Abstract
Production of alpha/beta interferon in response to viral double-stranded RNA (dsRNA) produced during viral replication is a first line of defense against viral infections. Here we demonstrate that the Erns glycoprotein of the pestivirus bovine viral diarrhea virus can act as an inhibitor of dsRNA-induced responses of cells. This effect is seen whether Erns is constitutively expressed in cells or exogenously added to the culture medium. The Erns effect is specific to dsRNA since activation of NF-κB in cells infected with Semliki Forest virus or treated with tumor necrosis factor alpha was not affected. We also show that Erns contains a dsRNA-binding activity, and its RNase is active against dsRNA at a low pH. Both the dsRNA binding and RNase activities are required for the inhibition of dsRNA signaling, and we discuss here a model to account for these observations.
Bovine viral diarrhea virus (BVDV) is a Pestivirus belonging to the family Flaviviridae and is a major pathogen of cattle throughout the world with an incidence of infection often in excess of 70% (9, 24). An unusual feature of pestiviruses is their ability to sustain a persistent infection after in utero infection of a fetus. BVDV exists as two biotypes according to their cytopathogenicity on cultured cells, and the ability to sustain a persistent infection is exclusively a property of the noncytopathogenic BVDV biotype. Persistently infected calves born after infection of the fetus during the early stages of pregnancy are held to be the source of acute virus infection and the subsequent generation of cytopathogenic BVDV variants isolated from animals with mucosal disease. Survival of the virus in the fetus requires avoiding immune responses; since the acquired immune response is not developed early in embryogenesis, innate immune responses are the main host defenses from infection. Evasion of innate immunity in the fetus is at least in part due to the lack of alpha/beta interferon (IFN-α/β) production after noncytopathogenic BVDV infection (6). We and others have shown that cultured cells infected with noncytopathogenic BVDV are refractory to the IFN-inducing effects of infection with heterologous virus or the addition of double-stranded RNA (dsRNA) to cells (3, 30). Thus, noncytopathogenic BVDV takes active measures to block the production of IFN-α/β. A key component of this inhibition operates at the level of inhibiting the DNA-binding activity of the IFN regulatory transcription factor IRF-3 induced by heterologous virus infection (2).
The viral gene products associated with the evasion of the IFN system have not been identified. The virus genome is positive-strand RNA that is translated to form a single virus polyprotein, which, through cleavage by both host and virus proteases, gives rise to either 11 or 12 mature viral proteins (NH2-Npro-C-Erns-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-COOH), dependent on the virus biotype (20). Erns, E1, and E2 are glycoproteins found on the surface of the viron (35). In addition to being a virion protein, a proportion of Erns is secreted from infected cells into the extracellular environment as a consequence of it lacking a typical transmembrane anchor domain (28). The biological significance of secreted Erns has not been determined. However, Erns has two defined activities. First, Erns has an RNase activity (10, 29, 37) that shows a strong preference for single-stranded RNA (ssRNA) (37). Second, Erns glycoproteins of both BVDV and a closely related pestivirus, classical swine fever virus (CSFV), have been shown to bind to cell surface glycosylaminoglycans (13, 14) through residues located close to their C termini (12, 15). The effects of inactivating the RNase activity on viral function are ambiguous: it has been reported that a genetically engineered strain of CSFV that lacks RNase activity induced cell death in swine kidney cells, but the parent virus did not (11); however, another mutant CSFV lacking RNase activity was not cytopathogenic (23).
In the present study we have investigated the properties of secreted BVDV Erns, and we show that this protein can bind to dsRNA-agarose and inactivate dsRNA-dependent signaling events initiated by the presence of dsRNA in the medium. Through the construction of mutated Erns by expression of plasmids subjected to site-directed mutagenesis at sites within the Erns-coding sequence (see Fig. 1), we show that this inhibition required the RNase activity and a region that influenced Erns binding to dsRNA.
FIG. 1.
Schematic representation of the primary amino acid sequence of the BVDV and CSFV glycoprotein Erns. Residues critical for RNase activity and for glycosaminoglycan binding (11, 12, 15, 23) and the mutants used in the present study are indicated.
MATERIALS AND METHODS
Cell culture and generation of cell lines.
Calf testes (CTe) cells were used between passages 4 and 7 and were cultured in Dulbecco modified Eagle medium (DMEM), supplemented with penicillin and streptomycin, and 10% fetal calf serum (FCS) free of both BVDV and anti-BVDV antibody. Human MG-63 cells were grown in DMEM supplemented with 10% FCS and penicillin and streptomycin. MDBK cells were cultured in Eagle minimal essential medium supplemented with 1% nonessential amino acids and 10% FCS, and insect (Schneider Drosophila S2) cells were obtained from Invitrogen and cultured at room temperature in Schneider's Drosophila medium (also from Invitrogen) containing 10% FCS.
The full-length Erns gene from noncytopathogenic BVDV strain pe515, preceded at the 5′ end by the CD33 signal peptide sequence in order to ensure efficient Erns secretion, was amplified from the previously cloned Signal/pIg/Fc plasmid (14) by using PCR, and cloned between the NheI and XbaI sites of the mammalian expression vector pcDNA6/V5-His (Invitrogen) to generate pcDNA6/CD33-Erns/V5-His. The forward primer used was 5′-ACCCAAGCTGCTAGCTCAGACATGCCG-3′, which corresponds to nucleotides 6267 to 6293 of the Signal/pIg/Fc plasmid (R&D Systems), incorporating an NheI restriction enzyme recognition site at the 5′ end of the region encoding the CD33 signal peptide. The reverse primer (pIgplus SEQ 3′ R&D Systems) sequence was 5′-ATGTGTGAGGTTTGTCACAAG-3′, which corresponds to nucleotides 101 to 121 of the Signal/pIg/Fc plasmid. An equivalent plasmid expressing a CD33-tagged BVDV E2 protein (pcDNA6/CD33-E2/V5-His) was generated in a similar manner. Stably transformed MDBK cell lines were established for constitutive transgene expression of Erns, E2, or β-galactosidase by transfection with pcDNA6/CD33-Erns/V5-His, pcDNA6/CD33-E2/V5-His, or pcDNA6/lacZ/V5-His (Invitrogen), respectively, by using FuGENE 6 (Roche Biochemicals) according to the manufacturer's instructions. The cells were treated with blasticidin (10 μg/ml), and pools of blasticidin-resistant cells were selected.
Construction of mutants of Erns.
The Erns gene was cloned previously into a Drosophila melanogaster expression vector to produce pMT/BiP/Erns/V5-His, which represented the full-length Erns cDNA of noncytopathogenic BVDV strain pe515 corresponding to amino acids 271 to 497 of the BVDV polyprotein (14). The construction of mutant ErnsΔ483-497 lacking amino acid residues 483 to 497 in the carboxy terminus of Erns and the construction of the mutants ErnsK409A, ErnsK410T, ErnsK412G, and ErnsK409K-K410T, containing specific substitutions of lysine residues for uncharged amino acids at positions 409, 410, and 412, respectively, in the Erns polypeptide, have been described previously (15). The schematic location of these Erns mutants is shown in Fig. 1. The RNase-negative mutant, ErnsH300K, was constructed from pMT/BiP/Erns/V5-His by PCR-mediated mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's guidelines. The resultant plasmid [pMT/BiP/Erns(H300K)/V5-His] was cotransfected with a hygromycin-resistant selection vector pCoHYGRO (Invitrogen) at a ratio of 19 [pMT/BiP/Erns(H300K)/V5-His] to 1 (pCoHYGRO) into insect (S2) cells by using a calcium phosphate transfection kit (Invitrogen) according to the manufacturer's instructions. Transfected cells were selected with hygromycin B (Invitrogen), and mutant Erns-expressing cell lines were established from clones selected by limiting dilution.
Expression and purification of Erns expressed in insect cells.
Wild-type and mutant Erns glycoproteins were expressed as described previously (14). Briefly, Drosophila S2 cells harboring the Erns gene were cultured in 150 ml of Schneider's Drosophila medium containing 10% FCS. After 5 to 6 days at room temperature the medium was replaced with 500 ml of Drosophila medium free of serum (Invitrogen) containing l-glutamine (90 mM), and expression of Erns was induced by the addition of 7.5 μM copper sulfate. At 4 to 5 days postinduction, the cells were harvested, and the culture supernatant was applied directly to a 10-ml chelating-Sepharose column (Amersham Biotech), which had previously been equilibrated with 20 mM phosphate buffer (pH 7.8) containing 300 mM NaCl. The column was washed with 20 mM phosphate buffer (pH 6.0) containing 1.0 M NaCl and 100 mM imidazole, and Erns was eluted with 20 mM phosphate buffer (pH 6.0) containing 1.0 M NaCl and 400 mM imidazole. Fractions containing Erns were pooled and dialyzed against phosphate-buffered saline (PBS). The purity of the glycoproteins was assessed by using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-10% PAGE), and the concentration of the purified proteins was determined by using the Coomassie Plus-200 protein assay reagent (Pierce). The stability of the wild type and mutant Erns was assessed after the addition of the purified glycoproteins to cell culture maintenance medium and incubation at 37°C for 18 h, followed by PAGE.
EMSAs.
Electrophoretic mobility shift assays (EMSAs) were carried out to analyze the inhibition of NF-κB activation by Erns. Confluent CTe and MG-63 cells were cultured in 9-cm dishes, and the cells were either mock treated or treated with polyriboinosinic acid-polyribocytidylic acid [poly(rI)-poly(rC); 5 μg/ml)], human tumor necrosis factor alpha (TNF-α; 10 ng/ml), or infected with Semliki Forest virus (SFV) at a multiplicity of infection of ∼5 PFU/cell in the presence or absence of Erns (1 μg/ml). Nuclear extracts were prepared as previously described (36), the protein concentrations were determined by a Bradford assay (Bio-Rad), and 10-μg aliquots of extracts were assayed by using the PRD II element from the human IFN-β promoter as a probe for NF-κB, as described by Visvanathan and Goodbourn (31).
Immunoblot analysis.
Induction of MxA protein by dsRNA was determined by immunoblot analysis. Confluent monolayers of CTe cells cultured in 24-well plates were either mock treated, treated with in vitro-transcribed dsRNA (5 μg/ml), or treated with poly(rI)-poly(rC) (5 μg/ml) and Erns (1 μg/ml). After 18 h at 37°C, the cells were washed once with PBS and treated with 100 μl of SDS-PAGE loading buffer (8 M urea, 10 mM Tris-HCl [pH 6.8], 2% SDS, 2% 2-mercaptoethanol, 0.1% bromophenol blue). The samples were boiled for 2 min, a 10-μl portion of the cell lysates was subjected to SDS-PAGE, and the separated proteins were electroblotted onto a polyvinylidene difluoride membrane (Amersham Biotech). After the membrane was blocked with 5% (wt/vol) skimmed milk in PBS containing 0.1% Tween 20 (PBS-T), the membrane was probed for MxA protein by using a rabbit antiserum raised against human MxA (serum no. 41; a generous gift from P. Staeheli, Freiburg, Germany) at a dilution of 1:2,000 and horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (Sigma) at a dilution of 1:5,000. The bound antibody was visualized by using an ECL detection kit (Amersham Biotech) according to the manufacturer's instructions.
IFN-specific RT-PCR.
Confluent monolayers of CTe cells cultured in 24-well plates were either mock treated or treated with poly(rI)-poly(rC) (100 μg/ml) and purified Erns (1 μg/ml). After 6 h at 37°C, the cells were washed once with PBS, and total RNA was extracted by using RNeasy kit (Qiagen) according to the manufacturer's protocol including a DNase treatment step. Reverse transcription (RT) was performed by using a cDNA synthesis kit (Promega) according to the manufacturer's instructions in a reaction volume of 30 μl containing 1 μg of total RNA, 0.75 μg of oligonucleotide dT, and 30 U of avian myeloblastosis virus reverse transcriptase and incubated at 42°C for 1 h, followed by 5 min at 99°C. Then, 2 μl of resulting cDNA was amplified by 25 cycles by using a standard PCR protocol with each cycle consisting of 45 s at 94°C, 45 s at 66°C, and 45 s at 72°C, followed by 10 min at 72°C. The primers used to amplify bovine IFN-β mRNA to produce a 571-bp product were 5′-TCACTCTGCAAACCCTTGAA-3′ (sense) and 5′-GTCCAGGCACACCTGTTGTA-3′ (antisense); the primers used for β-actin mRNA (270-bp product) were 5′-CCAGACAGCACTGTGTTGGC-3′ (sense) and 5′-GAGAAGCTGTGCTACGTCGC (antisense). The PCR for β-actin mRNA was performed as for IFN-β mRNA, but the annealing temperature was 55°C.
dsRNA-binding assays.
Poly(rI)-poly(rC)-agarose beads (200 μl) (Amersham Biotech and Sigma) were used in a 1-ml column (MoBiTec Columns; Biosciences Services, Cramlington, United Kingdom) and washed three times with binding buffer (20 mM sodium phosphate buffer [pH 7.2] containing 150 mM NaCl). A total of 2 μg of purified wild-type or mutant Erns glycoproteins diluted in 500 μl of binding buffer was applied to the columns. The columns were washed with binding buffer and eluted in 500-μl fractions with a stepwise gradient of increasing concentrations of NaCl (from 0.2 to 1.0 M) in 20 mM sodium phosphate buffer (pH 7.2). Dilutions of each fraction were coated onto a 96-well enzyme-linked immunosorbent assay (ELISA) plate at 4°C overnight and then blocked with 5% skimmed milk in PBS-T. Eluted Erns was detected by using an anti-V5 antibody (a generous gift from R. Randall, University of St. Andrew’s, St. Andrew’s, Scotland).
The binding of full-length and C-terminally truncated mutants of Erns to poly(rI)-poly(rC) was also analyzed by using radiolabeled recombinant proteins. [35S]methionine-labeled Erns was generated from pcDNA6/CD33-Erns/V5-His by using the T7 TNT-quick coupled transcription-translation system (Promega). In a standard reaction, 1 μg of template DNA and 1 μl of [35S]methionine (10 μCi/μl, >1,000 Ci/mmol [Redivue; Amersham]) was used in a final volume of 20 μl under conditions specified by the manufacturer. Truncations were generated by restriction enzyme digestion of aliquots of pcDNA6/CD33-Erns/V5-His with AflIII (Erns [residues 1 to 66], polyprotein residues 271 to 336), HincII (Erns [residues 1 to 104], polyprotein residues 271 to 374), ApoI (Erns [residues 1 to 142], polyprotein residues 271 to 412), BsgI (Erns [residues 1 to 165], polyprotein residues 271 to 435), AvaI (Erns [residues 1 to 207], polyprotein residues 271 to 477), or XbaI (full-length Erns [residues 1 to 227], polyprotein residues 271 to 497), and these proteins were similarly synthesized. Since the Erns protein synthesized from pcDNA6/CD33-Erns/V5-His contains an N-terminal CD33 tag, the effect of removing this sequence on dsRNA binding was also examined. To do this, a full-length Erns fragment (amino acids 1 to 227) was subcloned into pT7βplink (25); this plasmid was cut with all of the above restriction enzymes, and the resulting linearized plasmids were also expressed in the T7 TNT quick-coupled transcription-translation system. To prepare the poly(rI)-poly(rC)-agarose for binding assays, 250 μl of fresh beads was washed three times in 750 μl of binding buffer (100 mM KCl, 50 mM Tris [pH 7.4], 5 mM MgCl2, 0.1% NP-40, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 30 μg of leupeptin/ml, 5 μg of aprotinin/ml, 5 μg of pepstatin A/ml) and resuspended as a 50% slurry in binding buffer. To determine dsRNA binding, 2 μl of a TNT reaction mixture containing [35S]methionine-labeled recombinant protein was incubated with 10 μl of poly(rI)-poly(rC)-agarose bead slurry, followed by incubation at room temperature for 1 h in 100 μl of binding buffer plus 1 mM dithiothreitol with gentle mixing before three washes in 750 μl of binding buffer. Where indicated, the prewash, binding, and postbinding wash steps were performed in the presence of increased concentrations of KCl. Bound proteins were eluted from beads in 20 μl of SDS-PAGE buffer, separated by SDS-PAGE, and visualized by autoradiography.
RNase activity assay.
Purified wild-type and mutant Erns glycoproteins were diluted either in RNase assay buffer (20 mM sodium acetate buffer [pH 4.5]) or cell culture medium (DMEM containing 2% FCS). The glycoproteins in cell culture medium were further incubated at 37°C for 18 h, and the RNase activity was determined as described previously (15) with slight modification. Briefly, the assay mixture (25 μl in total) contained 0.2 μg of wild-type or mutant Erns, 12.5 μg of yeast 16-23S RNA (Worthington), and 40 U of RNase inhibitor (RNasin; Promega) in 20 mM sodium acetate buffer (pH 4.5). The mixture was incubated at 37°C for 45 min, and the enzyme reaction was stopped by acid precipitation with 5 μl of 25% (vol/vol) HClO4 containing 0.75% (wt/vol) uranyl acetate. After cooling on ice for 10 min, the reaction was centrifuged for 10 min at 10,000 × g, and the absorbance at 260 nm of the supernatant was measured.
Preparation of dsRNA.
Synthetic dsRNA poly(rI)-poly(rC) (Sigma or Amersham Biosciences) was prepared at 1 mg/ml in water or PBS. An alternative dsRNA preparation was made by using the Litmus 28i plasmid (New England Biolabs) containing only its polylinker site (140 nucleotides). Litmus 28i plasmid DNA was digested in two separate reactions with either BglII or StuI restriction enzymes, and transcription of equal amounts both linearized templates was carried out in a single reaction by using the RiboMAX system (Promega) according to the manufacturer's instructions. The resulting RNA was treated with RQ1 RNase-free DNase (Promega) at a concentration of 1 U per μg of template DNA for 15 min at 37°C. After incubation, the RNA was extracted with 1 volume of water-saturated phenol-chloroform-isoamyl alcohol (25:24:1) and then precipitated with 2.5 volumes of 100% ethanol. The resulting RNA pellet was washed with 70% ethanol, dried, and suspended in water. The transcribed RNA was incubated with S1 nuclease (Promega) at a concentration of 10 U per μg of RNA for 20 min at 25°C, and the dsRNA was recovered by ethanol precipitation.
dsRNA degradation assay.
Purified wild-type or mutant Erns glycoproteins (1 μg) were incubated with poly(rI)-poly(rC) (5 μg) or in vitro-synthesized dsRNA (5 μg) either in cell culture medium (DMEM containing 2% FCS) for 18 h at 37°C or in sodium acetate buffer (20 mM [pH 4.5] containing 150 mM NaCl) for 1 h at 37°C. The samples were analyzed on a 1.8% agarose gel, and the dsRNA was stained with ethidium bromide (0.2 μg/ml).
RESULTS
Inhibition of dsRNA-dependent gene expression by Erns.
In order to study a possible role for Erns in the induction of IFN and IFN-stimulated genes, stably transfected pools of MDBK cells expressing approximately similar levels of either wild-type Erns or control proteins E2 and β-galactosidase (data not shown) were established (see Materials and Methods). These pools were exposed to the synthetic dsRNA, poly(rI)-poly(rC), added directly to the medium at 5 μg/ml, and the expression levels of MxA were determined. As shown in Fig. 2A, MxA was inducible by poly(rI)-poly(rC) in cell pools expressing E2 or β-galactosidase; however, in contrast, no MxA was observed after treatment with poly(rI)-poly(rC) in cells that expressed Erns.
FIG. 2.
Effect of Erns on the induction of MxA by poly(rI)-poly(rC). (A) Pools of MDBK cells expressing Erns (lane1), E2 (lane 2), or β-galactosidase (lane 3) were treated with maintenance medium (DMEM plus 2% FCS) supplemented with 5 μg of the synthetic dsRNA, poly(rI)-poly(rC), per ml. After 18 h the cells were harvested in sample buffer and then subjected to SDS-PAGE and immunoblot analysis with rabbit antiserum against human MxA. (B) SDS-PAGE of purified Erns and GFP/V5-His. Prestained molecular mass standards (lane 1), 5 μg of Erns (lane 2), and 5 μg of GFP/V5-His (lane 3). The gel was stained with brilliant blue R (Sigma). (C) Confluent monolayers of CTe cells were rinsed and replaced with medium either supplemented with 5 μg of poly(rI)-poly(rC)/ml or not and then treated with 1 μg of control protein (GFP/V5-His)/ml (lane 1), 1 μg of Erns/ml (lane 2), or mock treated (lane 3). After 18 h, the cells were harvested in sample buffer and analyzed for MxA expression. Lanes marked “+” in panels A and C indicate treatment with poly(rI)-poly(rC).
We next examined whether the addition of exogenous recombinant Erns produced in insect cells and purified to ca. 95% homogeneity (Fig. 2B) could inhibit the expression of MxA induced by treatment of CTe cells with poly(rI)-poly(rC). The cells were treated with a mixture of poly(rI)-poly(rC) and Erns, and after overnight incubation at 37°C, cell lysates were examined for MxA expression. The results (Fig. 2C) demonstrated that the addition of Erns blocks the induction of MxA by poly(rI)-poly(rC), whereas the addition of green fluorescent protein (GFP) (a control protein) had no effect on MxA induction. A minimum of 0.25 μg of Erns/ml was required to inhibit MxA inducation by 5 μg of poly(rI)-poly(rC)/ml (data not shown). In addition, MxA induction by an alternative in vitro-transcribed dsRNA was also inhibited by the presence of Erns in the medium (see Fig. 9C). These results indicate that soluble Erns prevents MxA expression in response to dsRNA and that this effect is not cell type specific.
FIG. 9.
Effect of Erns on dsRNA stability at neutral and acidic pH conditions. (A) Synthetic dsRNA poly(rI)-poly(rC) (5 μg) was either mock treated or treated with wild-type Erns (0.2 μg) in 50 μl of cell culture medium (1× DMEM plus 2% FCS) at 37°C for 18 h. (B) In vitro-transcribed dsRNA (5 μg) was either mock treated or treated with wild-type Erns or the indicated Erns mutants (0.2 μg). The digestion was carried out either in 50 μl of cell culture medium (1× DMEM plus 2% FCS) at 37°C for 18 h (upper panel) or 50 μl of sodium acetate buffer (20 mM [pH 4.5], containing 150 mM NaCl) for 1 h at 37°C (lower panel). The samples were analyzed on a 1.8% agarose gel, and the RNA was stained with ethidium bromide. (C) The induction of MxA by dsRNA previously incubated with Erns. RNA was produced by in vitro transcription and incubated with Erns at 37°C for 18 h, followed by phenol-chloroform extraction and ethanol precipitation. CTe cells were rinsed, and the medium was replaced with maintenance medium (DMEM plus 2% FCS) with or without wild-type Erns (1 μg/ml) and equal amounts (5 μg/ml) of either Erns-pretreated in vitro-transcribed dsRNA or poly(rI)-poly(rC) not treated with Erns. The cells were incubated at 37°C for a further 18 h, and MxA levels were analyzed by immunoblotting. Lanes marked “+” indicate the samples incubated with both RNA and Erns.
Although MxA synthesis may be induced directly by virus infection, this activation appears to be inefficient compared to its induction by IFN-α/β (5, 34); thus, the Erns-mediated inhibition of the poly(rI)-poly(rC) effect is likely to be acting indirectly through the inhibition of the intermediate production of IFN-β. To establish this, RT-PCR was used to analyze IFN-β mRNA production. As shown in Fig. 3 (upper panel), cells stimulated with poly(rI)-poly(rC) alone showed a clear IFN-β mRNA signal in contrast to cells treated with poly(rI)-poly(rC) in the presence of Erns when no IFN-β signal was seen. Figure 3 (lower panel) shows that β-actin mRNA was detected in all RT reactions, indicating that all preparations contained similar amounts of RNA. These results clearly demonstrated that dsRNA induction of IFN-β mRNA synthesis was inhibited in the presence of Erns. Consistent with this, Erns also blocked the activation of an IFN-β promoter-luciferase reporter construct in response to the addition of poly(rI)-poly(rC) to the medium (data not shown).
FIG. 3.
Induction of IFN-β mRNA by dsRNA is inhibited by Erns. CTe cells were either treated with 100 μg of poly(rI)-poly(rC)/ml (lanes 1, 2, and 3) or mock treated (lane 4) in the presence (lane 1) or absence (lanes 2, 3, and 4) of 1 μg of Erns/ml. After 6 h RNA was extracted from the cells, reverse transcribed with oligonucleotide (dT) as a primer, and amplified by PCR with primers specific for bovine IFN-β mRNA (upper panel) or β-actin mRNA (lower panel). Lane 3 shows the absence of a PCR product from poly(rI)-poly(rC)-treated cells with no RT step. Lane 5 shows the PCR products of a bovine IFN-β cDNA clone or a bovine β-actin cDNA clone. The respective DNA mobilities of IFN-β and β-actin are indicated on the right-hand sides of each panel.
Inhibition of dsRNA-dependent NF-κB activation by Erns.
An early response of cells to treatment with dsRNA is to induce the nuclear translocation of the transcription factor NF-κB and promote its DNA-binding activity in nuclear extracts; these events are important for the activation of IFN-β gene transcription. The effect of purified Erns on NF-κB activation was examined in CTe cells and human MG-63 cells. As shown in Fig. 4 cells infected with SFV or treated with TNF-α clearly activated NF-κB in the presence or absence of Erns. In contrast, although cells treated with poly(rI)-poly(rC) activated NF-κB in the absence of Erns, the presence of Erns completely blocked NF-κB activation. These results show that the inhibition of activation of NF-κB by Erns is specific to dsRNA as the inducing signal. Furthermore, they indicate that the inhibition is not restricted to bovine cells.
FIG. 4.
Erns treatment prevents NF-κB activation by dsRNA. (A) CTe cells were either mock treated (lanes 1 and 2), infected with SFV (∼10 PFU/cell) (lanes 3 and 4), or treated with 5 μg/ml of poly(rI)-poly(rC) (lanes 5 and 6) for 2 h in the presence or absence of Erns. (B) MG-63 cells were either mock treated (lane 1) or treated with 5 μg of poly(rI)-poly(rC)/ml (lanes 2 and 3) or 10 ng of TNF/ml (lanes 4 and 5) for 2 h in the presence or absence of Erns. Nuclear extracts were prepared from each experiment and analyzed by EMSA with the PRD II probe from the human IFN-β gene (31). The mobilities of the DNA-protein complexes are indicated on the sides of each panel. Lanes marked “+” in panels A and B indicate treatment with Erns (1 μg/ml).
RNase activity of Erns is essential for the inhibition of dsRNA-dependent signaling.
Histidine 297 of the CSFV polyprotein (300 in BVDV; Fig. 1) has been shown to be essential for RNase activity of CSFV Erns (11, 23). In order to determine the importance of the RNase activity of BVDV glycoprotein Erns to its ability to inhibit the signaling by dsRNA, we engineered a histidine-to-lysine substitution at amino acid 300 in BVDV (ErnsH300K). We first established whether this mutated protein has lost the RNase activity of wild-type BVDV Erns. ErnsH300K was expressed in insect cells and purified to homogeneity as assessed by SDS-PAGE analysis (Fig. 5A). The RNase activity of the mutant glycoprotein was determined, and as shown in Fig. 5B, ErnsH300K is devoid of RNase activity. Next, we analyzed the stability and enzymatic activity of the wild-type and mutant proteins. Incubation of wild-type and mutant Erns in tissue culture medium at 37°C for 18 h altered neither the stability of mutant or wild-type protein (both remained intact after incubation in medium) nor the enzymatic activity of wild-type Erns (Fig. 5B).
FIG. 5.
Mutation of amino acid residue H300K impairs both the RNase activity of Erns and the inhibitory effect of Erns on dsRNA-induced MxA expression. (A) Stability of wild-type and mutant Erns as determined by SDS-PAGE. Prestained molecular mass standards (lane 1), 1 μg of wild-type Erns (lane 2), 1 μg of H300K mutant (lane 3), 1 μg of wild-type Erns in maintenance medium (lanes 4 and 5), and 1 μg of H300K mutant in maintenance medium (lanes 6 and 7) are shown. The samples in lanes 5 and 7 were incubated at 37°C for 18 h before electrophoresis. The gel was stained with Coomassie brilliant blue R250. (B) Stability of RNase activity of Erns. Purified wild-type and H300K mutant Erns (bars 1 and 4) and wild-type and mutant Erns incubated in cell culture medium (DMEM plus 2% FCS) for either 10 min at room temperature (bars 2 and 5) or for 18 h at 37°C (bars 3 and 6) were assayed for RNase activity at pH 4.5. The final assay mixture (25 μl) contained wild-type Erns and mutant H300K glycoproteins (0.2 μg), yeast 16-23S RNA (12.5 μg), and RNase inhibitor (RNasin; 40 U) as described in Materials and Methods. The release of acid-soluble RNA was determined by measuring the absorbance at 260 nm. (C) Comparison of dsRNA-induced MxA inhibition by wild-type Erns and H300K mutant. Confluent monolayers of cells were rinsed with maintenance medium and replaced with medium containing 5 μg of poly(rI)-poly(rC)/ml and 1 μg of wild-type Erns/ml (lane 1), 5 μg of poly(rI)-poly(rC)/ml and 1 μg of H300K mutant/ml (lane 2), and 5 μg of poly(rI)-poly(rC)/ml (lane 4). Lane 3, mock treated. Cells were incubated at 37°C for 18 h and subjected to immunoblot analysis for the MxA protein.
We next investigated whether ErnsH300K could block dsRNA-dependent signaling. When this protein was added to the medium of CTe cells, it was unable to block the induction of MxA in response to poly(rI)-poly(rC), in contrast to wild-type Erns (Fig. 5C). A similar failure of ErnsH300K to block dsRNA-dependent signaling was also seen for NF-κB induction (data not shown). We conclude that the histidine residue at position 300 in the Erns polypeptide, crucial for RNase activity, is critically required to block dsRNA-dependent signaling responses in cultured cells.
Erns binds to dsRNA-agarose.
Since Erns efficiently blocks dsRNA signaling, we sought to establish whether BVDV Erns can bind to dsRNA. Accordingly, Erns expressed in S2 cells was examined for its ability to bind to poly(rI)-poly(rC)-linked agarose. Erns was applied to a bed of poly(rI)-poly(rC)-agarose and bound Erns was eluted by stepwise increased concentrations of NaCl of up to 1 M. Eluted Erns was detected by ELISA. The results (Fig. 6A) show that Erns bound with greater affinity to poly(rI)-poly(rC)-agarose than it did to underivatized agarose, with a substantial proportion eluting at concentrations of ≥0.25 M NaCl.
FIG. 6.
Binding of Erns to poly(rI)-poly(rC)-agarose. (A) Purified H300K mutant Erns (2 μg) was applied to a 100-μl Minispin column containing either poly(rI)-poly(rC)-agarose (▪) or agarose (▧). The columns were equilibrated with 20 mM sodium phosphate buffer (pH 7.2) containing 150 mM NaCl, and the Erns was eluted with steps of increasing concentrations of NaCl (0.2 to 1 M) in 20 mM sodium phosphate buffer (pH 7.2). The Erns concentration in each fraction was determined by ELISA as described in Materials and Methods. (B) [35S]methionine-labeled full-length Erns (upper panel) or ErnsΔ375 (lower panel) was incubated with poly(rI)-poly(rC)-agarose beads and washed at increasing ionic strengths as described in Materials and Methods and as indicated on the top of each lane. The beads were then treated with polyacrylamide gel sample buffer, and released protein was separated by SDS-PAGE and visualized by autoradiography.
We also examined the ability of poly(rI)-poly(rC)-agarose to bind to recombinant Erns produced in reticulocyte lysates. Figure 6B shows that a significant proportion of Erns remained associated with the matrix at a concentration of at least 0.3 M NaCl (Fig. 6B, upper panel) in agreement with the result obtained with Erns expressed in insect cells.
Identification of amino acids involved in dsRNA binding by Erns.
We next took advantage of the ability of recombinant protein produced in reticulocyte lysates to bind to poly(rI)-poly(rC)-agarose to map regions of Erns involved in dsRNA binding. A series of C-terminal truncations were produced (see Materials and Methods) and examined for their ability to bind to poly(rI)-poly(rC)-agarose. Truncations that retained only 143 amino acids of Erns, terminating at K412 of the polyprotein, showed no loss of binding (data not shown), but only residual binding was seen with further truncation to produce a 104-amino-acid polypeptide, terminating at G375 (ErnsΔ375; Fig. 6B). In addition, we were able to show that the dsRNA-binding functions were not influenced by the presence of the CD33 tag at the N terminus of the Erns truncations (see Materials and Methods). These results indicate that the N-terminal region of Erns contains residues that influence dsRNA binding; however, when we tested the effects of KCl concentration on binding, it was clear that the ErnsΔ375 truncation bound less stably (Fig. 6B, lower panel) than full-length Erns. Thus, there are amino acids influencing binding to poly(rI)-poly(rC) resident in the C-terminal 123 amino acid residues of Erns.
Several dsRNA-binding proteins from a wide variety of organisms interact with dsRNA through basic amino acid residues (8, 32, 33). The Erns glycoprotein contains two clusters of basic amino acids: 409KKGK412 and 483KKLENKSK490. The latter has been shown to be responsible for binding to cell surface glycosaminoglycans (15). Both of these basic clusters are present in the C-terminal 123 amino acids of Erns, the region that was implicated above in modulating the affinity of binding to dsRNA. To investigate the contribution of these clusters to binding dsRNA by Erns, we examined a series of single (K409A, K410T, and K412G) amino acid substitutions and a double substitution (K409A-K410T) in wild-type Erns and the deletion mutant ErnsΔ483-497 from which the 483KKLENKSK490 motif had been removed (15). These proteins, expressed in Drosophila S2 cells and purified by chelating chromatography, were tested for dsRNA-binding activity by using poly(rI)-poly(rC)-agarose. Figure 7 shows that there are marked differences in the dsRNA-binding abilities of various mutant glycoproteins. ErnsΔ483-497 and the substitution mutants ErnsK410T, ErnsK412G, and ErnsH300K behave like wild-type Erns, whereas ErnsK409A and the double-substitution mutant ErnsK409A-K410T were deficient in dsRNA binding. The reduced ability of the mutant glycoprotein ErnsK409A to bind to dsRNA is consistent with the lowering of affinity for dsRNA associated with the deletion of these residues from the studies by using protein produced by in vitro translation. The cluster of basic amino acid residues at the C terminus (483KKLENKSK490), previously implicated in the binding of Erns to glycosylaminoglycans (15), appears not to contribute in dsRNA-Erns binding.
FIG. 7.
Binding of mutant Erns glycoprotein to poly(rI)-poly(rC)-agarose. Erns mutant glycoproteins expressed in Drosophila S2 cells were purifed by chelating chromatography (see Materials and Methods) and applied to poly(rI)-poly(rC)-agarose columns. The binding of mutant glycoproteins to poly(rI)-poly(rC)-agarose was analyzed as described in Fig. 6A legend.
Although these results indicated that Erns is able to bind to a matrix of dsRNA, we also sought to determine whether Erns was also able to bind to ssRNA or dsDNA. The results of experiments set up to examine binding of Erns to poly(C)-agarose or DNA-agarose were inconclusive; Erns bound strongly and irreversibly both to poly(rC)-agarose and to dsDNA-agarose and failed to elute when it was washed with 1 M NaCl in a batch procedure. This finding contrasted with binding to poly(rI)-poly(rC)-agarose, in which bound Erns can be eluted and none remained bound to the matrix. We cannot therefore be sure that binding to these other matrices represents a specific or nonspecific binding. We have also investigated the binding of Erns to ssRNA in a nitrocellulose filter-binding assay. Evidence for Erns binding to ssRNA was seen only for mutants that lacked RNase activity (H300K and K412G [see below]; also data not shown). We presume that the RNase activity of the Erns destroys the ssRNA. Our results on binding of Erns to poly(rI)-poly(rC)-agarose, however, show differences between mutants, and these mutants were investigated further.
Erns mutants lack the ability to block dsRNA-dependent signaling.
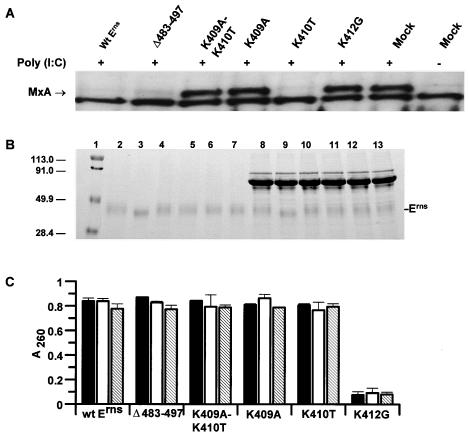
To determine the effects on dsRNA signaling of the substitutions in the two basic amino acid motifs, cells were exposed to dsRNA in the presence of wild-type or mutant forms of Erns. Figure 8A shows that Erns Δ483-497 and ErnsK410T behaved like wild-type Erns in inhibiting the induction of MxA by dsRNA. In contrast, the double substitution ErnsK409A-K410T, or the single substitutions ErnsK409A or ErnsK412G, were unable to block dsRNA-dependent signaling.
FIG. 8.
Functional analysis of Erns mutants. (A) Inhibition of MxA induction by dsRNA. CTe cells were either mock treated or treated with 5 μg of poly(rI)-poly(rC)/ml in maintenance medium in the presence or absence of wild-type or mutant Erns glycoproteins (1 μg/ml) as indicated. After incubation at 37°C for 18 h, MxA induction was analyzed by immunoblotting. Lanes marked “+” indicate that the cells were treated with poly(rI)-poly(rC). (B) Comparison of the protein stability of wild-type and mutant Erns. Wild-type and mutant Erns was purified and added to maintenance medium (DMEM plus 2% FCS) and incubated for 18 h at 37°C, and samples containing 1 μg of Erns were taken for electrophoresis. Lanes 2 to 7, purified Erns; lanes 8 to 13, Erns in maintenance medium following incubation. Lane 1, prestained markers; lanes 2 to 8, wild-type Erns; lanes 3 and 9, deletion mutant Δ483-497; lanes 4 and 10, double mutant K409A-K410T; lanes 5 and 11, mutant K409A; lanes 6 and 12, mutant K410T; and lanes 7 and 13, mutant K412G. Following electrophoresis the gel was stained with Coomassie brilliant blue R250. (C) RNase activity assays of purified wild-type (wt) Erns and Erns mutants with the indicated substitutions in the Erns sequence. The glycoproteins were either incubated in 20 mM sodium acetate buffer (pH 4.5) (▪), in cell culture medium (DMEM plus 2% FCS) for 10 min at room temperature (□), or in cell culture medium for 18 h at 37°C (▧). RNase assays were performed at pH 4.5 as described in Materials and Methods. Bars show the standard deviations of triplicate datum points.
The results presented above correlate Erns binding to poly(rI)-poly(rC)-agarose with its ability to block dsRNA-dependent signaling. However, they do not account for the properties of the ErnsK412G protein, since it is able to bind to poly(rI)-poly(rC)-agarose and yet has lost the ability to inhibit dsRNA-dependent signaling. Since we also saw a loss of dsRNA signaling inhibition associated with the RNase-deficient Erns H300K protein (Fig. 5B), we reasoned that the loss of signaling inhibition by the ErnsK412G protein could be due to a similar loss of RNase activity or, alternatively, the mutant Erns glycoproteins may not be stable. To test these possibilities, two series of experiments were carried out.
First, as described above for wild-type Erns, wild-type and mutant Erns was mixed with tissue culture maintenance medium and incubated for 18 h at 37°C. The results showed no degradation of Erns after incubation (Fig. 8B). Second, RNase assays were performed on each of the purified recombinant Erns proteins, either as purified Erns or as purified Erns added to tissue culture maintenance medium and incubated for 18 h at 37°C. The results showed that the mutant glycoproteins ErnsK409A-K410T and ErnsK409A and the deletion mutant ErnsΔ483-497 showed RNase activity similar to the wild-type Erns, which was not lost upon incubation at 37°C (Fig. 8C). In contrast, ErnsK412G was completely devoid of RNase activity (Fig. 8C), a finding comparable to the activity seen with the RNase-negative mutant glycoprotein ErnsH300K (Fig. 5B).
Erns degrades dsRNA at low pH.
The results presented above demonstrate that both the binding activity of Erns to dsRNA-agarose and the RNase activity of Erns correlate with the inhibition of dsRNA-dependent signaling. One explanation for the ability of Erns to inhibit dsRNA-dependent signaling is that the dsRNA in the cell culture medium is targeted for degradation by the RNase activity, although it should be stressed that previous reports indicate that ssRNA is the preferred substrate for the RNase activity of Erns, with dsRNA being degraded inefficiently (37). To test whether the dsRNA is degraded by Erns, poly(rI)-poly(rC) was incubated in the cell culture medium at 37°C for 1 h and analyzed by agarose gel electrophoresis. As shown in Fig. 9A, poly(rI)-poly(rC) in tissue culture medium was degraded by the addition of Erns. However, we considered that the poly(rI)-poly(rC) preparations may contain regions of ssRNA, which could be digested by Erns.
To investigate more carefully the degradation of dsRNA by Erns, we synthesized a 140-bp dsRNA by in vitro transcription and treated this product so as to remove all ssRNA structures (see Materials and Methods). The synthetic dsRNA retained the ability to induce dsRNA-dependent signaling events and is inactivated by the coaddition of Erns to the cell culture medium (Fig. 9C). When this dsRNA product was incubated with wild-type Erns in the cell culture medium for 18 h at 37°C, it remained intact (Fig. 9B, upper panel), unlike the result seen above with poly(rI)-poly(rC). From these results, we deduced that the integrity of dsRNA was not affected by Erns in the tissue culture medium. Furthermore, when this dsRNA was recovered from the medium after incubation with Erns and any associated proteins were removed by phenol extraction, this dsRNA retained its ability to stimulate dsRNA-dependent signaling in cells, and yet this activation was inhibited by the addition of Erns to the medium (Fig. 9C).
Although these results indicate that, under the conditions extant, Erns does not degrade dsRNA in the cell culture medium, we note that the conditions of the cell culture medium (pH 7.2 to 7.4) are not optimal for the RNase activity of Erns, pH 4.5 to 5.5 (8; unpublished results)—the pH range of several compartments of the cell. Experiments were carried out to examine whether Erns can degrade dsRNA at pH 4.5. The 140-bp synthetic dsRNA produced in vitro was mixed with wild-type Erns in either acidic conditions (pH 4.5) or in cell culture medium. Figure 9B (lower panel) shows that the synthetic dsRNA was completely degraded in acidic conditions (pH 4.5) by Erns in contrast to its stability in cell culture medium. We also investigated the dsRNase activities of mutant forms of Erns. In addition to the wild-type Erns, ErnsK409A-K410T, ErnsK409A, and ErnsK410T and the deletion mutant ErnsΔ483-497 retained dsRNase activity, whereas ErnsK412G and ErnsH300K were unable to degrade dsRNA under the conditions tested (Fig. 9B, lower panel). These results show that Erns can degrade dsRNA in acidic conditions and suggest that this activity may manifest in a low-pH compartment of the cell.
DISCUSSION
The innate immune response generated against viral infections is predominantly mediated by IFNs whose induction is triggered by dsRNA produced as a consequence of viral replication (16). Although most of this dsRNA is intracellular, it has been hypothesized that virally induced cell death can result in the release of dsRNA, and it is this material that is thought to be the ligand for Toll-like receptor 3 (2). However, direct evidence for the release of dsRNA from virus-infected cells is very sparse, influenza virus-infected cells being the only example known to the authors (21). Nevertheless, the peculiar property of pestiviruses to encode a secreted glycoprotein with RNase activity (the Erns product) stimulated us to examine whether it was involved in modulating cellular responses to dsRNA added directly to the cell culture medium. Our results show that bovine cells transfected with a plasmid encoding a secreted form of Erns no longer responded to the addition of dsRNA to the culture medium; similarly, dsRNA responses were blocked by the addition of recombinant Erns protein to the medium. The effect of Erns on the IFN system operates at the level of blocking the IFN-inducing signal since when Erns was present in the medium with the dsRNA, we saw no activation of NF-κB DNA-binding activity, which controls the expression of the IFN-β gene (19, 31), we saw no accumulation of IFN-β mRNA, and Erns blocked the activation of an IFN-β promoter-luciferase reporter construct in response to the addition of poly(rI)-poly(rC) to the medium.
The RNase activity is essential for the ability of Erns to block the dsRNA-mediated signaling as evinced by the effects of mutations at amino acids 300 and 412. Since other mutations blocked dsRNA-dependent signaling without affecting RNase activity there must be additional functions of Erns. We have shown that Erns also binds to dsRNA-agarose, and this property correlates with the inhibition of dsRNA-dependent signaling. In contrast, GAG-binding activity was not needed for inhibition of dsRNA-induced responses. It is probable that Erns degrades the dsRNA added to the cell culture medium. Although we saw no degradation of dsRNA at neutral pH, dsRNA was degraded by Erns at a lower pH. From these results we suggest that dsRNA is bound by Erns, but it is not degraded by Erns until the complex is present at a lower pH, such as a low-pH compartment of the cell. We have sought to test this hypothesis by altering the pH of cellular compartments by the addition of 10 μM chloroquine to the cells. The results were not conclusive since the addition of chloroquine inhibited the induction of MxA by dsRNA directly (data not shown). If Erns acts in, for example, an endosomal compartment of cells, then whether Erns can access this compartment becomes an issue. Recently, this issue has been addressed by Langedijk (18), who adduced that peptides corresponding to the C-terminal domain of Erns were able to translocate across cell membranes and the full-length glycosylated Erns was detected inside the cell, as well as on the cell surface. Thus, it is likely that Erns can access low-pH compartments inside the cell.
Pestiviruses that have been engineered to lose RNase activity are viable. One study with a CSFV virus which lacked RNase activity showed that, intriguingly, it was cytopathogenic (11). The reason for this was not clear. In light of our results, we can suggest that, because treatment of cells with dsRNA can lead to apoptosis (4, 17), it may be that the cells (SK6 cells) used in the study by Hulst et al. (11) were particularly sensitive to dsRNA from cellular debris, and thus the apoptosis may have been due to dsRNA released from cells into the medium. If so, supplementing this mutant with exogenous soluble Erns might have reduced cytopathogenicity.
Further studies carried out with a CSFV virus and a BVDV genotype 2 genetically modified to lack RNase activity (neither of which were reported to be cytopathogenic) showed that they were attenuated in animals (22, 23). In acute infections with these modified viruses either no viremia (in pigs infected with mutant CSFV) or markedly reduced viremia (in calves infected with a BVDV-2 mutant) was observed after infection, and infected animals showed reduced clinical signs. To our knowledge, however, there have been no reports on whether these mutants can initiate persistent infections. Taken as a whole, it seems likely that, during either an acute or a persistent infection, these mutants will be more susceptible to the IFN response or induce an altered innate immune response in vivo. The results of experiments to measure IFN responses after infection of animals with these mutants would be of considerable interest.
Pestiviruses may be unique in their ability to inhibit dsRNA signaling added to the exterior of the cell, but viruses generally have a wide range of ways to control the antiviral action of IFNs. For example, viruses encode proteins that inhibit the IFN signaling pathways; examples include the V protein of paramyxoviruses that target STATs for degradation (38) and poxviruses that encode secreted proteins to act as decoys for IFN and block its action (1, 7). It seems unlikely that pestiviruses take active steps to inhibit IFN signaling in cells: cells infected with BVDV produce MxA on incubation with IFN with the same kinetics as uninfected cells (3), and the antiviral activity of IFN is not inhibited in BVDV-infected cells (30). It is becoming clear that viruses also have mechanisms to limit the yield of IFN produced by an infected cell. A common way to do this is by producing dsRNA-binding proteins (for example, influenza virus NS1 protein [33] and reovirus sigma 3 protein [32; reviewed in reference 7]), although more specific pathways may also exist (for example, the E6 protein of human papillomavirus type 16 binds to and inhbits IRF-3 function [26]).
It is not likely that Erns is sufficient for the inhibition of all IFN-inducing signals produced by virus, since no effect was observed on the induction of IRF-3 DNA-binding activity in CTe cells infected with SFV in the presence of Erns (data not shown). There are additional signals, and there must be additional blocks to the induction of IFN by BVDV (3). Recently, the role of Npro has been associated with the ability of classical swine fever virus to inhibit IFN induction (27); whether Npro acts directly to inhibit IFN-β transcription or whether it acts through another virus polypeptide is not known.
It is probable that the ability of Erns to block signaling induced by the presence of extracellular dsRNA plays an important role in the ability of BVDV to sustain a persistent infection of the fetus where the innate immune response is likely to be the predominant antiviral defense of the host. During infection of the early fetus, secreted Erns may act to counteract any dsRNA released by dying infected cells and thus the host fetus would fail to mount an innate antiviral response by this route. In addition, the existence of a protein with the properties of Erns is strong evidence of the importance of the need for cells to respond to extracellular dsRNA in an antiviral response.
Acknowledgments
We thank J.-F. Valarcher for advice on IFN-β mRNA RT-PCR analysis, Sue Baigent for critically reviewing the manuscript, and P. Staeheli for the gift of anti-MxA antiserum. We thank Michael Clarke and Bryan Charleston for helpful discussions.
S.G. and E.P. are supported by The Wellcome Trust.
REFERENCES
- 1.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Trends Microbiol. 8:410-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexopoulou, L., A. C. Holt, R. Medzhitov, and R. A. Flavell. 2001. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 413:732-738. [DOI] [PubMed] [Google Scholar]
- 3.Baigent, S. J., G. Zhang, M. D. Fray, H. Flick-Smith, S. Goodbourn, and J. W. McCauley. 2002. Inhibition of beta interferon transcription by noncytopathogenic bovine viral diarrhea virus is through an interferon regulatory factor 3-dependent mechanism. J. Virol. 76:8979-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber, G. N. 2001. Host defense, viruses, and apoptosis. Cell Death Differ. 8:113-126. [DOI] [PubMed] [Google Scholar]
- 5.Bazzigher, L., J. Pavlovic, O. Haller, and P. Staeheli. 1992. Mx genes show weaker primary response to virus than other interferon-regulated genes. Virology 186:154-160. [DOI] [PubMed] [Google Scholar]
- 6.Charleston, B., M. D. Fray, S. Baigent, B. V. Carr, and W. I. Morrison. 2001. Establishment of persistent infection with non-cytopathic bovine viral diarrhoea virus in cattle is associated with a failure to induce type I interferon. J. Gen. Virol. 82:1893-1897. [DOI] [PubMed] [Google Scholar]
- 7.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 8.Green, S. R., and M. B. Mathews. 1992. Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev. 6:2478-2490. [DOI] [PubMed] [Google Scholar]
- 9.Houe, H. 1999. Epidemiological features and economical importance of bovine virus diarrhoea virus (BVDV) infections. Vet. Microbiol. 64:89-107. [DOI] [PubMed] [Google Scholar]
- 10.Hulst, M. M., G. Himes, E. Newbigin, and R. J. Moormann. 1994. Glycoprotein E2 of classical swine fever virus: expression in insect cells and identification as a ribonuclease. Virology 200:558-565. [DOI] [PubMed] [Google Scholar]
- 11.Hulst, M. M., F. E. Panoto, A. Hoekman, H. G. van Gennip, and R. J. Moormann. 1998. Inactivation of the RNase activity of glycoprotein Erns of classical swine fever virus results in a cytopathogenic virus. J. Virol. 72:151-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hulst, M. M., H. G. van Gennip, and R. J. Moormann. 2000. Passage of classical swine fever virus in cultured swine kidney cells selects virus variants that bind to heparan sulfate due to a single amino acid change in envelope protein Erns. J. Virol. 74:9553-9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hulst, M. M., H. G. van Gennip, A. C. Vlot, E. Schooten, A. J. de Smit, and R. J. Moormann. 2001. Interaction of classical swine fever virus with membrane-associated heparan sulfate: role for virus replication in vivo and virulence. J. Virol. 75:9585-9595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iqbal, M., H. Flick-Smith, and J. W. McCauley. 2000. Interactions of bovine viral diarrhoea virus glycoprotein Erns with cell surface glycosaminoglycans. J. Gen. Virol. 81:451-459. [DOI] [PubMed] [Google Scholar]
- 15.Iqbal, M., and J. W. McCauley. 2002. Identification of the glycosaminoglycan-binding site on the glycoprotein Erns of bovine viral diarrhoea virus by site-directed mutagenesis. J. Gen. Virol. 83:2153-2159. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs, B. L., and J. O. Langland. 1996. When two strands are better than one: the mediators and modulators of the cellular responses to double-stranded RNA. Virology 219:339-349. [DOI] [PubMed] [Google Scholar]
- 17.King, P., and S. Goodbourn. 1998. STAT1 is inactivated by a caspase. J. Biol. Chem. 273:8699-8704. [DOI] [PubMed] [Google Scholar]
- 18.Langedijk, J. P. 2002. Translocation activity of C-terminal domain of pestivirus Erns and ribotoxin L3 loop. J. Biol. Chem. 277:5308-5314. [DOI] [PubMed] [Google Scholar]
- 19.Lenardo, M. J., C. M. Fan, T. Maniatis, and D. Baltimore. 1989. The involvement of NF-κB in beta-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell 57:287-294. [DOI] [PubMed] [Google Scholar]
- 20.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 21.Majde, J. A., N. Guha-Thakurta, Z. Chen, S. Bredow, and J. M. Krueger. 1998. Spontaneous release of stable viral double-stranded RNA into the extracellular medium by influenza virus-infected MDCK epithelial cells: implications for the viral acute phase response. Arch. Virol. 143:2371-2380. [DOI] [PubMed] [Google Scholar]
- 22.Meyer, C., M. Von Freyburg, K. Elbers, and G. Meyers. 2002. Recovery of virulent and RNase-negative attenuated type 2 bovine viral diarrhea viruses from infectious cDNA clones. J. Virol. 76:8494-8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyers, G., A. Saalmuller, and M. Buttner. 1999. Mutations abrogating the RNase activity in glycoprotein Erns of the pestivirus classical swine fever virus lead to virus attenuation. J. Virol. 73:10224-10235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paton, D. J., K. H. Christiansen, S. Alenius, M. P. Cranwell, G. C. Pritchard, and T. W. Drew. 1998. Prevalence of antibodies to bovine virus diarrhoea virus and other viruses in bulk tank milk in England and Wales. Vet. Rec. 142:385-391. [DOI] [PubMed] [Google Scholar]
- 25.Pollock, R., and R. Treisman. 1990. A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res. 18:6197-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruggli, N., J. D. Tratschin, M. Schweizer, K. C. McCullough, M. A. Hofmann, and A. Summerfield. 2003. Classical swine fever virus interferes with cellular antiviral defense: evidence for a novel function of Npro. J. Virol. 77:7645-7654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rumenapf, T., G. Unger, J. H. Strauss, and H. J. Thiel. 1993. Processing of the envelope glycoproteins of pestiviruses. J. Virol. 67:3288-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider, R., G. Unger, R. Stark, E. Schneider-Scherzer, and H. J. Thiel. 1993. Identification of a structural glycoprotein of an RNA virus as a ribonuclease. Science 261:1169-1171. [DOI] [PubMed] [Google Scholar]
- 30.Schweizer, M., and E. Peterhans. 2001. Noncytopathic bovine viral diarrhea virus inhibits double-stranded RNA-induced apoptosis and interferon synthesis. J. Virol. 75:4692-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Visvanathan, K. V., and S. Goodbourn. 1989. Double-stranded RNA activates binding of NF-κB to an inducible element in the human beta-interferon promoter. EMBO J. 8:1129-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, Q., J. Bergeron, T. Mabrouk, and G. Lemay. 1996. Site-directed mutagenesis of the double-stranded RNA binding domain of bacterially expressed sigma 3 reovirus protein. Virus Res. 41:141-151. [DOI] [PubMed] [Google Scholar]
- 33.Wang, W., K. Riedel, P. Lynch, C. Y. Chien, G. T. Montelione, and R. M. Krug. 1999. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5:195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wathelet, M. G., P. M. Berr, and G. A. Huez. 1992. Regulation of gene expression by cytokines and virus in human cells lacking the type-I interferon locus. Eur. J. Biochem. 206:901-910. [DOI] [PubMed] [Google Scholar]
- 35.Weiland, F., E. Weiland, G. Unger, A. Saalmuller, and H. J. Thiel. 1999. Localization of pestiviral envelope proteins Erns and E2 at the cell surface and on isolated particles. J. Gen. Virol. 80:1157-1165. [DOI] [PubMed] [Google Scholar]
- 36.Whiteside, S. T., K. V. Visvanathan, and S. Goodbourn. 1992. Identification of novel factors that bind to the PRD I region of the human beta-interferon promoter. Nucleic Acids Res. 20:1531-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Windisch, J. M., R. Schneider, R. Stark, E. Weiland, G. Meyers, and H. J. Thiel. 1996. RNase of classical swine fever virus: biochemical characterization and inhibition by virus-neutralizing monoclonal antibodies. J. Virol. 70:352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]