Abstract
The doublesex and mab-3 related transcription factor 1 (Dmrt1) is a putative transcriptional regulator that is expressed exclusively in the gonads and is required for postnatal testis differentiation. Here we describe the transcriptional mechanisms regulating testis-specific expression of the Dmrt1 gene. Transient-transfection analysis identified a region of the promoter between kb −3.2 and −2.8 that is important for Sertoli cell-specific expression. DNase I footprinting revealed four sites of DNA-protein interaction within this region, three of which were prominent in primary Sertoli cells. Analysis of these sites, using electrophoretic mobility shift assays, revealed that Gata4 and another unknown factor bound within these regions. Further transient-transfection assays of various mutant promoters established the functional relevance of the Gata4-response and unknown factor-response elements, while studies of Dmrt1 expression in 13.5 days postcoitum Fog2 null gonads supported the in vivo importance of Gata4's regulation. As a whole, these studies identify Gata4 as an important regulator in the Dmrt1 transcriptional machinery that is responsible for robust expression of Dmrt1 in the testis.
The doublesex and mab-3 related transcription factor 1 (Dmrt1) is a member of a protein family that shares a novel DNA binding motif called the DM domain. This unique feature was first identified as a region of sequence similarity between two regions within the Caenorhabditis elegans protein mab-3 and the DNA binding motif of the Drosophila melanogaster transcription factor doublesex (32). Interestingly, both mab-3 and doublesex play important roles in sex determination and male-specific differentiation, providing the first genetic evidence that aspects of sex determination and differentiation are conserved across different phylogenetic groups (32). Identification of this conserved structural motif also suggested that the DM domain represents an evolutionarily conserved feature common among proteins in the sex determination pathway (32). Discovery of the mammalian DM-containing protein, Dmrt1, and determination of its testis-specific expression implicated this protein as a regulatory factor in the mammalian sex determination pathway (32). In support of this, human DMRT1 was shown to map to a cluster of DM-containing genes on chromosome 9p, with DMRT1 located less than 30 kb from a known breakpoint responsible for human 46 XY sex reversal (4, 31).
Dmrt1 has been cloned from a variety of vertebrates, including fish, alligator, turtle, and chicken (9, 15, 21, 27, 35). In each, embryonic expression of Dmrt1 occurred in a sexually dimorphic manner, where higher levels were observed in the developing male gonad prior to sexual differentiation (8, 9, 15, 21, 26, 29, 35). In mice, Dmrt1 expression was first detected in the indifferent urogenital ridge, but as differentiation proceeded it was gradually lost from the ovary (1, 8). In contrast, expression of Dmrt1 in the testis was up-regulated at 11.5 days postcoitum (dpc), and a high level was maintained after birth and throughout adulthood (8). This conserved expression pattern suggested that Dmrt1 is important during early stages of gonadogenesis and testis differentiation and that it has been functionally maintained during evolution of vertebrate sex determination (45).
Unexpectedly, elimination of Dmrt1 from the mouse genome did not give rise to any abnormal embryonic gonadogenesis or sex determination, indicating either that it does not function at the embryonic stage of gonadogenesis or that its role is functionally redundant. However, multiple defects did occur in postnatal testis differentiation (30). In these mice, postnatal Sertoli cells failed to complete their differentiation and overproliferated to fill the testicular cords. These immature cells eventually died, resulting in highly disorganized testes with few seminiferous tubules in the adult male. The germ cells in Dmrt1−/− testis failed to migrate to the periphery of the developing seminiferous tubules and began to die shortly after postnatal day 7. Hence, mouse Dmrt1 was critical for male-specific sterility by guiding both Sertoli cell and germ cell differentiation in the postnatal testis (30).
Dmrt1's expression pattern and role in testis differentiation places it at a pivotal position within the genetic pathway leading to formation and function of the adult testis. Thus, determination of the mechanism regulating Dmrt1 will contribute substantially to our understanding of this genetic pathway. Previous study of the rat Dmrt1 gene revealed that 5,000 bp of 5′-flanking sequence was transcriptionally active in primary Sertoli cells, and deletion analysis of this promoter identified two major regions that contributed to its transcriptional activity (19). These regions were located between bp −3280 and −2000 and downstream of bp −150 relative to the major transcriptional start site. Analysis of the first 150 bp of the promoter identified two elements that activate Dmrt1 transcription and two that repress it. Transcription factors Sp1, Sp3, and Egr1 bound the positive regulatory elements (19). While these factors clearly contribute to Dmrt1 expression, their global expression patterns suggest that additional factors are responsible for directing the specific expression pattern of Dmrt1. The present study focuses on the distal regulatory region between bp −3280 and −2000. Within this region, a series of essential response elements were discovered to contribute to testis-specific expression of Dmrt1. Additional analysis showed that among these elements three bound the transcription factor Gata4. Data from Fog2 knockout mice also support a role for Gata4 in Dmrt1 transcription. For the first time, we report herein the hierarchical relationship of Dmrt1 and Gata4, give explanations to their testicular expression and regulation, and have further determined aspects of their roles in testis development.
MATERIALS AND METHODS
Cell preparation and transfection analysis.
Preparation and transient transfection of day 15 primary rat Sertoli cells and mouse Sertoli cell lines MSC-1 and TM4 were as described elsewhere (10, 11, 19). Preparation of primary cultures of rat peritubular myoid cells was done as previously described (5). JEG3 cells, a human choriocarcinoma cell line, were cultured in Dulbecco's modified Eagle's medium (Cellgro Mediatech; Fisher Scientific, Pittsburgh, Pa.) supplemented with 5% fetal bovine serum. For transient transfection, myoid cells and JEG3 cells were seeded onto 24-well plates (Costar, Corning, N.Y.) at a density of 26,000 and 23,000 cells per well, respectively. Unless otherwise stated, myoid cells and JEG3 cells were transfected with 0.1 and 0.2 μg of promoterless control vector pGL3-Basic (Promega Corp., Madison, Wis.) or equimolar amounts of Dmrt1-luciferase constructs. Ten or 20 ng of pRL-TK (a control vector containing Renilla luciferase driven by the thymidine kinase promoter; Promega Corp.) was included to control for transfection efficiency by using 1 μl of Lipofectamine reagent (Life Technologies, Inc., Gaithersburg, Md.) in myoid cells or JEG3 cells, respectively. Sixty hours after transfection, cells were lysed and assayed for both firefly and Renilla luciferase activities using the Dual-Luciferase reporter assay system (Promega Corp.) as described previously (7, 19). The data are presented as the firefly/Renilla luciferase activity ratio of Dmrt1-luciferase constructs relative to the firefly/Renilla luciferase activity ratio of the promoterless control vector, pGL3-Basic. All plasmid DNAs were prepared from overnight bacterial cultures using a QIAwell 8 plasmid kit according to the supplier's recommendations (Qiagen, Valencia, Calif.). All buffers used for plasmid DNA preparation were endotoxin free.
RNase protection analysis.
RNase protection analysis was performed as previously described elsewhere (5).
Dmrt1 promoter clones.
If not otherwise indicated, all PCRs had the following cycle parameters: 30 cycles for 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C; PCRs were performed using Dmrt1(−3,280/+75)Luc as template and Bio-X-ACT DNA polymerase, according to the manufacturer's recommendations (Intermountain Scientific Corp., Kaysville, Utah). Cloning of rat Dmrt1 promoter-reporter constructs Dmrt1(−5,000/+75)Luc, Dmrt1(−3,280/+75)Luc, Dmrt1(−2,000/+75)Luc, and Dmrt1(−150/+75)Luc was described elsewhere (19). To generate Dmrt1(−2,240/+75)Luc, Dmrt1(−3,280/+75)Luc was digested with MluI and EcoRI, and following isolation from the bp −3,280/−2,240 DNA fragment the remaining vector was filled in by Klenow (NEN, Boston, Mass.) and religated. For Dmrt1(−3,100/+75)Luc, Dmrt1(−3,000/+75)Luc, Dmrt1(−2,900/+75)Luc, and Dmrt1(−2,800/+75)Luc, DNA sequences between −3100 and −2900 or between −2800 and −2240 were amplified, digested with MluI and EcoRI, and ligated upstream of Dmrt1(−2,240/+75)Luc using the EcoRI site at −2,240. Sequences for the 3′ primer (Dmrt1.26) and 5′ primers (Dmrt1.30 [2], Dmrt1.33, Dmrt1.29, and Dmrt1-2800), are listed in Table 1.
TABLE 1.
Sequences of primers used for chimeric and small mutant clones
| Clone | 5′ primera | 3′ primer |
|---|---|---|
| Dmrt1 (−3, 100/+75) Luc | Dmrt1.30 (2), GCGCACGCGTCAGGAGGATCAAAACTTCAAGGC | Dmrt1.26, AGTGAATTCAAAGCCAGCC |
| Dmrt1 (−3, 000/+75) Luc | Dmrt1.33, GCGCACGCGTGGGTCCCACAACACTTTTTTTTAATGCAC | Dmrt1.26, AGTGAATTCAAAGCCAGCC |
| Dmrt1 (−2, 900/+75) Luc | Dmrt1.29, GCGCACGCGTGCAGGCCCATTTTC | Dmrt1.26, AGTGAATTCAAAGCCAGCC |
| Dmrt1 (−2, 800/+75) Luc | Dmrt1-2800, GCGCACGCGTGCATGGTGTGTCCTCTCTG | Dmrt1.26, AGTGAATTCAAAGCCAGCC |
| Dmrt1 (−3, 280_−2, 985/ −150_+75) Luc | RV3, CTAGCAAAATAGGCTGTCCC | Dmrt1-2985, GCGCGGTACCCAGGAGGCA GTGACACAAATAC |
| Dmrt1 (−3, 000_−2, 735/ −150_+75) Luc | Dmrt1-3000, GCGCACGCGTCAGGAGGATCAAAACTTCAAGGC | Dmrt1-2375, GCGCAAGCTTCTAGGAAGT TATGAAAAAC |
| Dmrt1 (−2, 750_−2, 485/ −150_+75) Luc | Dmrt1-2750, GCGCGGTACCTTTCATAACTTCCTAGGTTTC | Dmrt1-2485, GCGCAAGCTTTAATTGGTCGATTAATTGGTCG |
| Dmrt1 (−2, 500_−2, 225/ −150_+75) Luc | Dmrt1-2500, GCGCGGTACCATTAATCGACCAATTAATTAATTTG | Dmrt1-2225, GCGCAAGCTTAGCTACAGAGAATTCAAGC |
| Dmrt1 usm5 | Dmrt1-usm5-up, CGCGTGCTAGCTAGTCAGTGAGCATGGACTATTTCTCATGTCA TATGTAAAGATCTGTGCCATCTC | Dmrt1-2225, GCGCAAGCTTAGCTACAGAGTGAATTCAAAGC |
| Dmrt1 usm4/5 | Dmrt1-usm4/5-up, CGCGTGCTAGCTAGTCAGTGAGCATGGACTATT TCTCATGTCATATGTAAAGATCTGT | Dmrt1-2225, GCGCAAGCTTAGCTACAGAGTGAATTCAAAGC |
The underlined sequences indicate the restriction endonuclease sites. Italic letters indicate that bases have been changed.
The chimeric constructs Dmrt1(−3,280_−2,985/−150_+75)Luc, Dmrt1(−3,000_−2,735/−150_+75)Luc, Dmrt1(−2,750_−2,485/−150_+75)Luc, and Dmrt1(−2,500_−2,225/−150_+75)Luc, containing a promoter fragment of ∼250 bp (between kb −3.2 and −2.8) cloned immediately upstream of the Dmrt1 proximal promoter (bp −150/+75). DNA sequences of upstream promoter fragments −3280 to −2985, −3000 to −2735, −2750 to −2485, and −2500 to −2225 were amplified with primer pairs RV3 and Dmrt1-2985, Dmrt1-3000 and Dmrt1-2735, Dmrt1-2750 and Dmrt1-2485, and Dmrt1-2500 and Dmrt1-2225, respectively (Table 1). The proximal promoter was amplified using the 5′ primer Dmrt1-150-HindIII (5′-GCGCAAGCTTCTAGGAAGTTATGAAAAAC-3′) and the 3′ primer Luc1. Amplified promoter fragments were digested with KpnI and HindIII, and the amplified proximal promoter was digested with HindIII and XhoI. The two promoter segments (upstream plus proximal promoter) were ligated into mpGL3-Basic Plus (19) digested with KpnI and XhoI. Dmrt1(−3,280/+75)Luc and Dmrt1(−2,240/+75)Luc were digested with KpnI and HindIII and yielded the upstream promoter fragments of bp −3,280/−2,000 and bp −2240/−1978, respectively. The isolated upstream promoter together with the proximal promoter (bp −150/+75 digested by KpnI and XhoI) were cloned into the XhoI and KpnI sites of mpGL3-Basic Plus to generate Dmrt1(−3,280_−2,000/−150_+75)Luc and Dmrt1(−2,240_−1,978/−150_+75)Luc. To generate Dmrt1(−3,280_−2,000/SV40)Luc, the proximal promoter of Dmrt1(−3,280_−2,000/−150_+75)Luc was replaced with the simian virus 40 (SV40) promoter.
Mutations in the regulatory region were generated in the context of Dmrt1(−3,280/+75)Luc. For usm1, usm2, and usm3, the sites of interest were changed into HindIII (for usm1 and usm3) or XhoI (for usm2) recognition sequences. The introduced recognition sequences were used as a cloning site for ligation of upstream and downstream promoter sequences. The primer pairs used to amplify the two Dmrt1 promoter fragments were Dmrt1-NheI and Dmrt1-usm1/2/3-up for the 5′ fragment and Dmrt1-usm1/2/3-down and Dmrt1-2225 for the 3′ fragment (Table 2). Amplified products of 5′ pieces were digested by NheI/HindIII (for usm1 and usm3) or XhoI (for usm2), while the 3′ pieces were digested by HindIII/EcoRI (for usm1 and usm3) or XhoI/EcoRI (for usm2). Finally, the digested 5′ and 3′ pieces were ligated into NheI and EcoRI sites of Dmrt1(−2,240/+75)Luc. The usm4 mutation was achieved by overlap extension as described previously (12). The primer pairs Dmrt1-NheI/Dmrt1-usm4-up and Dmrt1-usm4-down/Dmrt1-2225 (Table 2), with Dmrt1-usm4-up and Dmrt1-usm4-down containing the mutant sites, were used to amplify adjoining pieces with a 15-bp overlap. Annealed products of the sense strand of the 5′ piece and the antisense strand of the 3′ piece were used as templates to amplify the recombinant promoter. The recombinant promoter was digested by NheI and EcoRI and cloned into the same sites of Dmrt1(−2,240/+75)Luc. To generate usm5 and usm4/5, the region between bp −3280 and −2240 was amplified with the respective 5′ (Dmrt1-usm5-up and Dmrt1-usm4/5-up) and 3′ (Dmrt1-2225) primers (Table 1). The amplified fragment was digested with NheI and EcoRI and cloned into these sites in Dmrt1(−2,240/+75)Luc. For usm4/5, PCR of the upstream promoter was performed with usm4 as template.
TABLE 2.
Sequences of primers used for usm1, -2, -3, and -4 clones
| Dmrt1 clone | Upstream DNA sequence amplificationa
|
Downstream DNA sequence amplification
|
||
|---|---|---|---|---|
| 5′ primer | 3′ primer | 5′ primer | 3′ primer | |
| usm1 | Dmrt1-Nhe1, CGCGTGCTAGC TAGT CAGTGA | Dmrt1-usm1-up, CCTCTCAAGCTTTTGCGG TCTTTCCTGC | Dmrt1-usm1-down, CGCAAAAGCTTG AGAGGCTGAGTCTGTACCTC | Dmrt1-2225, AGCTACAGAGTGAATTCAAA |
| usm2 | Dmrt1-Nhe1, CGCGTGCTAGC TAGTCAGTGA | Dmrt1-usm2-up, TAAACAACTCGAG CGGGATTGTTCACTCTG | Dmrt1-usm2-down, ATCCCGCTCGAGTTGTTTATTTATGCA TTAAAAAAAA | Dmrt1-2225, AGCTACAGAGTGAATTCAAA |
| usm3 | Dmrt1-Nhe1, CGCGTGCTAGC TAGTCAGTGA | Dmrt1-usm3-up, CTGTATGAAGCTTTAAC CCTGGGAAG GCAGAGAC | Dmrt1-usm3-down, GGGTTAAAGCTTCATACAGTAATCCT ACAGGAAAAG | Dmrt1-2225, AGCTACAGAGTGAATTCAAA |
| usm4 | Dmrt1-Nhe1, CGCGTGCTAGC TAGTCAGTGA | Dmrt1-usm4-up, CATGTAAAGATCTGTTAA CTCTCTTTG GGGTCTTGAGAAAG | Dmrt1-usm4-down, ACAGATCTTTACATG ATTACTGAGAAATATG | Dmrt1-2225, AGCTACAGAGTGAATTCAAA |
The underlined sequences indicate the restriction endonuclease sites. Italic letters indicate that bases have been changed.
DNase I footprint analysis.
The preparation of nuclear extracts and the method to generate DNA probes were as described elsewhere (7, 19). To generate sense probes, the 5′ primers RV3, Dmrt1.30 (2), or Dmrt1.29 were radiolabeled by T4 polynucleotide kinase (New England Biolabs, Inc.) and used with the 3′ primer Dmrt1-2750 to amplify the upstream promoter region between bp −3280 and −2750, bp −3111 and −2750, or bp −2910 and −2750 (Table 1). For antisense probes, the 3′ primers Dmrt1-2985 and Dmrt1-2750 were radiolabeled and used with the 5′ primer RV3 or Dmrt1-3000 to amplify the upstream promoter region between bp −3280 and −2985 or between bp −3000 and −2750, respectively (Table 1). For both sense and antisense probes, PCR was performed using Dmrt1(−3,280/+75)Luc as template and Bio-X-ACT DNA polymerase, according to the manufacturer's recommendations (Intermountain Scientific Corp.). DNase I footprinting using nuclear extracts from primary cultures of either Sertoli cells or TM4 cells was performed as previously described (19).
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were performed as described elsewhere (7, 19). Competitors (Table 3) and antibodies, when included, were added to the reaction mixture immediately before the addition of nuclear extracts. Competitors were added at a concentration 100 times that of the probe, unless otherwise noted. Antibodies for Gata1, Gata2, Gata3, and Gata4 were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, Calif.). Antibodies were supplied as rat monoclonal (Gata-2 and Gata-3), goat polyclonal (Gata-1), and rabbit polyclonal (Gata-4) immunoglobulin G and used (1 or 2 μg per binding reaction mixture) directly as supplied by the manufacturer (Santa Cruz Biotechnology, Inc.).
TABLE 3.
Sequences of oligodeoxynucleotides used in EMSAs
| Oligodeoxynucleotide | Sequencea |
|---|---|
| Region IV | ACTCAGCCTCTCCTATCTTTGCG |
| Region IV μ1 | ACTCAGCCTCTCCagggTTTGCG |
| Region IV μ2 | ACTCAGaagaTCCTATCTTTGCG |
| Region V | TAATGCACAAATAAACAGATAGAGCGGG ATTGTTCACTCT |
| Region V μ1 | TAATGCACAAATAAACAaggcGAGCGGGAT TGTTCACTCT |
| Region V μ2 | TAATGCACAAATAAACAGATAGAGCGGGc ggaccCACTCT |
| Region VI | CCTGTAGGATTACTGTATTGGCCTTTAACC CTGGG |
| Region VI μ1 | CCTGTAGGAggcaTGTATTGGCCTTTAACCCT GGG |
| Region VI μ2 | CCTGTAGGATTACTGTATgagaCTTTAACCCT GGG |
| Region VII3′ | ATGTAAAGATCTGTGCCATCTCTTTGGGG |
| Region VII3′ μ1 | ATGTAActcgaTGTGCCATCTCTTTGGGG |
| Region VII3′ μ2 | ATGTAAAGATCTGTtaacTCTCTTTGGGG |
| Region VII3′ μ1/2 | ATGTAActcgaTGTtaacTCTCTTTGGGG |
| Region VII5′ | GGACATATTTCTCAGTAATCATGTAAAGA |
| Region VII5′ μ1 | GGACATATTTCTCAtgtcTCATGTAAAGA |
| Region VII5′ μ2 | GGACATATTTCTCAGTAATCATtgccAGA |
| Region VII5′ μ3 | GGACATATTTCTCAGTAcatATGTAAAGA |
| Region VII5′ μ4 | GGACcgcagTCTCAGTAATCATGTAAAGA |
| Gata consensus | CACTTGATAACAGAAAGTGATAACTCT |
| C/E BP consensus | TGCAGATTGCGCAATCTGCA |
| Sox9 consensus | GGGTTAACAGAACAATGGAATCTGGTAGA |
| Sp1 consensus | ATTCGATCGGGGCGGGGCGAGC |
| NS | CTAGAGTCGACCTGCAGGCATGCAAGCTT GGCATTC |
The lowercase letters indicate that bases have been changed.
Semiquantitative RT-PCR and Southern blotting.
mRNA levels of Dmrt1 were evaluated using semiquantitative reverse transcription-PCR (RT-PCR) as described elsewhere (7). Briefly, cDNA synthesis was performed using 0.5 μg of oligo(dT) and 2 μl of RNA template in 10-μl reaction volumes in the presence of reverse transcriptase Superscript II (Invitrogen, Carlsbad, Calif.). One microliter of the synthesized cDNA was used as template in an amplification reaction to test for the presence of mRNA of the ribosomal protein L7 using primer pairs L7.1 and L7.2 (L7.1, 5′-GGGGGAAGCTTCGAAAGGCAAGGAGGAAGCT-3′; L7.2, 5′-GGGGGGTCGACTCCTCCATGCAGATGATGCC-3′). For quantification of Dmrt1 expression, PCR was performed using 2 μl of cDNA and the Dmrt1-specific primer pair mDmrt1.1 and mDmrt1.2 (mDmrt1.1, 5′-CGAGCTCCTGGTCAAAAGAG-3′; mDmrt1.2, 5′-GGCCCGTAGTATGAGTGCAT-3′). The primer pair spans 458 bp from exon 2 to exon 4 within the Dmrt1 cDNA sequences and thus can distinguish between contaminating genomic DNA and processed mRNA. The PCR cycle numbers were determined to assure amplification in the linear ranges. Seventeen cycles were used for L7 (94°C for 30 s, 52°C for 30 s, and 72°C for 30 s), and 30 cycles were used for Dmrt1 (94°C for 30 s, 52°C for 30 s, and 72°C for 30 s). Products were resolved by agarose gel electrophoresis and analyzed by Southern blot assays. 32P-5′-end-labeled primers L7.1 and Dmrt1.1 (5′-CCTACTACAGCAG-3′, located between the amplification primers for Dmrt1) were used as probes to detect the amplified DNA products, and the signal strength was determined using phosphorimaging (PhosphorImager; Molecular Dynamics, Sunnyvale, Calif.).
In these studies, 17 gonads from 13.5-dpc Fog2−/− embryos and their littermates were obtained from Sergei Tevosian. Total RNA was isolated using an RNeasy mini kit, according to the manufacturer's recommendations (Qiagen). Among the 17 gonads, there were 5 XY mutant, 5 XY wild type or heterozygous, 1 XX mutant, and 3 XX wild type or heterozygous. Genotype was determined by the morphological characteristics of the gonads and confirmed by RT-PCR analysis for Fog2 using the primer pair Fog2 Forward (2) (5′-CCAGCAGTATTCATAGCTGTGGT-3′) and Fog2 Reverse (2) (5′-GAGACAGGGCTGCATCGG-3′), which are located in the deleted region of the mutant Fog2 gene. Wild-type transcripts resulted in a 536-bp DNA fragment. RT-PCR using the Sry-specific primers Sry forward (5′-AAGCGCCCCATGAATGCAT-3′) and Sry backward (5′-CGATGAGGCTGATATTTATA-3′) was performed to determine the chromosomal sex of the gonads, with males having a 218-bp amplified DNA product.
Three independent cDNA synthesis reactions were performed for each RNA sample. Semiquantitative RT-PCR using Dmrt1- and L7-specific primers and Southern blot analysis were performed with each set of cDNA. Each sample was represented as the Dmrt1 signal relative to the L7 signal. Data were calculated relative to the average value of the Fog2+/+/Fog2+/− XY group and then averaged across each independent amplification group.
RESULTS
Transcriptional activity of the 5-kb Dmrt1 promoter is highest in primary cultures of Sertoli cells.
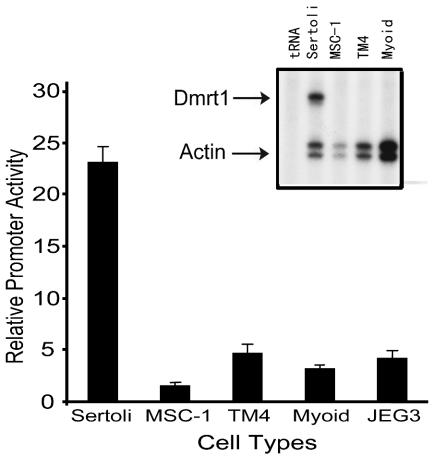
To evaluate the mechanisms regulating Dmrt1 transcription, a portion of the rat gene from bp +75 to approximately 5000 5′ to the transcriptional start site was cloned upstream of the firefly luciferase reporter gene [Dmrt1(−5,000/+75)Luc], and the activity of the promoter was evaluated in a variety of cell types using transient-transfection analysis. Since previous studies had shown that Dmrt1 was expressed in primary cultures of Sertoli cells, we used these as a positive cell type for the evaluation of Dmrt1 transcription (19, 29). In addition, the promoter construct was assayed in different nonexpressing cell types, as determined by RNase protection analysis (Fig. 1, inset). These included TM4 and MSC-1 cells, mouse cell lines derived from Sertoli cells, and myoid cells, another testicular cell type that was prepared as a primary culture. JEG3 is a human choriocarcinoma cell line and was included as a fourth nonexpressing cell type. The data showed that the activity of Dmrt1(−5,000/+75)Luc was significantly higher in primary Sertoli cells than in nonexpressing cell types (Fig. 1). Thus, transcriptional activity of Dmrt1(−5,000/+75)Luc appears to reflect the activity of the endogenous gene and suggests that the 5-kb fragment contains elements important for cell-specific regulation of the gene.
FIG. 1.
Dmrt1 promoter activity is highest in primary cultures of Sertoli cells. Dmrt1(−5,000/+75)Luc was transfected into various cell types together with pRL-TK. The data are represented as the firefly/Renilla luciferase activity ratio of Dmrt1(−5,000/+75)Luc relative to the firefly/Renilla luciferase activity ratio of pGL3-Basic. Transfections were done a minimum of three times. Error bars represent the standard errors of the means. The inset shows an RNase protection analysis for Dmrt1 mRNA. RNA samples were isolated from primary rat Sertoli cells (SC), Sertoli cell lines MSC-1 and TM4, and primary myoid cells. tRNA was added as a negative control.
The region between bp −3280 and −2750 is required for activity of the Dmrt1 promoter in primary cultures of Sertoli cells.
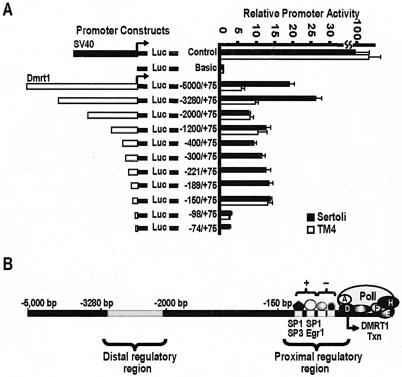
Sequential deletions of the 5-kb promoter were generated from Dmrt1(−5,000/+75)Luc, and the activities were compared by transient-transfection analysis in primary Sertoli cells and the cell line TM4. In both cell types, a dramatic drop in promoter activity was observed when the region between bp −150 and −98 was deleted (Fig. 2A). Previous characterization of this proximal promoter region showed that it contains two positive and two negative elements, and the positive elements bind the transcription factors Sp1, Sp3, and Egr1 (Fig. 2B) (19). The observed decrease in promoter activity in both cells types with deletion of the proximal regulatory region suggests that these elements are also functional in TM4 cells. Also, in both cell types the promoter activity of Dmrt1(−5,000/+75)Luc was higher than that of Dmrt1(−3,280/+75)Luc, suggesting a potential repressor element residing between bp −5000 and −3280 of the Dmrt1 promoter (Fig. 2A).
FIG. 2.
Two regulatory regions are important for Dmrt1 transcription. (A) Various 5′-deletion mutants were generated from Dmrt1(−5,000/+75)Luc and characterized by transient-transfection analysis in either primary Sertoli cells or TM4 cells. The data are represented as the firefly/Renilla luciferase activity ratio of each construct relative to the firefly/Renilla luciferase activity ratio of pGL3-Basic (black bars are for primary Sertoli cells and white bars are for TM4 cells). Control represents the pGL3-Control vector, which contains the SV40 promoter and enhancer sequences. The −400/+75, −300/+75, −221/+75, −179/+75, and −75/+75 constructs were not tested in TM4 cells. Transfections were done a minimum of three times. Error bars represent the standard errors of the means. (B) Schematic of the Dmrt1 promoter region. Results in panel A identified distal (bp −3280/−2000) and proximal (below bp −150) regulatory regions that contribute to activity of the Dmrt1 promoter in primary Sertoli cells.
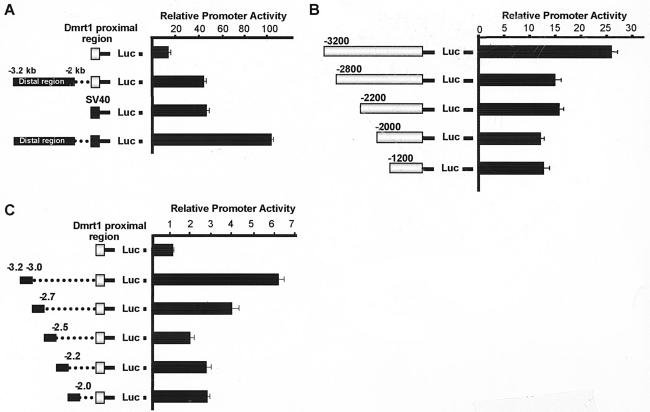
A second important promoter region was found between bp −3280 and −2000 (Fig. 2A). Interestingly, activity of this distal regulatory region was not apparent when tested in TM4 cells, indicating that important positive regulatory elements specific to the Dmrt1-expressing cells reside in the distal regulatory region (Fig. 2A). Further analysis of the distal regulatory region was performed by transient-transfection assays using refined sequential deletions and chimeric promoter constructs. The transcriptional activity of the distal regulatory region from kb −3.2 to −2.0 was tested when placed directly upstream of either the Dmrt1 proximal region (bp −150/+75) or the SV40 promoter. Transient-transfection analysis revealed a two- to threefold increase in promoter activity in the presence of the distal regulatory region, regardless of the nature of the proximal promoter (Fig. 3A). Thus, the context of the proximal promoter and sequences between bp −2.0 and −150 is not essential for activity of the distal regulatory region. Additional deletion mutagenesis showed that transcription was significantly reduced when the promoter was deleted from kb −3.2 to −2.8, indicating the presence of important regulatory elements within this region (Fig. 3B). Consistent with this finding, transient-transfection analysis of chimeric constructs containing overlapping sequence blocks (∼250 bp each) that span the distal regulatory region identified sequences between kb −3.2 and −3.0 and between kb −3.0 and −2.7 as containing the majority of the region's activity (Fig. 3C).
FIG. 3.
Activity of the distal regulatory region associates primarily with the region between bp −3280 and −2750. (A) Dmrt1 promoter sequences between bp −3280 and −2000 were cloned upstream of the Dmrt1 proximal promoter (bp −150/+75) or the SV40 promoter and assayed for promoter activity by transient-transfection analysis in primary Sertoli cells. (B) Additional 5′-deletion mutants were generated between bp −3280 and −1200 and assayed as above. (C) Sequential 250-bp blocks of the distal regulatory region were cloned upstream of the Dmrt1 proximal promoter (bp −150/+75) and assayed as above. Each 250-bp upstream sequence overlaps 15 bp of its neighbors. Relative promoter activity represents the luciferase/Renilla of each promoter construct relative to the luciferase/Renilla activity of Dmrt1(−150/+75)Luc. Transfections were done a minimum of three times. Error bars represent the standard errors of the means.
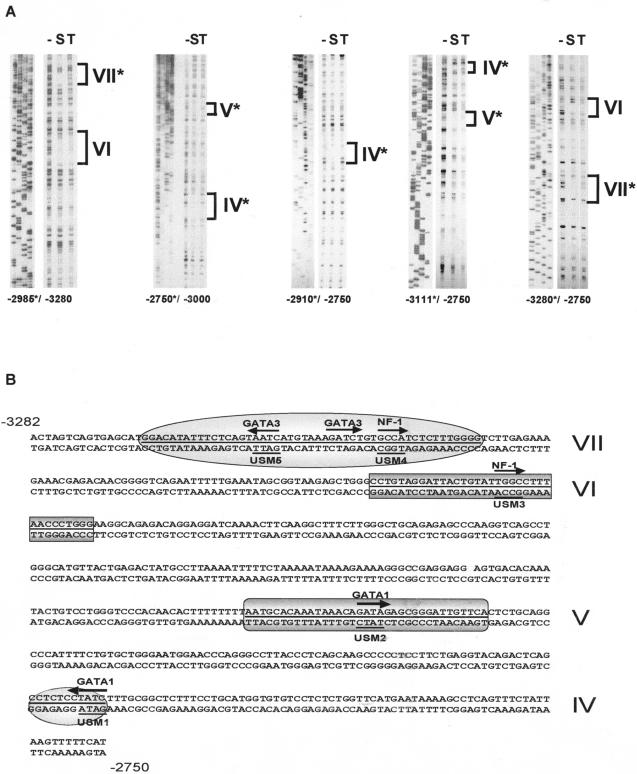
Four protein binding sites reside between bp −3280 and −2750.
To disclose regulatory elements within the distal regulatory region between bp −3280 and −2750, DNase I footprint analysis was preformed using nuclear extracts isolated from primary cultures of Sertoli cells or TM4 cells. Five end-labeled probes were synthesized to span the Dmrt1 promoter from bp −3280 to −2750. These data identified four regions that were protected from DNase I digestion in the presence of Sertoli cell nuclear extracts (Fig. 4A, regions IV to VII). Interestingly, three of the four footprinted regions were specific to primary Sertoli cells (Fig. 4A, regions IV, V, and VII). Sequence analysis using a transcription factor database (43) identified potential binding sites for Gata transcription factors and NF-1 (Fig. 4B).
FIG. 4.
Four prominent protein binding sites are located within the bp −3280 to −2750 region. (A) DNase I footprint analysis of the distal regulatory region of the Dmrt1 promoter. −2985*/−3280 and −2750*/−3000 represent antisense probes, while −2910*/−2750, −3111*/−2750, and −3280*/−2750 represent sense probes. The asterisks mark the radiolabeled primers. The probes were incubated either in the absence of nuclear extracts (−) or the presence of primary Sertoli (S) or TM4 (T) cell nuclear extracts and then digested with DNase I. Dideoxynucleotide sequencing ladders were generated using Dmrt1(−3,280/+75)Luc as a template and the same radiolabeled primer used in the associated footprint reactions. Regions protected from DNase I digestion are marked IV to VII. Regions protected in the presence of nuclear extracts from primary rat Sertoli cells but not TM4 cells have an asterisk. (B) Positions and sequences of the DNase I-protected sites within the Dmrt1 distal regulatory region from bp −3282 to −2750. Protected regions IV to VII are shaded, and the names of the potential binding sites are labeled on the top of sequences. Sites mutated to test the function of the potential binding sites are underlined and marked as usm1, usm2, usm3, usm4, and usm5.
Gata4 binds three functionally important DNA response elements within the distal regulatory region of the Dmrt1 promoter.
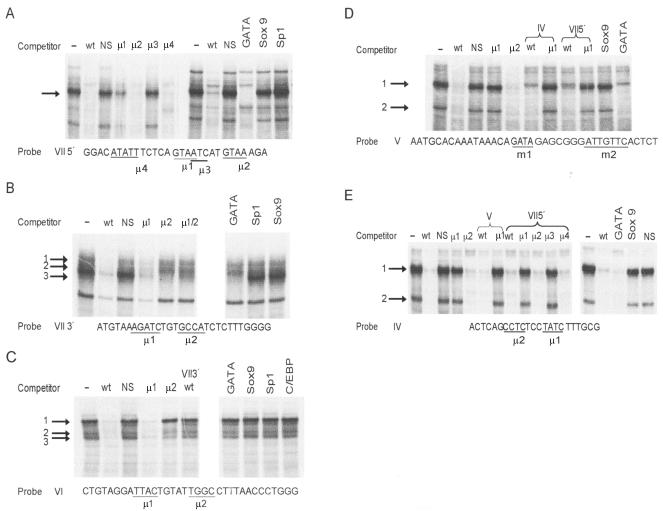
EMSAs were performed to further evaluate the proteins binding within the four protected regions. Specifically bound protein complexes were observed within each of the four regions, as revealed by elimination of the complexes with the addition of competitor DNAs containing homologous (wild-type) but not nonspecific sequences (Fig. 5, IV to VII). Studies on region VII utilized two overlapping oligodeoxynucleotides, VII5′ and VII3′. One major protein complex and several minor complexes bound to the VII5′ probe and were competed with an oligodeoxynucleotide containing a consensus Gata binding site, but not with consensus Sox9 or Sp1 binding sites (Fig. 5A). With the addition of various mutant oligodeoxynucleotides, differences were observed in their abilities to eliminate protein binding. Competitor oligodeoxynucleotides μ2 and μ4 fully competed for proteins binding to the probe, while μ1 and μ3 were less effective. Thus, bases mutated at sites of μ1 (GTAA) and μ3 (ATC, whose reverse complement is GAT) were required for the observed DNA-protein interaction. Three specific binding complexes bound the VII3′ probe, and the Gata consensus element competed for complex 3. In addition, competition analysis showed that the bases mutated within μ2 (GCCA) were needed for binding of complexes 1 and 2 (Fig. 5B). Three major binding complexes were also observed on the region VI probe, with complex 1 showing a requirement for bases in site μ2 (TGGC) for binding (Fig. 5C). The reverse complement of this binding site is GCCA, which is identical to the binding site for complexes 1 and 2 on the VII3′ probe. However, VII3′ wild type could not compete with the complexes formed on the region VI probe (Fig. 5C), suggesting that bases other than TGGC are required for protein binding.
FIG. 5.
Gata sequences are important for nuclear protein binding within the distal regulatory region of the Dmrt1 promoter. In EMSAs, oligodeoxynucleotides that span the footprinted regions were radiolabeled and used as probes together with nuclear extracts from primary Sertoli cells. Oligodeoxynucleotide sequences of each probe are shown underneath the respective EMSA, with sites mutated for competitor oligodeoxynucleotides underlined. Wild-type (wt), nonspecific (NS), and consensus oligodeoxynucleotides of C/EBP, GATA, Sp1, and Sox9Competitor oligodeoxynucleotides were included as competitors. The major DNA-protein complexes are indicated by arrows. (A) EMSAs using an oligodeoxynucleotide probe to the 5′ end of region VII (bp −3249 to −3221 bp). VII5′ μ1, μ2, μ3, and μ4 are mutated oligodeoxynucleotides used as competitors. (B) EMSAs using an oligodeoxynucleotide probe to the 3′ end of region VII (bp −3229 to −3201). VII3′ μ1, μ2, and μ1/2 are mutated oligodeoxynucleotides used as competitors. (C) EMSAs using an oligodeoxynucleotide probe to region VI (bp −3141 to −3107). VI μ1 and μ2 are mutated oligodeoxynucleotides used as competitors. VII3′ wt was also included as a competitor. (D) EMSAs using an oligodeoxynucleotide probe to region V (bp −2934 to −2896). V μ1 and μ2 are mutated oligodeoxynucleotides used as competitors. IV wt, IV μ1, VII5′ wt, and VII5′ μ1 were also included as competitors. (E) EMSAs using an oligodeoxynucleotide probe to region IV (bp −2821 to −2799). IV μ1 and μ2 are mutated oligodeoxynucleotides used as competitors. V wt, V μ1, and VII5′ μ1 to μ4 were also included as competitors.
The major complexes bound to region IV and V probes were competed with the Gata consensus sequence, and the required bases for each were within a Gata binding site (μ1, GATA for region V probe [Fig. 5D] and μ1, TATC, whose reverse complement is GATA, for region IV probe [Fig. 5E]). Thus, proteins binding regions IV, V, and VII5′ all interacted with Gata sequences within these regions. Furthermore, cross-competition of the sequences from these regions revealed that regions IV, V, and VII5′ were capable of binding the same protein complexes (Fig. 5D and E). Thus, Gata transcription factors are implicated in binding to three of the four footprinted regions, and two additional protein complexes interact with TGGC sequence, or its reverse complement GCCA, within regions VII3′ and VI. Interestingly, additional studies of regions IV, V, and VII5′ revealed that the major binding complexes were either diminished or absent from TM4 and myoid cell nuclear extracts (data not shown).
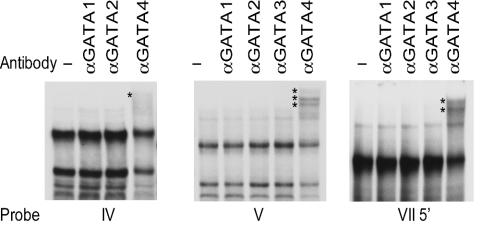
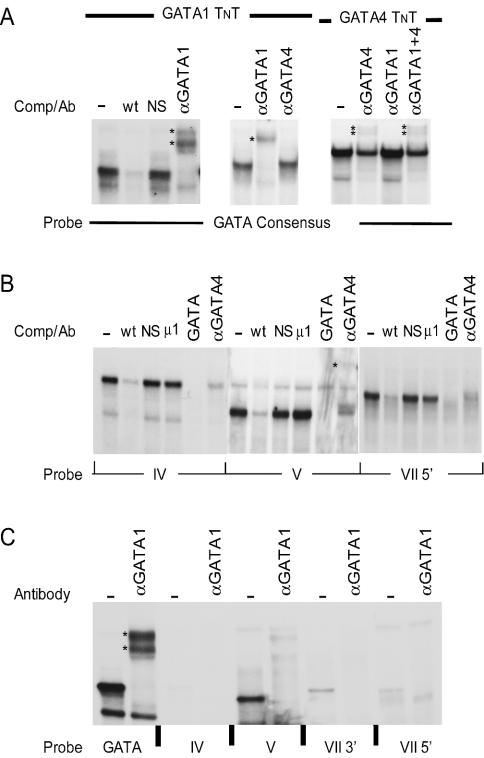
Antibodies specific for Gata1, Gata2, Gata3, and Gata4 were used to determine if members of the Gata family are present in the complexes observed in regions IV, V, and VII5′. In each region, only the antibody generated against Gata4 was able to diminish or supershift the major complexes in Sertoli cells (Fig. 6). These diminished or supershifted complexes were similar to reactions that included in vitro-transcribed-translated Gata4 protein and a Gata consensus probe (Fig. 7A). As a complementary approach, each of the probes was incubated with in vitro-transcribed-translated Gata1 or Gata4 protein. Gata4 protein interacted with regions IV, V, and VII5′ and required the same bases needed for interactions with the Sertoli cell proteins (Fig. 7B). In contrast, Gata1 protein appeared to bind less efficiently to probes against any of the regions, despite the presence of adequate Gata1 protein, as determined by binding to the Gata consensus probe (Fig. 7C). These studies indicated that Gata4, not Gata1, is the major protein binding to regions IV, V, and VII5′.
FIG. 6.
Sertoli cell complexes formed on regions IV, V, and VII5′ contain Gata4. EMSAs were performed using oligodeoxynucleotide probes of region IV, V, or VII5′ and nuclear extracts from primary rat Sertoli cells. When indicated, antibodies against the transcription factors Gata1 (αGATA1), Gata2 (αGATA2), Gata3 (αGATA3), or Gata4 (αGATA4) were added to the reactions. Asterisks mark the supershifted complexes.
FIG. 7.
In vitro-transcribed-translated (TNT) Gata4 binds to regions IV, V, and VII5′. (A) EMSAs using a consensus Gata probe with Gata1 or Gata4 TNT reactions. Where indicated, antibodies against the transcription factors Gata1 (αGATA1) or Gata4 (αGATA4), or a combination of αGATA1 and αGATA4 (αGATA1 + 4), were added to the reactions. Consensus Gata wild-type (wt) and nonspecific (NS) oligodeoxynucleotides were used as competitor. (B) EMSAs using oligodeoxynucleotide probes to regions IV, V, and VII5′ and Gata4 TNT reactions. Antibody against the transcription factor Gata4 (αGATA4) was added to the indicated reactions. Wild-type (wt) and mutant oligodeoxynucleotide μ1 for regions IV, V, or VII 5′ were used as competitors. For each probe, nonspecific (NS) and consensus Gata wild-type (GATA) oligodeoxynucleotides were also used as competitors. (C) EMSAs using a Gata consensus probe or oligodeoxynucleotide probes to regions IV, V, VII3′, and VII 5′ and Gata1 TNT reactions. Antibody against the transcription factor Gata1 (αGATA1) was added to the indicated reactions.
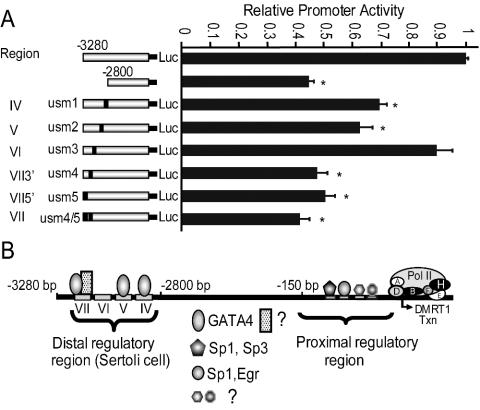
To determine the functional importance of these protein binding sites, the same mutations shown to disrupt protein binding were introduced into the Dmrt1(−3,280/+75)Luc construct and evaluated for their impact on transcriptional activity via transient-transfection analysis. Mutations usm1, usm2, usm4, and usm5 (Fig. 4B) significantly diminished Dmrt1 promoter activity in primary Sertoli cells, similar to that observed with deletion of the entire region between bp −3280 and −2800 (Fig. 8A). In contrast, the usm3 mutation did not have a significant impact on promoter activity (Fig. 8A). Furthermore, a double mutation of usm4 and usm5 had no greater effect on promoter activity than either mutation alone, suggesting proteins binding these elements act synergistically in their regulation of Dmrt1 (Fig. 8A). As a whole, the data showed that the nucleotides required for several of the described protein-DNA interactions are the same as those required for function of the response elements, implicating Gata4 as well as other factors in the regulation of Dmrt1 promoter activity (Fig. 8B).
FIG. 8.
Three Gata4 binding sites and a second site within region VII are important for transcriptional activity of the Dmrt1 promoter. (A) Promoter constructs containing mutations usm1, usm2, usm3, usm4, ums5, and usm4/5 (Fig. 4) were generated in the context of Dmrt1(−3,280/+75)Luc and examined by transient-transfection analysis in primary rat Sertoli cells. The data represent the firefly/Renilla luciferase activity ratio of each construct relative to the firefly/Renilla luciferase activity ratio of Dmrt1(−3,280/+75)Luc. Transfections were done a minimum of three times, and error bars represent the standard errors of the means. *, P < 0.05 by Student's t test. (B) Model of Dmrt1 transcription in Sertoli cells. The transcriptional apparatus of mRNA polymerase II is assembled at the transcriptional start site of Dmrt1 through the participation of transcription factors residing within the Dmrt1 promoter. Gata4 (oval) and another unidentified transcription factor (rectangle) regulate transcription through the distal regulatory region (bp −3280 to −2800) and Sp1, Sp3, and Egr1 function in the proximal regulatory region (19).
The Gata4 coactivator Fog2 is important for Dmrt1 expression in embryonic testes.
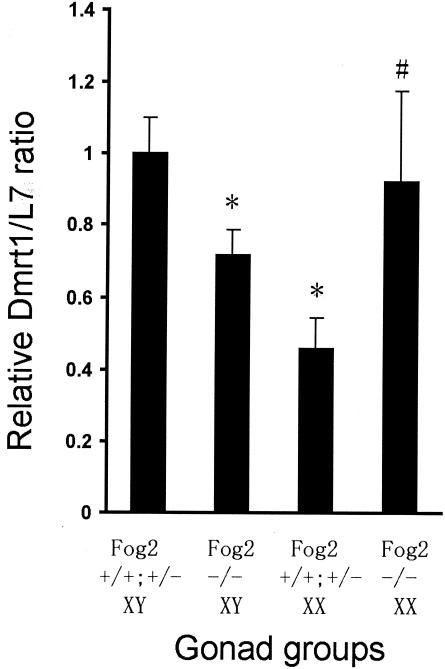
To help verify Gata4's role in the transcriptional regulation of Dmrt1 in vivo, Dmrt1 mRNA abundance was measured in mouse embryos lacking the essential Gata4 cofactor Fog2. Semiquantitative RT-PCR was used to measure Dmrt1 mRNA in 13.5-dpc gonads from Fog2−/− mice, and this was compared to that in their wild-type and heterozygous littermates. Chromosomal sex was determined by PCR using Sry-specific primers (data not shown). The amount of amplified Dmrt1 was standardized to the amount of amplified L7, revealing that Dmrt1 expression in Fog2 null XY gonads was significantly lower than in wild-type and Fog2 heterozygous XY gonads (Fig. 9, Fog2+/+/Fog2+/− and Fog2−/− XY). Since Fog2 is an essential cofactor for Gata4, this result suggests a difference in male Dmrt1 transcription in the presence and absence of Gata4. In contrast, Dmrt1 expression in the single XX mutant gonad was highly increased compared to that in the wild-type and Fog2 heterozygous XX gonads (Fig. 9, Fog2+/+/Fog2+/− and Fog2−/− XX). Despite the limited sample number for Fog2−/− XX gonads, the data suggest that the Gata4/Fog2 complex differentially regulates Dmrt1 transcription in male and female gonads.
FIG. 9.
Dmrt1 expression is decreased in embryonic testes from 13.5-dpc Fog2−/− mice. Semiquantitative RT-PCR was used to determine mRNA levels for Dmrt1 and ribosomal protein L7. The data are represented as the Dmrt1/L7 ratio of Fog2+/+ and Fog2+/− XY gonads (n = 5), Fog2−/− XY gonads (n = 5), Fog2+/+ and Fog2+/− XX gonads (n = 3), and Fog2−/− XX gonads (n = 1) relative to the average Dmrt1/L7 ratio in Fog2+/+ and Fog2+/− XY gonads. For each sample, semiquantitative RT-PCR was done three times. Error bars represent the standard errors of the means. *, P < 0.05 by Student's t test, relevant to Fog2+/+ and Fog2+/− XY gonads; #, P = 0.127 by Student's t test, relevant to Fog2+/+ and Fog2+/− XX gonads.
DISCUSSION
The process of sex determination in mammals is controlled by a hierarchy of genes that is initiated by the actions of the sex-determining region on Y (Sry), which induces formation of a testis from a bipotential gonad. Human mutations associated with sexual disorders or sex reversal and studies of induced mouse mutations that impair sex determination have identified several genes that participate in this process, including the DSS-AHC critical region on X, gene 1 (Dax-1), Dmrt1, Emx2, Gata4, Lim1, Lhx9, steroidogenic factor-1 (Sf-1), Sry-related HMG box-containing gene 9 (Sox9), and Wilms' tumor suppressor gene 1 (Wt1). Among these genes, Sf-1, Wt1, Lim1, Lhx9, and Emx2 are required for formation of the genital ridge and Sf-1, Wt1, Sox9, Dax-1, Gata4, and Dmrt1 are implicated in later stages of embryonic and postnatal testis differentiation (3, 17, 23, 30, 33, 34, 37). All of the above genes are transcription factors, and most of them, except Sry and Dmrt1, are involved in development of tissues other than the gonad. From studies of organogenesis, including gonadogenesis, there is an acknowledgment that the hierarchical control of developmentally important genes is the guidance for morphogenetic events that require specific temporal and spatial cues. However, details of this control are poorly defined in the testis. As a transcription factor that is solely expressed in the testis and implicated in the cascade of testis differentiation, Dmrt1 may hold a key position in this genetic pathway. Thus, studies of Dmrt1's transcriptional mechanism will not only reveal unique features of testicular transcriptional regulation but also will further our understanding of the mechanisms driving gonadogenesis. In this paper, we found that Gata4, an important transcription factor in testis determination and differentiation, regulates Dmrt1 expression in the testis. This discovery not only identified a critical component of the transcriptional regulatory mechanism for Dmrt1, but it also revealed that Gata4, through Dmrt1, is important in Sertoli cell differentiation and later testicular development.
The importance of Gata4 in heart morphogenesis and development of coronary vessels from epicardium was shown through studies of Gata4 null mice (6, 18, 25). Since these mice died prior to formation of the urogenital system, the function of Gata4 in gonadal morphogenesis was not detectable (18, 25). Fortunately, recent genetic evidence in mice null for Fog2 (Fog2−/−) and mice with a targeted mutation in Gata4 that disrupts binding to Fog2 (Gata4ki/ki) illustrated Gata4's functional role in the genetic pathway of mammalian sex determination (37, 39). Gata4 belongs to the evolutionary conserved family of Gata transcription factors, each of which contains a highly conserved two-zinc-finger DNA binding domain and is indispensable for embryonic development (16, 22, 24). Of this family, the subfamily of Gata4/5/6 plays an active role in the formation and differentiation of various mesoderm-derived tissues, while the subfamily of Gata1/2/3 predominantly regulates hematopoiesis (24, 28). In addition, Gata factors are often important for cell-specific gene expression but frequently require the cooperation of other more-restricted factors, such as with the cooperative interactions between Sf-1 and Gata4 in the transactivation of Mullerian inhibiting substance (Mis) (40).
Fog2 is a member of the friends of Gata (Fog) family and is a required partner of Gata4 as well as Gata5 and Gata6 (20, 38, 39, 41). Fog2 is expressed predominantly in heart, brain, and testis and interacts directly with the N-terminal fingers of Gata4 to regulate Gata4-dependent transcription, whereby it functions as either an activator or repressor, depending on the promoter and cell type (20, 36, 38). Deletion of Fog2 in mice resulted in mid-gestation lethality (14.5 dpc) due to cardiac defects (37). Since Fog2 regulates Gata4 and the same developmental abnormalities were observed in Gata4ki/ki and Fog2−/− mice, the Fog2−/− defects are likely due to loss of the functional Gata4-Fog2 interaction (6, 37, 39).
In Fog2−/− and Gata4ki/ki mice, differentiation of the testis failed (37, 39). With both mutations, the gonadal defect initiates with testis determination (11.5 dpc) and is marked by a significant decrease in Sry, supporting Gata4's regulatory importance in pre-Sertoli cells. In addition, the normal testis-specific induction of Sf1, Wt1, and Gata4 (or Gata4ki/ki) was not observed in 12.5-dpc XY gonads from the mutant mice. However, until 11.5 dpc, expression of these factors was similar to that in their wild-type littermates. Defects were most prominent in Sertoli cell differentiation, as noted by the apparent transcriptional block in the Sertoli cell-expressed genes Sox9, Mis, and desert hedgehog gene (Dhh) (37). Of these genes, only Mis has been identified as a direct target of Gata4's regulation (40). The loss of Mis expression in Fog2−/− testes reflected the importance of Fog2 modulation on Gata4's activation of Mis.
Given that Fog2 can function as either an activator or repressor, our observations of Dmrt1 expression in Fog2−/− mouse embryos and the contribution of the Gata4 binding sites to Dmrt1 promoter activity are particularly intriguing. The decreased Dmrt1 expression in the Fog2−/− testis indicates that Fog2 is acting as a transcriptional activator of Dmrt1 through Gata4. In contrast, the limited data set for the Fog2−/− ovary indicated elevated Dmrt1 expression, suggesting that Fog2 acts as a repressor in this cell type. Our favored interpretation of these results is that in the cells destined to be Sertoli cells, Gata4 binds the three Gata response elements within the Dmrt1 promoter and recruits Fog2, which acts as a transcriptional activator of the gene. However, if the same precursor cells in an XX background differentiate down the granulosa cell path, the different cellular context causes the Gata4/Fog2 complex to repress Dmrt1 transcription. This would lead to the sexually dimorphic expression pattern observed for Dmrt1. It will be interesting to see if other genes that demonstrate sexually dimorphic expression are similarly regulated by the Gata4/Fog2 complex.
Gata4's direct regulation of Dmrt1 is also supported by their similar expression patterns after Sry determines gonadal sex at 11.5 dpc and until the neonatal stage. Thus, Gata4 is expressed specifically in the developing Sertoli/granulosa cell lineage throughout embryonic gonadogenesis (42). In the ovary, Gata4 is dramatically down-regulated shortly after differentiation is initiated at 13.5 dpc, while in the testis expression is markedly higher (42). Notably, expression of Dmrt1 in embryonic mouse gonads begins to show this same dimorphic pattern that favors testis expression by 13.5 dpc (8, 29). Is the similar ontogeny of both genes in embryonic gonads a coincidence or a reflection of Gata4's regulation of Dmrt1? The study herein provides convincing evidence for the latter, as Gata4 binds three critical regulatory elements within the Dmrt1 promoter and Dmrt1 expression is significantly reduced in the absence of Gata4's required coregulator Fog2 in 13.5-dpc testis.
Gata4's direct regulation of Dmrt1 in postnatal testicular development is supported by recent studies by Mikko Anttonen et al., which showed that Gata4 expression in the postnatal testis reaches its peak at day 14 and is maintained throughout adulthood (2). This expression profile is in agreement with postnatal Dmrt1 expression, which is most robust at day 15 and persists evenly afterwards (5). However, there are also earlier reports that contradict the above findings regarding temporal Gata4 expression in postnatal testis. As reported in these papers, Gata4 expression is particularly intense in Sertoli cells of neonatal testis until day 4 and appears somewhat weaker or is entirely lost in pubertal and adult testes (13, 14, 42). However, in evaluating some of the data on Gata4 and Dmrt1 expression in the postnatal testis, changes in Sertoli cell levels cannot be easily deduced (from Northern or Western blot analyses) because of the significant dilution effect from induction of nonexpressing germ cells. Nonetheless, if this conflicting profile reflects the in vivo trend of Gata4 expression, the discrepancy between Gata4 and Dmrt1 expression suggests that there is a shift in the mechanism directing Dmrt1 transcription between embryonic and early postnatal stages and that in the adult. Thus, initial Dmrt1 expression through early puberty requires Gata4, while later stages are controlled by a Gata4-independent mechanism. Reports of Gata1 show that its expression pattern is similar to that of Dmrt1 in the postnatal testis (42, 44). This leads to the intriguing possibility that Gata1 replaces Gata4 in Dmrt1 expression at the latter stages of testicular development. However, our in vitro binding data indicate that Gata1 does not bind well to the upstream regulatory regions. So, if Gata1 regulation occurs, it is likely that Gata1 uses different regulatory elements than Gata4.
Unlike Dmrt1, Gata4 is expressed in many tissues and is involved in morphogenesis of multiple organs. Thus, it is doubtful that it acts alone to restrict expression of Dmrt1 to the testis. Rather, we hypothesize that Gata4 helps control temporal and spatial expression of Dmrt1 through its ability to interact with other spatially defining transcription factors and induce robust expression of Dmrt1 in the testis. Notably, we also identified a second protein binding site (usm4) within region VII that synergizes with a nearby Gata4 binding site (usm5). The nature of this interaction awaits further characterization but may reveal important insight into the role of Gata4 in Dmrt1 expression and its restriction to the testis.
Acknowledgments
We thank Jiang-kai Chen, Jeremy Presley, and Kaori Hornbaker for their preparations of Sertoli cells and technical assistance. We thank Sergei Tevosian for the generous gift of Fog2 null gonads.
This work was supported by the Madison and Lila Self Graduate Fellowship and the National Institute of Child Health and Development (grant HD41056 to L.L.H.).
REFERENCES
- 1.Albrecht, K. H., and E. M. Eicher. 2001. Evidence that Sry is expressed in pre-Sertoli cells and Sertoli and granulosa cells have a common precursor. Dev. Biol. 240:92-107. [DOI] [PubMed] [Google Scholar]
- 2.Anttonen, M., I. Ketola, H. Parviainen, A. K. Pusa, and M. Heikinheimo. 2003. FOG-2 and GATA-4 are coexpressed in the mouse ovary and can modulate mullerian-inhibiting substance expression. Biol. Reprod. 68:1333-1340. [DOI] [PubMed] [Google Scholar]
- 3.Birk, O. S., D. E. Casiano, C. A. Wassif, T. Cogliati, L. Zhao, Y. Zhao, A. Grinberg, S. Huang, J. A. Kreidberg, K. L. Parker, F. D. Porter, and H. Westphal. 2000. The LIM homeobox gene Lhx9 is essential for mouse gonad formation. Nature 403:909-913. [DOI] [PubMed] [Google Scholar]
- 4.Calvari, V., V. Bertini, A. De Grandi, G. Peverali, O. Zuffardi, M. Ferguson-Smith, J. Knudtzon, G. Camerino, G. Borsani, and S. Guioli. 2000. A new submicroscopic deletion that refines the 9p region for sex reversal. Genomics 65:203-212. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J. K., and L. L. Heckert. 2001. Dmrt1 expression is regulated by follicle-stimulating hormone and phorbol esters in postnatal Sertoli cells. Endocrinology 142:1167-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crispino, J. D., M. B. Lodish, B. L. Thurberg, S. H. Litovsky, T. Collins, J. D. Molkentin, and S. H. Orkin. 2001. Proper coronary vascular development and heart morphogenesis depend on interaction of GATA-4 with FOG cofactors. Genes Dev. 15:839-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daggett, M. A., D. A. Rice, and L. L. Heckert. 2000. Expression of steroidogenic factor 1 in the testis requires an E box and CCAAT box in its promoter proximal region. Biol. Reprod. 62:670-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Grandi, A., V. Calvari, V. Bertini, A. Bulfone, G. Peverali, G. Camerino, G. Borsani, and S. Guioli. 2000. The expression pattern of a mouse doublesex-related gene is consistent with a role in gonadal differentiation. Mech. Dev. 90:323-326. [DOI] [PubMed] [Google Scholar]
- 9.Guan, G., T. Kobayashi, and Y. Nagahama. 2000. Sexually dimorphic expression of two types of DM (Doublesex/Mab-3)-domain genes in a teleost fish, the tilapia (Oreochromis niloticus). Biochem. Biophys. Res. Commun. 272:662-666. [DOI] [PubMed] [Google Scholar]
- 10.Heckert, L. L. 2001. Activation of the rat follicle-stimulating hormone receptor promoter by steroidogenic factor 1 is blocked by protein kinase A and requires upstream stimulatory factor binding to a proximal e box element. Mol. Endocrinol. 15:704-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heckert, L. L., M. Sawadogo, M. A. Daggett, and J. Chen. 2000. The USF proteins regulate transcription of the follicle-stimulating hormone receptor but are insufficient for cell-specific expression. Mol. Endocrinol. 14:1836-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horton R. M. 1993. In vitro recombination and mutagenesis of DNA, p. 251-261. In B. A. White (ed.), Methods in molecular biology. Humana Press, Inc., Totowa, N.J. [DOI] [PubMed]
- 13.Ketola, I., M. Anttonen, T. Vaskivuo, J. S. Tapanainen, J. Toppari, and M. Heikinheimo. 2002. Developmental expression and spermatogenic stage specificity of transcription factors GATA-1 and GATA-4 and their cofactors FOG-1 and FOG-2 in the mouse testis. Eur. J. Endocrinol. 147:397-406. [DOI] [PubMed] [Google Scholar]
- 14.Ketola, I., N. Rahman, J. Toppari, M. Bielinska, S. B. Porter-Tinge, J. S. Tapanainen, I. T. Huhtaniemi, D. B. Wilson, and M. Heikinheimo. 1999. Expression and regulation of transcription factors GATA-4 and GATA-6 in developing mouse testis. Endocrinology 140:1470-1480. [DOI] [PubMed] [Google Scholar]
- 15.Kettlewell, J. R., C. S. Raymond, and D. Zarkower. 2000. Temperature-dependent expression of turtle Dmrt1 prior to sexual differentiation. Genesis 26:174-178. [PubMed] [Google Scholar]
- 16.Ko, L. J., and J. D. Engel. 1993. DNA-binding specificities of the GATA transcription factor family. Mol. Cell. Biol. 13:4011-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krreidberg, J. A., H. Sariola, J. M. Loring, M. Maeda, J. Pelletier, D. Housman, and R. Jaenisch. 1993. WT-1 is required for early kidney development. Cell 74:679-691. [DOI] [PubMed] [Google Scholar]
- 18.Kuo, C. T., M. L. Veselits, K. P. Barton, M. M. Lu, C. Clendenin, and J. M. Leiden. 1997. The LKLF transcription factor is required for normal tunica media formation and blood vessel stabilization during murine embryogenesis. Genes Dev. 11:2996-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei, N., and L. L. Heckert. 2002. Sp1 and Egr1 regulate transcription of the Dmrt1 gene in Sertoli cells. Biol. Reprod. 66:675-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu, J. R., T. A. McKinsey, H. Xu, D. Z. Wang, J. A. Richardson, and E. N. Olson. 1999. FOG2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol. Cell. Biol. 19:4495-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchand, O., M. Govoroun, H. D'Cotta, O. McMeel, J. Lareyre, A. Bernot, V. Laudet, and Y. Guiguen. 2000. DMRT1 expression during gonadal differentiation and spermatogenesis in the rainbow trout, Oncorhynchus mykiss. Biochim. Biophys. Acta 1493:180-187. [DOI] [PubMed] [Google Scholar]
- 22.Merika, M., and S. H. Orkin. 1993. DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol. 13:3999-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyamoto, N., M. Yoshida, S. Kuratani, I. Matsuo, and S. Aizawa. 1997. Defects of urogenital development in mice lacking Emx2. Development 124:1653-1664. [DOI] [PubMed] [Google Scholar]
- 24.Molkentin, J. D. 2000. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 275:38949-38952. [DOI] [PubMed] [Google Scholar]
- 25.Molkentin, J. D., Q. Lin, S. A. Duncan, and E. N. Olson. 1997. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 11:1061-1072. [DOI] [PubMed] [Google Scholar]
- 26.Moniot, B., P. Berta, G. Scherer, P. Sudbeck, and F. Poulat. 2000. Male specific expression suggests role of DMRT1 in human sex determination. Mech. Dev. 91:323-325. [DOI] [PubMed] [Google Scholar]
- 27.Nanda, I., Z. Shan, M. Schartl, D. W. Burt, M. Koehler, H. Nothwang, F. Grutzner, I. R. Paton, D. Windsor, I. Dunn, W. Engel, P. Staeheli, S. Mizuno, T. Haaf, and M. Schmid. 1999. 300 million years of conserved synteny between chicken Z and human chromosome 9. Nat. Genet. 21:258-259. [DOI] [PubMed] [Google Scholar]
- 28.Orkin, S. H. 1998. Embryonic stem cells and transgenic mice in the study of hematopoiesis. Int. J. Dev. Biol. 42:927-934. [PubMed] [Google Scholar]
- 29.Raymond, C. S., J. R. Kettlewell, B. Hirsch, V. J. Bardwell, and D. Zarkower. 1999. Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev. Biol. 215:208-220. [DOI] [PubMed] [Google Scholar]
- 30.Raymond, C. S., M. W. Murphy, M. G. O'Sullivan, V. J. Bardwell, and D. Zarkower. 2000. Dmrt1, a gene related to worm and fly sexual regulators, is required for mammalian testis differentiation. Genes Dev. 14:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raymond, C. S., E. D. Parker, J. R. Kettlewell, L. G. Brown, D. C. Page, K. Kusz, J. Jaruzelska, Y. Reinberg, W. L. Flejter, V. J. Bardwell, B. Hirsch, and D. Zarkower. 1999. A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators. Hum. Mol. Genet. 8:989-996. [DOI] [PubMed] [Google Scholar]
- 32.Raymond, C. S., C. E. Shamu, M. M. Shen, K. J. Seifert, B. Hirsch, J. Hodgkin, and D. Zarkower. 1998. Evidence for evolutionary conservation of sex-determining genes. Nature 391:691-695. [DOI] [PubMed] [Google Scholar]
- 33.Shawlot, W., and R. R. Behringer. 1995. Requirement for Lim1 in head-organizer function. Nature 374:425-430. [DOI] [PubMed] [Google Scholar]
- 34.Shen, W.-H., C. D. Moore, Y. Ikeda, K. L. Parker, and H. A. Ingraham. 1994. Nuclear receptor steroidogenic factor 1 regulates the Mullerian inhibiting substance gene: a link to the sex determination cascade. Cell 77:651-661. [DOI] [PubMed] [Google Scholar]
- 35.Smith, C. A., P. J. McClive, P. S. Western, K. J. Reed, and A. H. Sinclair. 1999. Conservation of a sex-determining gene. Nature 402:601-602. [DOI] [PubMed] [Google Scholar]
- 36.Svensson, E. C., G. S. Huggins, H. Lin, C. Clendenin, F. Jiang, R. Tufts, F. B. Dardik, and J. M. Leiden. 2000. A syndrome of tricuspid atresia in mice with a targeted mutation of the gene encoding Fog2. Nat. Genet. 25:353-356. [DOI] [PubMed] [Google Scholar]
- 37.Tevosian, S. G., K. H. Albrecht, J. D. Crispino, Y. Fujiwara, E. M. Eicher, and S. H. Orkin. 2002. Gonadal differentiation, sex determination and normal Sry expression in mice require direct interaction between transcription partners GATA4 and FOG2. Development 129:4627-4634. [DOI] [PubMed] [Google Scholar]
- 38.Tevosian, S. G., A. E. Deconinck, A. B. Cantor, H. I. Rieff, Y. Fujiwara, G. Corfas, and S. H. Orkin. 1999. FOG2: a novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc. Natl. Acad. Sci. USA 96:950-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tevosian, S. G., A. E. Deconinck, M. Tanaka, M. Schinke, S. H. Litovsky, S. Izumo, Y. Fujiwara, and S. H. Orkin. 2000. FOG2, a cofactor for GATA transcription factors, is essential for heart morphogenesis and development of coronary vessels from epicardium. Cell 101:729-739. [DOI] [PubMed] [Google Scholar]
- 40.Tremblay, J. J., and R. S. Viger. 1999. Transcription factor GATA-4 enhances Mullerian inhibiting substance gene transcription through a direct interaction with the nuclear receptor SF-1. Mol. Endocrinol. 13:1388-1401. [DOI] [PubMed] [Google Scholar]
- 41.Tsang, A. P., J. E. Visvader, C. A. Turner, Y. Fujiwara, C. Yu, M. J. Weiss, M. Crossley, and S. H. Orkin. 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90:109-119. [DOI] [PubMed] [Google Scholar]
- 42.Viger, R. S., C. Mertineit, J. M. Trasler, and M. Nemer. 1998. Transcription factor GATA-4 is expressed in a sexually dimorphic pattern during mouse gonadal development and is a potent activator of the Mullerian inhibiting substance promoter. Development 125:2665-2675. [DOI] [PubMed] [Google Scholar]
- 43.Wingender, E., X. Chen, R. Hehl, H. Karas, I. Liebich, V. Matys, T. Meinhardt, M. Pruss, I. Reuter, and F. Schacherer. 2000. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 28:316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yomogida, K., H. Ohtani, H. Harigae, E. Ito, Y. Nishimune, J. D. Engel, and M. Yamamoto. 1994. Developmental stage- and spermatogenic cycle-specific expression of transcription factor GATA-1 in mouse Sertoli cells. Development 120:1759-1766. [DOI] [PubMed] [Google Scholar]
- 45.Zarkower, D. 2002. Invertebrates may not be so different after all. Novartis Found. Symp. 244:115-126. [PubMed] [Google Scholar]