Abstract
The last ten years have witnessed an explosion of new techniques that can be used to probe the dynamic behavior of individual biological molecules, leading to discoveries that would not have been possible with more traditional biochemical methods. A common feature among these single-molecule approaches is the need for the biological molecules to be anchored to a solid support surface. This must be done under conditions that minimize nonspecific adsorption without compromising the biological integrity of the sample. In this review we highlight why surface attachments are a critical aspect of many single-molecule studies and we discuss current methods for anchoring biomolecules. Finally, we provide a detailed description of a new method developed by our laboratory for anchoring and organizing hundreds of individual DNA molecules on a surface, allowing “high-throughput” studies of protein–DNA interactions at the single-molecule level.
1. Introduction
Advances in single-molecule techniques have provided new information on aspects of biology that were largely inaccessible by bulk techniques, and as these techniques continue to improve they hold the promise of even greater insights.1–3 For example, groundbreaking studies in single-molecule visualization revealed many aspects of the dynamics of F1-ATPase rotary motor, including rotation rates and details of the mechanochemical reaction cycle.4–7 Several studies also probed the behavior of mechanoenzymes such as myosin, kinesin and dynein; most prominent were those techniques that measured the step size and force exerted by these proteins as they travel along actin filaments or microtubules.8–19 Early work in single-molecule bioscience also included measurements of individual transcribing molecules of bacterial RNA polymerase,20–22 and more recent studies with this enzyme were carried out to greater precision with instrumentation capable of making real time measurements with spatial resolution approaching one base pair.23–26 Single-molecule methods have also been used to examine other nucleic acid motors like bacteriophage lambda exonuclease,27,28 DNA polymerases, 29–32 Rad54 and Rdh54 DNA translocases,33–35 the chromatin remodeling complexes RSC and Swi/Snf,36,37 and several different helicases.38–40 A large volume of work has also focused on the conformational dynamics and physical properties of single molecules including the folding, unfolding and strain response of proteins, DNA, and RNA.41–48
This review is written for the nonspecialist who may be interested in understanding or pursuing single-molecule experiments. We outline the primary approaches that have been used for probing the properties of single biological molecules, including atomic force microscopy (AFM), tethered particle motion (TPM), optical and magnetic tweezers (OT and MT), and total internal reflection fluorescence microscopy (TIRFM). We briefly highlight some of the seminal studies that have utilized these techniques, discuss why each approach requires that the molecules under investigation be anchored to a solid support, and outline the general methods that are used for support surface design and molecule tethering. We conclude by describing a novel high-throughput approach to single-molecule TIRFM imaging of DNA molecules that has been developed in our laboratory. This method takes advantage of the unique properties of two-dimensional lipid bilayers, which allows us to organize and align hundreds of DNA molecules on a surface that closely mimics the natural environment encountered within a living cell.
2. Single-molecule methods and surface attachments
Many methods capable of making real time measurements of single-molecule behavior and dynamics require that the molecules under investigation be attached to a solid support surface. Typical attachment chemistries include covalent coupling, nonspecific adsorption, or specific attachments mediated by a noncovalent interaction. In the following sections, we briefly review some of the most common single-molecule techniques, and for each technique we provide examples of key studies and highlight why and how the molecules were attached to a surface.
2.1 Atomic force microscopy (AFM)
The most common use of AFM is to probe a sample by scanning a flexible cantilever tip across a surface. Any sample- induced deflection of the flexible cantilever is measured with a reflected laser beam that is centered on a quadrant photodetector, yielding a 3-dimensional profile of the sample 2,49 (Fig. 1A). For example, AFM has been used to explore the structure of RNA polymerase transcription complexes,50,51 the MRN (Mre11/Rad50/Nbs1) DNA damage-recognition proteins,52 and filaments of the homologous recombination protein Rad51.53 For these studies complexes were formed in solution, adsorbed nonspecifically onto a mica surface, and either dehydrated or imaged in a thin layer of buffer, to reveal individual protein DNA interactions. The use of AFM for scanning surface adsorbed molecules is best suited for gaining structural insights. It is difficult to make real time dynamic measurements of proteins or nucleic acids because behaviors may be perturbed by nonspecific contacts with the mica surface. In the past, AFM has been restricted by the rate at which the cantilever can be scanned across the sample, and for most instruments it currently takes on the order of at least 30 seconds to scan a region corresponding to the size of a typical protein and/or DNA molecule. However, this may become less of an impediment with the recent development of atomic force microscopes capable of imaging biological molecules at rates nearly approaching 10 frames per second.54
Fig. 1.
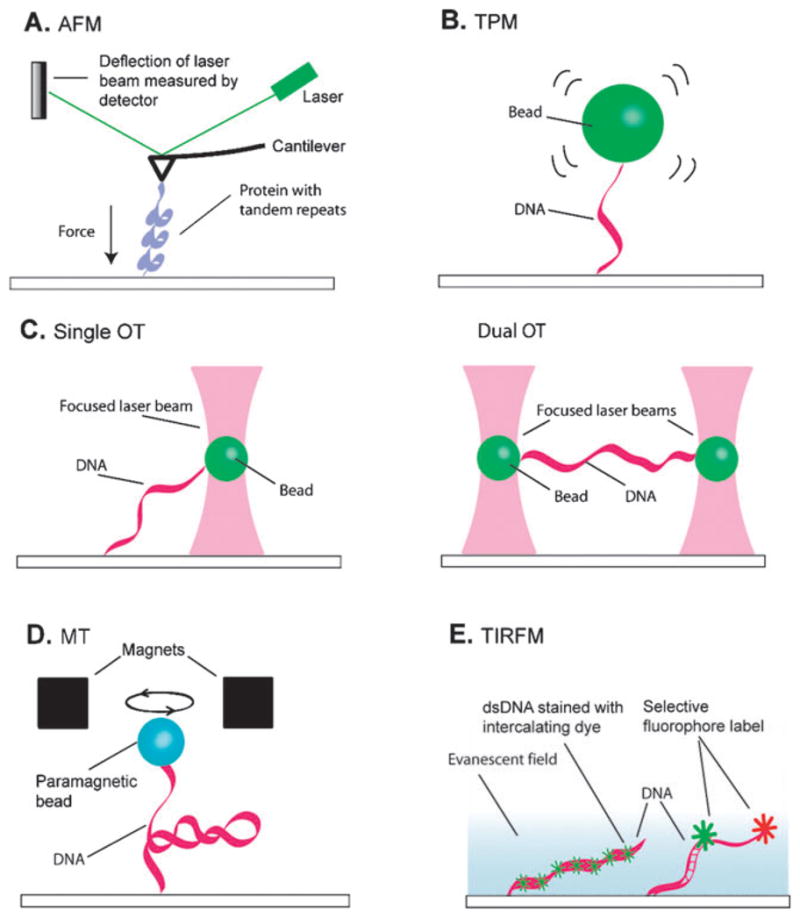
Single-molecule methods requiring surface attachment. (A) Atomic force microscopy (AFM). (B) Tethered particle motion (TPM). (C) Optical tweezers (OT). (D) Magnetic tweezers (MT). (E) Total internal reflection fluorescence microscopy (TIRFM). Details describing each technique are provided in the text.
AFM is not limited to imaging surface adsorbed molecules; it can also be used to make real time dynamic measurements of molecules that are stretched between the cantilever and a solid support (Fig. 1A).49 Measuring the dynamic responses of individual biomolecules placed under tension is technically demanding, but can yield detailed force–extension curves that reveal tension-dependent changes in molecular states. A challenging aspect of these studies is that one end of the biomolecule must be adhered to the cantilever tip while the other end of the molecule is linked to a solid support surface. Usually, the protein under investigation is nonspecifically adsorbed to a gold surface (Fig. 2A), then the cantilever probe tip is pushed against the surface so that protein molecules can attach to the tip. The support surface is then moved relative to the cantilever by means of piezo-electric actuators, exerting a force on the biomolecule tethered between the cantilever and the surface, and the resulting tension causes changes in the tethered molecule that are reflected as movement of the cantilever. AFM can be used to apply forces ranging from 5–10 000 pN (pico-newtons), with typical spatial limits on the order of 1 nm over millisecond to second time scales.2 Proteins comprised of tandem repeats, such as titin or poly-ubiquitin, are ideally suited to unfolding by AFM.41,42,55,56 The folding and unfolding reveals saw-tooth patterns in the force-extension curves where the gradual rise in force terminates upon the complete unfolding of one of the protein repeats, and this is repeated until all of the protein repeats have unfolded. A more recent AFM study achieved sub-angstrom spatial resolution sufficient to detect the rupture of single disulfide bonds in repeated arrays of thioredoxin.57
Fig. 2.
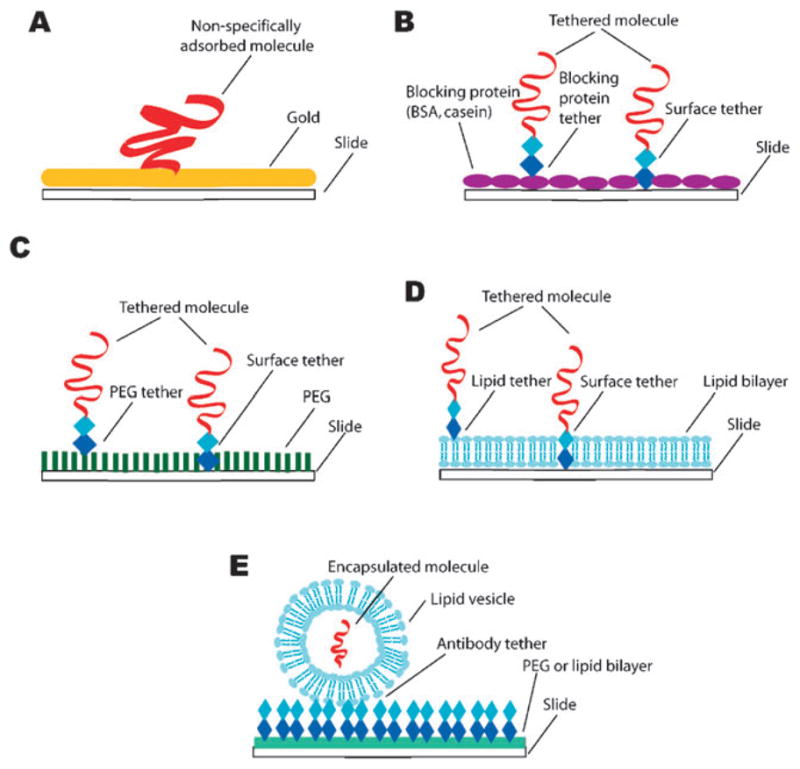
Attachment methods used in single-molecule studies. (A) Nonspecific adsorption of a protein to a gold surface is often used for AFM force-extension measurements. Specific tethering via biotin–streptavidin, or hapten–antibody interactions to surfaces coated with either (B) nonspecific blocking proteins, (C) polyethylene glycol (PEG), or (D) supported lipid bilayers are the most commonly used methods for single-molecule studies. Vesicle encapsulation (E) is another approach that can be used for single-molecule fluorescence studies. The general attachment methods shown in (A–D) can also be used to link molecules to beads for use in TPM, optical or magnetic tweezers.
2.2 Tethered particle motion (TPM)
Tethered particle motion relies on optical detection of a small bead anchored to a slide surface through a DNA tether (Fig. 1B). Changes in the length of the DNA tether are detected by analysis of the Brownian motion of the bead; shortening of the DNA restricts the motion of the bead and increases in DNA length allow for a greater freedom of motion. The beads themselves can be gold, silica, or polystyrene, ranging from a few tens of nanometres up to a few microns in diameter. These beads can be observed with video microscopy and the geometric center of each bead can be determined with 1 nm precision, allowing for accurate calculation of the DNA length.
This approach was developed by Landick and colleagues to study the movement of bacterial RNA polymerase (RNAP).20 For these studies a DNA molecule was attached to a 40 nm gold bead via a biotin–streptavidin interaction. RNA polymerase bound to the DNA was nonspecifically adsorbed to a glass cover slip, and appropriately tethered complexes were identified based upon the range of motion displayed by the gold beads. Transcription was initiated by addition of rNTPs and the length of the DNA tether increased as the polymerase moved along the template. The beads were eventually released from the surface once the polymerase reached the ends of the DNA. TPM has also been used to study the movement of the RecBCD helicase/nuclease.58 For this study, RecBCD was biotinylated in vivo by expressing the RecD subunit as a fusion with BCCP (biotin carboxyl carrier protein). RecBCD-bio was then coupled to strepavidin-coated polystyrene beads. These conjugates were then introduced into a flow chamber containing DNA molecules labeled at one end with a digoxigenin and anchored to a coverslip surface, which had been previously coated with nonspecifically adsorbed anti-digoxigenin antibodies. The bead-coupled RecBCD was observed as it bound to the exposed ends of tethered DNA molecules and the DNA translocation activity was monitored by the corresponding decrease in the length of the DNA tether as the enzyme degraded the DNA and moved towards the coverslip surface.
The studies with RNAP and RecBCD relied upon the chemo-mechanical properties of the enzymes to physically shorten the length of the DNA tether. However, TPM can also be used to observe reactions that involve changes in the end-to-end length of the DNA, but do not involve actively translocating proteins, such as DNA looping by lactose repressor protein.59 For this work the DNA molecules were labeled at one end with a biotin and at the other end with a digoxigenin. They were then linked to avidin-coated polystyrene beads and an anti-digoxigenin coated glass coverslip. The DNA molecules contained two lac repressor binding sites separated by a distance of 305 base pairs, such that concurrent binding of a lac repressor tetramer to both sites would result in shortening of the DNA tether. Using this approach the authors were able to assess the rate at which lac repressor formed and released the loops of DNA.
Most TPM measurements are done in the absence of any buffer flow. However, studies by van Oijen and colleagues have used flow-stretched DNA molecules.3,60 The force exerted on the DNA by the flowing buffer can be calculated based on both the end-to-end length of the DNA as well as the magnitude of the fluctuations exhibited by the beads. Because ssDNA and dsDNA exhibit distinct force–extension profiles, this flow-stretching approach to TPM is particularly well suited to studying systems that involve the conversion of dsDNA to ssDNA or vice versa, such as single molecules of λ exonuclease and single DNA replication forks.28,32
2.3 Optical tweezers (OT)
For optical tweezers a laser is tightly focused through a high numerical aperture objective lens to create a “trap” that can capture a small bead (Fig. 1C). The trap results from a combination of the scattering and gradient forces that are generated as momentum from the photons is transferred to the bead. The location of the bead relative to the focal position of the trap reflects the forces experienced by the bead and can be measured with video imaging or a quadrant photodetector. The beam intensity determines the magnitude of the forces exerted upon the bead and this can be exploited to force-clamp a system by modulating the beam intensity to maintain the bead at a fixed position.2,61,62
Optical tweezers can apply tension to molecules linked between a bead held within the trap and either a second bead or a solid support surface (Fig. 1C). For example, the energy landscape of the DNA helix was explored by unzipping dsDNA molecules.63 Digoxigenin and biotin tags on adjacent overhangs of dsDNA were used to attach one strand to a bead and the other to a slide surface, which was then moved away from the trap. The DNA unzipping force was measured by tracking bead displacement from the beam focus. This same approach has been used to measure the forces required to disrupt a variety of protein–DNA complexes, such as nucleosomes 64 and mismatch repair protein complexes.65 One recent study used this DNA unzipping configuration to study the DNA unwinding activity of the ring-shaped T7 helicase, and revealed that the helicase functioned through a mechanism involving protein induced separation of the DNA strands.39
Optical tweezers can also measure the forces exerted by molecules. For example, the first studies that measured the forces produced by a transcribing molecule of RNA polymerase relied upon optical tweezers.21 Forces exerted by different DNA translocating motor proteins have been measured.66,67 OT measurements have yielded insights into motor enzymes such as kinesin, myosin, and dynein, as well as DNA processing enzymes such as polymerases, nucleases, and helicases [reviewed in ref. 22].
Optical traps have also been used as single-molecule “stopped flow” devices to control the assembly and activity of different enzymes on single DNA molecules as exemplified by studies of the RecBCD exonuclease complex by Kowalczykowski and colleagues.68–71 These experiments utilize fluorescently tagged protein or DNA molecules and rely on a laminar flow system, which has input channels for multiple buffers that do not mix as they pass through the sample chamber. The experiments are conducted using the optical trap to capture a DNA molecule tethered to a polystyrene bead through a biotin–streptavidin interaction. Once captured, the DNA molecule can be moved from one buffer to the next to allow loading of RecBCD in one channel while initiating the exonuclease/translocation reaction with ATP in a second channel. Similar studies have been conducted on the RecA recombinase and the Rad54 and Rdh54 DNA translocases.34,35,72
2.4 Magnetic tweezers (MT)
Magnetic tweezers exploit the forces experienced by a paramagnetic bead placed within a magnetic field2,73 (Fig. 1D). Similar to TPM, magnetic tweezers require the macromolecule under investigation to be tethered between a bead and a solid supporting surface. The force experienced by a bead is proportional to the magnetic field gradient, but the changes in bead position during single-molecule experiments are relatively small. Consequently, the field is locally uniform and the force remains constant and can be calculated based on the Brownian fluctuations of the bead. The lateral positions of the beads in the x,y plane are determined by centroid tracking, and movement in the z-direction is analyzed through deconvolution of the interference patterns surrounding the images, yielding data at millisecond time scales with resolution limits on the order of 5–10 nm in all directions, while exerting forces on the order of 0.05–20 pN.
Protein–DNA interactions that lead to changes in DNA length are well-suited to analysis by magnetic tweezers. For example, the force dependence of chromatin assembly was examined with magnetic tweezers.74 These studies revealed that chromatin assembly rates decreased and disassembly rates increased as higher tensions were placed on the DNA molecules, and assembly was completely inhibited at forces greater than 10 pN.74 Similarly, the real time assembly/disassembly kinetics of Rad51 nucleoprotein filaments have been studied by magnetic tweezers.75 In addition, the conversion of ssDNA to dsDNA by DNA polymerases can be measured using magnetic tweezers and the rates of conversion in turn yield biochemical information about the polymerization reaction.29,30
A unique aspect of magnetic tweezers is that they can apply torque to an attached molecule. This requires that the molecules be anchored in a manner that prevents free rotation. For example, using magnetic tweezers Strick and colleagues were able to study the elastic properties of supercoiled dsDNA.76 This was accomplished by labeling one end of a DNA molecule with multiple molecules of digoxigenin and the other end with multiple biotins. Anti-digoxigenin was then nonspecifically adsorbed to a slide surface, which served as an anchor point for the DNA, and the other end of the DNA was attached to a streptavidin-coated bead. The torsionally constrained DNA molecule was then extended by the magnetic field and twisted by rotating the magnets, allowing characterization of the elasticity of the single, supercoiled DNA. This setup has proven remarkably versatile, and similar experiments with twisted molecules of DNA have now been used to characterize type I and type II topoisomerases,77,78 promotor escape by RNA polymerase,79 and DNA translocation by the RSC chromatin remodeling complex.36
Magnetic tweezers can also be used to manipulate other types of protein motors. In a remarkable series of experiments, Kinosita and colleagues were able to control the F1-ATPase motor by coupling the rotating γ-subunit to a magnetic bead and anchoring the stationary α3β3 subunit to a glass coverslip. 5,6 For these experiments it was crucial that the anchor points prevent free rotation of any component of the complex. To accomplish this the γ-subunit was engineered with two cysteines, which were then biotinylated and linked to a streptavidin-coated bead. The α3β3 was expressed with N-terminal histidine tags on the β-subunits, which were coupled to a coverslip covalently modified with a thiolated silane reagent followed by maleimide-Ni2+ NTA (nickel nitrilotriacetic acid). Once attached to the surface, the rotation speed and/or direction of the F1-ATPase motors was experimentally manipulated using an external magnetic field. These studies helped reveal how mechanical rotation is related to ATP hydrolysis when the motor rotates in one direction and its coupling to ATP synthesis when the direction of rotation is reversed.
2.5 Total internal reflection fluorescence microscopy (TIRFM)
Total internal reflection fluorescence microscopy (TIRFM) makes use of the evanescent field that is generated when light is reflected off the interface between two transparent materials of differing refractive indexes. If the angle of incidence exceeds a critical angle then the light reflects away from the interface, however an electromagnetic field persists in the region beyond the interface boundary. This is known as the evanescent wave and its intensity decays exponentially away from the interface. This illumination geometry eliminates background fluorescence typically associated with wide-field epifluorescence by selectively exciting a small volume located immediately adjacent to the reflective interface, thereby allowing observation of single fluorescent molecules. Because the evanescent field only penetrates a short distance into the sample chamber these studies typically rely on reactions that are coupled to a fused silica surface, most often through a biotin–streptavidin interaction.
One common application of TIRFM is the study of reactions using fluorescence resonance energy transfer between single pairs of donor and acceptor fluorophores (spFRET).1,80 FRET is strongly dependent upon the distance between the donor and acceptor fluorophores, and is typically useful for studying changes in distances in the 20–80Å range. Therefore spFRET is particularly well-suited for studying systems that undergo structural transitions within this distance regime. For example, the folding of an RNA molecule,81–83 structural transitions in DNA molecules,84,85 assembly and behavior of protein–DNA complexes86–88 and ribosomal structural transitions during translation have all been studied using spFRET.89–91
TIRFM studies have also been conducted with λ DNA (48 502 bp) since these long DNA substrates are particularly useful for studying protein–DNA interactions that occur at multiple, well-resolved locations or those that involve movement of the proteins along the DNA. The contour length of λ DNA (~16 μm) is considerably larger than the effective depth of the evanescent field and therefore the DNA must be confined parallel to the surface to allow visualization along its entire length. This can be accomplished by tethering the DNA to the surface at just one end (typically via a biotin– streptavidin linkage), in which case buffer flow is necessary to stretch the molecules, or by anchoring both ends in an extended configuration along the surface, which does not require continuous buffer flow.92,93 Using TIRFM and λ DNA our laboratory has studied the behaviors of DNA damage repair proteins such as the Rad51 recombinase, mismatch recognition proteins, and chromatin remodeling enzymes.33,92–94
3. Commonly used methods for making “bio-friendly” surfaces
It is generally appreciated that single-molecule measurements require specialized microscopy equipment, and in many cases these research tools are custom-built by specialists with training in physics and optics within the laboratories where the studies are conducted. However, one of the least appreciated factors in all of these experiments is the requirement for the molecules under investigation to be physically attached or confined to an inert surface. This requirement is a critical aspect of single-molecule experimental design, and both the attachment point and the immediate surroundings must be carefully designed such that they do not compromise the biological properties of the sample under investigation.
3.1 Nonspecific blocking proteins
Blocking with nonspecific proteins such as BSA (bovine serum albumin) is the most popular method for preventing nonspecific adsorption, and this can be accomplished by incubating the surfaces in a buffer preparation that contains the blocking protein for several minutes (Fig. 2B). In addition, BSA (or other nonspecific proteins) is often added to the reaction buffers to ensure that the surfaces remain blocked for the duration of the experiment. Although simple, the use of nonspecific blocking proteins is not applicable in many situations,95 and many biochemical systems require more complex surface modifications to prevent nonspecific adsorption.
3.2 PEGylation
Polyethylene glycol (PEG) is a commonly used linear polymer for generating inert surfaces (Fig. 2C), and it can often be much more effective than BSA alone.38,95 Amino-reactive PEG (e.g. N-hydroxysuccinimidyl PEG) is covalently coupled to a fused silica surface that has been coated with an aminosilane reagent (e.g. 2-aminopropyltriethoxysilane). The aminosilane treatment yields a surface that is densely coated with exposed primary amines, which then react with the succinimidyl PEG, yielding a surface with covalently attached PEG. The inclusion of a small fraction of biotinylated PEG provides anchor points for attaching molecules of interest. Most studies that use PEG also incorporate BSA in their reaction buffers to serve as an additional level of protection against nonspecific adsorption.
The density of the PEG coating on the surface is critical for preventing nonspecific protein adsorption. In an effort to maximize the PEG density one group has developed an extremely promising approach based on six-arm, star-shaped molecules of PEG, rather than linear PEG (reviewed in ref. 96). Functionalized versions of the star-shaped PEG can be intermolecularly crosslinked after application to a surface forming an extremely dense protective coating that is highly resistant to nonspecific surface adsorption of proteins.96
3.3 Lipid bilayers
Lipid bilayers provide an attractive option for single-molecule studies (Fig. 2D) because they provide a surface environment that is similar to that which might be encountered by a macromolecule inside a living cell.97,98 Therefore bilayers are compatible with many different proteins and/or nucleic acids, and will prevent any nonspecific interactions with the underlying surface. Bilayers can be modified through the incorporation of lipids with various functional moieties attached to the head groups, such as biotin or PEG (see section 4 for specific details). A practical advantage of lipid bilayers is the simplicity with which they can be prepared. Dehydrated phospholipids are suspended into an aqueous buffer and vesicles are made by sonicating the lipids or extruding them through a porous membrane. The resulting vesicles can be stored at 4 °C for up to several weeks. Bilayers are made by injecting the vesicles into a fused silica flow chamber. The vesicles spontaneously fuse with one another until they reach a critical size, at which time they rupture and form a bilayer that coats the entire surface. For experiments that require solid attachment points the flow chamber can first be modified through the deposition of a very sparse, nonspecifically adsorbed coating of neutravidin prior to laying down the bilayer, yielding isolated molecules of neutravidin that are then surrounded by the inert bilayer.98
3.4 Lipid vesicles
A promising new approach for anchoring molecules to a surface is to encapsulate them within a lipid vesicle and then tether the vesicle to a solid surface99,100 (Fig. 2E). The molecules inside the vesicles are free of potential perturbations that may be induced by direct attachment methods. This approach is especially advantageous for studying systems that are not amenable to direct coupling to a surface or bimolecular interactions that would otherwise require that two molecules be anchored in very close proximity to one another. The challenge of this approach lies in encapsulating the targets of interest into the vesicles. Cisse et al.101 describe novel methods of introducing pores into the lipid vesicles by either using a bacterial toxin or taking advantage of the melting characteristics of the lipids. Their FRET-based study encapsulated labeled DNA and the RecA protein into a small vesicle that is attached to the slide surface via a biotinylated PEG-neutravidin- biotin linkage, allowing them to probe the highly dynamic RecA filament.
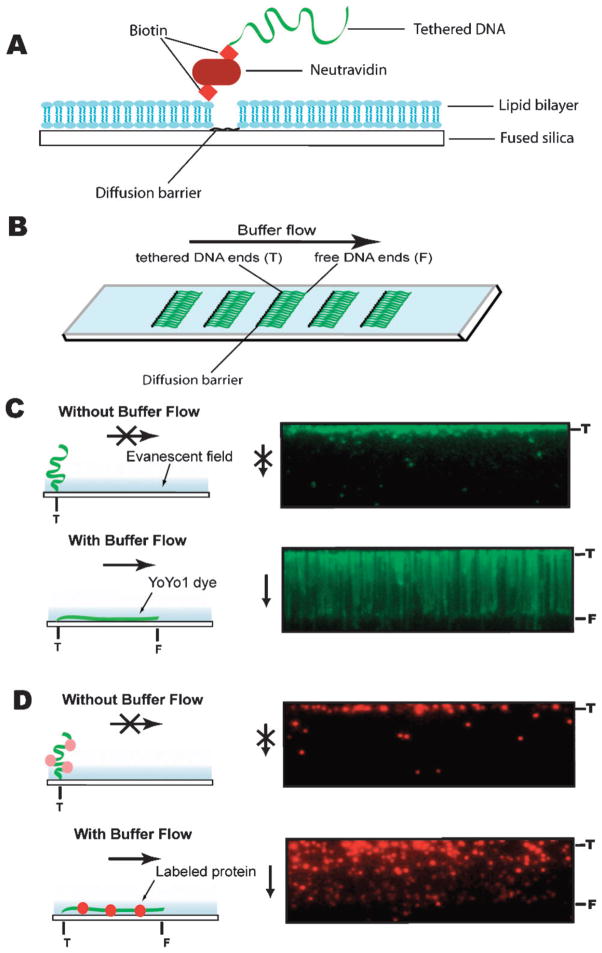
4. DNA curtains as a high-throughput approach for single-molecule analysis
For single molecule studies it is essential to use surfaces that minimize nonspecific interactions with the biomolecules under investigation and provide tethering points that do not affect the biological integrity of the sample. In addition, it is inherently difficult to collect statistically relevant information from methods designed to look at one reaction at a time. To help overcome these limitations we have developed a new technology referred to as “DNA curtains”. This assay allows simultaneous study of up to hundreds of individual DNA molecules anchored to an inert lipid bilayer and aligned with respect to one another within a single field-of-view with TIRFM33,92,94,98 (Fig. 3). These DNA curtains are similar to “belts” of DNA that can be aligned by dielectrophoresis on surfaces blocked with casein,102 but distinct in that the DNA molecules making up the curtain are aligned in the same direction with respect the long axis of the molecules and also with respect to the sequence of the DNA. Moreover, the DNA curtains developed in our group can be used to study a broad range of protein–DNA interactions because of the inert properties of the lipid bilayers.
Fig. 3.
Lipid bilayers and DNA curtains as tools for single-molecule bioscience. (a) Shows a side view illustrating the components that make up a DNA curtain and (b) shows a top view of a slide with multiple curtains. The images in (c) show a cartoon depicting the response of a tethered DNA molecule to changes in buffer flow, and the panels to the right show actual images of a DNA curtain with and without flow. The DNA is stained with the fluorescent dye YOYO1, and in the absence of buffer flow the molecules diffuse away from the surface and out of the evanescent field. This control is used to demonstrate that the DNA molecules are anchored by only one end and are not randomly stuck to the surface. (d) Shows an example of a DNA curtain bound by a fluorescently tagged protein. Here the DNA is not labeled, and the protein (the chromatin remodeling enzyme Rdh54) is labeled with a single quantum dot. Transiently turning buffer flow off causes the proteins (and the DNA) to diffuse out of the evanescent field, confirming that neither the DNA nor the proteins are stuck to the bilayer coated surface.
4.1 Microscale barriers to lipid diffusion
Molecules anchored to a bilayer are free to diffuse in two dimensions, and they will eventually diffuse out of the field-of-view, making it difficult to follow individual reactions over time. However, the fluidity of the bilayer can be used as a tool to organize molecules on the flowcell surface. For example, Boxer and colleagues have shown that lipids can not traverse imperfections on glass surfaces [reviewed in ref. 103]. These lipid barriers can be as simple as mechanical scratches on the glass surface, or they can be made with chrome, photoresist, aluminum oxide, gold, proteins or other materials deposited by photolithographic techniques, all of which function by disrupting the continuity of the bilayer. The inability of lipids to traverse these various barriers has been exploited to develop ways of controlling micropatterns of lipid-anchored molecules on solid support surfaces. For example, several studies have demonstrated that micrometre-scale barriers can be used to make corrals containing chemically distinct membrane patches or proteins coupled to lipids that can be manipulated through the application of an electric field [reviewed in ref. 103]. Our group has shown that DNA molecules tethered to lipids can be organized along the leading edges of microscale barriers using hydrodynamic force and we have exploited this as a tool for making curtains of DNA molecules that can be studied with TIRFM.98
4.2 How to make “DNA curtains”
To make DNA curtains a slide is first etched with a diamond-coated scribe, yielding microscale lipid diffusion barriers perpendicular to the direction in which buffer will flow. After assembly of the flowcell, the sample chamber is coated with a bilayer by injecting lipid vesicles (Avanti Polar Lipids, Inc.), which for most applications are comprised of a mixture of DOPC (1,2-dioleoyl-sn-glycero-phosphocholine), 0.5% biotinylated- DPPE (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl)), and 8–10% mPEG 550-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N- [methoxy-(polyethylene glycol)-550]), into the sample chamber. The biotinylated lipids serve as mobile attachment points for DNA molecules and the PEGylated lipids offer an additional level of protection against nonspecific binding. The vesicles are prepared by sonicating aqueous mixtures of these lipids and when injected into the sample chamber they spontaneously rupture on the surface and self-assemble into a planar bilayer. After the bilayer has formed, the excess vesicles are removed by flushing the chamber with fresh buffer. Neutravidin (Pierce Biotechnology, Inc.) is then applied to the surface and acts as a multivalent linker between the biotinylated-DPPE and any biotinylated DNA molecules that are subsequently injected into the chamber. Most of our experiments utilize λ-DNA (48 502 base pairs, ~16 μm in length; Invitrogen), which is biotinylated at one end with a oligonucleotide complementary to the 12-nucleotide ssDNA overhang present at the ends of the linear phage genome. This strategy yields DNA molecules that are anchored by one end to the bilayer through the neutravidin coupled to the lipids. For visual detection the DNA molecules can be labeled with a fluorescent dye such as YOYO-1 (Invitrogen).
The bilayer serves two purposes: (1) it renders the surface inert, and (2) it permits the DNA molecules to move in two dimensions while confining them near the surface. The barriers etched into the flowcell surface cannot be traversed by the lipids, and when buffer flow is applied the lipid-tethered DNA molecules are driven in the direction of flow and accumulate at the leading edges of the barriers (Fig. 3A, B and C). Application of buffer flow also extends the DNA molecules and confines them within the evanescent field. Pausing flow causes the DNA molecules to briefly diffuse out of the evanescent field, therefore any molecules that are nonspecifically bound to the surface remain within view and can be discounted from further analysis (Fig. 3C and D). Thousands of DNA molecules can be aligned at a given microscale diffusion barrier, and the density of the DNA can be varied to suit any specific experimental requirements. This is achieved by increasing or decreasing the amount of DNA injected into the sample chamber or by varying the distance between adjacent barriers on the surface. These DNA curtains greatly increase the throughput capacity of the TIRFM single-molecule experiments. Data analysis is also simplified because all of the DNA molecules are physically aligned with respect to one another, enabling straightforward molecule-to-molecule comparisons.
4.3 Visualizing and analyzing protein–DNA interactions
Using TIRFM and DNA curtains we have visualized the behavior of the molecular motor Rdh54, which is an Snf2-like chromatin remodeling protein involved in homologous DNA recombination.33 We have shown that Rdh54 can actively translocate along DNA at a mean velocity of ~80 base pairs per second, generates large loops in the DNA molecules as it travels along the double helix, and can spontaneously stop and reverse direction. Rdh54 interacts with Rad51, another protein required for homologous DNA recombination. Rad51 forms extended nucleoprotein filaments along DNA, and the DNA within this filament is stretched by approximately 50% relative to normal B-DNA.94 We have begun using TIRFM to visualize the assembly and disassembly of Rad51 filaments in real time, and we are moving towards using these methods to observe the interactions between Rad51, Rdh54, and other proteins involved in DNA repair. These ongoing investigations promise to yield new information regarding the interactions between these nucleoprotein filaments and other proteins that are critical for homologous recombination.
We have also used TIRFM to probe the mechanisms of post-replicative mismatch repair (MMR) protein complex Msh2-Msh6.92 Msh2-Msh6 is a damage sensing protein complex responsible for detecting and initiating the repair of mispaired bases that occur as a consequence of errors during DNA replication. It is still unclear precisely how MMR proteins function, but many of the remaining questions in the field concern how the various protein complexes travel along DNA at different stages of the repair reaction. We have found that Msh2-Msh6 travels passively along DNA by one-dimensional diffusion driven by thermal energy and our data suggest a model in which Msh2-Msh6 rotates around the DNA by tracking the phosphate backbone as it slides back and forth scanning for mispaired bases. Moreover, this protein complex stops moving at some sites along the molecules that function as traps in the energy landscape. Interestingly, when given ATP the “trapped” molecules of Msh2-Msh6 can continue diffusing along the DNA molecules. This nucleotide exchange dependent escape from the traps may reflect mechanistic steps leading to the downstream events in the repair pathway; further studies are now underway to probe these steps in the pathway.
5. Summary
Single-molecule methods have already demonstrated tremendous potential for biological research. An important consideration for all of these studies is the need for surfaces that are compatible with the biomolecules that are being studied. Lipid bilayers and DNA curtains offer the advantage of an inert surface compatible with a wide range of molecules and also offer the advantage of a high throughput approach to single-molecule fluorescence visualization using TIRFM. These new approaches to single-molecule research can potentially be applied to any protein that interacts with DNA molecules and can be used to learn where the proteins bind and what they do once they are bound.
Acknowledgments
We thank Dr Dana Moses and other members of this laboratory for careful reading of this manuscript and insightful discussions. The Greene laboratory is supported by funding from the National Institutes of Health, the National Science Foundation, the Susan G. Komen Foundation, and the Irma T. Hirschl Trust. We apologize to any colleagues whose work we may not have been able to cite due to length limitations.
Biographies

Mari-Liis Visnapuu grew up in Tallinn, Estonia and received her BA in Biochemistry from Vassar College, Poughkeepsie, NY in 2005. She joined the laboratory of Dr Eric Greene in September, 2006 as a graduate student in the Department of Biochemistry and Molecular Biophysics at Columbia University.

Daniel Duzdevich is a third-year biophysics BA candidate at Columbia University. Active involvement in research as a volunteer has spurred his interest in single-molecule studies of cellular machinery, specifically in elucidating the mechanisms by which DNA repair proteins interact with their substrates.

Eric C. Greene received his BS in biochemistry from the University of Illinois, his PhD from Texas A&M University, and he conducted postdoctoral studies at the National Institute of Health. He joined the Department of Biochemistry and Molecular Biophysics at Columbia University in 2004 as an assistant professor.
References
- 1.Ha T. Single-molecule fluorescence resonance energy transfer. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 2.Greenleaf WJ, Woodside MT, Block SM. High-resolution single-molecule measurments of biomolecular motion. Annu Rev Biophys Biomol Struct. 2007;36:171–190. doi: 10.1146/annurev.biophys.36.101106.101451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Oijen AM. Honey, I shrunk the DNA: DNA length as a probe for nucleic-acid enzyme activity. Biopolymers. 2007;85:144–53. doi: 10.1002/bip.20624. [DOI] [PubMed] [Google Scholar]
- 4.Noji H, Yasuda R, Yoshida M, Kinosita KJ. Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 5.Itoh H, Takahshi A, Adachi K, Noji H, Yasuda R, Yoshida M, Kinosita KJ. Mechanically driven ATP synthesis by F1-ATPase. Nature. 2004;427:465–468. doi: 10.1038/nature02212. [DOI] [PubMed] [Google Scholar]
- 6.Adachi K, Oiwa K, Nishizaka T, Furuike S, Noji H, Itoh H, Yoshida M, Kinosita KJ. Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell. 2007;130:309–321. doi: 10.1016/j.cell.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 7.Sambongi Y, Iko Y, Tanabe M, Omote H, Iwamoto-Kihara A, Ueda I, Tanagida T, Wada Y, Futai M. Mechanical rotation of the c subunit in ATP synthase (F0F1): Direct observation. Science. 1999;286:1722–1724. doi: 10.1126/science.286.5445.1722. [DOI] [PubMed] [Google Scholar]
- 8.Altman D, Sweeney HL, Spudich JA. The mechanism of myosin VI translocation and its load-inducing anchoring. Cell. 2004;116:737–749. doi: 10.1016/s0092-8674(04)00211-9. [DOI] [PubMed] [Google Scholar]
- 9.Kural C, Serpinskaya AS, Chou YH, Goldman RD, Gelfand VI, Selvin PR. Tracking melanosomes inside a cell to study molecular motors and their interaction. Proc Natl Acad Sci U S A. 2007;104:5378–82. doi: 10.1073/pnas.0700145104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kural C, Kim H, Syed S, Goshima G, Gelfand VI, Selvin PR. Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science. 2005;308:1469–72. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 11.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303:676–8. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 12.Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–5. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 13.Asbury CL, Fehr AN, Block SM. Kinesin moves by an asymmetric hand-over-hand mechanism. Science. 2003;302:2130–2134. doi: 10.1126/science.1092985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 15.Schnitzer MJ, Block SM. Kinesin hydrolyzes one ATP per 8-nm step. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 16.Svoboda K, Block SM. Force and velocity measured for single kinesin molecules. Cell. 1994;77:773–84. doi: 10.1016/0092-8674(94)90060-4. [DOI] [PubMed] [Google Scholar]
- 17.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–7. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 18.Kitamura K, Tokunaga M, Iwane AH, Yanagida T. A single myosin head moves along an actin filament with regular steps of 5.3 nanometres. Nature. 1999;397:129–34. doi: 10.1038/16403. [DOI] [PubMed] [Google Scholar]
- 19.Ishijima A, Kojima H, Funatsu T, Tokunaga M, Higuchi H, Tanaka H, Yanagida T. Simultaneous observation of individual ATPase and mechanical events by a single myosin molecule during interaction with actin. Cell. 1998;92:161–71. doi: 10.1016/s0092-8674(00)80911-3. [DOI] [PubMed] [Google Scholar]
- 20.Schafer DA, Gelles J, Sheetz MP, Landick R. Transcription by single molecules of RNA polymerase observed by light microscopy. Nature. 1991;352:444–448. doi: 10.1038/352444a0. [DOI] [PubMed] [Google Scholar]
- 21.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 22.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 23.Greenleaf WJ, Nlock SM. Single-molecule, motion-based DNA sequencing using RNA polymerase. Science. 2006;313:801. doi: 10.1126/science.1130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herbert KM, La Porta A, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–94. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–5. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–7. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perkins TT, Dalal RV, Mitsis PG, Block SM. Sequece-dependent pausing of single lambda exonuclease molecules. Science. 2003;301:1914–1918. doi: 10.1126/science.1088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Oijen AM, Blainey PC, Crampton DJ, Richardson CC, Ellenberger T, Xie XS. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science. 2003;301:1235–8. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 29.Wuite GJ, Smith SB, Young M, Keller D, Bustamante C. Single-molecule studies of the effect of template tension on T7 DNA polymerase activity. Nature. 2000;404:103–6. doi: 10.1038/35003614. [DOI] [PubMed] [Google Scholar]
- 30.Maier B, Bensimon D, Croquette V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc Natl Acad Sci U S A. 2000;97:12002–7. doi: 10.1073/pnas.97.22.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamdan SM, Johnson DE, Tanner NA, Lee JB, Qimron U, Tabor S, van Oijen AM, Richardson CC. Dynamic DNA helicase–DNA polymerase interactions assure processive replication fork movement. Mol Cell. 2007;27:539–49. doi: 10.1016/j.molcel.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 32.Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. DNA primase acts as a molecular brake in DNA replication. Nature. 2006;439:621–4. doi: 10.1038/nature04317. [DOI] [PubMed] [Google Scholar]
- 33.Prasad TK, Robertson RB, Visnapuu ML, Chi P, Sung P. E.C. A DNA-translocating Snf2 molecular motor: Saccharomyces cerevisiae Rdh54 displays processive translocation and extrudes DNA loops. J Mol Biol. 2007;369:940–53. doi: 10.1016/j.jmb.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amitani I, Baskin RJ, Kowalczykowski SC. Visualization of Rad54, a chromatin remodeling protein, translocating on single DNA molecules. Mol Cell. 2006;23:143–8. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Nimonkar AV, Amitani I, Baskin RJ, Kowalczykowski SC. Single molecule imaging of Tid1/Rdh54, a Rad54 homolog that translocates on duplex DNA and can disrupt joint molecules. J Biol Chem. 2007;282:30776–84. doi: 10.1074/jbc.M704767200. [DOI] [PubMed] [Google Scholar]
- 36.Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, Croquette V, Bensimon D, Owen-Hughes T. Direct observation of DNA distortion by the RSC complex. Mol Cell. 2006;21:417–25. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol Cell. 2006;24:559–68. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ha T, Rasnik I, Cheng W, Babcock HP, Gauss GH, Lohman TM, Chu S. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 2002;419:638–41. doi: 10.1038/nature01083. [DOI] [PubMed] [Google Scholar]
- 39.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dumont S, Cheng W, Serebrov V, Beran RK, Tinoco I, Pyle AM, Bustamante C. RNA translocation and unwinding mechanism of HCV NS3 helicase and its coordination by ATP. Nature. 2006;439:105–8. doi: 10.1038/nature04331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, LInke WA, Oberhauser AF, Carrion-Vazquez M, Kerkvliet JG, Lu H, Marszalek PE, Fernandez JM. Reverse engineering of the giant muscle protein titin. Nature. 2002;418:998–1002. doi: 10.1038/nature00938. [DOI] [PubMed] [Google Scholar]
- 42.Fernandez JM, Li H. Force-clamp spectroscopy monitors the folding trajectory of a single protein. Science. 2004;303:1674–8. doi: 10.1126/science.1092497. [DOI] [PubMed] [Google Scholar]
- 43.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: the elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–9. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 44.Gore J, Bryant Z, Nollmann M, Le MU, Cozzarelli NR, Bustamante C. DNA overwinds when stretched. Nature. 2006;442:836–839. doi: 10.1038/nature04974. [DOI] [PubMed] [Google Scholar]
- 45.Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 46.Bryant Z, Stone MD, Gore J, Smith SB, Cozzarelli NR, Bustamante C. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–41. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 47.Collin D, Ritort F, Jarzynski C, Smith SB, Tinoco I, Bustamante C. Verification of the Crooks fluctuation theorem and recovery of RNA folding free energies. Nature. 2005;437:231–4. doi: 10.1038/nature04061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liphardt J, Onoa B, Smith SB, Tinoco I, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–7. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 49.Basche T, Nie S, Fernandez JM. Single molecules. Proc Natl Acad Sci U S A. 2001;98:10527–10528. doi: 10.1073/pnas.191365898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rivetti C, Codeluppi S, Dieci G, Bustamante C. Visualizing RNA extrusion and DNA wrapping in transcription elongation complexes of bacterial and eukaryotic RNA polymerases. J Mol Biol. 2003;326:1413–26. doi: 10.1016/s0022-2836(03)00042-1. [DOI] [PubMed] [Google Scholar]
- 51.Guthold M, Zhu X, Rivetti C, Yang G, Thomson NH, Kasas S, Hansma HG, Smith B, Hansma PK, Bustamante C. Direct observation of one-dimensional diffusion and transcription by Escherichia coli RNA polymerase. Biophys J. 1999;77:2284–94. doi: 10.1016/S0006-3495(99)77067-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moreno-Herrero F, de Jager M, Dekker NH, Kanaar R, Wyman C, Dekker C. Mesoscale conformational changes in the DNA–repair complex Rad50/Mre11/Nbs1 upon binding DNA. Nature. 2005;437:440–3. doi: 10.1038/nature03927. [DOI] [PubMed] [Google Scholar]
- 53.Ristic D, Modesti M, van der Heijden T, van Noort J, Dekker C, Kanaar R, Wyman C. Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 2005;33:3292–302. doi: 10.1093/nar/gki640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kobayoshi M, Sumitomo K, Torimitsu K. Real-time imaging of DNA–streptavidin complex formation in solution using high-speed atomic force microscopy. Ultramicroscopy. 2007;107:184–190. doi: 10.1016/j.ultramic.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 55.Schlierf M, Li H, Fernandez JM. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc Natl Acad Sci U S A. 2004;101:7299–7304. doi: 10.1073/pnas.0400033101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sarkar A, Caamano S, Fernandez JM. The elasticity of individual titin PEVK exons measured by single molecule atomic force microscopy. J Biol Chem. 2005;280:6261–4. doi: 10.1074/jbc.C400573200. [DOI] [PubMed] [Google Scholar]
- 57.Wiita AP, Perez-Jimenez R, Walther KA, Gräter F, Berne BJ, Holmgren A, Sanchez-Ruiz JM, Fernandez JM. Probing the chemistry of thioredoxin catalysis with force. Nature. 2007;450:124–7. doi: 10.1038/nature06231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dohoney KM, Gelles J. X-sequence recognition and DNA translocation by single RecBCD helicase/nuclease molecules. Nature. 2001;409:370–374. doi: 10.1038/35053124. [DOI] [PubMed] [Google Scholar]
- 59.Finzi L, Gelles J. Measurement of lactose repressor-mediated loop formation and breakdown in single DNA molecules. Science. 1995;267:378–380. doi: 10.1126/science.7824935. [DOI] [PubMed] [Google Scholar]
- 60.van Oijen AM. Single-molecule studies of complex systems: the replisome. Mol Biosyst. 2007;3:117–25. doi: 10.1039/b612545j. [DOI] [PubMed] [Google Scholar]
- 61.Neuman KC, Block SM. Optical Trapping. Rev Sci Instrum. 2004;75:2787–2809. doi: 10.1063/1.1785844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lang MJ, Block SM. Resource letter: LBOT-1: Laser-based optical tweezers. Am J Phys. 2003;71:201–215. doi: 10.1119/1.1532323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bockelmann U, Thomen P, Essevaz-Roulet B, Viasnoff V, Heslot F. Unzipping DNA with optical tweezers: high sequence sensitivity and force flips. Biophys J. 2002;82:1537–53. doi: 10.1016/S0006-3495(02)75506-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shundrovsky A, Smith CL, Lis JT, Peterson CL, Wang MD. Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nat Struct Mol Biol. 2006;13:549–54. doi: 10.1038/nsmb1102. [DOI] [PubMed] [Google Scholar]
- 65.Jiang J, Bai L, Surtees JA, Gemici Z, Wang MD, Alani E. Detection of high-affinity and sliding clamp modes for MSH2-MSH6 by single-molecule unzipping force analysis. Mol Cell. 2005;20:771–81. doi: 10.1016/j.molcel.2005.10.014. [DOI] [PubMed] [Google Scholar]
- 66.Pease PJ, Levy O, Cost GJ, Gore J, Ptacin JL, Sherratt D, Bustamante C, Cozzarelli NR. Sequence-directed DNA translocation by purified FtsK. Science. 2005;307:586–90. doi: 10.1126/science.1104885. [DOI] [PubMed] [Google Scholar]
- 67.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–52. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 68.Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, Baskin RJ. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–8. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 69.Spies M, Amitani I, Baskin RJ, Kowalczykowski SC. RecBCD Enzyme Switches Lead Motor Subunits in Response to chi Recognition. Cell. 2007;131:694–705. doi: 10.1016/j.cell.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Handa N, Bianco PR, Baskin RJ, Kowalczykowski SC. Direct visualization of RecBCD movement reveals cotranslocation of the RecD motor after chi recognition. Mol Cell. 2005;17:745–50. doi: 10.1016/j.molcel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 71.Spies M, Bianco PR, Dillingham MS, Handa N, Baskin RJ, Kowalczykowski SC. A molecular throttle: the recombination hotspot chi controls DNA translocation by the RecBCD helicase. Cell. 2003;114:647–54. doi: 10.1016/s0092-8674(03)00681-0. [DOI] [PubMed] [Google Scholar]
- 72.Galletto R, Amitani I, Baskin RJ, Kowalczykowski SC. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–8. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 73.Zlatanova J, Leuba SH. Magnetic tweezers: a sensitive tool to study DNA and chormatin at the single-molecule level. Biochem Cell Biol. 2003;81:151–159. doi: 10.1139/o03-048. [DOI] [PubMed] [Google Scholar]
- 74.Leuba SH, Karymov MA, Tomschik M, Ramjit R, Smith P, Zlatanova J. Assembly of single chromatin fibers depends on the tension in the DNA molecule: magnetic tweezers study. Proc Natl Acad Sci U S A. 2003;100:495–500. doi: 10.1073/pnas.0136890100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.van der Heijden T, Seidel R, Modesti M, Kanaar R, Wyman C, Dekker C. Real-time assembly and disassembly of human RAD51 filaments on individual DNA molecules. Nucleic Acids Res. 2007;35:5646–57. doi: 10.1093/nar/gkm629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Strick TR, Allemand J-F, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 77.Strick TR, Croquette V, Bensimon D. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature. 2000;404:901–904. doi: 10.1038/35009144. [DOI] [PubMed] [Google Scholar]
- 78.Koster DA, Croquette V, Dekker C, Shuman S, Dekker NH. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 79.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ha T. Single-molecule fluorescence methods for the study of nucleic acids. Curr Opin Struct Biol. 2001;11:287–92. doi: 10.1016/s0959-440x(00)00204-9. [DOI] [PubMed] [Google Scholar]
- 81.Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc Natl Acad Sci U S A. 2002;99:4284–9. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Russell R, Zhuang X, Babcock HP, Millett IS, Doniach S, Chu S, Herschlag D. Exploring the folding landscape of a structured RNA. Proc Natl Acad Sci U S A. 2002;99:155–60. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhuang X, Bartley LE, Babcock HP, Russell R, Ha T, Herschlag D, Chu S. A single-molecule study of RNA catalysis and folding. Science. 2000;288:2048–51. doi: 10.1126/science.288.5473.2048. [DOI] [PubMed] [Google Scholar]
- 84.McKinney SA, Freeman AD, Lilley DM, Ha T. Observing spontaneous branch migration of Holliday junctions one step at a time. Proc Natl Acad Sci U S A. 2005;102:5715–20. doi: 10.1073/pnas.0409328102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McKinney SA, DÈclais AC, Lilley DM, Ha T. Structural dynamics of individual Holliday junctions. Nat Struct Biol. 2003;10:93–7. doi: 10.1038/nsb883. [DOI] [PubMed] [Google Scholar]
- 86.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–6. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Joo C, McKinney SA, Nakamura M, Rasnik I, Myong S, Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–27. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 88.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–5. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 89.Lee TH, Blanchard SC, Kim HD, Puglisi JD, Chu S. The role of fluctuations in tRNA selection by the ribosome. Proc Natl Acad Sci U S A. 2007;104:13661–5. doi: 10.1073/pnas.0705988104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11:1008–14. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 91.Blanchard SC, Kim HD, Gonzalez RL, Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A. 2004;101:12893–8. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gorman J, Chowdhury A, Surtees JA, Shimada J, Reichman DR, Alani E, Greene EC. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2-Msh6. Mol Cell. 2007;28:359–70. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Granéli A, Yeykal C, Robertson RB, Greene EC. Long-distance lateral diffusion of human Rad51 on double-stranded DNA. Proc Natl Acad Sci U S A. 2006;103:1221–1226. doi: 10.1073/pnas.0508366103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Prasad TK, Yeykal C, Greene EC. Visualizing the assembly of human Rad51 filaments on double-stranded DNA. J Mol Biol. 2006;363:713–728. doi: 10.1016/j.jmb.2006.08.046. [DOI] [PubMed] [Google Scholar]
- 95.Rasnik I, Myong S, Cheng W, Lohman TM, Ha T. DNA-binding orientation and domain conformation of the E. coli rep helicase monomer bound to a partial duplex junction: single-molecule studies of fluorescently labeled enzymes. J Mol Biol. 2004;336:395–408. doi: 10.1016/j.jmb.2003.12.031. [DOI] [PubMed] [Google Scholar]
- 96.Heyes CD, Groll J, Möller M, Neienhaus GU. Synthesis patterning and applications of star-shaped poly(ethylene glycol) biofunctionalized surfaces. Mol Biosyst. 2007;3:419–430. doi: 10.1039/b700055n. [DOI] [PubMed] [Google Scholar]
- 97.Sackmann E. Supported membranes: scientific and practical applications. Science. 1996;271:43–48. doi: 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- 98.Granéli A, Yeykal C, Prasad TK, Greene EC. Organized arrays of individual DNA molecules tethered to supported lipid bilayers. Langmuir. 2006;22:292–299. doi: 10.1021/la051944a. [DOI] [PubMed] [Google Scholar]
- 99.Rhoades E, Gussakovsky E, Haran G. Watching proteins fold one molecule at a time. Proc Natl Acad Sci U S A. 2003;100:3197–3202. doi: 10.1073/pnas.2628068100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Okumus B, Wilson TJ, Lilley DMJ, Ha T. Vesicle encapsulation studies reveal that single molecule ribozyme heterogeneities are intrinsic. Biophys J. 2004;87:2798–2806. doi: 10.1529/biophysj.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cisse I, Okumus B, Joo C, Ha T. Fueling protein-DNA interactions inside porous nanocontainers. Proc Natl Acad Sci U S A. 2007;104:12646–12650. doi: 10.1073/pnas.0610673104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kabata H, Kurosawa O, Arai I, Washizu M, Margarson SA, Glass RE, Shimamoto N. Visualization of single molecules of RNA polymerase sliding along DNA. Science. 1993;262:1561–1563. doi: 10.1126/science.8248804. [DOI] [PubMed] [Google Scholar]
- 103.Groves J, Boxer S. Micropattern formation in supported lipid membranes. Acc Chem Res. 2002 Mar;35:149–57. doi: 10.1021/ar950039m. [DOI] [PubMed] [Google Scholar]