Abstract
Elucidating the mechanism of liver regeneration could lead to life-saving therapy for a large number of patients, especially the elderly, after segmental liver transplantation or resection of liver tumors. The forkhead box m1b (Foxm1b) transcription factor is required for normal liver regeneration and overexpression of Foxm1b restores the regenerative capacity of aging livers. Here, we demonstrated that Foxm1b is a direct farnesoid X receptor (FXR) target gene involved in cell cycle regulation. An inverted repeat (IR-0) of FXR response element (FXRE), acting as an enhancer in intron 3 of the Foxm1b gene was identified by a combination of transcriptional reporter assays, electrophoretic mobility shift and chromatin immunoprecipitation assays. Interestingly, FXR binding to the IR-0 element was dramatically diminished in aging regenerating livers, resulting in defective induction of Foxm1b, which contributes to defective liver regeneration. However, FXR activation by a novel ligand in aging livers induced Foxm1b expression and restored hepatocyte DNA replication to 70–80% of levels found in young regenerating livers, which was significantly suppressed by hepatic expression of an anti-Foxm1b short hairpin RNA. Our results highlight FXR as a potential drug target to induce Foxm1b expression and promote aging liver regeneration.
Introduction
Segmental liver transplantation from living donors has emerged as an alternative strategy to counteract the lack of available organs for transplant into the increasing number of patients with end-stage liver disease 1, 2. In addition, over the past decades, liver resection has become a common treatment for liver tumors in many patients. However, after surgery, the remnant livers in about 20% of patients fail to regenerate because of insufficient liver mass 3–6. For elderly patients in particular, the aging liver has a dramatically reduced ability to proliferate normally after partial hepatectomy (PH), resulting in greater mortality 7, 8. This reduced proliferative response has been mainly attributed to decreased expression of Foxm1b transcription factor 9–11 and to the failure of aging liver to diminish the age-specific C/EBPα-Brm complex after PH 12, 13.
Forkhead box m1b (Foxm1b; previously known as HFH-11B, Trident, WIN, MPP2) is a proliferation-specific member of the forkhead box family of transcription factors. It regulates chromosome segregation and hepatocyte proliferation through upregulating the expression of cell cycle genes that stimulate cyclin-dependent kinase 2 (Cdk2) and Cdk1 activity, which is essential for hepatocyte entry into mitosis during liver development, regeneration, and liver cancer 14–16. Interestingly, it has been demonstrated that elevated hepatic expression of Foxm1b is sufficient to restore the regenerative potential of aging liver10, 11, 17. Therefore, identifying the regulatory mechanisms that underlie Foxm1b transcriptional activation is crucial for further elucidating the liver regenerative process and the development of novel approaches to improve liver regeneration.
The farnesoid X receptor (FXR, NR1H4) is also involved in liver regeneration. FXR, a central metabolic regulator in bile acid homeostasis as well as lipid and glucose metabolism, is a member of the nuclear hormone receptor superfamily of ligand-activated transcription factors 18, 19. Many target genes of FXR have been identified, and these genes are involved in diverse metabolic pathways, including bile acid, lipid and glucose metabolisms 18, 19. However, no genes involved in cell cycle regulation have been reported among these direct target genes of FXR. Our recent work suggests that FXR is required for normal liver regeneration 20. In the current study, we identified Foxm1b as the first known direct FXR target gene that is a cell cycle regulator. We also determined that diminished binding of FXR to the IR-0 response element of Foxm1b could be an intrinsic defect in aging regenerating livers. Furthermore, activation of FXR alone was able to induce Foxm1b expression and alleviate age-related liver regeneration defects.
Materials and Methods
Reagents and plasmids
Cell transfection and analysis of reporter expression
Transient transfection of human hepatoblastoma cells (HepG2) and the determination of the luciferase activity were performed as previously described 21. Twenty-four hours after transfection, cells were treated with FXR ligands, CDCA (50 μM), GW4064 (2μM) or 6ECDCA (3μM), for 24 h and then harvested for the determination of luciferase activity.
Animals, adenovirus, PH, and BrdU assay
Young (8-week-old) and old (13–16 month-old, obtained from the National Institutes of Aging) male C57Bl/6 mice and young male FXR−/− mice (8-week-old) were housed and handled at the City of Hope Animal Research Facility in accordance with NIH guidelines. Eight-week-old WT and FXR−/− mice were injected intraperitoneally with a single dose of CCl4 (750μl/kg in corn oil 1:4 [v/v]). At 0, 1, 2 and 3 days after injection, mice were sacrificed, and livers were removed for gene expression, eletrophoretic mobility shift assay (EMSA) and ChIP assays. Young and old WT mice were subjected to PH. At 0, 1, 2, 3 days and 40 h after PH, mice were sacrificed, and livers were removed for gene expression and EMSA assay. Young WT and FXR−/− mice were injected in the tail vein with 1×109 plaque-forming units per mouse of either Ad-VP16 or Ad-FXRα2-VP16. After 7 days, their livers were harvested for RNA isolation. Old WT mice were injected with either Ad-VP16 or Ad-FXRα2-VP16. After 5 days, these mice were subjected to PH, and their livers were isolated at 40 and 44 h after PH for EMSA, BrdU staining and RNA extraction. The mice were then given Px20350 (17.5 mg/kg) or vehicle (10% HPBCD in 500 mM phosphate pH 7.0) by gavage twice daily for 2 days prior to PH. At 40 hours after PH, these mice were sacrificed, and livers were harvested for EMSA, gene expression analysis and BrdU staining. In parallel experiments, hydrodynamic transfections of 10 μg anti-Foxm1b or negative control shRNA plasmids in 1.8 ml PBS buffer to livers of mice treated with adenovirus or Px20350 were carried out as described previously 22, 23. Adenovirus purification, PH, and BrdU staining were performed as described previously 20, 21.
Primary hepatocyte culture
Mouse primary hepatocytes from 8-week-old mice were prepared as described previously 21. hepatocytes were treated with CDCA (50μM) and 6ECDCA (3μM) for 24 h prior to RNA isolation.
RNA isolation and quantitative real-time PCR
Total RNA was isolated from mouse livers or primary hepatocytes, and quantitative real-time PCR was performed as described previously 24.
EMSA assay
Mouse FXR and RXRα proteins were synthesized in vitro using a TNT® quick coupled transcription/translation system according to the manufacturer’s protocol (Promega, Madison, WI). EMSAs were performed by incubating in vitro translated FXR and RXRα (1 μl each) or the liver nuclear extract (5 μg) with 32P-labeled oligonucleotide duplex under standard binding conditions, and the DNA-protein complex was electrophoretically resolved on a 6% nondenaturing gel. A 32P-labeled bona fide IR-1 consensus oligo-duplex (FXRE, 5′-ctcaagAGGTCAtTGACCTttttg-3′) 25 was used as a positive control. For competition of the EMSA complex with the recombinant FXR and RXRα, the unlabeled homologous or heterologous duplex oligonucleotides were added at 100-and 500-fold molar excess during pre-incubation. When the liver nuclear extract was used in the EMSAs, the competing wild-type and mutant oligonucleotides were used at 200-fold molar excess. The nuclear extract was pre-incubated with antibodies against FXR (sc-1204X, Santa Cruz Biotechnology, Santa Cruz, CA) and/or RXRα (sc-774X, Santa Cruz Biotechnology, Santa Cruz, CA) for 20 min before addition of the DNAprobe in the supershift assay.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays for untreated and CCl4 treated WT livers were performed according to the protocol of a kit (Upstate, Lake Placid, NY). The polyclonal anti-FXR antibody (sc-1204X) was used in the immunoprecipitations.
Statistics
All data represent at least three independent experiments and are expressed as the mean ± SD. Student’s t test was used to calculate P values. P less than 0.05 was considered significant.
Results
FXR activation induces Foxm1b gene expression in vivo and in vitro
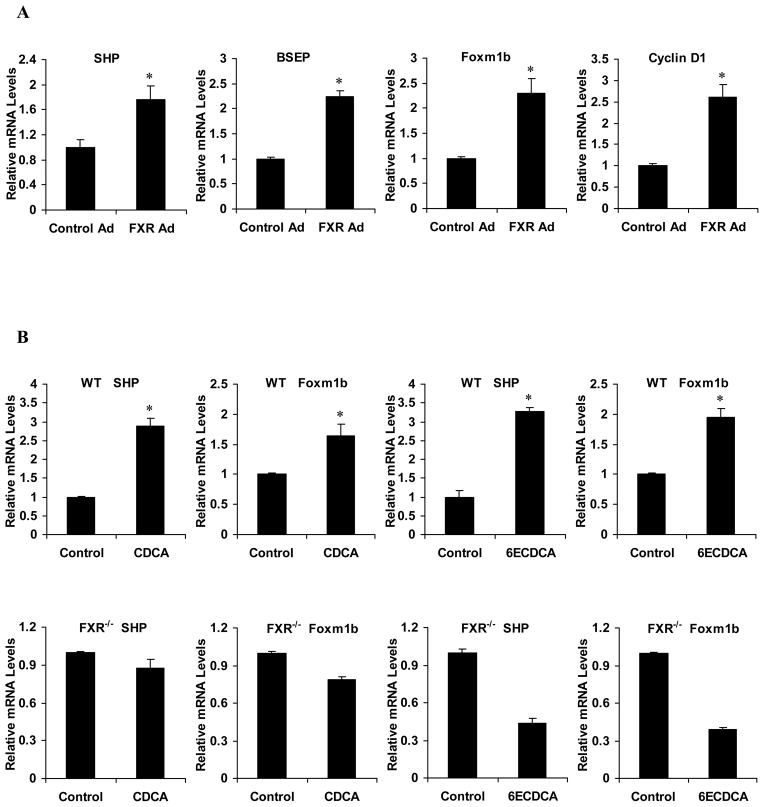
We previously showed that WT but not FXR null mice fed a diet containing 0.2% cholic acid (CA), an endogenous FXR agonist, increased Foxm1b gene expression after PH20. To determine whether FXR directly regulates Foxm1b gene expression, we injected 8-week-old WT mice with adenovirus that expressed murine FXRα2 fused to VP16 (Ad-FXRα2-VP16), the transactivation domain of herpes simplex virus, or VP16 only (Ad-VP16) as a control. FXRα2-VP16 is constitutively active in the absence of FXR ligands 21, 26. As controls, mRNA levels of the well-characterized FXR target genes, small heterodimer partner (SHP) 27, 28 and bile salt export pump (BSEP) 29, as well as the direct Foxm1b target Cyclin D111 were measured. Hepatic expression of FXRα2-VP16 in WT mice resulted in a 1.8-fold induction of SHP mRNA, a 2.2-fold induction of BSEP, a 2.3-fold induction of Foxm1b and a 2.6-fold induction of Cyclin D1 (Fig. 1A). Quantitative real-time PCR analysis of total RNA from mouse primary hepatocytes that were treated with chenodeoxycholic acid (CDCA) (50 −M), a natural ligand of FXR, or 6α-ethylchenodeoxycholic acid (6ECDCA) (3 μM), a synthetic highly specific FXR agonist, confirmed the up-regulation of Foxm1b (1.7-fold to 2-fold) and SHP (about 3-fold) in WT but not FXR−/− hepatocytes (Fig. 1B), further indicating that FXR activation can induce Foxm1b expression.
Fig. 1. Activation of FXR induces expression of Foxm1b mRNA in vitro and in vivo.
(A) Relative mRNA levels of the indicated genes in 8-week-old WT mice (6–8 mice per group) infected with adenovirus encoding VP16 (Control Ad) or the FXRα2-VP16 fusion protein (FXR Ad). Seven days after transfection, total hepatic RNA was extracted and mRNA levels were measured by real-time quantitative PCR. (B) Relative mRNA levels of primary hepatocytes from 8-week-old WT and FXR−/− mice treated with 50 μM CDCA or 3 μM 6ECDCA for 24 h. mRNA levels were measured by real-time quantitative PCR. *P < 0.05 versus control group.
FXR regulates Foxm1b transcriptional activation by binding to an IR-0 element in intron 3 of Foxm1b
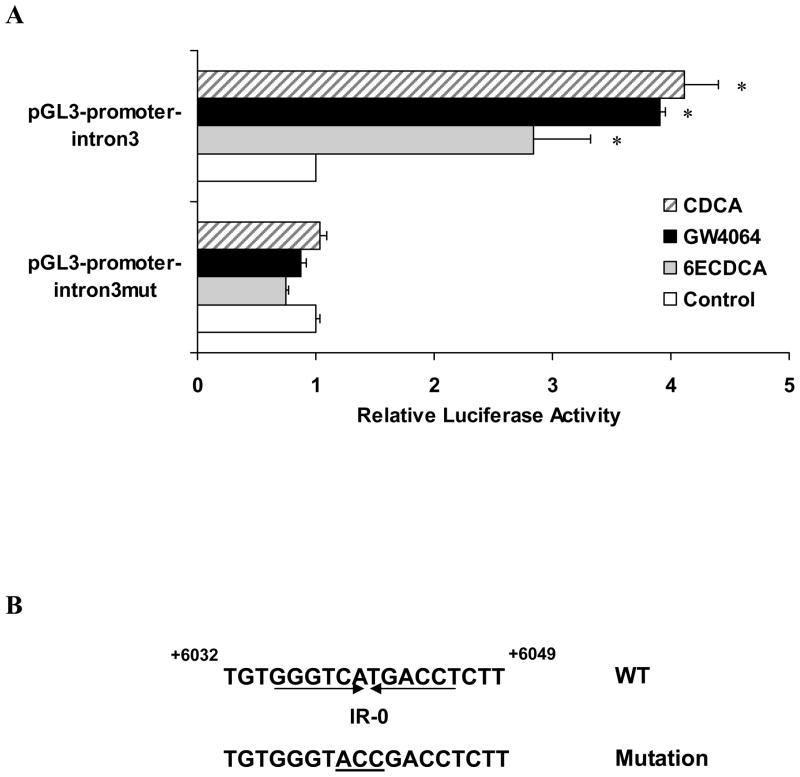
We then analyzed the mouse Foxm1b gene for potential FXR binding sites. To determine the region within the mouse Foxm1b gene that confers transcriptional responsiveness to FXR, a series of progressively deleted Foxm1b promoter and intron fragments were amplified by using a BAC clone containing the entire Foxm1b locus. These fragments were inserted into a pGL3-promoter luciferase vector. These constructs were transiently co-transfected with FXR and RXRα expression plasmids into HepG2 cells. We failed to detect any induction of luciferase from constructs containing only Foxm1b promoter regions after activation of FXR by FXR ligands (CDCA or GW4064) (Supporting Fig. 1). However, an approximately 4-fold increase in luciferase expression was detected after activation of FXR by CDCA, GW4064 or 6ECDCA in cells transfected with pGL3-promoter-intron3 (inserted DNA fragment position from +5604 to +6300 relative to the transcription start site), as compared to cells that were not exposed to FXR ligands (Fig. 2A). The pGL3-promoter-intron3 contains a putative FXR response element (FXRE) (IR-0 element, sequence, 5′-GGGTCA TGACCT-3′) with only a single nucleotide change in comparison to a consensus, nonclassical FXRE, IR-0 element (5′-GGGTCA TGAACT-3′) 30. We then performed site-directed mutagenesis of the IR-0 element (changing ‘CAT’ to ‘ACC’) (Fig. 2B), which blocked the induction of reporter expression by FXR activation (Fig. 2A).
Fig. 2. Intron 3 of Foxm1b intron 3 contains a putative FXRE and is transactivated by FXR/RXRα in HepG2 cells.
(A) Relative luciferase activity of the pGL3-promoter-intron3 and pGL3-promoter-intron3mut reporter constructs when co-transfected into HepG2 cells with FXR and RXRα expression constructs and exposed to the indicated FXR ligands. Data were calculated as fold induction relative to empty vector (pGL3-promoter luciferase vector). *P < 0.05 versus control group. (B) Schematic of the WT and mutant IR-0 elements. The three IR-0 nucleotides that were altered to form the mutant construct are underlined.
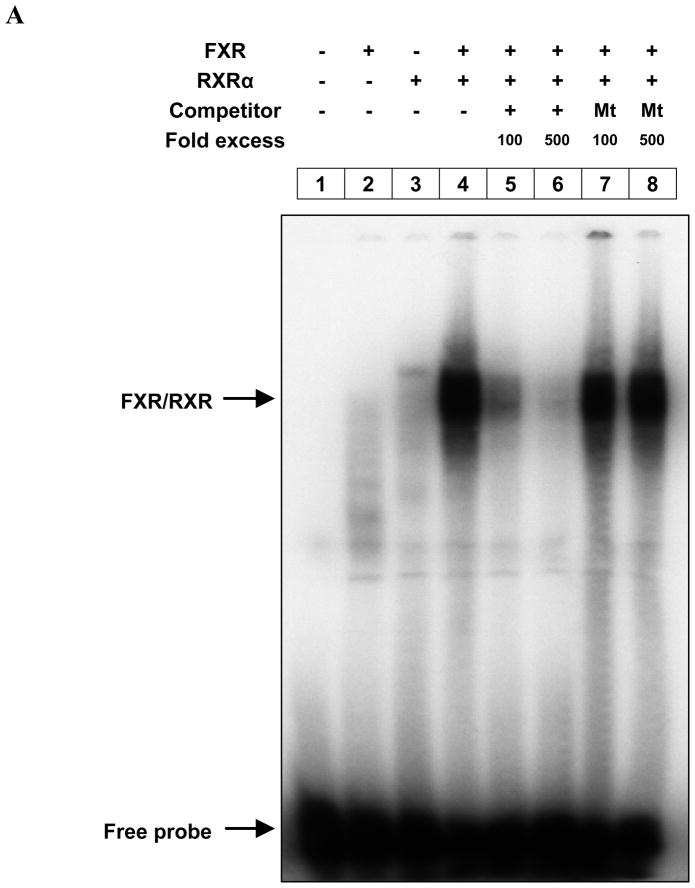
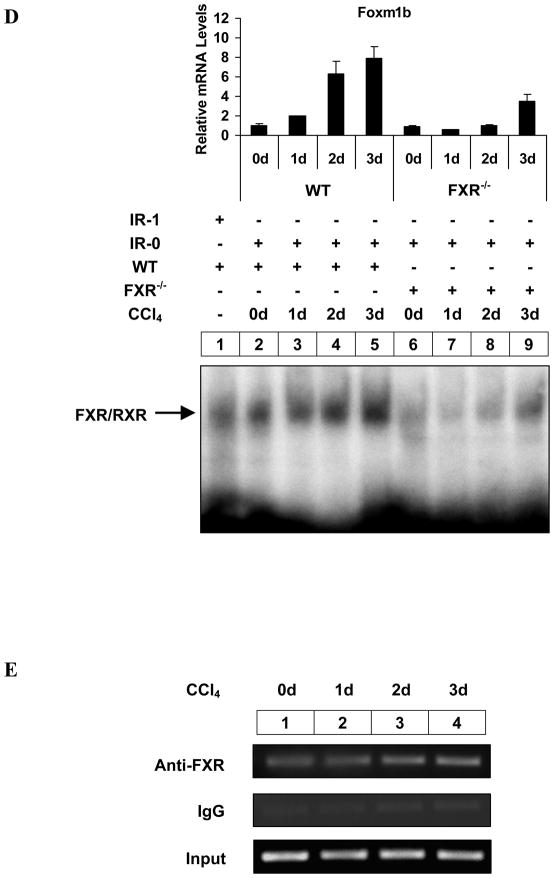
The direct interaction of FXR with the IR-0 element was further substantiated by EMSA. A mixture of recombinant FXR and RXRα, produced by in vitro transcription and translation of the corresponding cDNAs, bound to 32P-labeled 6028/6052 oligo-duplex containing the IR-0 element (Fig. 3A, lane 4). The binding of this complex was specifically competed out by the unlabeled homologous element (Fig. 3A, lanes 5 and 6) but not by the mutant IR-0 element (‘CAT’ to ‘ACC’) (Fig. 3A, lanes 7 and 8). Furthermore, we performed EMSAs using liver nuclear extract and either the oligo-duplexes containing a bona fide IR-1 consensus oligonucleotide (FXRE) (as a control) 25 or the IR-0 element. Upon incubation of the 32P-labeled oligo-duplex with the liver nuclear extract, increased protein binding specifically to both the IR-0 and IR-1 was visible (Fig. 3B, lane 1; Fig. 3C, lane 1; Fig. 3D, lanes 1 and 2). The DNA-protein complex was out competed by a 200-fold molar excess of the unlabeled homologous oligo-duplex (Fig. 3B, lane 2) but not by the mutant oligo-duplex (lane 3), thus confirming the specificity of the protein-DNA complex. Antibody supershift assay showed that the protein-DNA complex contained both FXR and RXRα (Fig. 3C, lanes 2–4). To confirm these results, we performed EMSAs with liver nuclear extract from carbon tetrachloride (CCl4)-treated mice, since CCl4 administration is a good model for inducing Foxm1b gene expression in vivo 31. CCl4 treatment induced both Foxm1b mRNA expression and protein binding to the IR-0 element (Fig. 3D). This increase in protein binding and Foxm1b expression upon CCl4 treatment further supports the hypothesis that increased protein binding to the IR-0 element induces Foxm1b transcription.
Fig. 3. Identification of the FXR binding site in intron 3 of Foxm1b.
(A) EMSA performed with in vitro synthesized mouse FXR/RXRα and 32P-labeled 6028/6052 oligo-duplex containing the IR-0 motif from intron 3 of mouse Foxm1b. The positions of the shifted FXR/RXRα complex and free probes are indicated. Mt, Mutant. (B) EMSA performed with liver nuclear extract from WT and FXR−/− mice. Mt, Mutant. (C) EMSA and super shift assay performed with liver nuclear extract from WT mice. Anti-FXR (2μl; lanes 2 and 3) and anti-RXR (2μl; lanes 2 and 4) antibodies were used. Due to long electrophoretic run, the free probe migrated out of the gel. (D) Relative liver Foxm1b mRNA expression and EMSA performed with liver nuclear extracts from 8-week-old WT and FXR−/− mice injected intraperitoneally with a single dose of CCl4 (750 μl/kg). (E) ChIP assay performed on soluble formaldehyde-cross-linked chromatin isolated from untreated and CCl4 treated WT livers with polyclonal anti-FXR antibodies (Anti-FXR) or control IgG. The final DNA extraction was PCR amplified using primer pair that covered the IR-0 sequence in intron 3 of Foxm1b. Total extract (input) was used as a positive PCR control.
To further demonstrate that FXR binds to the intronic enhancer in vivo, we performed ChIP assay on soluble formaldehyde-cross-linked chromatin isolated from untreated and CCl4 treated livers with a polyclonal anti-FXR antibody. FXR bound to the intronic enhancer in vivo (Fig. 3E, lanes 1–4), and acute CCl4 injury displayed increased association of FXR with the intronic enhancer from 0 to 3 days after treatment, which is consistent with the EMSA results shown in Fig. 3D. Together, both in vitro and in vivo results demonstrated that Foxm1b is upregulated by FXR activation through binding to the IR-0 element in intron 3 of Foxm1b.
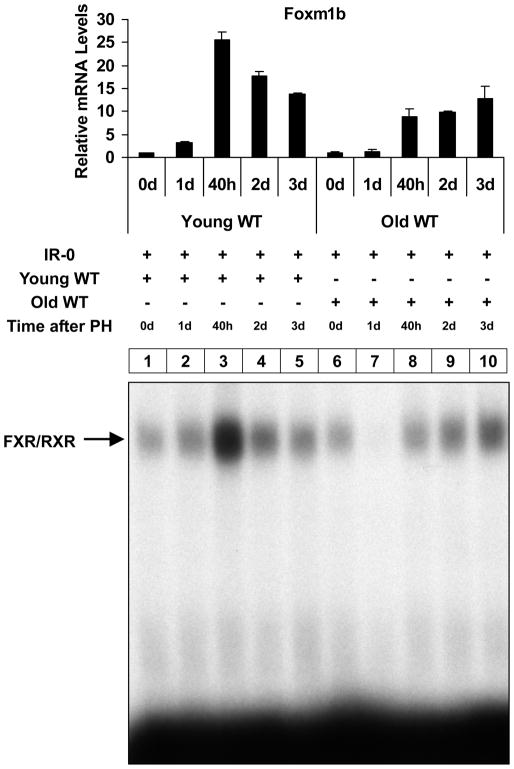
Binding of FXR to the IR-0 element is diminished in aging regenerating livers
Aging livers have dramatically reduced ability to proliferate normally after PH 12, 17. This decline in the regenerative capacity of aging liver has been mainly attributed to decreased expression of Foxm1b and other transcription factors 9–11. Because our data indicated that Foxm1b is a direct target gene of FXR, we speculated that defective activation of FXR might occur in aging regenerating livers, which could result in defective induction of Foxm1b leading to defective liver regeneration. Therefore, we examined FXR binding to the IR-0 element in regenerating livers of both young (8-week-old) and aging (13–16-month-old) mice. Compared with livers of young mice, aging regenerating livers had largely diminished FXR binding to the IR-0 element and defective induction of Foxm1b expression (Fig. 4). Especially on the first day post-PH, very little FXR bound to the IR-0 element in aging regenerating livers. The reduced FXR binding is not due to the lower FXR protein levels in aging livers (Supporting Fig. 2), suggesting a defective activation of FXR but not reduced FXR expression in aging regenerating livers may account for the defective proliferation in aging livers.
Fig. 4. Diminished FXR binding to the IR-0 element in regenerating aging livers.
Relative liver Foxm1b mRNA expression and EMSA performed with liver nuclear extracts from young (8-week-old) and old (13–16-month-old) WT mice subjected to PH. Mice were euthanized, and livers were removed and processed for the above analyses at the following times after PH: 0, 1, 2, 3 days and 40 h.
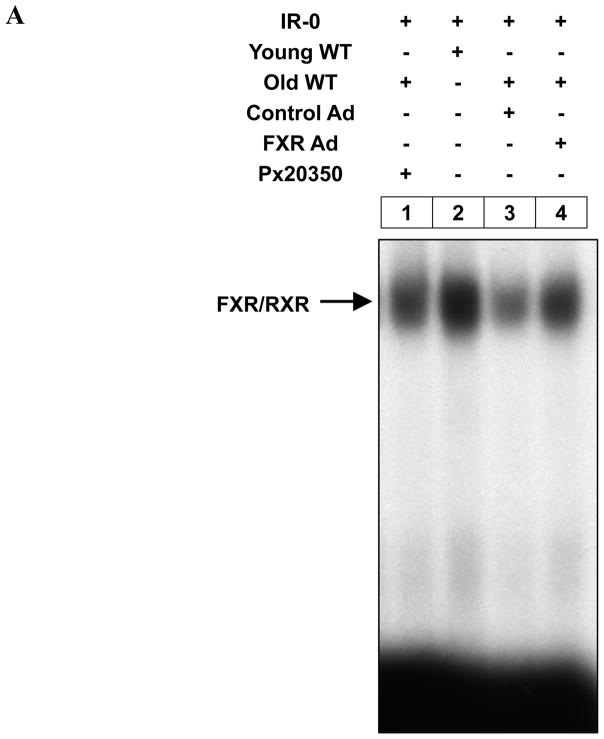
Activation of FXR alleviates aging-related liver regeneration defects
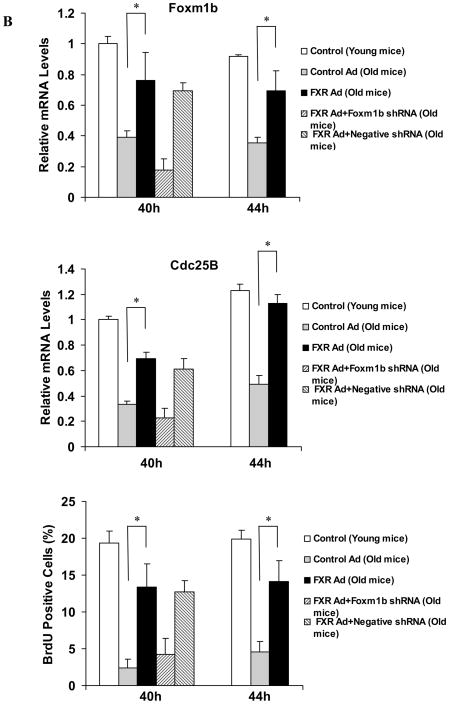
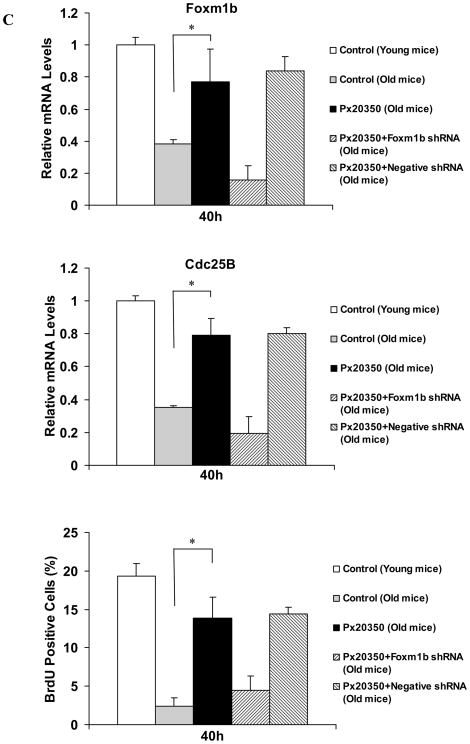
If reduced activation of FXR is an intrinsic defect in aging regenerating livers, resulting in defective liver regeneration, activation of FXR should correct or alleviate aging-related liver regeneration defects. To test this hypothesis, we examined the efficacy of liver regeneration in Ad-VP16 alone and Ad-FXRα2-VP16-infected aging mice (13–16 months). To avoid the initial acute phase response to viral infection, which subsides within 36 h following adenovirus infection 32, we performed PH on the fifth day after adenovirus infection. Regenerating livers from young mice exhibit a sharp S phase peak at 40–44 h after surgery, therefore, for these experiments, we isolated livers at 40 and 44 h after PH. As expected, hepatic expression of FXRα2-VP16 in aging mice enhanced FXR binding to the IR-0 element (Fig. 5A). Ad-FXRα2-VP16 infection of aging mice also resulted in the up-regulation of Foxm1b and Cdc25B expression (about 2-fold) and an increase in hepatocyte BrdU incorporation (a measure of DNA synthesis). Both Foxm1b and Cdc25B expression and BrdU incorporation were 70–80% of levels found in young regenerating livers (Fig. 5B). Compared with negative controls, hepatic expression of anti-Foxm1b short hairpin RNA (shRNA) specifically blocked the induction of Foxm1b and Cdc25B expression, and dramatically reduced BrdU incorporation in hepatocytes of mice infected with Ad-FXRα2-VP16 (Fig. 5B; see also Supporting Fig. 3A). Overall, these data indicate that FXR promotes regeneration of aging liver, mainly through activating Foxm1b transcription.
Fig. 5. Activation of FXR promotes regeneration of aging livers in a Foxm1b-dependent manner.
Young (8-week-old) and old WT (13–16-month-old) mice were injected with either Ad-VP16 or Ad-FXRα2-VP16. After 5 days, these mice were subjected to PH and their livers were isolated at 40 and 44 h after PH for EMSA, gene expression analysis and BrdU staining. In addition, old WT mice were fed Px20350 (17.5 mg/kg) or vehicle (10% HPBCD in 500 mM phosphate pH 7.0) by oral gavage twice daily for 2 days prior to PH. At 40 hours after PH, these livers were harvested for the same analyses. Shown are EMSA results (A) at 40 h after PH, and mean mRNA levels of Foxm1b and Cdc25B, and hepatocyte BrdU incorporation for Ad-VP16 or Ad-FXRα2-VP16 treated mice (B) or Px20350 treated mice (C). *P < 0.05 versus control group of old mice. To test whether FXR-dependent Foxm1b signaling is required to restore regeneration of aging livers, hydrodynamic transfections of 10 μg anti-Foxm1b or negative control shRNA plasmids to the livers of mice treated with adenovirus or Px20350 were carried out. These mice were sacrificed at 40 h after PH, and relative gene expression (B) and percentage of BrdU labeling (C) are shown. Representative images of BrdU immunostaining of these liver sections are shown in Supporting Fig. 2.
We then asked whether ligand-activated FXR could also restore the regenerative potential of aging liver regenerative. Previously, the treatment of GW4064 was not good to stimulate proliferation probably due to its poor bioavailability20. We test the effects of a novel FXR ligand, Px20350 that is a new synthetic, highly specific FXR ligand. Expression of he FXR target genes, SHP and Cyp7a1, was specifically regulated by Px20350 in WT but not FXR−/− mice, which confirmed that Px20350 is a potent and highly specific FXR agonist for in vivo study (Supporting Fig. 4). Measurement of serum alanine aminotransferase levels and histological staining indicated that administering Px20350 to mice by gavage did not induce toxic effects (data not shown). Therefore, we examined whether administration of Px20350 by gavage could restore the reduced regenerative potential of aging livers. As expected, ligand-activated FXR strongly promoted aging liver regeneration, as demonstrated by increased FXR binding (Fig. 5A), up-regulation of Foxm1b expression and increased BrdU incorporation (Fig. 5C). In shRNA experiments, elevated liver regeneration was observed in negative control, but not anti-Foxm1b shRNA, treated groups (Fig. 5C; see also Supporting Fig. 3B). These results indicate that FXR activation is sufficient to promote aging liver regeneration.
Discussion
Foxm1b is required for the timely expression of many cell cycle regulators essential for G1/S and G2/M progression and chromosome stability and segregation 33, 34. Loss of Foxm1b function in livers of young mice (8 week-olds) results in a significant reduction in hepatocyte DNA replication and inhibition of mitosis after PH 9, and overexpression of Foxm1b in regenerating livers of old mice (12-months-old) is sufficient to restore hepatocyte DNA replication and expression of necessary cell cycle regulatory genes, all of which are Foxm1b target genes, to levels seen in young animals 10, 11, 17. In the present work, we demonstrate that Foxm1b is a direct target gene of FXR, and elevated hepatic expression of Foxm1b through FXR activation can largely alleviate age-related liver regeneration defects.
To date, many target genes of FXR have been identified, which are involved in diverse metabolic pathways 18, 19. However, until this identification of Foxm1b as an FXR target genes, none of the reported direct target genes of FXR had been directly involved in cell cycle regulation. Foxm1b gene expression is negatively regulated in quiescent or terminally differentiated cells, while its expression is induced when cells are stimulated to enter the cell cycle after PH 34. It has been recently reported that, Foxm1b protein is actively degraded during exit from mitosis through active proteolysis involving the anaphase-promoting complex 35. However, it is currently unclear how Foxm1b gene expression is transiently induced after PH. Our results suggest that FXR signaling could be required for controlling the timely induction of Foxm1b for proper liver proliferation during liver regeneration. We found that defective activation of FXR occurred in aging regenerating livers (Fig. 4). This result suggests that insufficient activation of FXR, resulting in defective liver regeneration, could be an intrinsic defect in aging regenerating livers. Compared with young mice, we found that aging mice did not have altered protein levels of FXR (Supporting Fig. 2). Therefore, aging may affect the levels of endogenous FXR ligands such as bile acids or other liver-specific metabolites 19, which could result in defective activation of FXR during liver regeneration. On the other hand, we can not exclude other possibility that aging may affect FXR activation, further research is required to fully understand the defective FXR activation in aging livers.
Previous experiments with serum-starved cells showed that a 300 bp fragment of the proximal Foxm1b promoter is sufficient for the serum-induced expression of Foxm1b36. This 300 bp region contains multiple binding elements for c-Myc and E2F. Recent ChIP assays with chromatin isolated from untreated and 1,4-bis [2-(3,5-dichloropyridyloxy)] benzene (TCPOBOP) treated livers suggested that Foxm1b may be a direct transcriptional target of c-Myc and that binding of c-Myc to the Foxm1 promoter only occurs in response to TCPOBOP treatment 37. Studies from Timchenko’s group demonstrated that aging induces the binding of histone deacetylase 1 (HDAC1)-C/EBPα-E2F-Rb complex to Foxm1b promoter, which inhibits activation of Foxm1b promoter in quiescent old livers and after PH 38. However, our ChIP analyses showed that FXR regulated Foxm1b in a C/EBPα-independent manner (Supporting Fig. 4). Although regulation of the Foxm1b gene probably involves multiple transcription factors, FXR may direct a unique pathway for Foxm1b induction during normal liver regeneration.
In conclusion, our results have revealed Foxm1b as the first known direct FXR target gene involved in cell cycle regulation, and demonstrated that defective activation of FXR could be an intrinsic defect in aging regenerating livers. Activation of FXR by small molecules is able to largely alleviate age-related liver regeneration defects. These findings suggest that FXR agonist ligands are potential therapeutic candidates for prevention and treatment of insufficient liver regeneration after segmental liver transplantation or resection of liver tumors.
Supplementary Material
Acknowledgments
We thank Dr. Maria Hahn, Akio Kruoda, and Qiang Lu for plasmids; and Dr. Peter A. Edwards (University of California, Los Angeles, CA) for adenovirus that expressed VP16 alone (Ad-VP16) or murine FXRα2 fused to VP16. Px20350 was provided by Dr. Claus Kremoser from Phenex Pharmaceuticals AG (Ludwigshafen, Germany). We appreciate Keely Walker at City of Hope for proofreading and editing the manuscript. W.H. is supported by Sidney Kimmel Foundation and Concern Foundation.
Abbreviations
- CA
cholic acid
- 6ECDCA
6α-ethylchenodeoxycholic acid
- ChIP
chromatin immunoprecipitation
- Foxm1b
forkhead box m1b
- FXR
farnesoid X receptor
- PH
partial hepatectomy
Footnotes
Disclosures: All authors have nothing to disclose.
References
- 1.Biglarnia AR, Lorant T, Lee HS, Tufveson G, Totsch M, Malago M. Liver regeneration is impaired by FK778 in partially hepatectomized rats, while supplemental uridine restores both liver growth and hepatocyte proliferation. Hepatol Res. 2009;39:86–92. doi: 10.1111/j.1872-034X.2008.00402.x. [DOI] [PubMed] [Google Scholar]
- 2.Broelsch CE, Emond JC, Thistlethwaite JR, Rouch DA, Whitington PF, Lichtor JL. Liver transplantation with reduced-size donor organs. Transplantation. 1988;45:519–24. doi: 10.1097/00007890-198803000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Clavien PA. Liver regeneration: a spotlight on the novel role of platelets and serotonin. Swiss Med Wkly. 2008;138:361–70. doi: 10.4414/smw.2008.12231. [DOI] [PubMed] [Google Scholar]
- 4.Clavien PA, Petrowsky H, DeOliveira ML, Graf R. Strategies for safer liver surgery and partial liver transplantation. N Engl J Med. 2007;356:1545–59. doi: 10.1056/NEJMra065156. [DOI] [PubMed] [Google Scholar]
- 5.Helling TS. Liver failure following partial hepatectomy. HPB (Oxford) 2006;8:165–74. doi: 10.1080/13651820510035712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dahm F, Georgiev P, Clavien PA. Small-for-size syndrome after partial liver transplantation: definition, mechanisms of disease and clinical implications. Am J Transplant. 2005;5:2605–10. doi: 10.1111/j.1600-6143.2005.01081.x. [DOI] [PubMed] [Google Scholar]
- 7.Fortner JG, Lincer RM. Hepatic resection in the elderly. Ann Surg. 1990;211:141–5. doi: 10.1097/00000658-199002000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koperna T, Kisser M, Schulz F. Hepatic resection in the elderly. World J Surg. 1998;22:406–12. doi: 10.1007/s002689900405. [DOI] [PubMed] [Google Scholar]
- 9.Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The Forkhead Box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci U S A. 2002;99:16881–6. doi: 10.1073/pnas.252570299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Krupczak-Hollis K, Tan Y, Dennewitz MB, Adami GR, Costa RH. Increased hepatic Forkhead Box M1B (FoxM1B) levels in old-aged mice stimulated liver regeneration through diminished p27Kip1 protein levels and increased Cdc25B expression. J Biol Chem. 2002;277:44310–6. doi: 10.1074/jbc.M207510200. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Quail E, Hung NJ, Tan Y, Ye H, Costa RH. Increased levels of forkhead box M1B transcription factor in transgenic mouse hepatocytes prevent age-related proliferation defects in regenerating liver. Proc Natl Acad Sci U S A. 2001;98:11468–73. doi: 10.1073/pnas.201360898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iakova P, Awad SS, Timchenko NA. Aging reduces proliferative capacities of liver by switching pathways of C/EBPalpha growth arrest. Cell. 2003;113:495–506. doi: 10.1016/s0092-8674(03)00318-0. [DOI] [PubMed] [Google Scholar]
- 13.Wang GL, Shi X, Salisbury E, Sun Y, Albrecht JH, Smith RG, Timchenko NA. Growth hormone corrects proliferation and transcription of phosphoenolpyruvate carboxykinase in livers of old mice via elimination of CCAAT/enhancer-binding protein alpha-Brm complex. J Biol Chem. 2007;282:1468–78. doi: 10.1074/jbc.M608226200. [DOI] [PubMed] [Google Scholar]
- 14.Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM, Tindal DJ, Chen J. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol. 2008;10:1076–82. doi: 10.1038/ncb1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalinichenko VV, Major ML, Wang X, Petrovic V, Kuechle J, Yoder HM, Dennewitz MB, Shin B, Datta A, Raychaudhuri P, Costa RH. Foxm1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–50. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7:126–36. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 17.Krupczak-Hollis K, Wang X, Dennewitz MB, Costa RH. Growth hormone stimulates proliferation of old-aged regenerating liver through forkhead box m1b. Hepatology. 2003;38:1552–62. doi: 10.1016/j.hep.2003.08.052. [DOI] [PubMed] [Google Scholar]
- 18.Wang YD, Chen WD, Huang W. FXR, a target for different diseases. Histol Histopathol. 2008;23:621–7. doi: 10.14670/HH-23.621. [DOI] [PubMed] [Google Scholar]
- 19.Wang YD, Chen WD, Moore DD, Huang W. FXR: a metabolic regulator and cell protector. Cell Res. 2008;18:1087–95. doi: 10.1038/cr.2008.289. [DOI] [PubMed] [Google Scholar]
- 20.Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, Dong B, Huang X, Moore DD. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312:233–6. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- 21.Wang YD, Chen WD, Wang M, Yu D, Forman BM, Huang W. Farnesoid X receptor antagonizes nuclear factor kappaB in hepatic inflammatory response. Hepatology. 2008;48:1632–43. doi: 10.1002/hep.22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu F, Song Y, Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–66. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 23.Zhang G, Budker V, Wolff JA. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–7. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]
- 24.Wang YD, Yang F, Chen WD, Huang X, Lai L, Forman BM, Huang W. Farnesoid X receptor protects liver cells from apoptosis induced by serum deprivation in vitro and fasting in vivo. Mol Endocrinol. 2008;22:1622–32. doi: 10.1210/me.2007-0527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laffitte BA, Kast HR, Nguyen CM, Zavacki AM, Moore DD, Edwards PA. Identification of the DNA binding specificity and potential target genes for the farnesoid X-activated receptor. J Biol Chem. 2000;275:10638–47. doi: 10.1074/jbc.275.14.10638. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci U S A. 2006;103:1006–11. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–26. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 28.Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–15. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- 29.Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem. 2001;276:28857–65. doi: 10.1074/jbc.M011610200. [DOI] [PubMed] [Google Scholar]
- 30.Song CS, Echchgadda I, Baek BS, Ahn SC, Oh T, Roy AK, Chatterjee B. Dehydroepiandrosterone sulfotransferase gene induction by bile acid activated farnesoid X receptor. J Biol Chem. 2001;276:42549–56. doi: 10.1074/jbc.M107557200. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Hung NJ, Costa RH. Earlier expression of the transcription factor HFH-11B diminishes induction of p21(CIP1/WAF1) levels and accelerates mouse hepatocyte entry into S-phase following carbon tetrachloride liver injury. Hepatology. 2001;33:1404–14. doi: 10.1053/jhep.2001.24666. [DOI] [PubMed] [Google Scholar]
- 32.Benihoud K, Salone B, Esselin S, Opolon P, Poli V, Di Giovine M, Perricaudet M, Saggio I. The role of IL-6 in the inflammatory and humoral response to adenoviral vectors. J Gene Med. 2000;2:194–203. doi: 10.1002/(SICI)1521-2254(200005/06)2:3<194::AID-JGM102>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 33.Costa RH. FoxM1 dances with mitosis. Nat Cell Biol. 2005;7:108–10. doi: 10.1038/ncb0205-108. [DOI] [PubMed] [Google Scholar]
- 34.Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. doi: 10.1016/j.bbcan.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 35.Laoukili J, Alvarez-Fernandez M, Stahl M, Medema RH. FoxM1 is degraded at mitotic exit in a Cdh1-dependent manner. Cell Cycle. 2008;7:2720–6. doi: 10.4161/cc.7.17.6580. [DOI] [PubMed] [Google Scholar]
- 36.Korver W, Roose J, Heinen K, Weghuis DO, de Bruijn D, van Kessel AG, Clevers H. The human TRIDENT/HFH-11/FKHL16 gene: structure, localization, and promoter characterization. Genomics. 1997;46:435–42. doi: 10.1006/geno.1997.5065. [DOI] [PubMed] [Google Scholar]
- 37.Blanco-Bose WE, Murphy MJ, Ehninger A, Offner S, Dubey C, Huang W, Moore DD, Trumpp A. C-Myc and its target FoxM1 are critical downstream effectors of constitutive androstane receptor (CAR) mediated direct liver hyperplasia. Hepatology. 2008;48:1302–11. doi: 10.1002/hep.22475. [DOI] [PubMed] [Google Scholar]
- 38.Wang GL, Salisbury E, Shi X, Timchenko L, Medrano EE, Timchenko NA. HDAC1 cooperates with C/EBPalpha in the inhibition of liver proliferation in old mice. J Biol Chem. 2008;283:26169–78. doi: 10.1074/jbc.M803544200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.