and CYP2A5 colocalize to the same zones in the livers of the ethanol-fed KI mice such that areas of the liver where CYP2E1 is highly expressed are the areas where CYP2A5 is highly expressed (Fig. 2C).
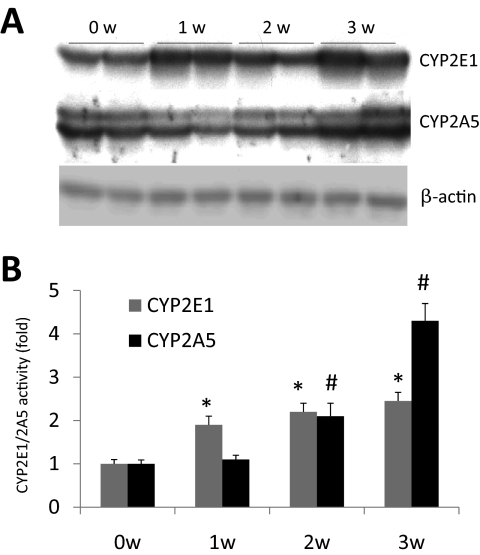
Time Course for Induction of CYP2E1 and CYP2A5 by Ethanol.
Because ethanol elevated both CYP2E1 and CYP2A5 and the ethanol elevation of CYP2A5 was blunted in the absence of CYP2E1 (CYP2E1 KO mice), we compared the time course for induction of the two P450s by ethanol. Immunoblot analysis showed that CYP2E1 was elevated after 1 week of ethanol feeding and then slowly increased further after 2 and 3 weeks of ethanol feeding (Fig. 3A). Likewise, CYP2E1 catalytic activity was also elevated after 1 week of ethanol feeding and slightly increased further at 2 and 3 weeks of ethanol feeding (Fig. 3B). Levels and activity of CYP2A5 were not increased after 1 week of ethanol feeding but began to increase after 2 and especially after 3 weeks of ethanol feeding (Fig. 3, A and B). Thus, ethanol induction of CYP2E1 occurs before ethanol induction of CYP2A5.
Fig. 3.
Time course for the induction of CYP2E1 and CYP2A5 by ethanol. SV/129 WT mice (n = 4 in each group) were fed the ethanol liquid diet for 0, 1, 2, or 3 weeks (w). A, Western blot analysis for levels of CYP2E1 and CYP2A5 protein. B, PNP oxidation as a reflection of CYP2E1 activity and coumarin 7-hydroxlase activity as a reflection of CYP2A5 activity. Results from mice before initiation of the ethanol feeding (0 week) are also shown. *, P < 0.05 compared with the 0-week CYP2E1; #, P < 0.05, compared with the 0-week CYP2A5.
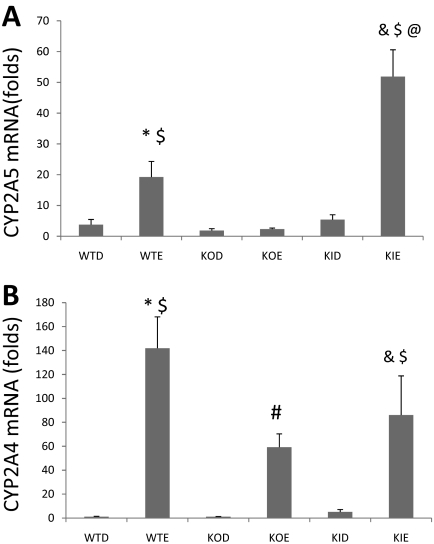
Ethanol Elevates CYP2A5 (and CYP2A4) mRNA Levels.
The above studies show that chronic ethanol consumption elevates CYP2A5 protein levels and activity in WT mice and CYP2E1 KI mice but not in CYP2E1 KO mice. We evaluated whether such increases in CYP2A5 by ethanol are associated with increases in CYP2A5 mRNA. Indeed, the CYP2A5 mRNA was elevated by ethanol in WT mice and CYP2E1 KI mice but not in CYP2E1 KO mice (Fig. 4B). The increase in CYP2A5 mRNA was highest in the CYP2E1 KI mice fed ethanol, analogous to the high protein levels and activity of CYP2A5 in these mice. Ethanol did not elevate levels of CYP2E1 mRNA in the WT or the CYP2E1 KI mice (data not shown) confirming that ethanol induction of CYP2E1 is largely post-transcriptional (Song et al., 1986). Ethanol increased CYP2A4 mRNA levels in all three mouse genotypes (Fig. 4A). The increase in CYP2A4 mRNA in the CYP2E1 KO mice by ethanol does indicate that these mice are responsive to induction of at least some members of the CYP2A family, e.g., CYP2A4 but not CYP2A5.
Fig. 4.
Ethanol elevates CYP2A4 (A) and CYP2A5 mRNA (B) levels. WT, KO, and KI mice (n = 4 in each group) were fed the dextrose control or the ethanol liquid diet for 3 weeks. Isolated RNA was reverse-transcribed to produce cDNA, and a real-time quantitative PCR assay for the expression of CYP2A5 and CYP2A4 mRNA was performed as described under Materials and Methods. *, P < 0.05 compared with the WTD group; #, P < 0.05 compared with the KOD group; P < 0.05 compared with the KID group; $, P < 0.05 compared with the KOE group; @, P < 0.05 compared with the WTE group. WTD, wild-type dextrose control; WTE, wild-type mice fed ethanol; KOD, knockout mice fed dextrose; KOE, knockout mice fed ethanol; KID, knockin mice fed dextrose; KIE, knockin mice fed ethanol.
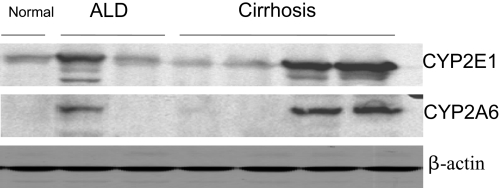
Comparison of CYP2E1 and CYP2A6 Levels in Human Liver.
Samples of human liver (no information on age, gender, medications, and possible polymorphisms was available) were obtained from the Department of Pathology at Mount Sinai Medical Center, and levels of CYP2E1 and CYP2A6 (the human ortholog of the mouse CYP2A5) were determined. The CYP2A5 antibody recognizes CYP2A6. Levels of CYP2E1 varied widely, being elevated in one of two patients with alcoholic liver disease and in two of four patients with cirrhosis (Fig. 5). Note that the levels of CYP2A6 varied widely as well but were reflective of the levels of CYP2E1 in that CYP2A6 was high in the one patient with alcoholic liver disease who displayed high CYP2E1 and in the two patients with cirrhosis who displayed elevated CYP2E1 but were low in those livers that displayed low CYP2E1 (Fig. 5).
Fig. 5.
Levels of CYP2E1 and CYP2A6 (the human ortholog of mouse CYP2A5) in human livers. Western blots were performed on liver extracts from two patients with alcoholic liver disease (ALD), from four patients with liver cirrhosis, and from a control healthy liver. Diagnoses and liver samples were provided by the Department of Surgical Pathology at Mount Sinai.
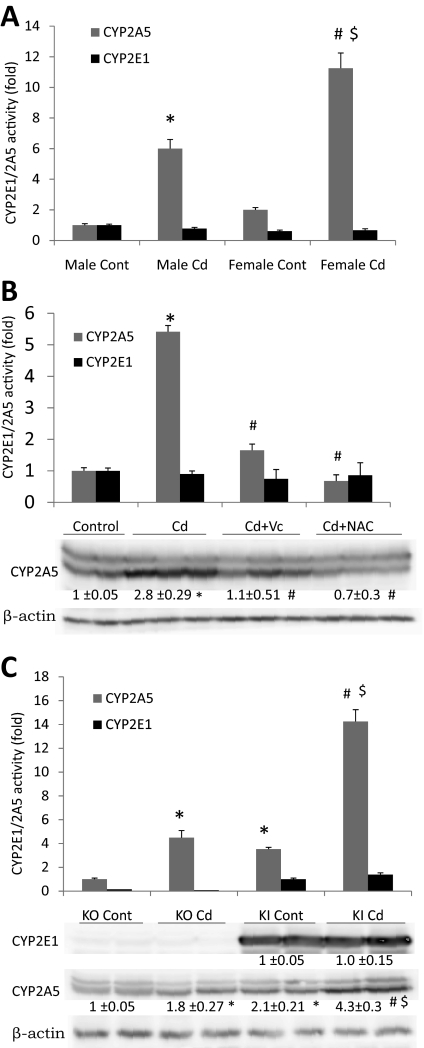
Cadmium Induction of CYP2A5.
The above results suggest that ethanol induction of CYP2A5 is largely CYP2E1-dependent, because ethanol also induces CYP2E1. We evaluated whether other CYP2A5 inducers that do not induce CYP2E1 also induce CYP2A5 in a CYP2E1-dependent way. CYP2A5 can be induced by heavy metals; induction of CYP2A5 by cadmium was shown to be dependent on Nrf2 in liver (Abu-Bakar et al., 2004, 2007; Lämsä et al., 2010). The ability of cadmium to induce CYP2A5 was evaluated in WT, CYP2E1 KO, and CYP2E1 KI mice. Figure 6A verifies that cadmium elevated CYP2A5 catalytic activity in SV129 male and female WT mice, under conditions in which it had no effect on CYP2E1 catalytic activity (Fig. 6A). To evaluate whether ROS are important for the elevated induction of CYP2A5 found in the presence of cadmium plus a basal level of CYP2E1, the effect of two antioxidants on the induction of CYP2A5 by cadmium in WT mice was determined. Results in Fig. 6B shows that administration of vitamin C or N-acetylcysteine to cadmium-treated WT mice blunted the elevation of CYP2A5 activity and content. The two antioxidants had no effect on the activity of CYP2E1 (Fig. 6B).
Fig. 6.
Induction of CYP2A5 by cadmium in WT, CYP2E1 KO, and CYP2E1 KI mice. Mice were given intraperitoneal injections of cadmium chloride (3 mg/kg) or saline as control, and 18 h later, mice were sacrificed. PNP oxidation as a reflection of CYP2E1 activity and coumarin 7-hydroxlase activity as a reflection of CYP2A5 activity were determined or levels of CYP2E1 or CYP2A5 were determined by immunoblots. A, CYP2A5 but not CYP2E1was induced by cadmium in female and male WT mice (n = 3 in each group). *, P < 0.05 compared with the male control group; #, P < 0.05 compared with the female control group; $, P < 0.05 compared with the male cadmium group. B, Vc and NAC blocked cadmium induction of CYP2A5 in WT mice. WT mice were pretreated with saline (n = 3), Vc (n = 3), or NAC (n = 3) before cadmium treatment as described under Materials and Methods. Controls received saline intraperitoneally (n = 4). Catalytic activities and Western blots are shown. *, P < 0.05 compared with the control group; #, P < 0.05 compared with the cadmium group. C, induction of CYP2A5 by cadmium in female CYP2E1 KO and CYP2E1 KI mice (n = 4 in each group). Treatments are identical to those for the female WT mice in A. Catalytic activities and Western blots are shown. *, P < 0.05 compared with the KO control group; #, P < 0.05 compared with the KI control group; $, P < 0.05 compared with the KO cadmium group. Cont, control; Cd, cadmium.
Unlike ethanol, cadmium treatment increased CYP2A5 catalytic activity in CYP2E1 KO mice (Fig. 6C). The cadmium-induced CYP2A5 catalytic activity was much higher in CYP2E1 KI mice compared with CYP2E1 KO mice, but considering the difference in basal levels of CYP2A5 in the KO and KI mice, the extent of increase in CYP2A5 activity by cadmium was similar (4.5-fold in KO versus 4.1-fold in KI) (Fig. 6C). Immunoblot analyses showed that cadmium increased the level of CYP2A5 in both CYP2E1 KI mice and to a less extent in CYP2E1 KO mice. The treatment with cadmium had no effect on CYP2E1 levels in the KI mice (Fig. 6C). Thus, unlike the chronic ethanol treatment, cadmium elevated CYP2A5 under conditions in which it did not alter CYP2E1 activity or content, and cadmium could induce CYP2A5 in the CYP2E1 KO mice.
Discussion
Although much of the focus on ethanol induction of P450s has been on CYP2E1, chronic ethanol consumption can elevate levels of other P450s such as CYP2B and CYP3A (Johansson et al., 1988; Niemelä et al., 1998). Levels of CYP2E1, CYP3A, and CYP2A6 were elevated in livers of patients with alcoholic and nonalcoholic liver diseases (Niemelä et al., 2000). Induction of CYP2A5 has been suggested to occur in response to liver injury (Su and Ding, 2004). Results in the current report show that feeding mice an ethanol liquid diet results in an increase in CYP2A5 levels and catalytic activity compared with those in pair-fed dextrose controls. Under these conditions, the ethanol feeding produces fatty liver and an increase in triglyceride levels, but liver injury is minimal; e.g., transaminase levels are increased 2- to 3-fold and no necroinflammation is observed (Lu et al., 2008b). Ethanol elevated CYP2A5 mRNA levels, which probably explains the elevated CYP2A5 protein and activity; however, possible post-transcriptional mechanisms contributing to the elevation of CYP2A5 remain to be evaluated. Ethanol induction of CYP2E1 is mainly post-transcriptional, reflecting stabilization of CYP2E1 against proteosome-mediated degradation (Song et al., 1986; Roberts, 1997; Gonzalez, 2007), although increases in CYP2E1 mRNA can occur at very high blood ethanol levels (Ronis et al., 1993). Ethanol is a ligand and substrate for CYP2E1, which explains its ability to stabilize and increase the half-life of CYP2E1 (Eliasson et al., 1988; Lieber, 1999). Whether ethanol is a ligand and/or substrate for CYP2A5 is currently under investigation.
Whether the increase in CYP2A5 mRNA by ethanol reflects transcriptional activation of the CYP2A5 gene or stabilization of the CYP2A5 mRNA remains to be evaluated. Both mechanisms are possible; e.g., pyrazole increases CYP2A5 by a post-transcriptional mechanism involving binding of heterogenous nuclear ribonucleic protein A1 to the 3′-untranslated region of CYP2A5 mRNA (Glisovic et al., 2003). The oxidant-sensitive transcription factor Nrf2 has been shown to regulate the CYP2A5 gene (Abu-Bakar et al., 2004, 2007; Lämsä et al., 2010). Two putative stress response elements were localized to positions −2514 to −2502 and −2386 to −2377 of the CYP2A5 promoter with the more proximal sequence specifically binding Nrf2 (Abu-Bakar et al., 2007). Pyrazole and heavy metals were ineffective in inducing CYP2A5 in Nrf2 knockout mice (Lu et al., 2008a; Lämsä et al., 2010). Chronic ethanol consumption increases oxidative stress in the liver and elevates Nrf2 mRNA (Gong and Cederbaum, 2006), which may contribute to the ethanol elevation of CYP2A5 mRNA levels. In the future, assays of nuclear run-on experiments, mRNA stability, and chromatin immunoprecipitation analyses of Nrf2 binding to the CYP2A5 promoter and investigation into whether ethanol induces CYP2A5 in Nrf2 knockout mice would help determine mechanisms by which ethanol elevates CYP2A5 mRNA levels.
We believe the most novel information in this study is the close association between levels of CYP2E1 and CYP2A5 and the fact that the induction of CYP2A5 by ethanol seems to be potentiated by the induction of CYP2E1. Ethanol induction of CYP2A5 was blunted in CYP2E1 KO mice but restored in CYP2E1 KI mice. Immunohistochemical staining showed that the distribution of the elevated CYP2E1 and CYP2A5 in the liver produced by the ethanol feeding overlapped. Time course experiments showed that the elevation of CYP2E1 by ethanol occurs before the elevation of CYP2A5, perhaps suggesting that elevation of CYP2E1 by ethanol is important for the subsequent induction of CYP2A5. In patients with alcoholic liver disease or cirrhosis of the liver, those samples showing high levels of CYP2E1 also had high levels of CYP2A6, and patients with low levels of CYP2E1 had low levels of CYP2A6. Nrf2 is increased in livers from mice and rats treated with pyrazole and in HepG2 hepatoma cells overexpressing CYP2E1 (Gong and Cederbaum, 2006). These increases may be due to increases in the production of ROS by the elevated levels of CYP2E1. Chronic ethanol feeding elevated production of thiobarbituric acid-reactive substances, an assay for lipid peroxidation, and lowered GSH levels in the liver of the WT and the CYP2E1 KI mice but not the CYP2E1 KO mice (Lu et al., 2010). Thus, ethanol increased oxidative stress in the WT and the KI mice but not in the KO mice. We speculate that ethanol induction of CYP2E1 followed by increases in production of ROS and then activation and translocation of Nrf2 to the nucleus are important steps in the mechanism by which ethanol induces CYP2A5. We are currently studying the effect of antioxidants such as N-acetylcysteine, ascorbate, and α-tocopherol on the ethanol induction of CYP2A5, as was done with cadmium induction of CYP2A5 (Fig. 6B).
In view of data in the literature that Nrf2 is an important transcription factor for induction of CYP2A5, the role of CYP2E1 in the mechanism by which chronic ethanol induces CYP2A5 is hypothesized to be due to induction of CYP2E1 and enhanced ROS production, followed by activation of Nrf2. More surprising is the role of CYP2E1 in contributing to cadmium induction of CYP2A5 under conditions in which cadmium, unlike ethanol, does not elevate CYP2E1. The decline in cadmium induction of CYP2A5 by vitamin C and N-acetylcysteine suggests that ROS play a role in the induction mechanism. We speculate that both cadmium and CYP2E1 separately produce ROS; however, the levels of ROS are insufficient for maximal, efficient activation of CYP2A5. Thus, WT mice with high levels of CYP2E1 but without cadmium or CYP2E1 KO mice without CYP2E1 but with cadmium do not generate sufficient ROS for maximal CYP2A5 induction. However, the combination of cadmium plus CYP2E1 in WT or CYP2E1 KI mice results in sufficient ROS to induce CYP2A5. Hence, basal levels of CYP2E1 in combination with cadmium are sufficient to induce CYP2A5. In the case of chronic ethanol induction, the ethanol elevation of CYP2E1 in the WT or KI mice is necessary to generate sufficient ROS in the absence of cadmium to activate CYP2A5; i.e., basal levels of CYP2E1 are not sufficient. Whether CYP2E1 plays a role in induction of CYP2A5 by other inducers, e.g., phenobarbital, is not known.
Although this study has focused on CYP2A5, chronic ethanol treatment also elevated levels of CYP2A4 mRNA. This increase appears to be independent of CYP2E1 because the increase occurs in the CYP2E1 KO mice fed ethanol, and the increase was not higher in the CYP2E1 KI mice with levels of CYP2E1 higher than those of the WT mice. The increase in CYP2A4 does indicate that the CYP2E1 KO mice are responsive to inducers of certain CYP2A family members by ethanol. The mechanism for this CYP2E1-independent ethanol induction of CYP2A4 is currently unknown.
Ethanol induction of CYP2A5 may be of toxicological significance in view of the ability of CYP2A5 to metabolize hepatocarcinogens (Su and Ding, 2004) and drugs such as nicotine (Yamazaki et al.,1999; Benowitz et al., 2006; Zhou et al., 2010). The latter is of special interest because most alcoholics are also smokers (Wall et al., 2007). The possible contribution of CYP2A5 to the metabolic and toxicological effects of ethanol, if any, would be important to evaluate; e.g., does CYP2A5, in addition to CYP2E1, contribute to the increase in ROS production by chronic ethanol consumption or to ethanol-induced fatty liver? Possible approaches to evaluate this question are to study the effect of CYP2A5 inhibitors such as 8-methoxysalen, analogous to the use of the CYP2E1 inhibitor chlormethiazole, on ethanol-induced oxidative stress, steatosis, or hepatotoxicity. CYP2A5 knockout mice have recently been developed and used to show that CYP2A5 is the major nicotine and cotinine oxidase in mouse liver (Zhou et al., 2010). It would be interesting to feed ethanol to CYP2A5 KO mice and assay for parameters such as induction of oxidant stress, liver injury, and steatosis. Is CYP2E1 induced by ethanol in CYP2A5 KO mice? Induction of CYP2A5 by ethanol was decreased in the CYP2E1 KO mice, suggesting interaction between these two P450s. Although CYP2E1 has been implicated in some of the toxicological and metabolic effects of ethanol, the possibility that induction of CYP2E1 is permissive for the induction of CYP2A5 may reflect a new contribution of CYP2E1 in the actions of ethanol.
This work was supported by the National Institutes of Health National Institute on Alcohol Abuse and Alcoholism [Grants R01-AA017425, AA018790, P20-AA017067].
Article, publication date, and citation information can be found at http://dmd.aspetjournals.org.
doi:10.1124/dmd.110.035691.
- KO
- knockout
- KI
- knockin
- WT
- wild-type
- Vc
- vitamin C
- NAC
- N-acetyl-cysteine
- PCR
- polymerase chain reaction
- PNP
- p-nitrophenol
- ROS
- reactive oxygen species.
Authorship Contributions
Participated in research design: Lu, Zhuge, and Cederbaum.
Conducted experiments: Lu, Zhuge, and Wu.
Contributed new reagents or analytic tools: Lu, Zhuge, and Wu.
Performed data analysis: Lu, Zhuge, and Cederbaum.
Wrote or contributed to the writing of the manuscript: Lu, Zhuge, and Cederbaum.
References
- Abu-Bakar A, Satarug S, Marks GC, Lang MA, Moore MR. (2004) Acute cadmium chloride administration induces hepatic and renal CYP2A5 mRNA, protein and activity in the mouse: involvement of transcription factor NRF2. Toxicol Lett 148:199–210 [DOI] [PubMed] [Google Scholar]
- Abu-Bakar A, Lämsä V, Arpiainen S, Moore MR, Lang MA, Hakkola J. (2007) Regulation of CYP2A5 gene by the transcription factor nuclear factor (erythroid-derived 2)-like 2. Drug Metab Dispos 35:787–794 [DOI] [PubMed] [Google Scholar]
- Benowitz NL, Swan GE, Jacob P, 3rd, Lessov-Schlaggar CN, Tyndale RF. (2006) CYP2A6 genotype and the metabolism and disposition kinetics of nicotine. Clin Pharmacol Ther 80:457–467 [DOI] [PubMed] [Google Scholar]
- Cederbaum AI, Lu Y, Wu D. (2009) Role of oxidative stress in alcohol-induced liver injury. Arch Toxicol 83:519–548 [DOI] [PubMed] [Google Scholar]
- Cheung C, Yu AM, Ward JM, Krausz KW, Akiyama TE, Feigenbaum L, Gonzalez FJ. (2005) The CYP2E1-humanized transgenic mouse: role of CYP2E1 in acetaminophen hepatotoxicity. Drug Metab Dispos 33:449–457 [DOI] [PubMed] [Google Scholar]
- De-Oliveira AC, Da-Matta AC, Paumgartten FJ. (2006) Plasmodium berghei (ANKA): infection induces CYP2A5 and 2E1 while depressing other CYP isoforms in the mouse liver. Exp Parasitol 113:256–261 [DOI] [PubMed] [Google Scholar]
- Eliasson E, Johansson I, Ingelman-Sundberg M. (1988) Ligand-dependent maintenance of ethanol-inducible cytochrome P-450 in primary rat hepatocyte cell cultures. Biochem Biophys Res Commun 150:436–443 [DOI] [PubMed] [Google Scholar]
- Gilmore WJ, Kirby GM. (2004) Endoplasmic reticulum stress due to altered cellular redox status positively regulates murine hepatic CYP2A5 expression. J Pharmacol Exp Ther 308:600–608 [DOI] [PubMed] [Google Scholar]
- Glisovic T, Ben-David Y, Lang MA, Raffalli-Mathieu F. (2003) Interplay between hnRNP A1 and a cis-acting element in the 3′ UTR of CYP2A5 mRNA is central for high expression of the gene. FEBS Lett 535:147–152 [DOI] [PubMed] [Google Scholar]
- Goldberg L, Rydberg U. (1969) Inhibition of ethanol metabolism in vivo by administration of pyrazole. Biochem Pharmacol 18:1749–1762 [DOI] [PubMed] [Google Scholar]
- Gong P, Cederbaum AI. (2006) Nrf2 is increased by CYP2E1 in rodent liver and HepG2 cells and protects against oxidative stress caused by CYP2E1. Hepatology 43:144–153 [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ. (2007) The 2006 Bernard B. Brodie Award Lecture. CYP2e1. Drug Metab Dispos 35:1–8 [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Autio S, Juvonen R, Raunio H, Gelboin HV, Park SS, Pelkonen O, Lang MA. (1988) Pyrazole is different from acetone and ethanol as an inducer of the polysubstrate monooxygenase system in mice: evidence that pyrazole-inducible P450Coh is distinct from acetone-inducible P450ac. Arch Biochem Biophys 267:589–598 [DOI] [PubMed] [Google Scholar]
- Honkakoski P, Negishi M. (1997) The structure, function, and regulation of cytochrome P450 2A enzymes. Drug Metab Rev 29:977–996 [DOI] [PubMed] [Google Scholar]
- Institute of Laboratory Animal Resources (1996) Guide for the Care and Use of Laboratory Animals, 7th ed, Institute of Laboratory Animal Resources, Commission on Life Sciences, National Research Council, Washington, DC [Google Scholar]
- Johansson I, Ekström G, Scholte B, Puzycki D, Jörnvall H, Ingelman-Sundberg M. (1988) Ethanol-, fasting-, and acetone-inducible cytochromes P-450 in rat liver: regulation and characteristics of enzymes belonging to the IIB and IIE gene subfamilies. Biochemistry 27:1925–1934 [DOI] [PubMed] [Google Scholar]
- Jounaïdi Y, Bonfils C, Périn F, Negishi M, Lange R. (1994) Overexpression of a cytochrome P-450 of the 2a family (CYP2a-5) in chemically induced hepatomas from female mice. Eur J Biochem 219:791–798 [DOI] [PubMed] [Google Scholar]
- Juvonen RO, Kaipainen PK, Lang MA. (1985) Selective induction of coumarin 7-hydroxylase by pyrazole in D2 mice. Eur J Biochem 152:3–8 [DOI] [PubMed] [Google Scholar]
- Lämsä V, Levonen AL, Leinonen H, Ylä-Herttuala S, Yamamoto M, Hakkola J. (2010) Cytochrome P450 2A5 constitutive expression and induction by heavy metals is dependent on redox-sensitive transcription factor Nrf2 in liver. Chem Res Toxicol 23:977–985 [DOI] [PubMed] [Google Scholar]
- Lee SS, Buters JT, Pineau T, Fernandez-Salguero P, Gonzalez FJ. (1996) Role of CYP2E1 in the hepatotoxicity of acetaminophen. J Biol Chem 271:12063–12067 [DOI] [PubMed] [Google Scholar]
- Lieber CS. (1999) Microsomal ethanol-oxidizing system (MEOS): the first 30 years (1968–1998)—a review. Alcohol Clin Exp Res 23:991–1007 [PubMed] [Google Scholar]
- Lieber CS, DeCarli LM. (1972) The role of the hepatic microsomal ethanol oxidizing system (MEOS) for ethanol metabolism in vivo. J Pharmacol Exp Ther 181:279–287 [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Lu Y, Cederbaum AI. (2006) Enhancement by pyrazole of lipopolysaccharide-induced liver injury in mice: role of cytochrome P450 2E1 and 2A5. Hepatology 44:263–274 [DOI] [PubMed] [Google Scholar]
- Lu Y, Cederbaum AI. (2008) CYP2E1 and oxidative liver injury by alcohol. Free Radic Biol Med 44:723–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Gong P, Cederbaum AI. (2008a) Pyrazole induced oxidative liver injury independent of CYP2E1/2A5 induction due to Nrf2 deficiency. Toxicology 252:9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wu D, Wang X, Ward SC, Cederbaum AI. (2010) Chronic alcohol-induced liver injury and oxidant stress are decreased in cytochrome P4502E1 knockout mice and restored in humanized cytochrome P4502E1 knock-in mice. Free Radic Biol Med 49:1406–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Zhuge J, Wang X, Bai J, Cederbaum AI. (2008b) Cytochrome P450 2E1 contributes to ethanol-induced fatty liver in mice. Hepatology 47:1483–1494 [DOI] [PubMed] [Google Scholar]
- Nichols KD, Kirby GM. (2008) Microarray analysis of hepatic gene expression in pyrazole-mediated hepatotoxicity: identification of potential stimuli of Cyp2a5 induction. Biochem Pharmacol 75:538–551 [DOI] [PubMed] [Google Scholar]
- Niemelä O, Parkkila S, Juvonen RO, Viitala K, Gelboin HV, Pasanen M. (2000) Cytochromes P450 2A6, 2E1, and 3A and production of protein-aldehyde adducts in the liver of patients with alcoholic and non-alcoholic liver diseases. J Hepatol 33:893–901 [DOI] [PubMed] [Google Scholar]
- Niemelä O, Parkkila S, Pasanen M, Iimuro Y, Bradford B, Thurman RG. (1998) Early alcoholic liver injury: formation of protein adducts with acetaldehyde and lipid peroxidation products, and expression of CYP2E1 and CYP3A. Alcohol Clin Exp Res 22:2118–2124 [DOI] [PubMed] [Google Scholar]
- Roberts BJ. (1997) Evidence of proteosome-mediated cytochrome P-450 degradation. J Biol Chem 272:9771–9778 [DOI] [PubMed] [Google Scholar]
- Ronis MJ, Huang J, Crouch J, Mercado C, Irby D, Valentine CR, Lumpkin CK, Ingelman-Sundberg M, Badger TM. (1993) Cytochrome P450 CYP 2E1 induction during chronic alcohol exposure occurs by a two-step mechanism associated with blood alcohol concentrations in rats. J Pharmacol Exp Ther 264:944–950 [PubMed] [Google Scholar]
- Sipowicz MA, Chomarat P, Diwan BA, Anver MA, Awasthi YC, Ward JM, Rice JM, Kasprzak KS, Wild CP, Anderson LM. (1997) Increased oxidative DNA damage and hepatocyte overexpression of specific cytochrome P450 isoforms in hepatitis of mice infected with Helicobacter hepaticus. Am J Pathol 151:933–941 [PMC free article] [PubMed] [Google Scholar]
- Song BJ, Gelboin HV, Park SS, Yang CS, Gonzalez FJ. (1986) Complementary DNA and protein sequences of ethanol-inducible rat and human cytochrome P450s. Transcriptional and post-transcriptional regulation of the rat enzyme. J Biol Chem 261:16689–16697 [PubMed] [Google Scholar]
- Su T, Ding X. (2004) Regulation of the cytochrome P450 2A genes. Toxicol Appl Pharmacol 199:285–294 [DOI] [PubMed] [Google Scholar]
- Wall TL, Schoedel K, Ring HZ, Luczak SE, Katsuyoshi DM, Tyndale RF. (2007) Differences in pharmacogenetics of nicotine and alcohol metabolism: review and recommendations for future research. Nicotine Tobacco Res 9 (Suppl 3):S459–S474 [DOI] [PubMed] [Google Scholar]
- Winters DK, Cederbaum AI. (1992) Time course characterization of the induction of cytochrome P-450 2E1 by pyrazole and 4-methylpyrazole. Biochim Biophys Acta 117:15–24 [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Inoue K, Hashimoto M, Shimada T. (1999) Roles of CYP2A6 and CYP2B6 in nicotine C-oxidation by human liver microsomes. Arch Toxicol 73:65–70 [DOI] [PubMed] [Google Scholar]
- Yang CS, Yoo JS, Ishizaki H, Hong JY. (1990) Cytochrome P450IIE1: roles in nitrosamine metabolism and mechanisms of regulation. Drug Metab Rev 22:147–159 [DOI] [PubMed] [Google Scholar]
- Zhou X, Zhuo X, Xie F, Kluetzman K, Shu YZ, Humphreys WG, Ding X. (2010) Role of CYP2A5 in the clearance of nicotine and cotinine: insights from studies on a Cyp2a5-null mouse model. J Pharmacol Exp Ther 332:578–587 [DOI] [PMC free article] [PubMed] [Google Scholar]