Abstract
Sialyl Lewisx (SLex, Siaα2–3Galβ1–4(Fucα1–3)GlcNAcβOR) is an important sialic acid-containing carbohydrate epitope involved in many biological processes such as inflammation and cancer metastasis. In the biosynthetic process of SLex, α2–3-sialyltransferase-catalyzed sialylation generally proceeds prior to α1–3-fucosyltransferase-catalyzed fucosylation. For the chemoenzymatic synthesis of SLex containing different sialic acid forms, however, it would be more efficient if diverse sialic acid forms are transferred in the last step to the fucosylated substrate Lewisx (Lex). An α2–3-sialyltransferase obtained from myxoma virus-infected European rabbit kidney RK13 cells (viral α2–3-sialyltransferase (vST3Gal-I)) was reported to be able to tolerate fucosylated substrate Lex. Nevertheless, the substrate specificity of the enzyme was only determined using partially purified protein from extracts of cells infected with myxoma virus. Herein we demonstrate that a previously reported multifunctional bacterial enzyme Pasteurella multocida sialyltransferase 1 (PmST1) can also use Lex as an acceptor substrate, although at a much lower efficiency compared to nonfucosylated acceptor. In addition, N-terminal 30-amino-acid truncated vST3Gal-I has been successfully cloned and expressed in Escherichia coli Origami™ B(DE3) cells as a fusion protein with an N-terminal maltose binding protein (MBP) and a C-terminal His6-tag (MBP-Δ30vST3Gal-I-His6). The viral protein has been purified to homogeneity and characterized biochemically. The enzyme is active in a broad pH range varying from 5.0 to 9.0. It does not require a divalent metal for its α2–3-sialyltransferase activity. It has been used in one-pot multienzyme sialylation of Lex for the synthesis of SLex containing different sialic acid forms with good yields.
Keywords: cloning, sialic acid, Lewisx, sialyltransferase, sialyl Lewisx
Introduction
Sialic acids belong to an important family of negatively charged monosaccharides. They have a common nine-carbon backbone of a polyhydroxy α-keto acid nature and are usually presented as the terminal residues on cell surface glycoconjugates of higher eukaryotes (Schauer 2000; Angata and Varki 2002; Chen and Varki 2010). The carboxyl group of sialic acids is normally deprotonated under physiological pH (Vimr et al. 2004), which makes them bear a net negative charge that affects their properties. Aside from their common features, sialic acids are structurally diverse in nature with more than 50 different sialic acid forms that have been identified. The diversity includes modifications at the carbon 5 which can link to an acetamido group providing N-acetylneuraminic acid (Neu5Ac), the most abundant sialic acid broadly presented in humans, animals, bacteria and protozoa. A hydroxyacetamido group on carbon 5 leads to N-glycolylneuraminic acid (Neu5Gc), a nonhuman sialic acid form, and a hydroxyl group at carbon 5 gives 2-keto-3-deoxy-nonulosonic acid (Kdn). Furthermore, additional modifications include single or multiple acetylation at hydroxyl groups at C4, C5, C7, C8 and/or C9, sulfation at C8-OH, phosphorylation or lactylation at C9-OH and methylation at C8-OH, C5-OH (for Kdn), or N-glycolyl hydroxyl group (for Neu5Gc) (Angata and Varki 2002; Schauer 2004; Chen and Varki 2010). Owing to their negative charge, exposed position and diversity, sialic acids play important roles in either masking recognition sites or facilitating cell recognition and adhesion. These biological processes are normally carried out through carbohydrate and protein interactions. Both sialic acids and underlying sugars are involved in determining the binding characteristics of the interactions. Among sialic acid-containing carbohydrates, sialyl Lewisx (SLex) Neu5Acα2–3Galβ1–4(Fucα1–3)GlcNAcβOR is one of the most well-studied structures. SLex is involved in inflammation (Rosen 2004). Its interaction with E-, P-, and L-selectins (Lowe et al. 1990; Phillips et al. 1990; Polley et al. 1991; Foxall et al. 1992; Lasky 1992) facilitates the recruitment of leukocytes to inflammation sites. SLex is also a well-known tumor-associated carbohydrate antigen that is involved in adhesion and metastasis of cancer cells (Takada et al. 1993; Ugorski and Laskowska 2002).
Both chemical and enzymatic methods have been developed to study the biological functions of SLex and its potential therapeutic applications such as cancer vaccine candidates. Chemical sialylation in the synthesis of SLex is usually low yielding and complicated by tedious protection and deprotection processes, steric hindrance of sialic acid for sialylation as well as difficulties in choosing suitable protecting groups and controlling stereospecificity (Boons and Demchenko 2000; Halcomb and Chappell 2002; Muthana et al. 2009). Chemically or enzymatically synthesized sialyloligosaccharides have been used as building blocks to improve the synthetic yields of SLex (Hayashi et al. 1996; Hanashima et al. 2007; Cao et al. 2008). Glycosyltransferase-catalyzed enzymatic methods have been developed with or without the regeneration of sugar nucleotides (Ichikawa et al. 1992; DeFrees et al. 1993, 1995; Lin et al. 1995; Duffels et al. 2000; Koeller and Wong 2000; Huang et al. 2006; Soriano del Amo et al. 2010; Zhang et al. 2010). Nevertheless, only Neu5Ac, the most abundant sialic acid form, was the sialic acid form in previous SLex synthetic targets.
In order to study the importance of naturally occurring sialic acid forms in SLex, instead of following its natural biosynthetic pathway in which sialylation takes place before fucosylation, a more efficient enzymatic synthetic approach would be to carry out the fucosylation before the final sialylation process to introduce diverse sialic acid forms. Although most bacterial α1–3-fucosyltransferases can tolerate both sialylated and nonsialylated N-acetyllactosamine (LacNAc) as acceptors (Lin et al. 2006; Soriano del Amo et al. 2010; Zhang et al. 2010), α2–3-sialyltransferases are usually restricted to nonfucosylated galactoside acceptors. Therefore, the key issue for efficient chemoenzymatic synthesis of SLex-containing diverse sialic acid forms is to find an α2–3-sialyltransferase that can tolerate fucosylated galactoside Lex as acceptor substrate.
Among reported α2–3-sialyltransferases, a viral α2–3-sialyltransferase (vST3Gal-I) was shown to be able to tolerate fucosylated acceptors (Jackson et al. 1999; Sujino et al. 2000). The sialyltransferase was classified together with mammalian sialyltransferases in Carbohydrate-Active enZymes (CAZy, http://www.cazy.org/) glycosyltransfersae family 29 (GT 29; Coutinho et al. 2003; Cantarel et al. 2009). This vST3Gal-I, encoded by myxoma virus gene MST3N, was obtained from RK13 cells (European rabbit kidney cells) infected with myxoma virus (Jackson et al. 1999; Sujino et al. 2000). Partially purified extracts of the cells were used to test the acceptor substrate specificity of vST3Gal-I. It was shown that the enzyme sialylated both Lewisx and Lewisa with relative Vmax of 30 and 40%, respectively, compared to that of LacNAc (Sujino et al. 2000). Nevertheless, the vST3Gal-I has not been purified to homogeneity and hence the detailed kinetics data were not available.
Herein we report the cloning, expression (in Escherichia coli), purification and characterization of an N-terminal truncated vST3Gal-I as a fusion protein with an N-terminal maltose binding protein (MBP) and a C-terminal His6-tag (MBP-Δ30vST3Gal-I-His6). Its activities toward fucosylated LacNAc (Lewisx) and nonfucosylated lactoside were compared. In addition, the tolerance of fucosylated LacNAc as an acceptor by a previously reported multifunctional Pasteurella multocida sialyltransferase 1 (PmST1 or tPm0188Ph; Yu et al. 2005; Ni et al. 2006, 2007) was demonstrated and compared to the recombinant vST3Gal-I. Partially purified recombinant vST3Gal-I was used in one-pot multienzyme sialylation of Lex for the synthesis of SLex containing different sialic acid forms with good yields.
Results
Sequence comparison of vST3Gal-I and mammalian ST3Gal-IVs
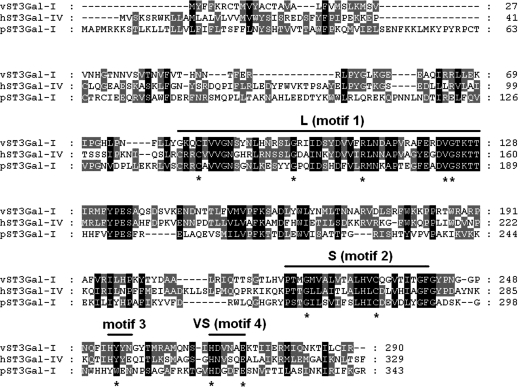
The amino-acid sequence of vST3Gal-I shares high identity to mouse ST3Gal-IV (37%) (Sujino et al. 2000), human ST3Gal-IV (38%) (Sasaki et al. 1993; Kitagawa and Paulson 1994), human ST3Gal-III (36%) (Kitagawa and Paulson 1993) and porcine ST3Gal-I (26%) whose X-ray crystal structure was recently reported (Rao et al. 2009). As shown in Figure 1, vST3Gal-I has all four conserved sialyl motifs including large (L), small (S), motif 3 and very small (VS) motifs identified previously (Datta and Paulson 1995; Geremia et al. 1997; Datta et al. 1998, 2001; Jeanneau et al. 2004). In addition, it has conserved amino-acid residues (shown by asterisks in Figure 1) including a conserved catalytic base H268 in sialyl motif VS identified in the porcine ST3Gal-I X-ray crystal structures (Rao et al. 2009).
Fig. 1.
Alignment of vST3Gal-I, hST3Gal-IV (GenBank accession number NP_006269) and pST3Gal-I (GenBank accession number AAA31125.1).
Cloning, expression and purification of recombinant protein
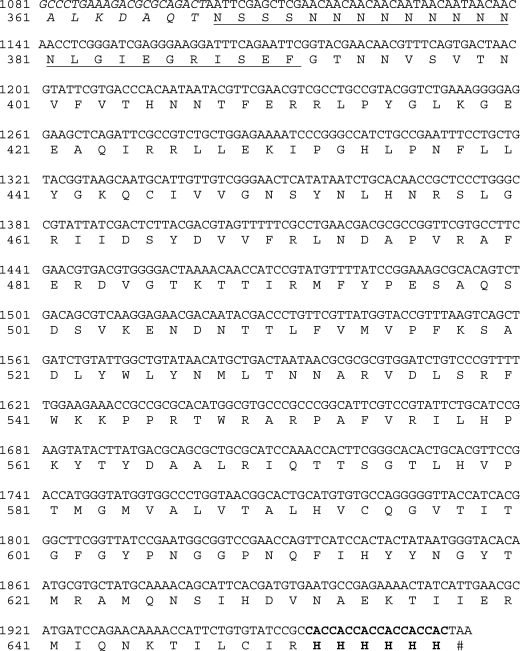
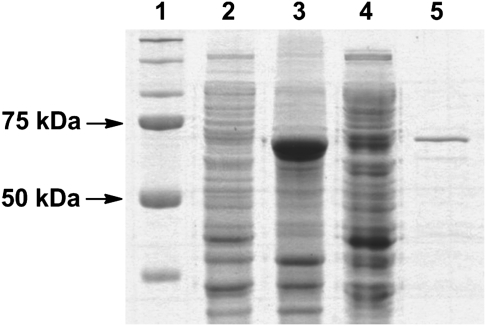
To obtain a soluble and active recombinant vST3Gal-I in E. coli expression system, a truncated protein with the deletion of an N-terminal cytoplasmic domain (1–6 aa) and a noncleavable signal-transmembrane sequence (7–30 aa) (Jackson et al. 1999) was cloned from a synthetic gene with codons optimized for E. coli expression. As shown in Figure 2, the codon-optimized Δ30vST3Gal-I gene contained 24% adenine, 27% cytosine, 25% guanine and 24% thymine as compared to the original sequence containing 28% adenine, 25% cytosine, 23% guanine and 24% thymine (GenBank accession number U46578.1). The recombinant protein was obtained as a fusion protein with an N-terminal MBP and a C-His6-tag. The MBP tag was introduced by using pMAL-c4X vector, while the C-His6-tag was introduced by including the His6-tag codons in the 3′-primer used for cloning. Optimal expression was achieved by incubating E. coli Origami™ B(DE3) cells at 20°C for 24 h with vigorous shaking (250 rpm) after the addition of isopropyl-1-thio-β-d-galactopyranoside (IPTG) (0.5 mM) for induction. Sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS–PAGE) analysis (Figure 3) indicated that the recombinant protein was overexpressed which constituted about 70% of the total protein in the whole-cell extraction. Nevertheless, only a small portion of the recombinant protein was seen in the cell lysate, the soluble portion of the cell extraction. Ni2+-NTA column purification of the fusion protein using an ÄKTATM fast protein liquid chromatography (FPLC) system yielded the purified protein, showing a molecular weight of around 72 kDa in the SDS–PAGE. The molecular weight was similar to that calculated (72.5 kDa) for the MBP-Δ30vST3Gal-I-His6.
Fig. 2.
Gene and protein sequences of the codon-optimized MBP-Δ30vST3Gal-I-His6. Only the C-terminal sequence of MBP is shown (in italics). The multiple cloning sites of pMAL-c4X vector are underlined. The six histidine residues introduced at the C-terminus during cloning are in bold.
Fig. 3.
SDS–PAGE analysis for MBP-Δ30vST3Gal-I expression and purification. Lanes: 1, protein standard; 2, whole-cell extraction before induction; 3, whole-cell extraction after induction; 4, cell lysates after induction; 5, Ni-column purified fraction.
pH profile of MBP-Δ30vST3Gal-I-His6
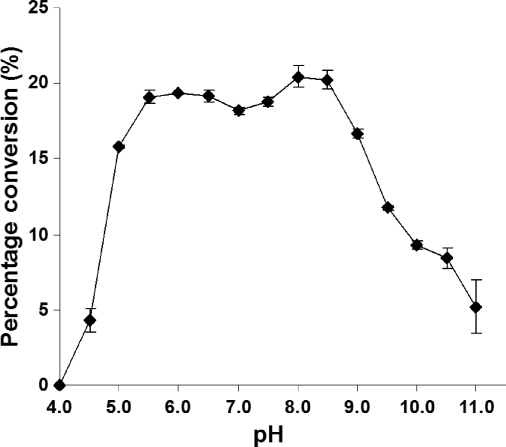
High-performance liquid chromatography (HPLC)-based pH profile study using a 4-methylumbelliferyl (MU)-labeled lactose (LacβMU or Galβ1–4GlcβMU) as an acceptor substrate demonstrated that MBP-Δ30vST3Gal-I-His6 was active in a wide pH range varying from 5.5 to 8.5 with minimum variation of activity (Figure 4). About 80% of the optimum activity was observed at pH 5.0 and 9.0. The activity decreased drastically when the pH was below 5.0 or above 9.0. Only about 20% of the optimum activity was observed at pH 4.5 and 11.0.
Fig. 4.
The pH profile of MBP-D30vST3Gal-I-His6. Activity was measured at indicated pH at 37°C for 30 min. Buffers (200 mM) used: sodium acetate (pH 4.5), MES (pH 5.0–6.5), HEPES (pH 7.0–7.5), Tris–HCl (pH 8.0–9.0), 3-(cyclohexylamino)-2-hydroxy-1-propanesulfonic acid (pH 9.5–11.0).
Kinetics
Both fluorophore-tagged lactoside (LacβMU) and Lewisx (LexβMU) were tolerable acceptor substrates by the MBP-Δ30vST3Gal-I-His6. As shown in Table I, LacβMU was a preferred acceptor for MBP-Δ30vST3Gal-I-His6 compared to LexβMU. Compared to LexβMU as an acceptor (kcat/Km = 1.5 mM−1 min−1), the catalytic efficiency of the recombinant viral enzyme was 19 times greater when LacβMU was used as an acceptor (kcat/Km = 28 mM−1 min−1). The difference was contributed by both a lower Km and a higher kcat value for LacβMU. Nevertheless, when LacβMU was used as an acceptor, the α2–3-sialyltransferase activity of MBP-Δ30vST3Gal-I-His6 was much weaker (about 70-fold difference) than a previously reported multifunctional P. multocida α2–3-sialyltransferase (PmST1; Yu et al. 2005; Ni et al. 2006, 2007). Quite interestingly, LexβMU was also a tolerable acceptor substrate for PmST1. When LexβMU was used as an acceptor, the catalytic efficiency of PmST1 (kcat/Km = 23 mM−1 min−1) was actually 15 times higher than that of MBP-Δ30vST3Gal-I-His6 (kcat/Km = 1.5 mM−1 min−1). Nonetheless, this PmST1 activity with LexβMU acceptor was 87-fold less efficient compared to the PmST1 activity with LacβMU acceptor due to a much (about 13-fold) higher Km value and a much (7-fold) lower kcat value for LexβMU.
Table I.
Apparent kinetic parameters of recombinant MBP-Δ30vST3Gal-I-His6 and PmST1
| Enzyme | Substrate | Km (mM) | kcat (min−1) | kcat/Km (mM−1 min−1) |
|---|---|---|---|---|
| MBP-Δ30vST3Gal-I-His6 | CMP-Neu5Ac | 1.4 ± 0.2 | 32 ± 2 | 23 |
| LacβMU | 1.2 ± 0.1 | 33 ± 1 | 28 | |
| LexβMU | 3.3 ± 0.5 | 4.7 ± 0.2 | 1.5 | |
| PmST1 | CMP-Neu5Aca | 0.44 | 1.9 × 103 | 4.4 × 103 |
| LacβMUa | 1.4 | 2.8 × 103 | 2.0 × 103 | |
| LexβMU | 18 ± 2 | (4.0 ± 0.2) × 102 | 23 |
Confirmation of the α2–3-sialyltransferase activity of MBP-Δ30vST3Gal-I-His6 in using Lex acceptor by one-pot two-enzyme synthesis of SLex Neu5Acα2–3Galβ1–4(Fucα1–3)GlcNAcβProN3
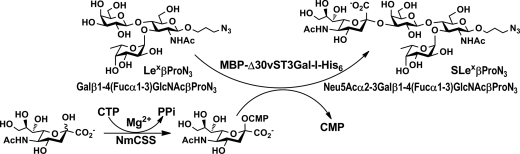
In order to confirm that Lex was indeed a tolerable acceptor for the α2–3-sialyltransferase activity of MBP-Δ30vST3Gal-I-His6, a preparative-scale synthesis of SLex tetrasaccharide Neu5Acα2–3Galβ1–4(Fucα1–3)GlcNAcβProN3 was carried out from Lex trisaccharide Galβ1–4(Fucα1–3)GlcNAcβProN3, Neu5Ac and cytidine 5′-triphosphate (CTP) using a one-pot two-enzyme reaction (Figure 5) containing partially purified recombinant sialyltransferase MBP-Δ30vST3Gal-I-His6 and a recombinant Neisseria meningitidis CMP-sialic acid synthetase (NmCSS; Yu et al. 2004). The presence of NmCSS allowed in situ synthesis of CMP-Neu5Ac, the sugar nucleotide donor for MBP-Δ30vST3Gal-I-His6, from inexpensive, commercially readily available Neu5Ac and CTP. SLex Neu5Acα2–3Galβ1–4(Fucα1–3)GlcNAcβProN3 was obtained in a yield of 61%. The product was confirmed by nuclear magnetic resonance (NMR) and high-resolution mass spectrometry (HRMS) studies. As shown in Table II, comparing the 13C NMR chemical shifts of the purified product and the starting trisaccharide Lex indicated a significant downfield shift (72.58 ppm to 75.82 ppm) of the C-3 signal of the galactose (Gal) residue in the product and a moderate upfield shift (71.16 ppm to 69.43 ppm) of the neighboring C-4 signal in the Gal. This confirmed that the sialylation took place at the C-3 of the Gal in Lex. High resolution mass spectrometry electrospray ionization (HRMS ESI) spectrum obtained showed the desired m/z of 926.3309 for C34H57N5O23Na (M+Na) (calculated: 926.3342).
Fig. 5.
Schematic illustration for the one-pot two-enzyme preparative scale synthesis of sialyl LexβProN3 Neu5Acα2–3Galβ1–4(Fucα1–3)GlcNAcβProN3 from Neu5Ac, CTP and LexβProN3 Galβ1–4(Fucα1–3)GlcNAcβProN3. Enzymes used: MBP-Δ30vST3Gal-I-His6 and a recombinant NmCSS.
Table II.
13C NMR chemical shift assignments of LexβProN3 Galβ1–4(Fucα1–3)GlcNAcβProN3 and SLexβProN3 including Neu5Acα2–3Galβ1–4(Fucα1–3)GlcNAcβProN3, Kdnα2–3Galβ1–4(Fucα1–3)GlcNAcβProN3 and Neu5AcN3α2–3Galβ1–4(Fucα1–3)GlcNAcβProN3
| Residue | Carbon atom | Chemical shift (ppm) |
|||
|---|---|---|---|---|---|
| βDGlcNAc | C | LexβProN3 | Neu5Acα2–3LexβProN3 | Kdnα2–3LexβProN3 | Neu5AcN3α2–3LexβProN3 |
| 1 | 101.09 | 101.15 | 101.09 | 101.17 | |
| 2 | 55.94 | 55.99 | 55.91 | 55.98 | |
| 3 | 75.05 | 75.07 | 75.02 | 75.08 | |
| 4 | 75.47 | 75.43 | 75.35 | 75.40 | |
| 5 | 73.49 | 73.52 | 73.43 | 73.47 | |
| 6 | 59.88 | 59.83 | 59.73 | 59.79 | |
| C=O | 174.40 | 174.41 | 174.17 | 174.43 | |
| CH3 | 22.36 | 22.40 | 22.31 | 22.38 | |
| βDGal(1–4) | 1 | 101.96 | 101.79 | 101.72 | 101.75 |
| 2 | 71.16 | 69.43 | 69.34 | 69.43 | |
| 3 | 72.58 | 75.82 | 75.70 | 75.79 | |
| 4 | 68.47 | 68.48 | 69.26 | 68.35 | |
| 5 | 75.10 | 74.99 | 74.93 | 74.99 | |
| 6 | 61.63 | 61.65 | 61.60 | 61.67 | |
| αLFuc(1–3) | 1 | 98.78 | 98.77 | 98.74 | 98.81 |
| 2 | 67.82 | 67.88 | 67.79 | 67.86 | |
| 3 | 69.33 | 69.36 | 69.84 | 69.33 | |
| 4 | 72.03 | 72.03 | 71.99 | 72.06 | |
| 5 | 66.85 | 66.85 | 66.79 | 66.86 | |
| CH3 | 15.43 | 15.44 | 15.37 | 15.44 | |
| αDNeu5Ac(2–3) | 1 | 174.05 | |||
| 2 | 99.84 | ||||
| 3 | 39.95 | ||||
| 4 | 67.48 | ||||
| 5 | 51.87 | ||||
| 6 | 73.08 | ||||
| 7 | 68.29 | ||||
| 8 | 72.07 | ||||
| 9 | 62.78 | ||||
| C=O | 175.20 | ||||
| CH3 | 22.21 | ||||
| αDKdn(2–3) | 1 | 174.36 | |||
| 2 | 99.76 | ||||
| 3 | 39.51 | ||||
| 4 | 67.28 | ||||
| 5 | 70.32 | ||||
| 6 | 74.04 | ||||
| 7 | 67.28 | ||||
| 8 | 72.26 | ||||
| 9 | 62.73 | ||||
| αDNeu5AcN3(2–3) | 1 | 174.09 | |||
| 2 | 99.82 | ||||
| 3 | 39.97 | ||||
| 4 | 67.44 | ||||
| 5 | 51.92 | ||||
| 6 | 72.74 | ||||
| 7 | 68.19 | ||||
| 8 | 72.10 | ||||
| 9 | 62.72 | ||||
| C=O | 171.36 | ||||
| NHCOCH2N3 | 52.06 | ||||
| ProN3 | OCH2CH2CH2N3 | 67.32 | 67.37 | 67.36 | 67.35 |
| OCH2CH2CH2N3 | 28.24 | 28.28 | 28.21 | 28.28 | |
| OCH2CH2CH2N3 | 47.88 | 47.94 | 47.84 | 47.91 | |
One-pot three-enzyme synthesis of SLex analogs containing different sialic acid forms
To demonstrate the application of MBP-Δ30vST3Gal-I-His6 in enzymatic synthesis of SLex containing different sialic acid forms from Lex, partially purified MBP-Δ30vST3Gal-I-His6 was used with NmCSS (Yu et al. 2004) and a recombinant P. multocida sialic acid aldolase (Li et al. 2008) in a one-pot three-enzyme system (Yu et al. 2005, 2006) for the preparative-scale synthesis of SLex tetrasaccharides Kdnα2–3Galβ1–4(Fucα1–3)GlcNAcβProN3 and Neu5AcN3α2–3Galβ1–4(Fucα1–3)GlcNAcβProN3 from mannose and N-azidoacetylmannosamine (ManNAcN3; Yu et al. 2005), respectively, in the presence of Lex, sodium pyruvate, CTP and MgCl2. In this system, aldolase catalyzed the formation of Kdn and N-azidoacetylneuraminic acid (Neu5AcN3) from mannose and ManNAcN3 in the presence of an excess amount of pyruvate. These sialic acids were converted into their activated CMP-sialic acid by NmCSS which were used by the viral sialyltransferase to transfer sialic acids to Lex for the formation of SLex containing different sialic acid forms. SLex tetrasaccharides Kdnα2–3Galβ1–4(Fucα1–3)GlcNAcβProN3 and Neu5AcN3α2–3Galβ1–4(Fucα1–3)GlcNAcβProN3 were formed with 64% and 58% yields, respectively. The products were confirmed by NMR as shown in Table II and HRMS studies. Similar to that observed for Neu5Acα2–3Galβ1–4(Fucα1–3)GlcNAcβProN3, a significant downfield shift of the C-3 signal of the Gal residue in the product and a moderate upfield shift of the neighboring C-4 signal in the Gal were observed in the 13C NMR spectra of both products when compared to that of Lex starting material, indicating the formation of a 2–3-sialyl linkage. HRMS (ESI) spectrum obtained showed the desired m/z of 885.3103 for Kdn-containing SLex C32H54N4O23Na (M+Na) (calculated: 885.3077) and 967.396 for Neu5AcN3-containing SLex C34H56N8O23Na (calculated: 967.3356).
Discussion
Synthesizing SLex with different sialic acid forms is crucial to understand the importance of structural diversity of naturally occurring sialic acids and to generate nonnatural carbohydrate probes. Variation of sialic acid structures, especially at the C-5 position, has been demonstrated to enhance the immunogenicity of some carbohydrate vaccines (Livingston 1995; Liu et al. 2000; Lemieux and Bertozzi 2001; Krug et al. 2004; Zou et al. 2004; Pan et al. 2005). We demonstrate here that SLex oligosaccharides containing natural and nonnatural sialic acid forms can be directly synthesized by enzyme-catalyzed sialylation of fucose-containing acceptor Lex. Although viral vST3Gal-I and bacterial PmST1 do not share significant protein sequence homology and belong to different CAZy glycosyltransferase families (GT 29 and GT 80, respectively), both have α2–3-sialyltransferase activity and can tolerate nonfucosylated and fucosylated galactosides as acceptors. Nonetheless, their activities toward fucosylated acceptors are much lower than the nonfucosylated acceptor substrates. We have successfully demonstrated here that the flexible acceptor substrate specificity of vST3Gal-I can be applied in a previously described one-pot three-enzyme sialylation system containing a sialic acid aldolase, a CMP-sialic acid synthetase and a suitable sialyltransferase (Yu et al. 2005, 2006) to introduce different sialic acid structures onto Lex to generate structurally diverse SLex and analogs. In comparison, the application of PmST1 in sialylation of LexβMU for the formation of Neu5Acα2–3LexβMU was unsuccessful as less than 5% yields were observed by HPLC assays due to its strong donor hydrolysis activity (data not shown). Nevertheless, the relatively low expression level of soluble and active vST3Gal-I hampers its application in large-scale synthesis of SLex. Due to the high expression level and high activity of PmST1, generating PmST1 mutants with increased activity toward fucosylated acceptors may be a preferred approach to achieve efficient synthesis of SLex with diverse sialic acid forms.
Compared to the common biosynthetic route for the formation of SLex structure with α2–3-sialylation followed by α1–3-fucosylation, enzymatic synthesis with an alternative glycosylation sequence of α1–3-fucosylation followed by α2–3-sialylation has a great advantage for introducing different sialic acid forms in the last step. Obtaining a sialyltransferase that can tolerate fucosylated acceptor substrate (e.g. Lex), such as the recombinant vST3Gal-I described here, provides an efficient catalyst to do so. With the availability of MBP-Δ30vST3Gal-I-His6 by expression in E. coli system, SLex with diverse naturally occurring and nonnatural sialic acid forms can be obtained from a common acceptor substrate Lex and a sialic acid precursor such as mannose, ManNAc or their derivatives, using an efficient one-pot three-enzyme system containing the sialyltransferase, a sialic acid aldolase and a CMP-sialic acid synthetase.
vST3Gal-I has high sequence homology with mouse ST3Gal-IV (Kono et al. 1997) and human ST3Gal-IV (Sasaki et al. 1993; Kitagawa and Paulson 1994). Similar to human placenta hST3Gal-IV expressed in COS-1 cells which showed marginal activity toward Lex with the relative rate of the reaction 33-fold less than that of type-II acceptor (Kitagawa and Paulson 1994), the recombinant MBP-Δ30vST3Gal-I-His6 shows a 19-fold less efficiency in its sialyltransferase activity toward Lex acceptor (LexβMU) compared to an acceptor (LacβMU) closely resembling type-II glycan.
As shown in Figures 1 and 2, vST3Gal-I has several potential N-linked glycosylation sites at 34, 45, 95, 147 and 283 (Jackson et al. 1999) with conserved NxS/T where x is any amino acid other than a proline. We demonstrate in this report that similar to pST3Gal-I (Rao et al. 2009), glycosylation of vST3Gal-I is not essential for its activity in vitro.
Materials and methods
Bacterial strains, plasmids and materials
E. coli electrocompetent cells DH5α were from Invitrogen (Carlsbad, CA) and chemically competent Origami™ B(DE3) from Novagen (Madison, WI). EcoRI and SalI restriction enzymes as well as vector plasmid pMAL-c4X were purchased from New England BioLabs (Beverly, MA). Ni2+-NTA agarose (nickel-nitrilotriacetic acid agarose), QIAprep Spin Miniprep Kit and QIAquick Gel Extraction Kit were from Qiagen (Valencia, CA). Herculase-enhanced DNA polymerase was from Stratagene (La Jolla, CA). T4 DNA ligase and 1 kb DNA ladder were from Promega (Madison, WI). Bicinchoninic acid (BCA) Protein Assay Kit was from Pierce Biotechnology, Inc. (Rockford, IL). CTP, d-N-acetylmannosamine (ManNAc) and sodium pyruvate were purchased from Sigma (St. Louis, MO). Full-length vST3Gal-I synthetic gene with the codons optimized for E. coli expression system was custom-synthesized by Codon Devices, Inc. (Cambridge, MA).
Synthesis of CMP-Neu5Ac
CMP-Neu5Ac was synthesized enzymatically from ManNAc, sodium pyruvate and CTP by a one-pot two-enzyme system using a recombinant sialic acid aldolase cloned from E. coli K12 and a recombinant NmCSS as described previously (Yu et al. 2004).
Synthesis of 4-methylumbelliferyl β-d-lactoside (LacβMU) and Galβ1–4GlcNAcβProN3 (LacNAcβProN3)
LacβMU was synthesized from lactose through a hepta-O-acetyllactosyl trichloroacetimidate intermediate as reported by Yu et al. (2005). The synthesis of LacNAcβProN3 was carried out as described previously (Chokhawala et al. 2008).
Synthesis of LexβProN3 and LexβMU as the acceptors for MBP-Δ30vST3Gal-I-His6
Both LexβProN3 and LexβMU were synthesized from the corresponding LacNAc derivatives LacNAcβProN3 and LacNAcβMU using a one-pot two-enzyme system containing a recombinant N-His6-tagged C-terminal truncated Helicobacter pylori α1–3-fucosyltransferase (Hp1-3FTΔ66; Sun et al. 2007) and a recombinant bifunctional L-fucokinase/GDP-fucose pyrophosphorylase (FKP) from Bacteroides fragilis (Yi et al. 2009). To synthesize LewisxβProN3, LacNAcβProN3 (47 mg, 0.10 mmol), L-fucose (25 mg, 0.15 mmol), adenosine 5'-triphosphate (ATP) (84 mg, 0.15 mmol), guanosine 5'-triphosphate (GTP) (79 mg, 0.15 mmol) and MgCl2 (41 mg, 0.20 mmol) were dissolved in H2O (5 mL). A stock solution of Tris–HCl buffer (1 M, pH 7.5, 1 mL) was added. After the addition of a recombinant FKP (3.4 mg) which catalyzes the synthesis of GDP–fucose from L-fucose, ATP and GTP via a fucose-1-phosphate intermediate and Hp1–3FTΔ66 (2.2 mg), water was added to bring the volume of the reaction mixture to 10 mL. The reaction mixture was then incubated in an incubator shaker at 37°C for 48 h. The reaction was stopped by adding cold EtOH (10 mL). The mixture was incubated on ice for 30 min and was centrifuged to remove precipitates. The supernatant was concentrated by rotavaporation. A Bio-Gel P-2 filtration column and a silica gel column (eluted with EtOAc:MeOH:H2O = 7:2:1, v/v/v) were used to purify the product LexβProN3 Galβ1–4(Fuc1–3)GlcNAcβProN3 (40 mg, 65% yield). The LexβMU Galβ1–4(Fucα1–3)GlcNAcβMU was synthesized similarly with a 59% yield.
Cloning
To clone MBP-Δ30vST3Gal-I-His6 in pMAL-c4X vector, the forward primer used was 5′-CCGGAATTCGGTACGAACAACGTTTCAG-3′ (EcoRI restriction site is underlined) and the reverse primer used was 5′-ACGCGTCGACTTAGTGGTGGTGGTGGTGGTGGCGGATACACAGAATG-3′ (SalI restriction site is underlined and the sequence encoding the hexahistidine tag is italicized). Polymerase chain reactions (PCRs) for amplifying the target gene were performed in a 50 μL of reaction mixture containing plasmid DNA (10 ng), forward and reverse primers (0.2 μM each), 10× Herculase buffer (5 μL), dNTP mixture (0.2 mM) and 5 U (1 μL) of Herculase-enhanced DNA polymerase. The reaction mixture was subjected to 30 cycles of amplification at an annealing temperature of 53°C. The resulted PCR product was purified and double-digested with EcoRI and SalI restriction enzymes. The digested PCR product was purified, ligated with the predigested pMAL-c4X vector and transformed into electrocompetent E. coli DH5α cells. Selected clones were grown for minipreps and characterization by restriction mapping. DNA sequencing was performed by the Davis Sequencing Facility in the University of California-Davis.
Expression
The plasmids of positive clones were selected and transformed into E. coli Origami™ B(DE3) chemically competent cells. The plasmid-bearing E. coli strain was cultured in an LB-rich medium (10 g L−1 tryptone, 5 g L−1 yeast extract and 10 g L–1 NaCl) supplemented with ampicillin (100 μg mL–1). Overexpression of the target protein was achieved by inducing the E. coli culture with 0.5 mM IPTG when the OD600 nm of the culture reached 1.0 followed by incubating the induced culture at 20°C for 24 h with vigorous shaking at 250 rpm in a C25KC incubator shaker (New Brunswick Scientific, Edison, NJ).
Purification
His6-tagged target protein was purified from cell lysates. To obtain the cell lysate, cell pellets harvested by centrifugation at 3452 × g for 2 h were resuspended in lysis buffer (pH 8.0, 100 mM Tris–HCl containing 0.1% Triton X-100) (20 mL for cells obtained from 1 L of cell culture). Lysozyme (50 μg mL–1) and DNaseI (3 μg mL–1) were then added and the mixture was incubated at 37°C for 60 min with vigorous shaking. Cell lysates were obtained as the supernatant after centrifugation at 11,000 rpm for 20 min. Purification of His6-tagged proteins from the lysate was achieved using an AKTA FPLC system (GE Healthcare) equipped with a HisTrap™ FF 5 mL column. The column was pre-equilibrated with 10 column volumes of binding buffer (5 mM imidazole, 0.5 M NaCl, 50 mM Tris–HCl, pH 7.5) before the lysate was loaded. After washing with 10 column volumes of binding buffer, stepwise gradient elution was carried out sequentially with 10 column volumes of elute buffers containing 34.25, 102.5 and 200 mM imidazole, respectively, in Tris–HCl buffer (50 mM, pH 7.5, 0.5 M NaCl). The fractions containing the purified enzyme were collected and stored at 4°C.
Quantification of purified protein
The concentration of purified enzyme was obtained in a 96-well plate using a BCA Protein Assay Kit (Pierce Biotechnology, Rockford, IL) with bovine serum albumin as a protein standard. The absorbance of each sample was measured at 562 nm by a BioTek Synergy™ HT Multi-Mode Microplate Reader.
Sodium dodecylsulfate–polyacrylamide gel electrophoresis
SDS–PAGE was performed in a 12% Tris–glycine gel using a Bio-Rad Mini-protein III Cell Gel Electrophoresis Unit (Bio-Rad) at a DC voltage of 120 V. Bio-Rad Precision Plus Protein Standards (10–250 kDa) were used as molecular weight standards. Gels were stained with Coomassie Blue.
pH profile by HPLC
Assays were performed in a total volume of 10 μL in a buffer (200 mM) with pH varying from 4.5 to 11.0 containing CMP-Neu5Ac (1 mM), LacβMU (1 mM) and MBP-Δ30vST3Gal-I-His6 (0.3 μg). The buffers used were: sodium acetate, pH 4.5; MES, pH 5.0–6.5; HEPES, pH 7.0–7.5; Tris–HCl, pH 8.0–9.0; 3-(cyclohexylamino)-2-hydroxy-1-propanesulfonic acid, pH 9.5–11.0. Reactions were allowed to proceed at 37°C for 30 min before being quenched by adding 10 μL of ethanol. An aliquot (2 μL) of the mixture was then added to 138 μL of 25% acetonitrile to make 70-fold dilutions. The samples were then kept on ice until aliquots of 8 μL were injected and analyzed by a Shimadzu LC-6AD system equipped with a membrane on-line degasser, a temperature control unit and a fluorescence detector (Shimadzu RF-10AXL). A reversed-phase Premier C18 column (250 × 4.6 mm i.d., 5 μm particle size, Shimadzu) protected with a C18 guard column cartridge was used. The mobile phase was 25% acetonitrile. The fluorescent compounds LacβMU and Neu5Acα2–3LacβMU were detected by excitation at 325 nm and emission at 372 nm (Yu et al. 2005). All assays were carried out in duplicates.
Expression and purification of the recombinant PmST1
The recombinant PmST1 was expressed as a His6-tag protein and purified using an ÄKTATM FPLC system (GE Healthcare) equipped with a HisTrap™ FF 5 mL column as previously described (Yu et al. 2005).
Kinetics by HPLC assay
The enzymatic assays were carried out in a total volume of 10 μL in a Tris–HCl buffer (200 mM, pH 8.0) containing CMP-Neu5Ac, acceptor substrates (LacβMU or LexβMU) and the recombinant proteins (MBP-Δ30vST3Gal-I-His6 or PmST1). All reactions for the MBP-Δ30vST3Gal-I-His6 were allowed to proceed at 37°C for 60 min. The PmST1 reactions with LexβMU acceptor were allowed to proceed for 7 min at 37°C. Apparent kinetic parameters of CMP-Neu5Ac for MBP-Δ30vST3Gal-I-His6 (0.2 μg was used) were obtained by varying the CMP-Neu5Ac concentrations (0.1, 0.2, 0.4, 1.0, 2.0 and 4.0 mM), while the concentration of LacβMU was fixed at 1 mM. Kinetic parameters of LacβMU (concentrations used were 0.1, 0.2, 0.4, 1.0, 2.0, 4.0, 6.0 and 8.0 mM) and LexβMU (concentrations used were 0.1, 0.2, 0.4, 1.0, 2.0, 4.0, 8.0 and 14.0 mM) for MBP-Δ30vST3Gal-I-His6 (0.3 and 0.4 μg were used for determining the parameters for LacβMU and LexβMU, respectively) were obtained with a fixed concentration of CMP-Neu5Ac (1 mM). The same concentration of CMP-Neu5Ac (1 mM) was used for the kinetic study of PmST1 (40 ng was used) on LexβMU (concentrations used were 1.0, 5.0, 10.0, 15.0, 25.0 and 35.0 mM). Apparent kinetic parameters were obtained by fitting the experimental data (the average values of duplicate assay results) into the Michaelis–Menten equation using Grafit 5.0.
One-pot two-enzyme synthesis of Neu5Acα2–3LexβProN3 from LexβProN3 using MBP-Δ30vST3Gal-I-His6
For the synthesis of Neu5Acα2–3LexβProN3, LexβProN3 (21 mg, 0.03 mmol), Neu5Ac (16 mg, 0.05 mmol), CTP (29 mg, 0.05 mmol) and MgCl2 (22 mg, 0.11 mmol) were dissolved in H2O (2.5 mL). A stock solution of Tris–HCl buffer (1 M, pH 8.5, 0.5 mL) was added. After the addition of a recombinant N. meningitidis CMP–sialic acid synthetase (1.1 mg) (Yu et al. 2004) and Ni2+-NTA column purified MBP-Δ30vST3Gal-I-His6 (262 μg, estimated), water was added to bring the volume of the reaction mixture to 5 mL. The reaction was carried out by incubating the solution in an incubator shaker at 37°C for overnight. The reaction was stopped by adding cold EtOH (5 mL) followed by incubation on ice for 30 min. The mixture was then centrifuged to remove precipitates. The supernatant was concentrated by rotavaporation. A BioGel P-2 filtration column and a silica gel column (eluted with EtOAc:MeOH:H2O = 5:2:1, v/v/v) were used to purify the product SLexβProN3Neu5Acα2–3Galβ1–4(Fucα1–3)GlcNAcβProN3 (19 mg, 61% yield).
One-pot three-enzyme synthesis of Kdnα2–3LexβProN3 and Neu5AcN3α2–3LexβProN3 from LexβProN3 using MBP-Δ30vST3Gal-I-His6
For the synthesis of Kdnα2–3LexβProN3 and Neu5AcN3α2–3LexβProN3, LexβProN3 (20 mg, 0.03 mmol), mannose (9 mg, 0.05 mmol) or ManNAcN3 (Yu et al. 2005) (12 mg, 0.05 mmol), sodium pyruvate (18 mg, 0.16 mmol), CTP (28 mg, 0.05 mmol) and MgCl2 (22 mg, 0.11 mmol) were dissolved in H2O (2.5 mL). A stock solution of Tris–HCl buffer (1 M, pH 8.5, 0.5 mL) was added. After the addition of a recombinant P. multocida sialic acid aldolase (Li et al. 2008) (1.7 mg), a recombinant N. meningitidis CMP–sialic acid synthetase (Yu et al. 2004) (1.1 mg), and Ni2+-NTA column purified MBP-Δ30vST3Gal-I-His6 (262 μg, estimated), water was added to bring the volume of the reaction mixture to 5 mL in both reactions. The reactions were carried out by incubating the solution in an incubator shaker at 37°C for overnight. The reactions were stopped by adding cold EtOH (5 mL) followed by incubation on ice for 30 min. The mixtures were then centrifuged to remove precipitates. The supernatants were concentrated by rotavaporation. A BioGel P-2 filtration column and a silica gel column (eluted with EtOAc:MeOH:H2O = 5:2:1, v/v/v) were used to purify the product SLexβProN3Kdnα2–3LexβProN3 (18 mg, 64% yield) and Neu5AcN3α2–3LexβProN3 (18 mg, 58% yield).
Supplementary data
Supplementary data are available at Glycobiology online (http://glycob.oxfordjournals.org/).
Funding
This work was supported by NIH grant R01GM076360. X.C. is an Alfred P. Sloan Research Fellow, a Camille Dreyfus Teacher-Scholar and a UC-Davis Chancellor's Fellow.
Conflict of interest statement
None declared.
Abbreviations
BCA, bicinchoninic acid; CAZy, Carbohydrate-Active enZymes; CMP, cytidine 5′-monophosphate; CTP, cytidine 5′-triphosphate; Hp1–3FTΔ66, Helicobacter pylori α1–3-fucosyltransferase; FKP, bifunctional L-fucokinase/GDP-fucose pyrophosphorylase; FPLC, fast protein liquid chromatography; Gal, galactose; HPLC, high-performance liquid chromatography; HRMS, high-resolution mass spectrometry; IPTG, isopropyl-1-thio-β-d-galactopyranoside; Kdn, 2-keto-3-deoxy-nonulosonic acid; LacβMU, 4-methylumbelliferyl-β-d-lactoside; LexβMU, 4-methylumbelliferyl-β-d-Lewisx; ManNAc, d-N-acetylmannosamine; ManNAcN3, N-azidoacetylmannosamine; MBP, maltose binding protein; MES, 2-(N-morpholino)ethanesulfonic acid; MU, 4-methylumbelliferyl; Neu5Ac, N-acetylneuraminic acid; Neu5AcN3, N-azidoacetylneuraminic acid; Neu5Gc, N-glycolylneuraminic acid; NmCSS, Neisseria meningitidis CMP-sialic acid synthetase; NMR, nuclear magnetic resonance; PCR, polymerase chain reactions; PmST1, Pasteurella multocida sialyltransferase 1; SDS–PAGE, sodium dodecylsulfate–polyacrylamide gel electrophoresis; SLex, sialyl Lewisx; vST3Gal-I, viral α2–3-sialyltransferase.
References
- Angata T, Varki A. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem Rev. 2002;102:439–469. doi: 10.1021/cr000407m. doi:10.1021/cr000407m. [DOI] [PubMed] [Google Scholar]
- Boons GJ, Demchenko AV. Recent advances in o-sialylation. Chem Rev. 2000;100:4539–4566. doi: 10.1021/cr990313g. doi:10.1021/cr990313g. [DOI] [PubMed] [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. doi:10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Huang S, Cheng J, Li Y, Muthana S, Son B, Chen X. Chemical preparation of sialyl Lewis x using an enzymatically synthesized sialoside building block. Carbohydr Res. 2008;343:2863–2869. doi: 10.1016/j.carres.2008.06.020. doi:10.1016/j.carres.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Varki A. Advances in the biology and chemistry of sialic acids. ACS Chem Biol. 2010;5:163–176. doi: 10.1021/cb900266r. doi:10.1021/cb900266r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokhawala HA, Huang S, Lau K, Yu H, Cheng J, Thon V, Hurtado-Ziola N, Guerrero JA, Varki A, Chen X. Combinatorial chemoenzymatic synthesis and high-throughput screening of sialosides. ACS Chem Biol. 2008;3:567–576. doi: 10.1021/cb800127n. doi:10.1021/cb800127n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho PM, Deleury E, Davies GJ, Henrissat B. An evolving hierarchical family classification for glycosyltransferases. J Mol Biol. 2003;328:307–317. doi: 10.1016/s0022-2836(03)00307-3. doi:10.1016/S0022-2836(03)00307-3. [DOI] [PubMed] [Google Scholar]
- Datta AK, Chammas R, Paulson JC. Conserved cysteines in the sialyltransferase sialylmotifs form an essential disulfide bond. J Biol Chem. 2001;276:15200–15207. doi: 10.1074/jbc.M010542200. doi:10.1074/jbc.M010542200. [DOI] [PubMed] [Google Scholar]
- Datta AK, Paulson JC. The sialyltransferase “sialylmotif” participates in binding the donor substrate CMP-NeuAc. J Biol Chem. 1995;270:1497–1500. doi: 10.1074/jbc.270.4.1497. doi:10.1074/jbc.270.4.1497. [DOI] [PubMed] [Google Scholar]
- Datta AK, Sinha A, Paulson JC. Mutation of the sialyltransferase S-sialylmotif alters the kinetics of the donor and acceptor substrates. J Biol Chem. 1998;273:9608–9614. doi: 10.1074/jbc.273.16.9608. doi:10.1074/jbc.273.16.9608. [DOI] [PubMed] [Google Scholar]
- DeFrees SA, Gaeta FCA, Lin Y-C, Ichikawa Y, Wong C-H. Ligand recognition by E-selectin: Analysis of conformation and activity of synthetic monomeric and bivalent sialyl Lewis X analogs. J Am Chem Soc. 1993;115:7549–7550. doi:10.1021/ja00069a083. [Google Scholar]
- DeFrees SA, Kosch W, Way W, Paulson JC, Sabesan S, Halcomb RL, Huang D-H, Ichikawa Y, Wong C-H. Ligand recognition by E-selectin: Synthesis, inhibitory activity, and conformational analysis of bivalent sialyl Lewis x analogs. J Am Chem Soc. 1995;117:66–79. doi:10.1021/ja00106a008. [Google Scholar]
- Duffels A, Green LG, Lenz R, Ley SV, Vincent SP, Wong C-H. Chemoenzymatic synthesis of l-galactosylated dimeric sialyl Lewis X structures employing α-1,3-fucosyltransferase V. Bioorg Med Chem. 2000;8:2519–2525. doi: 10.1016/s0968-0896(00)00187-5. doi:10.1016/S0968-0896(00)00187-5. [DOI] [PubMed] [Google Scholar]
- Foxall C, Watson SR, Dowbenko D, Fennie C, Lasky LA, Kiso M, Hasegawa A, Asa D, Brandley BK. The three members of the selectin receptor family recognize a common carbohydrate epitope, the sialyl Lewis(x) oligosaccharide. J Cell Biol. 1992;117:895–902. doi: 10.1083/jcb.117.4.895. doi:10.1083/jcb.117.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geremia RA, Harduin-Lepers A, Delannoy P. Identification of two novel conserved amino acid residues in eukaryotic sialyltransferases: Implications for their mechanism of action. Glycobiology. 1997;7:v–vii. doi: 10.1093/glycob/7.2.161. [DOI] [PubMed] [Google Scholar]
- Halcomb RL, Chappell MD. Recent developments in technology for glycosylation with sialic acid. J Carbohydr Chem. 2002;21:723–768. doi:10.1081/CAR-120016488. [Google Scholar]
- Hanashima S, Castagner B, Esposito D, Nokami T, Seeberger PH. Synthesis of a sialic acid α(2–3) galactose building block and its use in a linear synthesis of sialyl Lewis X. Org Lett. 2007;9:1777–1779. doi: 10.1021/ol0704946. doi:10.1021/ol0704946. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Tanaka M, Itoh M, Miyauchi H. A convenient and efficient synthesis of SLeX analogs. J Org Chem. 1996;61:2938–2945. doi: 10.1021/jo960125f. doi:10.1021/jo960125f. [DOI] [PubMed] [Google Scholar]
- Huang KT, Wu BC, Lin CC, Luo SC, Chen C, Wong C-H. Multi-enzyme one-pot strategy for the synthesis of sialyl Lewis X-containing PSGL-1 glycopeptide. Carbohydr Res. 2006;341:2151–2155. doi: 10.1016/j.carres.2006.04.047. doi:10.1016/j.carres.2006.04.047. [DOI] [PubMed] [Google Scholar]
- Ichikawa Y, Lin Y-C, Dumas DP, Shen G-J, Garcia-Junceda E, Williams MA, Bayer R, Ketcham C, Walker LE, Paulson JC, et al. Chemical-enzymic synthesis and conformational analysis of sialyl Lewis X and derivatives. J Am Chem Soc. 1992;114:9283–9298. doi:10.1021/ja00050a007. [Google Scholar]
- Jackson RJ, Hall DF, Kerr PJ. Myxoma virus encodes an alpha2,3-sialyltransferase that enhances virulence. J Virol. 1999;73:2376–2384. doi: 10.1128/jvi.73.3.2376-2384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeanneau C, Chazalet V, Auge C, Soumpasis DM, Harduin-Lepers A, Delannoy P, Imberty A, Breton C. Structure–function analysis of the human sialyltransferase ST3Gal I: Role of N-glycosylation and a novel conserved sialylmotif. J Biol Chem. 2004;279:13461–13468. doi: 10.1074/jbc.M311764200. doi:10.1074/jbc.M311764200. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Paulson JC. Cloning and expression of human Gal β1,3(4)GlcNAc α2,3-sialyltransferase. Biochem Biophys Res Commun. 1993;194:375–382. doi: 10.1006/bbrc.1993.1830. doi:10.1006/bbrc.1993.1830. [DOI] [PubMed] [Google Scholar]
- Kitagawa H, Paulson JC. Cloning of a novel alpha 2,3-sialyltransferase that sialylates glycoprotein and glycolipid carbohydrate groups. J Biol Chem. 1994;269:1394–1401. [PubMed] [Google Scholar]
- Koeller KM, Wong CH. Chemoenzymatic synthesis of sialyl-trimeric-Lewis x. Chemistry. 2000;6:1243–1251. doi: 10.1002/(sici)1521-3765(20000403)6:7<1243::aid-chem1243>3.3.co;2-a. doi:10.1002/(SICI)1521-3765(20000403)6:7<1243::AID-CHEM1243>3.3.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Kono M, Ohyama Y, Lee Y-C, Hamamoto T, Kojima N, Tsuji S. Mouse β-galactoside α2,3-sialyltransferases: Comparison of in vitro substrate specificities and tissue specific expression. Glycobiology. 1997;7:469–479. doi: 10.1093/glycob/7.4.469. doi:10.1093/glycob/7.4.469. [DOI] [PubMed] [Google Scholar]
- Krug LM, Ragupathi G, Ng KK, Hood C, Jennings HJ, Guo Z, Kris MG, Miller V, Pizzo B, Tyson L, et al. Vaccination of small cell lung cancer patients with polysialic acid or N-propionylated polysialic acid conjugated to keyhole limpet hemocyanin. Clin Cancer Res. 2004;10:916–923. doi: 10.1158/1078-0432.ccr-03-0101. doi:10.1158/1078-0432.CCR-03-0101. [DOI] [PubMed] [Google Scholar]
- Lasky LA. Selectins: Interpreters of cell-specific carbohydrate information during inflammation. Science. 1992;258:964–969. doi: 10.1126/science.1439808. doi:10.1126/science.1439808. [DOI] [PubMed] [Google Scholar]
- Lemieux GA, Bertozzi CR. Modulating cell surface immunoreactivity by metabolic induction of unnatural carbohydrate antigens. Chem Biol. 2001;8:265–275. doi: 10.1016/s1074-5521(01)00008-4. doi:10.1016/S1074-5521(01)00008-4. [DOI] [PubMed] [Google Scholar]
- Li Y, Yu H, Cao H, Lau K, Muthana S, Tiwari VK, Son B, Chen X. Pasteurella multocida sialic acid aldolase: a promising biocatalyst. Appl Microbiol Biotech. 2008;79:963–970. doi: 10.1007/s00253-008-1506-2. doi:10.1007/s00253-008-1506-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C-H, Shimazaki M, Wong C-H, Koketsu M, Juneja LR, Kim M. Enzymatic synthesis of a sialyl Lewis X dimer from egg yolk as an inhibitor of E-selectin. Bioorg Med Chem. 1995;3:1625–1630. doi: 10.1016/0968-0896(95)00150-6. doi:10.1016/0968-0896(95)00150-6. [DOI] [PubMed] [Google Scholar]
- Lin S-W, Yuan T-M, Li J-R, Lin C-H. Carboxyl terminus of Helicobacter pylori α1,3-fucosyltransferase determines the structure and stability. Biochemistry. 2006;45:8108–8116. doi: 10.1021/bi0601297. doi:10.1021/bi0601297. [DOI] [PubMed] [Google Scholar]
- Liu T, Guo Z, Yang Q, Sad S, Jennings HJ. Biochemical engineering of surface α2–8 polysialic acid for immunotargeting tumor cells. J Biol Chem. 2000;275:32832–32836. doi: 10.1074/jbc.C000573200. doi:10.1074/jbc.C000573200. [DOI] [PubMed] [Google Scholar]
- Livingston PO. Approaches to augmenting the immunogenicity of melanoma gangliosides: From whole melanoma cells to ganglioside-KLH conjugate vaccines. Immunol Rev. 1995;145:147–166. doi: 10.1111/j.1600-065x.1995.tb00080.x. doi:10.1111/j.1600-065X.1995.tb00080.x. [DOI] [PubMed] [Google Scholar]
- Lowe JB, Stoolman LM, Nair RP, Larsen RD, Berhend TL, Marks RM. ELAM-1-dependent cell adhesion to vascular endothelium determined by a transfected human fucosyltransferase cDNA. Cell. 1990;63:475–484. doi: 10.1016/0092-8674(90)90444-j. doi:10.1016/0092-8674(90)90444-J. [DOI] [PubMed] [Google Scholar]
- Muthana S, Cao H, Chen X. Recent progress in chemical and chemoenzymatic synthesis of carbohydrates. Curr Opin Chem Biol. 2009;13:573–581. doi: 10.1016/j.cbpa.2009.09.013. doi:10.1016/j.cbpa.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L, Chokhawala HA, Cao H, Henning R, Ng L, Huang S, Yu H, Chen X, Fisher AJ. Crystal structures of Pasteurella multocida sialyltransferase complexes with acceptor and donor analogues reveal substrate binding sites and catalytic mechanism. Biochemistry. 2007;46:6288–6298. doi: 10.1021/bi700346w. doi:10.1021/bi700346w. [DOI] [PubMed] [Google Scholar]
- Ni L, Sun M, Yu H, Chokhawala H, Chen X, Fisher AJ. Cytidine 5′-monophosphate (CMP)-induced structural changes in a multifunctional sialyltransferase from Pasteurella multocida. Biochemistry. 2006;45:2139–2148. doi: 10.1021/bi0524013. doi:10.1021/bi0524013. [DOI] [PubMed] [Google Scholar]
- Pan Y, Chefalo P, Nagy N, Harding C, Guo Z. Synthesis and immunological properties of N-modified GM3 antigens as therapeutic cancer vaccines. J Med Chem. 2005;48:875–883. doi: 10.1021/jm0494422. doi:10.1021/jm0494422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips ML, Nudelman E, Gaeta FC, Perez M, Singhal AK, Hakomori S, Paulson JC. ELAM-1 mediates cell adhesion by recognition of a carbohydrate ligand, sialyl-Lex. Science. 1990;250:1130–1132. doi: 10.1126/science.1701274. doi:10.1126/science.1701274. [DOI] [PubMed] [Google Scholar]
- Polley MJ, Phillips ML, Wayner E, Nudelman E, Singhal AK, Hakomori S, Paulson JC. CD62 and endothelial cell-leukocyte adhesion molecule 1 (ELAM-1) recognize the same carbohydrate ligand, sialyl-Lewis x. Proc Natl Acad Sci USA. 1991;88:6224–6228. doi: 10.1073/pnas.88.14.6224. doi:10.1073/pnas.88.14.6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao FV, Rich JR, Rakic B, Buddai S, Schwartz MF, Johnson K, Bowe C, Wakarchuk WW, DeFrees S, Withers SG, Strynadka NC. Structural insight into mammalian sialyltransferases. Nat Struct Mol Biol. 2009;16:1186–1188. doi: 10.1038/nsmb.1685. doi:10.1038/nsmb.1685. [DOI] [PubMed] [Google Scholar]
- Rosen SD. Ligands for l-selectin: Homing, inflammation, and beyond. Annu Rev Immunol. 2004;22:129–156. doi: 10.1146/annurev.immunol.21.090501.080131. doi:10.1146/annurev.immunol.21.090501.080131. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Watanabe E, Kawashima K, Sekine S, Dohi T, Oshima M, Hanai N, Nishi T, Hasegawa M. Expression cloning of a novel Gal beta(1–3/1–4) GlcNAc alpha2,3-sialyltransferase using lectin resistance selection. J Biol Chem. 1993;268:22782–22787. [PubMed] [Google Scholar]
- Schauer R. Achievements and challenges of sialic acid research. Glycoconj J. 2000;17:485–499. doi: 10.1023/A:1011062223612. doi:10.1023/A:1011062223612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer R. Victor Ginsburg's influence on my research of the role of sialic acids in biological recognition. Arch Biochem Biophys. 2004;426:132–141. doi: 10.1016/j.abb.2004.03.008. doi:10.1016/j.abb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Soriano del Amo D, Wang W, Besanceney C, Zheng T, He Y, Gerwe B, Seidel RD, III, Wu P. Chemoenzymatic synthesis of the sialyl Lewis X glycan and its derivatives. Carbohydr Res. 2010;345:1107–1113. doi: 10.1016/j.carres.2010.03.032. doi:10.1016/j.carres.2010.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sujino K, Jackson RJ, Chan NW, Tsuji S, Palcic MM. A novel viral α2,3-sialyltransferase (v-ST3Gal I): Transfer of sialic acid to fucosylated acceptors. Glycobiology. 2000;10:313–320. doi: 10.1093/glycob/10.3.313. doi:10.1093/glycob/10.3.313. [DOI] [PubMed] [Google Scholar]
- Sun H-Y, Lin S-W, Ko T-P, Pan J-F, Liu C-L, Lin C-N, Wang A-H, Lin C-H. Structure and mechanism of Helicobacter pylori fucosyltransferase: A basis for lipopolysaccharide variation and inhibitor design. J Biol Chem. 2007;282:9973–9982. doi: 10.1074/jbc.M610285200. [DOI] [PubMed] [Google Scholar]
- Takada A, Ohmori K, Yoneda T, Tsuyuoka K, Hasegawa A, Kiso M, Kannagi R. Contribution of carbohydrate antigens sialyl Lewis A and sialyl Lewis X to adhesion of human cancer cells to vascular endothelium. Cancer Res. 1993;53:354–361. [PubMed] [Google Scholar]
- Ugorski M, Laskowska A. Sialyl Lewis(a): a tumor-associated carbohydrate antigen involved in adhesion and metastatic potential of cancer cells. Acta Biochim Pol. 2002;49:303–311. [PubMed] [Google Scholar]
- Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 2004;68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. doi:10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Liu X, Li Y, Li J, Xia C, Zhou G, Zhang W, Zhao W, Chen X, Wang PG. Remodeling bacterial polysaccharides by metabolic pathway engineering. Proc Natl Acad Sci USA. 2009;106:4207–4212. doi: 10.1073/pnas.0812432106. doi:10.1073/pnas.0812432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chokhawala HA, Huang S, Chen X. One-pot three-enzyme chemoenzymatic approach to the synthesis of sialosides containing natural and non-natural functionalities. Nat Protoc. 2006;1:2485–2492. doi: 10.1038/nprot.2006.401. doi:10.1038/nprot.2006.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Chokhawala H, Karpel R, Yu H, Wu B, Zhang J, Zhang Y, Jia Q, Chen X. A multifunctional Pasteurella multocida sialyltransferase: A powerful tool for the synthesis of sialoside libraries. J Am Chem Soc. 2005;127:17618–17619. doi: 10.1021/ja0561690. doi:10.1021/ja0561690. [DOI] [PubMed] [Google Scholar]
- Yu H, Yu H, Karpel R, Chen X. Chemoenzymatic synthesis of CMP-sialic acid derivatives by a one-pot two-enzyme system: Comparison of substrate flexibility of three microbial CMP-sialic acid synthetases. Bioorg Med Chem. 2004;12:6427–6435. doi: 10.1016/j.bmc.2004.09.030. doi:10.1016/j.bmc.2004.09.030. [DOI] [PubMed] [Google Scholar]
- Zhang L, Lau K, Cheng J, Yu H, Li Y, Sugiarto G, Huang S, Ding L, Thon V, Wang PG, et al. Helicobacter hepaticus Hh0072 gene encodes a novel alpha1–3-fucosyltransferase belonging to CAZy GT11 family. Glycobiology. 2010;20:1077–1088. doi: 10.1093/glycob/cwq068. doi:10.1093/glycob.cwq068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Borrelli S, Gilbert M, Liu T, Pon RA, Jennings HJ. Bioengineering of surface GD3 ganglioside for immunotargeting human melanoma cells. J Biol Chem. 2004;279:25390–25399. doi: 10.1074/jbc.M402787200. doi:10.1074/jbc.M402787200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.