Abstract
The Dpp and Fat-Hippo signaling pathways both regulate growth in Drosophila. Dpp is a BMP family ligand, and acts via a Smad family DNA-binding transcription factor, Mad. Fat-Hippo signaling acts via a non-DNA-binding transcriptional co-activator protein, Yorkie. Here we show that these pathways are directly interlinked. They act synergistically to promote growth, in part via regulation of the micro RNA gene bantam, and their ability to promote growth is mutually dependent. Yorkie and Mad physically bind each other, and we identify a 410 bp minimal enhancer of bantam that responds to Yorkie:Mad in vivo and in cultured cells, and show that both Yorkie and Mad associate with this enhancer in vivo. Our results indicate that in promoting the growth of Drosophila tissues, Fat-Hippo and Dpp signaling contribute distinct subunits of a shared transcriptional activation complex, Yorkie:Mad.
INTRODUCTION
Metazoan development relies on the reiterative deployment of multiple, conserved intercellular signaling pathways. The developing Drosophila wing has served as a model for identifying and characterizing pathways that regulate growth and patterning. For example, the Decapentaplegic (Dpp), Wingless (Wg), Notch, Hedgehog, Epidermal Growth Factor Receptor, and Fat-Hippo pathways all play important roles during wing development, and key components of each pathway were first identified through studies in the wing. The action of these pathways must be coordinated to ensure proper development. Here, we describe an unexpected intertwining of the Fat-Hippo and Dpp signaling pathways that regulates the growth of Drosophila tissues.
The Dpp pathway is named for its ligand, Dpp, a member of the BMP family. Dpp signals through type I (Thickveins, Tkv) and type II (Punt) receptors to promote phosphorylation of an R-Smad transcription factor, Mad (reviewed in Affolter and Basler, 2007). Mad can then act in conjunction with a co-Smad, Medea (Med), to activate the transcription of downstream genes. Alternatively, Mad and Med can activate downstream genes through a de-repression mechanism, acting in concert with Schnurri to repress the expression of the transcriptional repressor Brinker (Brk).
The Fat-Hippo pathway plays a conserved role in growth control and oncogenesis (reviewed in Reddy and Irvine, 2008). Fat-Hippo signaling controls growth by regulating transcription, and the critical mediator of this is a transcriptional co-activator protein, known as Yorkie (Yki) in Drosophila and Yap in vertebrates (reviewed in Oh and Irvine, 2010). Multiple upstream branches of Fat-Hippo signaling have been identified, but they all act via a kinase, Warts, which inhibits Yki activity by phosphorylating it. Several downstream genes that contribute to organ growth have been identified as targets of Yki in Drosophila, including the micro RNA gene bantam (ban), Cyclins B and E, E2f1, and the inhibitor of apoptosis Diap1 (encoded by thread). Yki does not bind DNA itself, but instead acts in conjunction with DNA-binding partner proteins. Several DNA-binding partners for Yap were identified in mammals before Yap was linked to Hippo signaling (reviewed in Bertini et al., 2009; Oh and Irvine, 2010). One of these, Sd/Tead, was subsequently identified as a key Yki/Yap partner, contributing to the influence of Fat-Hippo signaling on growth in both Drosophila and vertebrates (Goulev et al., 2008; Ota and Sasaki, 2008; Wu et al., 2008; Zhang et al., 2008; Zhao et al., 2008). However, because sd is only expressed and required in a fraction of the cells where yki is required, the identification of Sd as a Yki partner raised the question of how Yki controls growth in areas where sd appears to play no role. More recently, Hth was identified as a second Yki partner in Drosophila, promoting growth in the anterior eye in conjunction with Yki, in part by regulating ban (Peng et al., 2009). However, Hth also exhibits spatially restricted expression and genetic requirements, and so its identification still did not provide a general solution to the question of how Yki acts to promote growth in diverse places.
The role of Dpp signaling in controlling growth and patterning has been extensively studied (reviewed in Affolter and Basler, 2007). As its name suggests, Dpp is broadly required for growth in all of the imaginal discs. In the wing, Dpp is secreted by a stripe of cells along the anterior-posterior (A-P) compartment boundary, and then spreads from its site of synthesis, forming a morphogen gradient. The Dpp gradient plays an important role in wing patterning, and genes expressed in distinct domains in response to different thresholds of Dpp pathway activity have been identified. Much of the downstream patterning and growth control by Dpp in the wing is thought to be regulated by establishment of an inverse gradient of Brk expression, although some genes are also directly activated by Mad-Med transcription complexes.
The observations that both the Dpp and the Fat-Hippo pathways are required for growth regulation in Drosophila raises the question of how they are related. One link between them was identified through studies which established that Dpp signaling helps to establish gradients of expression of regulators of Fat-Hippo signaling, including the Fat ligand Dachsous (Ds) and the Fat/Ds kinase Four-jointed (Fj) (Rogulja et al., 2008). In Fat signaling, the gradient, rather than just the absolute levels, of Fat regulators is critical for pathway regulation, presumably because their ability to polarize cells impinges on Warts regulation (Rogulja et al., 2008; Willecke et al., 2008). The role of the Fj and Ds gradients in wing growth thus helped to explain how cell proliferation could be evenly distributed in the developing wing, despite being promoted by a gradient of Dpp pathway activity (Rogulja and Irvine, 2005). However, studies of Dpp signaling have established that the Dpp pathway also possesses an autonomous ability to promote wing growth (Martín et al., 2004; Martin-Castellanos and Edgar, 2002; Rogulja and Irvine, 2005; Schwank et al., 2008). In this manuscript, we show that this autonomous growth promotion involves a second intertwining with Fat-Hippo signaling. Remarkably, we find that the transcription factors at the end of each pathway, Yki and Mad, can interact directly to form a transcription factor complex that regulates the expression of downstream target genes crucial for growth, including ban.
RESULTS
Co-regulation of growth by Yki and Mad
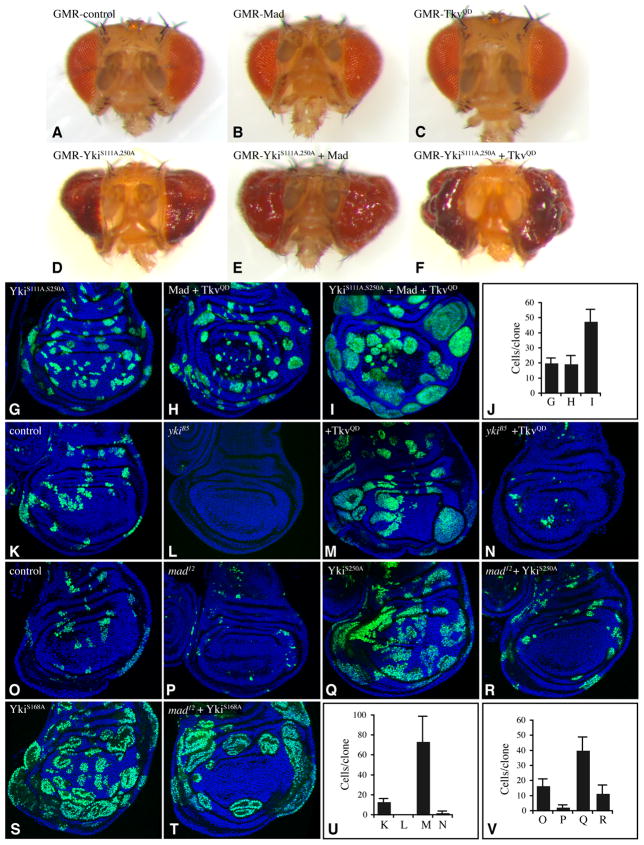
In a test of the functional relationship between Fat-Hippo and Dpp signaling, we explored the consequences of simultaneous elevation of the activity of both pathways. For Fat-Hippo, this was achieved by expressing isoforms of Yki activated by mutations in one or more Warts phosphorylation sites (Dong et al., 2007; Oh and Irvine, 2008, 2009). For Dpp, this was achieved either by expressing an activated form of the Tkv receptor, TkvQD (Nellen et al., 1996), or by over-expressing Mad. Transgenes encoding these proteins were expressed in vivo under UAS-Gal4 control. When GMR-Gal4 was used to drive expression in the eye disc, activated forms of Yki (YkiS168A:V5, or YkiS111A,S250A:V5) (Oh and Irvine, 2009) induce enlarged and irregular eyes (Figs 1D, S1A), whereas TkvQD or Mad did not significantly increase eye size (Fig. 1B,C). However, when activated-Yki was combined with Mad or TkvQD, the eye overgrowth phenotype was enhanced (Figs 1E,F, S1B,C). In a second assay, clones of cells expressing transgenes in the developing wing disc were examined. A mildly activated form of Yki, YkiS111A,S250A:V5, induces only mild growth enhancement (Fig. 1G), and Mad and TkvQD on their own modestly enhances the growth of clones in lateral regions (Fig. 1H). However, when they were all co-expressed, a stronger overgrowth phenotype was observed (Fig. 1I,J). Thus, Fat-Hippo and Dpp signaling can act cooperatively to promote growth in both eyes and wings.
Fig. 1. Yki and Tkv/Mad act cooperatively to promote growth.
A–F) Heads of adult females from GMR-Gal4 and A) control B) UAS-Flag:Mad, C) UAS-TkvQ235D, D) UAS-YkiS111,250A:V5, E) UAS-YkiS111,250A:V5 UAS-Flag:Mad, F) UAS-YkiS111,250A:V5 UAS- TkvQ235D. (G–I) Wing discs with clones (AyGal4) marked by UAS-GFP (green) and expressing G) UAS-YkiS111A,250A:V5, H) UAS-Flag:Mad UAS-TkvQ235D, I) UAS-YkiS111A,S250A:V5 UAS-Flag:Mad UAS-TkvQ235D, grown for 48 h, nuclei are labeled with Hoechst (blue). J) Average cells/clone in genotypes shown in panels G–I, as indicated. (K–N) Wing discs with MARCM clones expressing UAS-GFP, grown for 72 h. K) control, L) mutant for ykiB5, M) expressing UAS-TkvQ235D, N) expressing UAS-TkvQ235D and mutant for ykiB5. (O–T) Wing discs with MARCM clones grown for 72 h, expressing UAS-GFP. O) control, P) mutant for mad12, Q) expressing UAS-YkiS250A:V5, R) expressing UAS-YkiS250A:V5 and mutant for mad12, S) expressing UAS-YkiS168A:V5, T) expressing UAS-YkiS168A:V5 and mutant for mad12. U) Average cells/clone in genotypes shown in panels K–N, as indicated. V Average cells/clone in genotypes shown in panels O–R, as indicated. Error bars show standard deviation. See also Fig S1.
To further investigate this cooperativity, we created clones of cells in which one pathway was activated while the other was inactivated. Mutation of yki suppressed the ability of TkvQD to stimulate growth in the wing disc (Fig. 1M,N,U), which indicates that autonomous growth promotion by Dpp signaling requires Yki. In complementary experiments, the overgrowth that would normally be induced by expressing activated-Yki (Fig. 1Q,S) was partially suppressed by loss of function mutations in mad (Figs 1R,T,V, S1). With high level Yki activation (YkiS168A:V5), suppression of growth by mutation of mad was evident in the distal wing, but not in the proximal wing (Fig. 1S,T). With low level activation of Yki (YkiS250A:V5), suppression of growth was evident throughout the wing disc (Fig 1Q,R,V). These observations imply that Yki and Mad act in parallel to promote growth. In principle, this could occur either because they each independently regulate distinct downstream genes required for growth, or because they cooperate to regulate one or more common downstream genes. As described below, we identified a downstream gene essential for normal growth, ban, which is directly regulated by the concerted action of Yki and Mad.
ban is a common downstream target of Yki and Mad
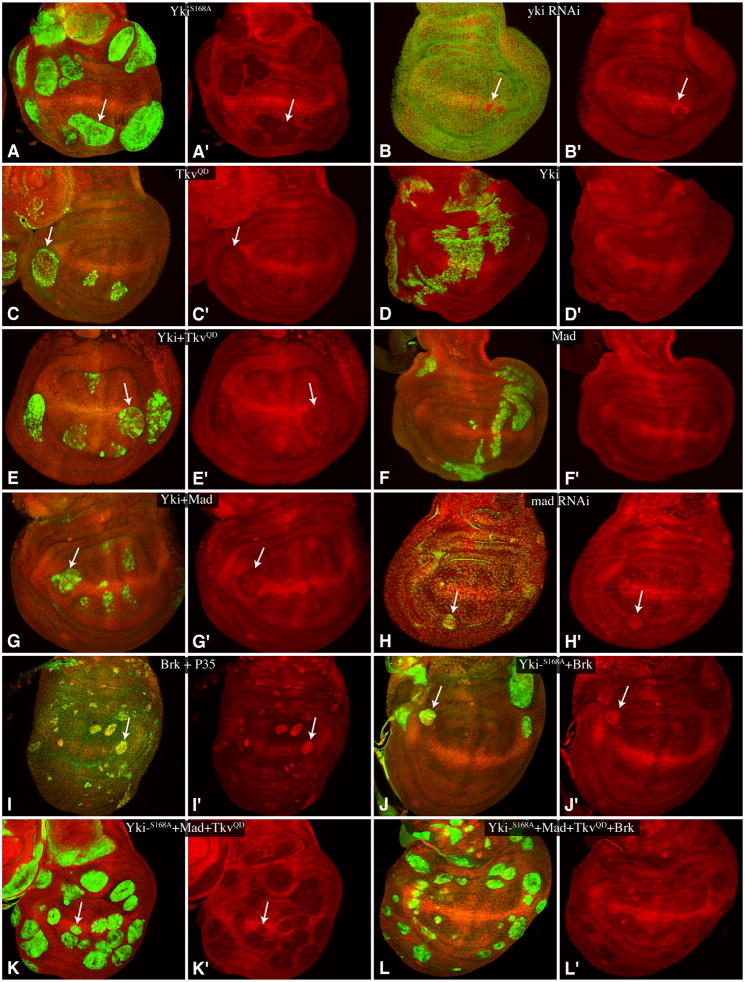
ban is a micro RNA gene that affects growth in developing tissues: loss of ban reduces growth, whereas over-expression of ban enhances growth (Brennecke et al., 2003; Hipfner et al., 2002). Expression of ban can be detected using a GFP-ban sensor, which inversely reports ban levels by virtue of a ban recognition sequence in the 3′ UTR of a GFP-expressing transgene (Brennecke et al., 2003). ban is a downstream target of Yki (Nolo et al., 2006; Thompson and Cohen, 2006), and consistent with published reports, GFP-ban sensor expression was decreased in clones expressing activated-Yki, and increased in clones in which yki levels were decreased by RNAi (Fig. 2A,B). Most clones of cells expressing TkvQD in wing discs did not noticeably influence ban sensor expression, although a slight decrease was observed in a fraction of clones in lateral regions of the wing disc, where TkvQD clones overgrow (Fig. 2C), consistent with an earlier report (Martín et al., 2004). However, when co-expressed, Tkv and Yki acted synergistically to decrease GFP-ban sensor expression. Thus, while expression of wild type Yki:V5 alone didn’t significantly influence ban sensor expression (Fig. 2D), and TkvQD clones only rarely influenced ban sensor expression, co-expression of TkvQD and wild-type Yki:V5 consistently decreased GFP-ban sensor expression (Fig. 2E). A similar synergy was observed when wild-type Mad and wild-type Yki were co-expressed: neither transcription factor significantly decreased GFP-ban sensor expression on its own (Fig. 2D,F), but when co-expressed they decreased GFP-ban sensor expression (Fig. 2G). Moreover, mad RNAi, like yki RNAi, increased GFP-ban sensor expression (Fig. 2H). These observations indicate that Fat-Hippo and Dpp signaling co-regulate ban through the key downstream transcription factors in each pathway, Yki and Mad.
Fig. 2. Regulation of ban by Yki, Mad/Tkv and Brk.
Wing discs expressing GFP-ban sensor (red) with A) Flip-out clones expressing UAS-YkiS168A:V5 (marked by anti-V5, green), B) UAS-RNAi-yki (anti-Yki, green), C) UAS- TkvQ235D (UAS-lacZ:NL (β-gal), green), D) UAS-Yki:V5 (anti-V5, green), E) UAS-Yki:V5 UAS-TkvQ235D (anti-V5, green), F) UAS-Flag:Mad (anti-Flag, green), G) UAS-Yki:V5 UAS-Flag:Mad (anti-V5, green) (G and G′), H) UAS-RNAi-mad UAS-Dcr2 (anti-Dcr2, green), I) UAS-Brk UAS-p35 (anti-Brk, green), J) UAS-YkiS168A:V5 UAS-Brk (anti-V5, green), K) UAS-YkiS168A:V5 UAS-Flag:Mad UAS-TkvQ235D (anti-V5, green), L) UAS-YkiS168A:V5 UAS-Flag:Mad UAS-TkvQ235D UAS-Brk (anti-V5, green). Panels marked prime show the GFP-ban sensor alone. In A–K arrows highlight clones with visible increases or decreases in GFP-ban sensor expression. Late third instar larvae were dissected either 48 h (B, H, I, K and L) or 72 h (A, C, D, E, F, G, and J) after clone induction.
It has been reported that yki mutant clones can be partially rescued by over-expression of ban, consistent with the inference that ban is a key downstream target of Yki (Nolo et al., 2006; Thompson and Cohen, 2006). To investigate whether ban could also rescue mad mutant clones, we used the MARCM technique to make clones of cells mutant for mad and over-expressing ban. Both mad10 and mad12 mutant clones were partially rescued by ban over-expression (Fig. S1).
Brk represses ban expression and Yki-Mad dependent growth
The results described above indicate that Yki and Mad act synergistically to promote growth at least in part by regulating ban. However, prior studies of growth regulation by Dpp indicated that Mad regulates growth principally through repression of expression of the transcriptional repressor Brk, which is a negative regulator of growth (Martín et al., 2004; Muller et al., 2003; Schwank et al., 2008). The disparate observations that Mad regulates growth through Brk, and that Mad cooperates with Yki to regulate growth by promoting ban expression, could be reconciled if Brk also regulates ban. Indeed, evidence for regulation of GFP-ban sensor expression by Brk has been reported (Martín et al., 2004). We performed a series of experiments to confirm and extend characterization of ban regulation by Brk.
Clones of cells over-expressing Brk grow poorly and are normally not recovered in wing imaginal discs. However, when the apoptosis inhibitor P35 was co-expressed with Brk, clones could be recovered, and these clones upregulated GFP-ban sensor expression (Fig. 2I). The growth and survival of Brk-expressing clones could also be partially rescued by co-expression of activated-Yki (Figs 2J,3C). Remarkably, even though activated-Yki upregulated ban expression (indicated by repression of GFP-ban sensor expression) (Fig. 2A), clones of cells co-expressing Brk and activated Yki often repressed ban expression (Fig. 2J). Thus Brk is able to repress ban even in the presence of Yki. Co-expression of Mad and TkvQD with Yki counteracted this repressive effect of Brk (Fig. 2L).
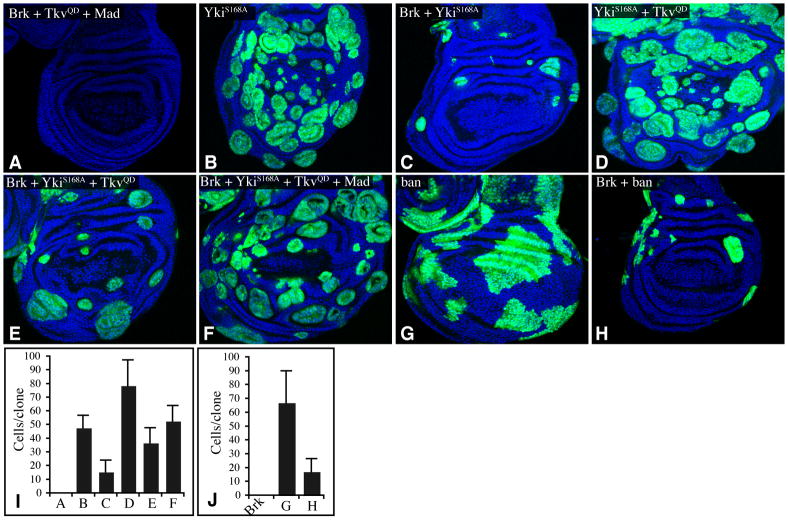
Fig. 3. Brinker represses ban and Yki-Mad dependent growth.
A–H) Wing discs (nuclei labeled by Hoechst, blue) with Flip-out clones (marked by UAS-GFP, green) expressing A) UAS-brk UAS-TkvQ235D UAS-Flag:Mad, B) UAS-ykiS168A:V5, C) UAS-brk UAS-ykiS168A:V5, D) UAS-ykiS168A:V5 UAS-TkvQ235D, E) UAS-brk UAS-ykiS168A:V5 UAS-TkvQ235D, F) UAS-brk UAS-ykiS168A:V5 UAS-TkvQ235D UAS-Flag:Mad, G) GS-ban, H) GS-ban UAS-brk. Third instar larvae were dissected 48h (A–F) or 72h (G,H) after heat-shock. I,J) Average cells/clone in genotypes shown in panels A–H, as indicated. See also Fig S2.
We also examined relationships among Brk, Yki, and ban in growth control. Co-expression of Mad and TkvQD was not sufficient to reverse the failure of Brk-expressing clones to survive (Fig. 3A). However, the rescue of Brk-expressing clones by co-expression of activated Yki was enhanced by expression of TkvQD, resulting in some clone overgrowth (Fig. 3E), and when TkvQD and Mad were co-expressed together with YkiS168A, strong growth of Brk-expressing clones was observed (Fig. 3F,I). Thus, Yki and Mad can act together to oppose growth repression by Brk. To investigate the opposing effects of Brk and Yki-Mad on growth in another context, we also examined eyes of animals expressing different combinations of transgenes under GMR-Gal4 control. Expression of Brk results in smaller eyes, and this small eye phenotype was suppressed by co-expression of Flag:Mad and TkvQD (Fig. S2). The small eye phenotype of Brk over-expression was also reversed by expression of activated-Yki, and co-expression of Yki and TkvQD together completely overcame the effect of Brk, resulting in overgrown eyes (Fig. S2). Because Brk was under a heterologous promoter when expressed in clones or under GMR-Gal4 control, the growth-promoting activity of Tkv and Mad in this context cannot be ascribed to their repression of Brk, but instead must reflect parallel influences of these genes on growth, which our results suggest are at least in part due to regulation of ban, in concert with Yki. Consistent with the importance of ban in growth regulation downstream of Brk, forced expression of ban could also partially rescue the growth of Brk-expressing clones (Fig. 3H,J), and Brk-expressing eyes (Fig. S2).
Yki and Mad co-regulate a ban enhancer
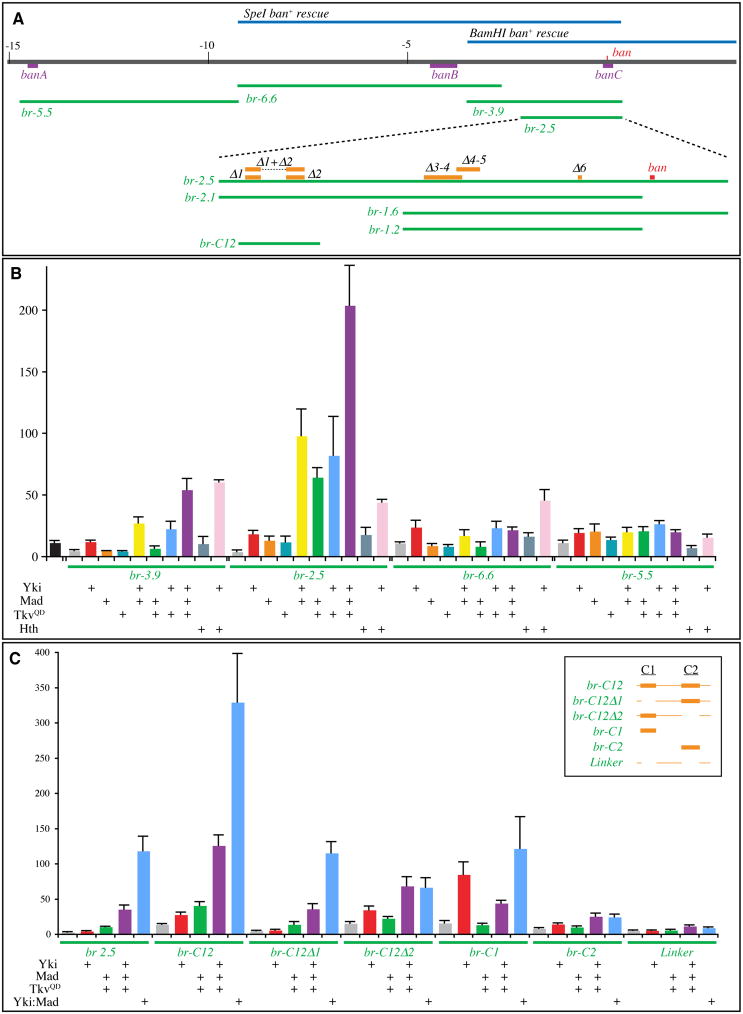
To determine whether the synergistic activation of ban expression by Yki and Mad reflects direct co-regulation, we sought to identify and characterize ban regulatory sequences. Two overlapping ban genomic rescue constructs (SpeI-ban+ and BamHI-ban+) together define a 3.9 kb region, which should thus contain all sequences essential for ban function (Brennecke et al., 2003), although we note that a Yki and Hth binding site was recently identified 14.5 kb upstream of the ban miRNA (Fig. 4A) (Peng et al., 2009). A reporter construct (br-3.9), including the entire 3.9 kb present in the overlap of ban rescue constructs, was created by cloning this DNA upstream of luciferase. In addition, we created a smaller ban reporter construct (br-2.5) from sequences within br-3.9, and two reporter constructs containing sequences farther upstream (br-5.5 and br-6.6, Fig. 4A). Their transcriptional activity was assayed by measuring Luciferase in transfected Drosophia S2 cells in the presence or absence of co-transfected transcription factors. Each of these ban reporters was stimulated to some degree by Yki (Fig. 4B). The greatest stimulation was observed using the br-2.5 reporter, which suggests that the 1.4 kb region that differs between br-3.9 and br-2.5 might contain elements that repress transcription. To investigate the influence of Dpp signaling, combinations of Yki, TkvQD and Mad were co-transfected together with the ban reporters (Fig. 4B). TkvQD or Mad, or both in combination, enhanced transcription from the br-2.5 reporter. Interestingly, they did not significantly enhance transcription from the br-3.9 or br-6.6 reporters, and the br-5.5 reporter was only modestly affected. Notably, when Yki was co-transfected together with Mad and/or TkvQD, a synergistic enhancement of transcription was detected from the br-3.9 and br-2.5 reporters, but not the br-6.6 or br-5.5 reporters. These observations support the conclusion from in vivo studies that the Fat-Hippo and the Dpp pathways can cooperate to promote ban expression, and define a 2.5 kb region that is sufficient to mediate this cooperative effect. For comparison, we also characterized the responsiveness of these enhancers to co-transfection with Hth; the br-2.5, br-3.9, and br-6.6 reporters were all modestly stimulated by co-transfection of Yki and Hth (Fig. 4B).
Fig. 4. Identification of ban enhancers.
A) Features of the ban locus. Blue lines: genomic rescue constructs (Brennecke et al., 2003), purple lines: Hth/Yki binding regions (Peng et al., 2009), green lines: ban reporter constructs, orange lines: deletions of sequences conserved among Drosophila species, red line: ban miRNA. B, C) Histograms depicting average ratios of firefly luciferase (experimental)/Renilla luciferase (control) from triplicate experiments; error bars indicate standard deviation. B) Expression from the indicated ban luciferase reporters in lysates of S2 cells transfected to express Yki, Mad, TkvQD and/or Hth, as indicated. Black bar represents a firefly luciferase reporter with a minimal Hsp70 promoter. C) Expression from the indicated ban luciferase reporters in lysates of S2 cells transfected to express Yki, Mad, TkvQD, and/or Yki:Mad, as indicated. Inset indicates the DNA present in br-C12 and subfragments, where the thick lines are the C1 and C2 regions identified in Fig S4. See also Figs S3,S4.
To further narrow down cis-regulatory elements responsible for Yki-Mad mediated activation, we next made three smaller reporter constructs (br-2.1, br-1.6, br-1.2) by deleting portions of the br-2.5 reporter (Fig. 4A). The br-1.6 and br-1.2 reporters failed to respond to Yki and Mad (Fig. S3A and data not shown), implicating the 900 bp region between the 5′ ends of br-1.6 and br-2.5 as essential for Yki-Mad mediated transcriptional activation. However, as the br-2.1 reporter was less active than br-2.5, we suspected that there might be multiple Yki-Mad responsive elements. To identify potential regulatory elements, we compared the sequence of the br-2.5 region among several Drosophila species (D.erecta, yakuba, simulans, sechellia, melanogaster, persimilis and pseudoobscura), and identified 7 conserved stretches of nucleotides, comprising the stem-loop region of the ban miRNA and 6 upstream regions (Figs 4A, S4, and not shown). To evaluate requirements for these conserved sequences, reporter constructs in which one or two of them were deleted within the context of br-2.5 were examined. Among the individual conserved motif deletions, Δ1 and Δ2 significantly lowered transcriptional activation, whereas Δ3, Δ4, Δ5 or Δ6 had no effect (Fig. S3A and data not shown). Among three double deletion constructs (Δ1-2, Δ3-4, Δ4-5) Δ1-2 almost completely eliminated the responsiveness of br-2.5 to Yki and Mad, whereas Δ3-4 and Δ4-5 had no effect (Fig. S3A). To investigate whether this region is not only necessary, but also sufficient, for Yki-Mad regulation, a 410 bp region including conserved sequences 1 and 2 (br-C12, Fig 4,S4) was assayed. Because multimerization of minimal transcription elements is sometimes needed for effective responses, we also made multimers of this enhancer. Both br-C12, and an 8x-br-C12 multimer, responded robustly to Yki and Mad (Figs 4, S3). Thus, the 410 bp br-C12 contains sequence elements sufficient for Yki-Mad-activated transcription. Since activation of br-C12 was even higher than that of br-2.5, br-2.5 might also include sequences that direct repression.
To delineate responsive elements with the C12 region, we characterized smaller subfragments, but these failed to fully recapitulate the Yki-Mad responsiveness of br-C12 (Fig. 4C). For example, br-C12 expression was induced 9-fold by co-expression of Yki, Mad and TkvQD, whereas br-C1 or br-C2 expression were induced only 3-fold, and expression regulated by the less conserved “Linker” sequences was induced only 2-fold. br-C1 was also strongly induced by Yki alone, which suggests that this region contain an element that responds to Yki independently of Mad. Addition of the Linker region to C1 (br-C12Δ2) slightly increased (to 4-fold) responsiveness to Yki, Mad and TkvQD. Combining the Linker region and C2 (br-C12Δ1) recreated a more robust (7-fold) responsiveness, although the absolute levels of expression were substantially lower than for br-C12 (Fig. 4C). These observations indicate that multiple sequence elements are required for effective co-responsiveness to Yki and Mad.
Two other DNA-binding proteins have previously been identified as partners for Yki in Drosophila: Sd and Hth (Goulev et al., 2008; Peng et al., 2009; Wu et al., 2008; Zhang et al., 2008). Previously mapped Hth binding sites at the ban locus lie outside br-C12 (Peng et al., 2009). There are no consensus Sd binding site sequences within the br-C12 sequence, and when Sd expression was reduced in S2 cells by RNAi, no effect on activation of br-C12 was observed (Fig. S3D), even though the same RNAi treatment reduced expression of the 3xsd-luciferase and Diap1-3.5-luciferase reporters (Fig. S3E,F).
Activity of Yki-Mad responsive ban enhancers in vivo
To examine the activity and regulation of ban enhancers in vivo, we first created transgenic flies that expressed a lacZ reporter under br-2.5 control. br2.5-lacZ expression was barely detectable with one copy of the reporter, although as a homozygote br2.5-lacZ expression could be detected, especially in the proximal, medial wing (Fig. 5A). Importantly, expression of activated forms of Yki or Tkv substantially increased br2.5-lacZ expression. Activated Yki was most effective in the medial, proximal wing disc (Fig. S5A,C), whereas TkvQD could increase br2.5-lacZ expression in lateral regions (Fig. S5B), but to a variable degree. Co-expression of Yki and TkvQD was more effective at inducing br2.5-lacZ, both medially and laterally (Fig. 5C). The preferential expression of br2.5-lacZ in medial regions, and its induction there by Yki, is consistent with an influence of Dpp signaling, because this is where endogenous pathway activity is highest.
Fig. 5. In vivo analysis of ban enhancers.
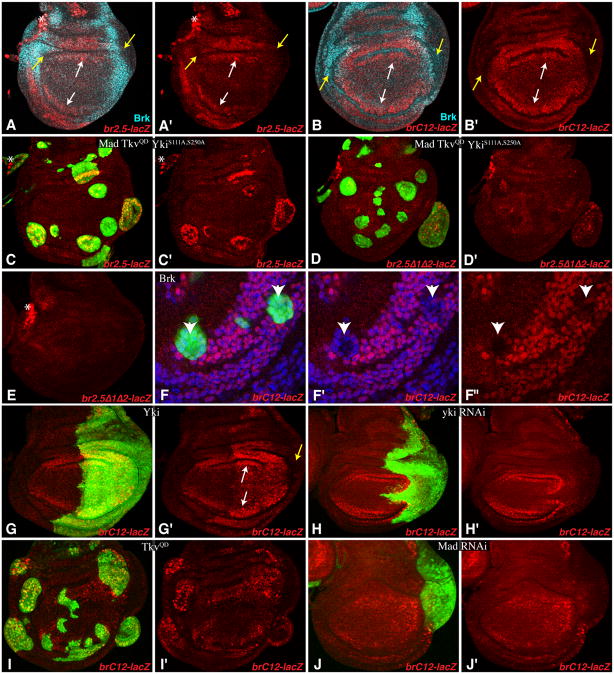
Wing discs from flies expressing ban-lacZ reporters. Panels marked prime show individual channel for β-gal (red); expression of activated or RNAi transgenes is marked by co-expression of GFP (green). Non-specific staining in the trachea (asterisk) is sometimes visible. Yellow arrows point to lateral regions with low expression, white arrows point to medial regions with higher expression. Brk expression is shown in cyan (A,B), and nuclei (F) in blue. A) With two copies of br-2.5-lacZ, expression is visible in the proximal wing, and is complementary to Brk. B) With one copies of br-C12-lacZ, expression is visible throughout the medial wing, and is complementary to Brk. C,D) Expression of one copy of br2.5-lacZ (C) and br2.5–Δ/Δ2-lacZ (D) in wing discs with Flip-out clones (marked by UAS-GFP, green) expressing UAS-Flag:Mad UAS- TkvQ235D UAS-YkiS111, 250A:V5. E) Even with two copies of br-2.5Δ1Δ2-lacZ, no expression is detected. F) Flip-out clones expressing UAS-brk UAS-p35 repress br-C12 expression (arrowheads), nuclei (marked by Hoechst, blue) are shown here to indicate that lack of staining is not an artefact of loss or movement of nuclei. G–J) Expression of br-C12-lacZ in cells with transgenes activating or depleting Yki or Mad are marked by GFP (green). G) en-Gal4 UAS-GFP UAS-Yki:V5 H) en-Gal4 UAS-GFP UAS-yki RNAi I) Ay-Gal4 UAS-GFP UAS-TkvQ235D J) en-Gal4 UAS-GFP UAS-Mad RNAi. See also Fig. S5.
To confirm the importance of the C12 region in vivo, we created and characterized two additional transgenes, one comprising br-C12 driving lacZ expression, and the other comprising the br-2.5-lacZ reporter with C1 and C2 deleted. Transgenic flies containing insertions of these reporters at the same cytological location as br2.5-lacZ were examined. By contrast to br2.5-lacZ, br2.5Δ1-Δ2-lacZ was not detectably expressed even as a homozygote, nor was its expression induced by activation of Yki or Tkv (Figs 5D,E, S5D). Similar to br-2.5-lacZ, br-C12-lacZ was expressed most strongly in the medial, proximal wing, but its expression was at a higher level, as it was readily detected with only one copy of the transgene (Fig. 5B). The observation that br-C12-lacZ expression was strongest in the medial area of the wing disc, and lowest at the lateral edges, is consistent with regulation by Dpp signaling (Fig. 5B). Moreover, br-C12-lacZ expression was elevated by activation of Yki or Tkv (Fig. 5G,I) and reduced by RNAi of yki or Mad (Fig. 5H,J), confirming its regulation by Yki and Mad in vivo.
In S2 reporter assays, Brk only reduced Yki-Mad mediated activation of br-C12 by half (Fig. S3B). However, in wing discs, br2.5-lacZ and br-C12-lacZ reporter expression were partially complementary to Brk expression (Fig. 5A,B), and their expression was repressed by Brk over-expression, and increased by brk RNAi (Figs 5F,S5 and not shown), indicating that they respond to Brk.
Independently-regulated downstream targets of Yki and Mad
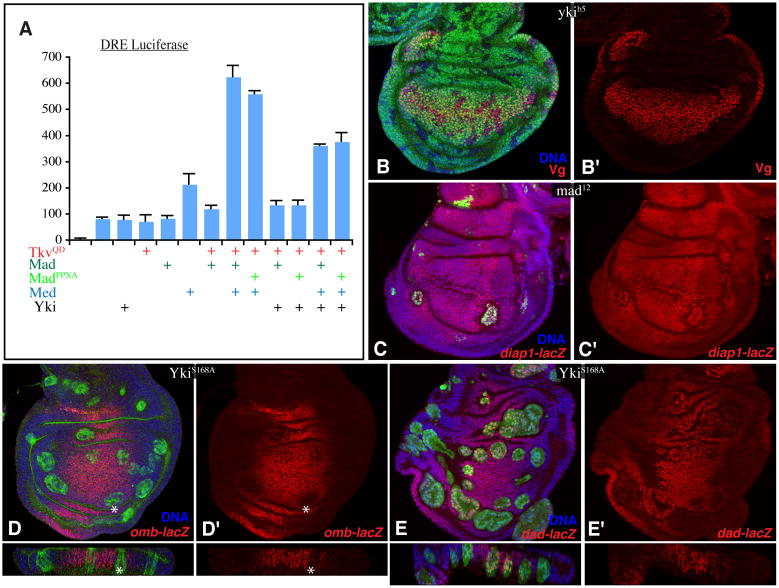
To investigate how generally Yki and Mad require each other to effect transcriptional activation, we examined their influence on previously identified downstream genes. A Dpp-responsive element (DRE) within an enhancer of the Ubx gene has been characterized previously (Kirkpatrick et al., 2001). In luciferase reporter assays in S2 cells, Yki did not stimulate DRE-mediated transcription, nor did it enhance the effects of Mad and TkvQD on the DRE (Fig. 6A). Regulation of the DRE and br-2.5 also differed in other respects. For the DRE, exogenous expression of the co-SMAD Med stimulated activation (Fig. 6A), and depletion of Med reduced activation (Fig. S6B). Conversely, addition or depletion of Med had little effect on br-2.5 (Figs 7A, S6A).
Fig. 6. Independently regulated targets of Yki and Mad.
A) Histogram of luciferase assay results (average firefly/Renilla ratio from triplicate experiments, error bars indicate standard deviation) using Ubx DRE reporter in S2 cells transfected to express Yki, TkvQ235D, Mad, MadPPxA, and/or Medea (Med) as indicated. B–E) Horizontal (square panels) and vertical (thin rectangles) sections of wing discs, with nuclei labeled by Hoechst (blue). Panels marked by prime symbols show stains without the clone marker. B) Vg (anti-Vg, red) expression in ykiB5 clones (marked by lack of β-gal, green). C) diap1-lacZ expression (marked by β-gal, red) in mad12 MARCM clones (marked by presence of GFP, green). D,E) Flip-out clones marked by UAS-GFP (green) expressing UAS-ykiS168A:V5 and stained for D) omb-lacZ or E) dad-lacZ (β-gal, red), asterisk marks decreased expression. Third instar larvae were dissected 48 h after heat-shock. See also Fig. S6.
Fig. 7. Interaction between Yki and Mad and association with ban enhancers.
A) Histogram of luciferase assay results (average firefly/Renilla ratio from triplicate experiments, error bars indicate standard deviation) using br-2.5 in S2 cells transfected to express Yki, Yki-WW, TkvQ235D, Mad, MadPPxA, Med, Yki:Mad, Yki: MadPPxA, and/or Yki-WW:Mad, as indicated. Inset (A–i) shows Western blot (anti-FLAG) on lysates used for luciferase assays; Flag:Mad, Flag:MadPPxA, Yki: Flag:Mad and Yki:Flag:MadPPxA were detected using anti-Flag antibodies. B) Co-immunoprecipitation of Yki:V5 and FLAG:Mad expressed in S2 cells. Upper panels (Input) show Western blots (anti-V5, anti-FLAG) on lysates, lower panel (IP) shows Western blot (anti-FLAG) on material precipitated using anti-V5 beads. C) EMSA of ban enhancer fragments, using the indicated amounts of Mad or Brk proteins. D) ChIP analysis of ban enhancers. Chromatin from eye discs of late 3rd instar larvae expressing UAS-YkiS168A:V5 and UAS-Flag:Mad under GMR-Gal4 control was immunoprecipitated using anti-V5, anti-Flag or mouse IgG (control) and amplified by PCR using primers for Pka (negative control), banA (positive control for Yki) and ban C12 region. E) Summary Model. Fat-Hippo signaling regulates the activity of Yki and Dpp signaling regulates the activity of Mad (black arrows). Three different DNA binding partners for Yki have been identified: Sd, Hth, and Mad. Several downstream targets of Yki have been identified, but only Diap1 and ban have been shown to be direct. Yki activates (gray arrows) Diap1 expression with Sd, and ban expression with Hth or Mad. Mad and Med activate some targets directly (eg dad), and others indirectly through repression (gray block lines) of brk. Production of Brk protein (dashed red line) generates a repressor of Dpp pathway targets, some of which are also regulated by Mad complexes (eg dad, ban), others of which are regulated only by Brk (eg omb). Both pathways control growth in part through regulation of ban. See also Fig. S7.
We also characterized downstream targets of Dpp signaling in wing discs (reviewed in Affolter and Basler, 2007). Four genes that are positively regulated by Dpp signaling (vestigial, vg, optomotorblind, omb, daughters against dpp, dad, spalt-related, salr), and one gene that is negatively regulated (brinker, brk) were examined. By contrast to the upregulation of ban, expression of activated-Yki did not lead to detectable increases in the expression of omb, salr or dad, nor decreases in the expression of brk (Fig. 6, S6). In fact, expression of omb appeared to be slightly decreased by expression of activated Yki. Vg was upregulated, but only in medial, proximal clones (Fig. S6D). Because this is a region where Vg can promote its own expression, and Vg can also act as a partner for Sd (Halder et al., 1998; Simmonds et al., 1998), we suspected that this activation of Vg might reflect an ability of over-expressed Yki to effect a Vg-like activity, rather than a normal role for Yki as a partner for Mad in activating Vg (Alarcon et al., 2009). Consistent with this hypothesis, yki mutant clones have no effect on endogenous Vg expression (Fig. 6B). In sum, while Yki functions as a Mad co-regulator for some genes (e.g. ban), Mad regulation of other genes is Yki-independent.
We also investigated the potential requirement for Mad in regulating previously identified direct targets of Yki. A 3.5 kb enhancer from the Diap1 (thread) gene is directly regulated by Sd and Yki (Zhang et al., 2008). However, a Diap1-3.5-GFP reporter was not regulated by TkvQD, either alone or in combination with YkiS250A, even though TkvQD and YkiS250A exert synergystic effects on growth (Fig. S6). To confirm the lack of responsiveness of this Diap1-3.5 Kb enhancer to Dpp signaling, we also assayed a Diap1-3.5-luciferase construct in S2 cells. Diap1-3.5-luciferase, as well as another direct target of Yki and Sd (3x-sd-luciferase) (Wu et al., 2008), were activated by Yki in a Sd-dependent fashion (Fig. S3). However, they were not stimulated by Mad or TkvQD (Fig. S3E,F). Moreover, an enhancer trap insertion in Diap1 (thread-lacZ) was unaffected by mad mutant clones in vivo (Fig. 6C). Thus, Diap1 is a Yki:Sd target, but not a Yki:Mad target. Altogether, our observations indicate that Yki and Mad co-regulate one or more target genes important for growth control, including ban, but they also each independently regulate distinct sets of downstream target genes in combination with other factors.
Yki and Mad interact directly to regulate a ban enhancer
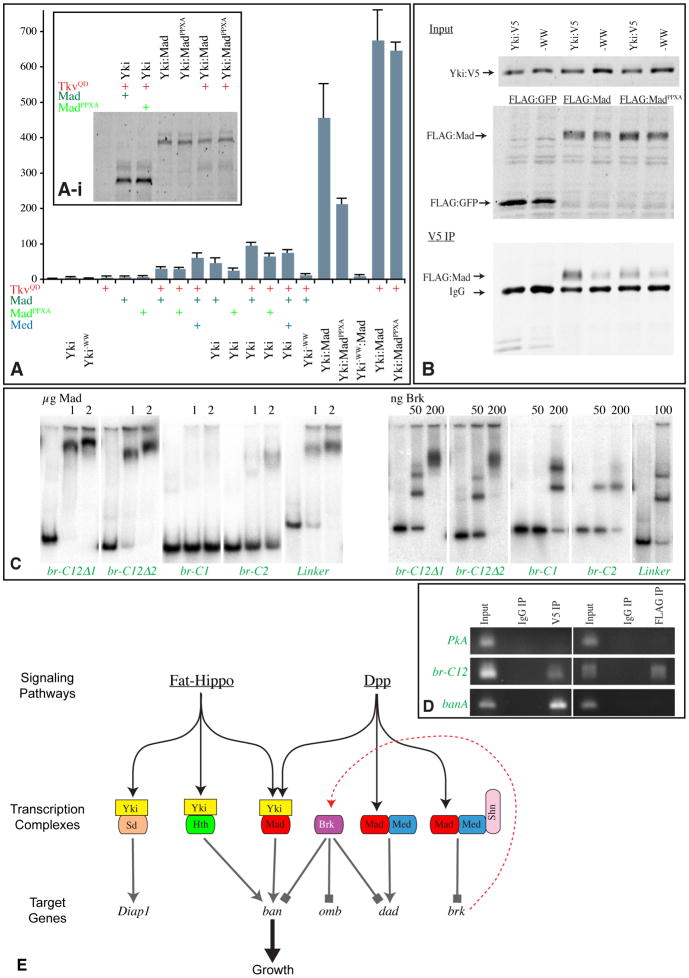
The co-regulation of ban enhancers by Yki and Mad could in principle be achieved by independent, parallel action. However, we considered the possibility that it might be mediated by direct association between Mad and Yki, acting as subunits of a shared transcriptional activation complex. Yki contains two WW domains, which can associate with PPXY motifs in other proteins to mediate direct binding (Macias et al., 2002). Mad has a PPXY motif in the linker region between its two Mad homology domains. To assay for direct interaction between Mad and Yki, S2 cells were co-transfected with FLAG:Mad and Yki:V5. Co-immunopreciptitation experiments confirmed Yki-Mad binding (Fig. 7B). Mutations in either the WW-domains of Yki, or in the PPXY motif of Mad, reduced, but did not abolish, Yki-Mad binding, which suggests that WW domain-PPXY interactions contribute to, but do not completely explain, the physical association between these proteins. Our detection of Yki-Mad binding is consistent with a recent study (Alarcon et al., 2009).
Since WW domain-PPXY interactions contribute to Yki-Mad binding, we investigated whether they contribute to Yki-Mad-mediated transcriptional activation of ban. When the PPXY motif of Mad was mutated (MadPPXA), the ability of Mad to enhance Yki-dependent activation of br-2.5 was reduced but not abolished (Fig. 7A), consistent with the effect of this mutation on Yki-Mad binding. MadPPXA was similar to wild type Mad on the Mad-Med responsive DRE (Fig. 6A). Mutation of the WW domains of Yki (Yki-WW) abolished its ability to stimulate ban reporter expression, either alone or in combination with Tkv and Mad (Fig. 7A), consistent with observations that the WW domains of Yki are essential for its ability to promote transcriptional activation (Oh et al., 2009; Zhang et al., 2009). To further investigate the significance of Yki-Mad binding, we created transgenes that expressed Yki:Mad fusion proteins. A wild-type Yki:Mad fusion was a potent activator of br-2.5 expression, significantly more active than the Yki, TkvQD and Mad combination, which indicates that direct interaction between Yki and Mad promotes ban transcription (Figs 4C,7A). A Yki:MadPPXA fusion protein was also an activator of the br-2.5 reporter, but Yki-WW:Mad was not (Fig. 7A), which implies that the requirement for the WW domains is not related to Mad recruitment.
Yki, Mad and Brk bind the ban C12 enhancer
The observations that Yki and Mad directly bind to each other, and act synergistically to regulate ban through the C12 enhancer, suggest that they can act together in a transcriptional activation complex that directly regulates ban via this enhancer. This hypothesis predicts that Mad and Yki should be co-associated with ban enhancer DNA in vivo. To test this, we performed chromatin immunoprecipitation (ChIP) experiments. Sheared genomic DNA was immunoprecipitated, using anti-V5 and anti-FLAG antibodies, from eye imaginal discs expressing YkiS168A:V5 and Flag:Mad under GMR-Gal4 control (The br-C12-lacZ reporter also responds to Yki and Tkv in eye discs, Fig. S5G,H). The 410 bp C12 region was amplified by PCR from DNA immunoprecipitated using either anti-V5 or anti-FLAG (Fig. 7D). As negative controls, DNA was precipitated with non-specific mouse IgG, or primers for an unrelated locus (protein kinase A) were employed; in neither case was a PCR band amplified from the precipitated material (Fig. 7D). Thus, Yki and Mad co-localize to the C12 region of ban in vivo. We also confirmed the association of Yki with a previously identified Yki-Hth site (banA) (Peng et al., 2009). Mad was not detected at banA (Fig. 7D), which suggests that distinct Yki-Mad and Yki-Hth complexes regulate ban through separate enhancers. Although antibodies suitable for ChIP of endogenous Mad were not available, we confirmed the association of endogenous Yki with the br-C12 enhancer in wild-type wing discs by ChIP using anti-Yki sera (Fig. S7A).
To confirm that Mad can directly associate with br-C12, we performed electrophoretic mobility shift assays (EMSA), using a Mad polypeptide including the DNA binding domain. In this assay, Mad also bound to br-C12, and not to banA (Fig. S7B). When subfragments of br-C12 were assayed, the strongest binding was detected to the Linker regions, and Mad also bound detectably to C2, but not C1 (Fig. 7C). Thus, br-C12 contains multiple Mad binding sites, despite the absence of sequences matching previously identified binding sites within the conserved C1 and C2 sequences. Notably, the regions that bind Mad in vitro (C2 and the Linker) together direct robust Yki:Mad responsiveness in S2 cells (Fig. 4C). Brk could also bind directly to br-C12 in vitro (Figs 7C, S7B), suggesting that its repression of ban is direct. Brk and Mad binding sites are related, and in some instances they compete for binding to these overlapping sequences (Affolter and Basler, 2007). However, Brk did not detectably compete for Mad binding within br-C12 (Fig. S7C), which suggests that Brk represses ban expression through distinct sites within this enhancer.
DISCUSSION
While substantial progress has been made towards identifying the regulatory pathways that control organ growth during development, elucidating how these pathways interact with each other to achieve proper growth control remains a significant challenge. Here, we have described an unanticipated intertwining of the Dpp and Fat-Hippo pathways that is required for growth control during imaginal development in Drosophila. Our observations argue that this involves a shared transcription factor complex, Yki-Mad, which includes subunits that are the key downstream transcriptional effectors for their respective pathways. The link between Yki and Mad is supported by several observations: The ability of each transcription factor to promote growth in vivo is partially dependent upon the other. They act synergistically to promote growth, and to regulate the expression of ban, a key regulator of growth in Drosophila. They bind directly to each other, and a mutation that decreases binding decreases their transcriptional activity, while directly fusing them together dramatically enhances it. And they regulate ban through the same minimal enhancer, to which they both bind in vivo, and to which Mad can bind in vitro.
An interaction between Yap and an inhibitory Smad, Smad7, was first reported several years ago (Ferrigno et al., 2002), and more recently interactions between Yap and Smad1, between Yki and Mad, and between Taz (a Yap homologue) and Smad2/3-4, have been reported (Alarcon et al., 2009; Varelas et al., 2008). However, the relevance of these interactions to either Fat-Hippo or Dpp signaling was not established. An obstacle to divining the general relevance of interactions between Smads and Yap family members has been their promiscuity – over a dozen partners for Yap have been identified previously (Bertini et al., 2009), and over fifty partners have been identified for Smads (Feng and Derynck, 2005; Ross and Hill, 2008). Importantly then, our observations establish Mad and Yki as critical partners for growth regulation in Drosophila, which is a crucial biological function of their respective pathways in imaginal discs.
Transcriptional regulation by Dpp signaling
Smads are DNA binding proteins, but on their own they generally bind with relatively low specificity (Feng and Derynck, 2005; Ross and Hill, 2008). This has led to a paradigm in which Smads bind regulatory sequences in conjunction with one or more co-factors. Dozens of co-factors have been identified in vertebrates, but far fewer in Drosophila. Instead, it has been concluded that most Dpp signaling in Drosophila is effected through transcriptional repression of Brk (Affolter and Basler, 2007). Repression of Brk is effected by a complex that includes Mad, Med, and the DNA-binding transcriptional repressor Schnurri, which together bind to “silencer elements”. However, genes that are directly activated by Dpp signaling have also been identified (Affolter and Basler, 2007). The best known of these is dad, which is activated by Dpp signaling via an activating element that binds a Mad-Medea complex without Schnurri (Weiss et al., 2010). The identification of Yki as a transcriptional co-activator for Mad raises the question of whether it is also a co-activator for other Dpp target genes. The lack of effect of Yki on dad expression, and on other Dpp pathway targets, argues that Yki is not a universal Mad co-activator, but rather acts as a co-activator for a subset of Mad target genes.
Although Smad-mediated transcription generally appears to involve complexes that include both R-Smads and co-Smads, some instances of Smad complexes composed only of R-Smads have been described in vertebrates (Ross and Hill, 2008). Our studies suggest that a Yki:Mad complex need not include Med. For example, in cultured cells, addition or depletion of Med did not significantly affect expression of br-2.5, but did affect the Ubx-DRE.
The role of Brk in growth control
Our delineation of a role for a Yki:Mad complex in growth control has to be squared with prior observations, which led to the conclusion that Dpp signaling regulates growth via repression of Brk. We propose that normal growth regulation by Dpp signaling is effected by two, parallel mechanisms – direct activation of downstream targets by Yki:Mad complexes, and de-repression of downstream targets by repression of Brk (Fig. 7E). This model fits studies of ban expression, as both Brk and Yki:Mad regulate ban and bind the C12 enhancer. Importantly, because Yki:Mad can influence growth and ban expression even when Brk is expressed under the control of a heterologous promoter, its effects can’t be ascribed simply to transcriptional repression of Brk: Yki-Mad and Brk must compete to regulate ban. This parallels studies of salm and dad regulation by Dpp signaling, for which Mad activates transcription both directly, and indirectly, via Brk (Affolter and Basler, 2007). In the parallel regulation model, the regulatory links could be partially redundant. Thus, the lack of requirement for Mad for growth in the absence of Brk could be explained by de-repression of Yki-Mad target genes like ban.
The sharing of components (e.g. Mad) between the direct (Yki-Mad) and indirect (Brk) growth regulatory pathways complicates assessment of their respective contributions, and indeed we would predict that their relative contributions could vary in time and space (e.g., depending on the level of Yki activation). However, if Yki-Mad activation is independent of Med, then med mutant clones might be expected to have weaker effects on growth than mad mutant clones, because Med would only participate in the Brk pathway. Notably, just such a difference has been reported (Wisotzkey et al., 1998). Moreover, Brk is evolutionarily derived, as thus far Brk homologues have only been found in insects. Thus, while in Drosophila, the Brk derepression pathway plays a major role in growth regulation, in other arthropods, direct activation by Yki-Mad might be more important.
Growth regulation by Yki and its DNA-binding partners
One of the puzzles of growth regulation by Yki in Drosophila has been the restricted distribution of its DNA-binding partners, Sd and Hth. Mad, by contrast, is expressed ubiquitously, and is required for growth in all imaginal discs. Thus, it is well positioned to be a general partner for Yki in growth regulation.
Since Smads often form complexes with other transcription factors, a Yki-Mad complex might form in conjunction with other proteins, and indeed our observation that the linker region of br-C12 exhibits the strongest binding to Mad, and was necessary but not sufficient for strong Yki-Mad-mediated activation, suggests that this might be the case. RNAi of sd did not significantly decrease the expression of ban reporters in S2 cells, which implies that Sd is not an obligate partner for Yki-Mad. RNAi of Hth also did not influence ban reporters (unpublished observations), although this is subject to the caveat that we lacked Hth-responsive reporters with which to compare. However, as co-regulators with Yki in over-expression experiments, Hth exhibited different profiles from TkvQD and Mad in terms of which ban enhancers were most sensitive. Thus, we favor the hypothesis that distinct Yki-Hth and Yki-Mad complexes independently regulate ban, acting through distinct enhancers. This could also explain the observations that mutation of mad did not block the overgrowth of YkiS168A-expressing clones in the proximal wing, where Hth expression is high, and did not block growth of expanded mutant clones in the eye imaginal disc, which upregulate Hth (Tyler and Baker, 2007); in these cases Hth might act in place of Mad to promote ban expression with Yki. Finally, we emphasize that while ban is clearly a key downstream target of Yki-Mad, there are also likely to be other targets important for growth control. Regardless of their number, our studies reveal that Drosophila tissues directly monitor the status of both the Dpp and Fat-Hippo pathways through a requirement for a shared transcription factor complex.
EXPERIMENTAL PROCEDURES
Histology and imaging
Imaginal discs were fixed and stained as described previously (Cho and Irvine, 2004), using as primary antibodies rabbit anti-Yki (1:400), mouse anti-V5 (1:400, Invitrogen), mouse anti-Flag (1:2000, Sigma), rabbit anti-Vg (1:200, gift of S. Carroll), rabbit anti-Dcr2 (1:200, abcam), goat anti-β-gal (1:500, Biogenesis), and guinea pig anti-Brk (1:2000, gift of G. Morata). Fluorescent stains were captured on a Leica TCS SP5 confocal microscope.
Clone sizes were measured by counting cell numbers (using nuclear staining) because in some instances clones induce folding which makes area measurements inaccurate, and cell size is not significantly affected by activation of Yki or Tkv. At least 30 clones, from multiple discs, were counted for the histograms in Fig 1, and at least 20 clones were counted for the histograms in Fig 7.
S2 cell assays and co-immunoprecipitation
S2 cells were cultured with Schneider’s Drosophila medium (Invitrogen) and 10% FBS (Sigma). For co-immunoprecipitation, transient transfections were performed with equal amounts of DNA (1 μg per construct) using Cellfectin (Invitrogen) in 6 well plates according to the manufacturer’s protocol. Co-immunoprecipitation assays were performed as described previously (Chen et al., 2003). For Western blotting mouse anti-V5 (1:5000, Invitrogen), IR dye 680 conjugated anti-mouse (goat,1:10000) (Odyssey) and IRdye 800 conjugated anti-Flag (1:100000, rabbit) were used. Blots were scanned and analyzed using the Odyssey Infrared Imaging system (LiCor biosciences).
Luciferase reporter assays were performed using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer’s instructions. S2 cells were transfected in triplicate with Luciferase-reporter constructs (25 ng) and copia-renilla luciferase (8 ng) reporters in 48-well plates together with 25 ng of each plasmid in a pAc5.1 vector (Invitrogen) and incubated for 48 hrs after transfection. For RNAi experiments, templates for in vitro transcription were amplified by PCR which have T7 promoter at each end, then used for in vitro transcription using MEGAscript Kit (Ambion) according to the manufacturer’s instructions. RNAi was performed as described previously (Clemens et al., 2000), but modified in order to combine RNAi and transfection. Briefly, 24 hrs after dsRNA was added to the cell, cells were washed once with serum-free medium and then transfection was performed and the cells were incubated for 48 hrs for luciferase assays.
Chromatin Immunoprecipitation (ChIP)
Late 3rd instar larvae, wild type or overexpressing YkiS168A:V5 and Flag:Mad under the control of GMR-Gal4, were dissected and wing or eye imaginal discs were collected in PBS on ice. Chromatin preparation and immunoprecipitation were performed as described in the protocol of EZ ChIP Chromatin Immunoprecipitation Kit (Millipore). About 50 discs were used for an IP reaction. Rabbit normal IgG (1 μg) (Millipore), rabbit anti-Yki (1:400), Mouse normal IgG (1 μg) (Millipore), mouse anti-V5 (1 μg) (Invitrogen), and mouse anti-Flag (0.5 μg) (Sigma) were used for immunoprecipitation. For PCR pairs of primers from Pka (5′-agccgcactcgcgcttctac/5′-caatcagcagattctccggct), banA (5′-aatccaaacgtgcagacggc/5′-agcggtgtctaagcacagcg) (Peng et al., 2009) and C12 region of bantam (5′-gcgactgagcgtgggtttttg/5′-cgactctcaacattctaaactta) were used.
Electrophoretic Mobility Shift Assay (EMSA)
As in earlier studies, we used the DNA binding domains of Brk and Mad rather than full length proteins, and we observed that Brk has significantly higher affinity for DNA than Mad (Kim et al., 1997; Kirkpatrick et al., 2001; Pyrowolakis et al., 2004; Weiss et al., 2010). DNA binding domains of Mad (MadPvuII, first 303 aa: MHI + Linker)(Kim et al., 1997) and Brk (Brk:DBD, first 99 aa) were amplified by PCR and inserted into SmaI site of pGEX-3X vector and transformed into BL21(DE3)pLysS to be induced by 0.5 mM IPTG. Purification of GST-MadPvuII and Brk:DBD were performed using B-PER GST Fusion Protein Spin Purification Kit (Thermo Scientific) according to manufacturer’s instruction. Radioactive-labeled probes were generated by PCR followed by T4 PNK treatment in the presence of [γ-32P] ATP. Binding reactions were carried out in 20 μl of 100 mM KCl, 10 mM Tris (pH 7.5), 1 mM DTT, 5 mM MgCl2, 0.05 % NP-40, 1 μg poly(dIdC), 2.5% Glycerol, 1 mM EDTA, 0.3 % BSA containing 10,000 cpm probes for 20 minutes at room temperature and then separated in 5 % polyacrylamide gels containing 1X TBE (Bio-rad) followed by autoradiography using phosphoimager.
Additional Methods
Standard methods were employed for Drosophila genetics and plasmids construction; a detailed description is in the Supplementary Material.
Supplementary Material
Acknowledgments
We thank K. Basler, S. Carroll, R. Mann, G. Morata, R. Padgett, C. Rushlow, Flybase, the DSHB and the Bloomington Stock Center for antibodies, plasmids, bioinformatics and Drosophila stocks, K. Malanga for technical assistance, and M. Affolter, K. Basler, and C. Rauskolb for comments on the manuscript. This research was supported by HHMI, NIH grant GM078620 to KDI, and NIH post-doctoral fellowship 1F32GM079817 to H.O.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Affolter M, Basler K. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat Rev Genet. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- Alarcon C, Zaromytidou AI, Xi Q, Gao S, Yu J, Fujisawa S, Barlas A, Miller AN, Manova-Todorova K, Macias MJ, et al. Nuclear CDKs drive Smad transcriptional activation and turnover in BMP and TGF-beta pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini E, Oka T, Sudol M, Strano S, Blandino G. YAP: at the crossroad between transformation and tumor suppression. Cell Cycle. 2009;8:49–57. doi: 10.4161/cc.8.1.7259. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Hipfner D, Stark A, Russell R, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- Cho E, Irvine KD. Action of fat, four-jointed, dachsous and dachs in distal-to-proximal wing signaling. Development. 2004;131:4489–4500. doi: 10.1242/dev.01315. [DOI] [PubMed] [Google Scholar]
- Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, Dixon JE. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A. 2000;97:6499–6503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Ferrigno O, Lallemand F, Verrecchia F, L’Hoste S, Camonis J, Atfi A, Mauviel A. Yes-associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF-beta/Smad signaling. Oncogene. 2002;21:4879–4884. doi: 10.1038/sj.onc.1205623. [DOI] [PubMed] [Google Scholar]
- Goulev Y, Fauny JD, Gonzalez-Marti B, Flagiello D, Silber J, Zider A. SCALLOPED interacts with YORKIE, the nuclear effector of the hippo tumor-suppressor pathway in Drosophila. Curr Biol. 2008;18:435–441. doi: 10.1016/j.cub.2008.02.034. [DOI] [PubMed] [Google Scholar]
- Halder G, Polaczyk P, Kraus ME, Hudson A, Kim J, Laughon A, Carroll S. The Vestigial and Scalloped proteins act together to directly regulate wing-specific gene expression in Drosophila. Genes Dev. 1998;12:3900–3909. doi: 10.1101/gad.12.24.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hipfner D, Weigmann K, Cohen SM. The bantam gene regulates Drosophila growth. Genetics. 2002;161:1527–1537. doi: 10.1093/genetics/161.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Johnson K, Chen HJ, Carroll S, Laughon A. Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature. 1997;388:304–308. doi: 10.1038/40906. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick H, Johnson K, Laughon A. Repression of dpp targets by binding of brinker to mad sites. J Biol Chem. 2001;276:18216–18222. doi: 10.1074/jbc.M101365200. [DOI] [PubMed] [Google Scholar]
- Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- Martín FA, Pérez-Garijo A, Moreno E, Morata G. The brinker gradient controls wing growth in Drosophila. Development. 2004;131:4921–4930. doi: 10.1242/dev.01385. [DOI] [PubMed] [Google Scholar]
- Martin-Castellanos C, Edgar BA. A characterization of the effects of Dpp signaling on cell growth and proliferation in the Drosophila wing. Development. 2002;129:1003–1013. doi: 10.1242/dev.129.4.1003. [DOI] [PubMed] [Google Scholar]
- Muller B, Hartmann B, Pyrowolakis G, Affolter M, Basler K. Conversion of an extracellular Dpp/BMP morphogen gradient into an inverse transcriptional gradient. Cell. 2003;113:221–233. doi: 10.1016/s0092-8674(03)00241-1. [DOI] [PubMed] [Google Scholar]
- Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895–1904. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. In vivo analysis of Yorkie phosphorylation sites. Oncogene. 2009;28:1916–1927. doi: 10.1038/onc.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Irvine KD. Yorkie: the final destination of Hippo signaling. Trends in Cell Biol. 2010;20:410–417. doi: 10.1016/j.tcb.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Reddy BV, Irvine KD. Phosphorylation-independent repression of Yorkie in Fat-Hippo signaling. Dev Biol. 2009;335:188–197. doi: 10.1016/j.ydbio.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota M, Sasaki H. Mammalian Tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of Hippo signaling. Development. 2008;135:4059–4069. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- Peng HW, Slattery M, Mann RS. Transcription factor choice in the Hippo signaling pathway: homothorax and yorkie regulation of the microRNA bantam in the progenitor domain of the Drosophila eye imaginal disc. Genes Dev. 2009;23:2307–2319. doi: 10.1101/gad.1820009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyrowolakis G, Hartmann B, Müller B, Basler K, Affolter M. A simple molecular complex mediates widespread BMP-induced repression during Drosophila development. Developmental Cell. 2004;7:229–240. doi: 10.1016/j.devcel.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Reddy BV, Irvine KD. The Fat and Warts signaling pathways: new insights into their regulation, mechanism and conservation. Development. 2008;135:2827–2838. doi: 10.1242/dev.020974. [DOI] [PubMed] [Google Scholar]
- Rogulja D, Irvine KD. Regulation of cell proliferation by a morphogen gradient. Cell. 2005;123:449–461. doi: 10.1016/j.cell.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Rogulja D, Rauskolb C, Irvine KD. Morphogen control of wing growth through the Fat signaling pathway. Dev Cell. 2008;15:309–321. doi: 10.1016/j.devcel.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S, Hill CS. How the Smads regulate transcription. Int J Biochem Cell Biol. 2008;40:383–408. doi: 10.1016/j.biocel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Schwank G, Restrepo S, Basler K. Growth regulation by Dpp: an essential role for Brinker and a non-essential role for graded signaling levels. Development. 2008;135:4003–4013. doi: 10.1242/dev.025635. [DOI] [PubMed] [Google Scholar]
- Simmonds AJ, Liu X, Soanes KH, Krause HM, Irvine KD, Bell JB. Molecular interactions between Vestigial and Scalloped promote wing formation in Drosophila. Genes Dev. 1998;12:3815–3820. doi: 10.1101/gad.12.24.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BJ, Cohen SM. The Hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in Drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Tyler DM, Baker NE. Expanded and fat regulate growth and differentiation in the Drosophila eye through multiple signaling pathways. Dev Biol. 2007;305:187–201. doi: 10.1016/j.ydbio.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X, Sakuma R, Samavarchi-Tehrani P, Peerani R, Rao BM, Dembowy J, Yaffe MB, Zandstra PW, Wrana JL. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- Weiss A, Charbonnier E, Ellertsdóttir E, Tsirigos A, Wolf C, Schuh R, Pyrowolakis G, Affolter M. A conserved activation element in BMP signaling during Drosophila development. Nat Struct Mol Biol. 2010;17:69–76. doi: 10.1038/nsmb.1715. [DOI] [PubMed] [Google Scholar]
- Willecke M, Hamaratoglu F, Sansores-Garcia L, Tao C, Halder G. Boundaries of Dachsous Cadherin activity modulate the Hippo signaling pathway to induce cell proliferation. Proc Natl Acad Sci U S A. 2008;105:14897–14902. doi: 10.1073/pnas.0805201105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisotzkey RG, Mehra A, Sutherland DJ, Dobens LL, Liu X, Dohrmann C, Attisano L, Raftery LA. Medea is a Drosophila Smad4 homolog that is differentially required to potentiate DPP responses. Development. 1998;125:1433–1445. doi: 10.1242/dev.125.8.1433. [DOI] [PubMed] [Google Scholar]
- Wu S, Liu Y, Zheng Y, Dong J, Pan D. The TEAD/TEF family protein Scalloped mediates transcriptional output of the Hippo growth-regulatory pathway. Dev Cell. 2008;14:388–398. doi: 10.1016/j.devcel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ren F, Zhang Q, Chen Y, Wang B, Jiang J. The TEAD/TEF family of transcription factor Scalloped mediates Hippo signaling in organ size control. Dev Cell. 2008;14:377–387. doi: 10.1016/j.devcel.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Milton CC, Humbert PO, Harvey KF. Transcriptional output of the Salvador/warts/hippo pathway is controlled in distinct fashions in Drosophila melanogaster and mammalian cell lines. Cancer Res. 2009;69:6033–6041. doi: 10.1158/0008-5472.CAN-08-4592. [DOI] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.