Abstract
Hepatitis E virus (HEV) is a major human pathogen in much of the developing world. It is a plus-strand RNA virus with a 7.2-kb polyadenylated genome consisting of three open reading frames, ORF1, ORF2, and ORF3. Of these, ORF2 encodes the major capsid protein of the virus and ORF3 encodes a small protein of unknown function. Using the yeast three-hybrid system and traditional biochemical techniques, we have studied the RNA binding activities of ORF2 and ORF3, two proteins encoded in the 3′ structural part of the genome. Since the genomic RNA from HEV has been postulated to contain secondary structures at the 5′ and 3′ ends, we used these two terminal regions, besides other regions within the genome, in this study. Experiments were designed to test for interactions between the genomic RNA fusion constructs with ORF2 and ORF3 hybrid proteins in a yeast cellular environment. We show here that the ORF2 protein contains RNA binding activity. The ORF2 protein specifically bound the 5′ end of the HEV genome. Deletion analysis of this protein showed that its RNA binding activity was lost when deletions were made beyond the N-terminal 111 amino acids. Finer mapping of the interacting RNA revealed that a 76-nucleotide (nt) region at the 5′ end of the HEV genome was responsible for binding the ORF2 protein. This 76-nt region included the 51-nt HEV sequence, conserved across alphaviruses. Our results support the requirement of this conserved sequence for interaction with ORF2 and also indicate an increase in the strength of the RNA-protein interaction when an additional 44 bases downstream of this 76-nt region were included. Secondary-structure predictions and the location of the ORF2 binding region within the HEV genome indicate that this interaction may play a role in viral encapsidation.
Hepatitis E virus (HEV), the causative agent of hepatitis E, is transmitted via the fecal-oral route, predominantly through contaminated water, and is responsible for sporadic infections as well as large epidemics of acute viral hepatitis in developing countries (3, 4, 11, 12, 13, 21, 22, 23, 36). The HEV genome organization resembles that of many alphaviruses, with nonstructural genes at the 5′ end and structural genes at the 3′ end (13, 23), and is currently classified in a separate unclassified group of hepatitis E-like viruses (2). It has a single-stranded positive-sense RNA genome of 7.2 kb with three forward open reading frames (ORF1, ORF2, and ORF3) encoding three different proteins (Fig. 1A) (13, 24, 29, 31). The 5′ untranslated region (UTR) is 28 nucleotides (nt) in length, and bases 2 through 26 can potentially assume a complex secondary structure or hairpin (7). Bases 150 to 208 form a sequence that is homologous to the 51-nt sequence, conserved across alphaviruses. The 3′ UTR is 68 nt long and has been postulated to form stem-loop structures (29).
FIG. 1.
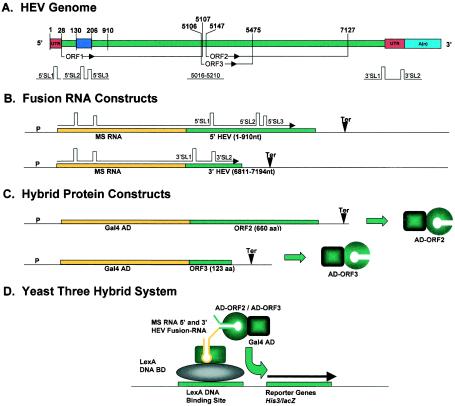
HEV genome and yeast three-hybrid constructs used to study RNA-protein interactions. (A) Genes and genome organization of HEV. Start sites for all three ORFs and both 3′ and 5′ UTRs are shown in red. A(n) represents the poly(A) tail (blue). Predicted stem-loop structures (SL) are shown schematically and numbered across the HEV genome. (B) Fusion RNA constructs designed to express fusion transcripts within the yeast cell. The MS2 RNA coding region (yellow) was cloned with two different HEV genomic regions (green). Predicted mRNA stem-loop structures are shown schematically as fusion transcripts. (C) Hybrid protein constructs to test the RNA binding activity of the ORF2 and ORF3 proteins of HEV using the yeast three-hybrid system. The schematic diagram shows the Gal4 Gal4 AD fused in frame to the ORF2 and ORF3 genes of HEV, thus expressing fusion proteins in yeast cells. (D) Schematic diagram of the yeast three-hybrid system showing the different fusion-RNA and hybrid-protein constructs being examined. P, promoter; Ter, terminator; SL, stem-loop structure.
In the absence of a reliable culture system for HEV, fundamental studies of its replication and expression strategy have been restricted to foreign gene expression systems. ORF2 and ORF3 have been expressed in Escherichia coli, animal cells, baculovirus, yeast and in vitro in a coupled transcription-translation system (6, 8, 9, 15, 19).
ORF1, the putative nonstructural gene, begins 28 nt from the 5′ end of the HEV genome, spans 5,079 bases before terminating at nt 5107, and codes for a polypeptide of 1,693 amino acid residues. ORF2 starts from nt 5147, extends 1,980 bases before ending at nt 7127, and codes for the major structural protein of 660 amino acids (88 kDa) that is expressed intracellularly as well as on the cell surface. It is synthesized as a precursor and is processed through signal sequence cleavage into the mature protein, which is capable of self-association and glycosylation (10, 30, 32). The third positive-polarity reading frame of 369 bases (ORF3) overlaps ORF1 at its 5′ end by 1 nt, significantly overlaps ORF2 (in a different frame), and codes for a protein of 123 amino acids (13.5 kDa). ORF3 encodes a phosphoprotein that is expressed intracellularly, fractionates with the cytoskeletal and membrane fractions, and shows no major processing (1, 20, 37). The ORF3 protein dimerizes using a 43-amino-acid interaction domain, interacts with proteins containing SH3 domains, and activates mitogen-activated protein kinase (14, 32). Recently, we have shown that the phosphorylated form of the ORF3 protein interacts with the nonglycosylated form of the major capsid protein, ORF2 of HEV (34).
Since HEV is a plus-strand RNA virus, it is expected that either one of the two proteins encoded in the structural part of the genome, ORF2 or ORF3, would fulfill the role of genomic RNA binding for viral encapsidation leading to headfull packaging of new HEV particles in the infected hepatocyte. This fundamental aspect of the HEV life cycle has not yet been elucidated.
We have used the yeast three-hybrid system (26) to detect the RNA binding properties of these two proteins of HEV and map the interaction domains for the interacting protein and genomic RNA. The yeast three-hybrid system is a genetic assay in which specific RNA-protein interactions can be detected rapidly in yeast, in a fashion that is independent of the biological role of the RNA or protein. This approach is based on the yeast two-hybrid system, in principle, which detects protein-protein interactions. The three-hybrid system allows simple phenotypic properties of yeast, such as the ability to grow or to metabolize a chromogenic compound, to be used to detect and analyze an RNA-protein interaction. In the cotransformed yeast cell, a fusion RNA molecule bridges two hybrid proteins, one containing a DNA binding domain and the other containing a transcriptional activation domain, resulting in the transcriptional activation of HIS3 and lacZ reporter genes downstream of the binding site for the DNA binding domain. To apply this system to HEV, we designed constructs fusing MS2-RNA with the 5′ HEV (nt 1 to 910) genome and the 3′ HEV (nt 6807 to 7184) genome in two separate constructs (Fig. 1B). The HEV ORF2 and ORF3 genes were cloned in-frame with the Gal4 activation domain (Gal4 AD) (Fig. 1C) in two separate constructs.
From the two ORFs tested, using the yeast three-hybrid assay, we found the ORF2 protein to be capable of binding to the 5′ end of the HEV genome. These results were verified by electrophoretic mobility shift assays (EMSA). Further, we have mapped the interaction domains of the genomic RNA and the interacting protein, ORF2. Deletion analysis of ORF2 showed that the RNA binding activity of the protein was lost when deletions were made beyond amino acid 111 from the N terminal, suggesting that its RNA binding activity is secondary-structure dependent rather than sequence specific. On the other hand, finer mapping of the 5′ genomic RNA revealed that a 76-nt region was responsible for this interaction. Although this 76-nt region was the smallest identifiable interaction domain that binds ORF2, genomic sequences up to 44 nt downstream of this region contributed to the strengthening of the RNA protein interaction. A detailed RNA secondary-structure model for the interaction domain has been postulated, and the functional significance of this interaction during the viral life cycle has been discussed.
MATERIALS AND METHODS
Growth media, yeast strains, and plasmids.
Yeast cells were grown either in synthetic media lacking the indicated nutrients or in rich media (25). The components for the three-hybrid selection were provided by the laboratory of M. Wickens (University of Wisconsin, Madison), and the selection was performed essentially as described elsewhere (26, 27, 38). The yeast strains, plasmids, and plasmid constructs used in this study are listed in Table 1.
TABLE 1.
Yeast strains, plasmids, and recombinant plasmid constructs used in this study
| Strain, plasmid, or constructa | Genotype and description |
|---|---|
| Strain | |
| L40-coat | MATaura3-52 leu2-3,112 his3Δ200 trp1Δ1 ade2 LYS::(LexA op)-HIS3 ura3::(LexA-op)-LacZ, LexA-MS2 coat (TRP1) |
| Plasmids | |
| pACT2 | GAL4 AD vector [GAL4(768-881)]; LEU2, 2μm, Ampr |
| pIII/MS2-2 | Derived from pIIIEx426RPR |
| Constructs | |
| pAS2-ORF2 | pMT-ORF2 digested with NcoI and BamHI, fragment ligated |
| pACT-ORF2 | pAS2-ORF2 digested with NcoI and BamHI, fragment ligated |
| pSG112-660 ORF2 | Described in reference 8 |
| pACT-ORF2 112-660 | pSG-ORF2 1-110 digested with EcoRI and BamHI, fragment religated |
| pACT-ORF2 1-226 | pACT-ORF2 digested with EcoRI, vector religated |
| pACT2-ORF2 1-358 | pACT2-ORF2 digested with NcoI and ScaI, fragment religated |
| pACT2-ORF2 1-586 | pACT2-ORF2 digested with BstXI and NcoI and religated |
| pSG-ORF2 | Described in reference 8 |
| pSG-ORF2 1-226 | pSG-ORF2 digested with EcoRI and PstI, fragment religated |
| pSG-ORF2 1-358 | pSG-ORF2 digested with ScaI and PstI, fragment religated |
| pSG-ORF2 1-586 | pSG-ORF2 digested with BstXI and PstI, fragment religated |
| pSGIb | pSGI vector digested with EcoRI and HindIII, blunt ended, and vector religated |
| pSG-ORF2b | pSG-ORF2 digested with PstI and BamHI, fragment religated into pSGIb |
| pSG-ORF2 228-660 | pSG-ORF2b digested with BstEII and EcoRI, blunt ended, and vector religated |
| pIIIMS2-2 10-910 HEV | Full-length HEV genome digested with SmaI and NdeI, fragment religated |
| pIIIMS2-1-250 HEV | Fragment from bp 1 to 250 PCR amplified and cloned into SmaI site of vector |
| pIIIMS2-2 1-130 HEV | Fragment from bp 1 to 130 PCR amplified and cloned into SmaI and ShpI site of vector |
| pIIIMS2-2 1-120 HEV | Fragment from bp 130 to 250 PCR amplified and cloned into SmaI and ShpI sites of vector |
| pGEMTeasy 1-250 HEV | Fragment from bp 1 to 250 PCR amplified and cloned using TA overhang ligation |
| pGEMT easy 130-250 HEV | pGEMT easy 1-250 digested with StuI and PstI, fragment religated |
| pGEMT 3′ 6807-7184 HEV | Fragment of 377 bp PCR amplified from pSG-ORF2 and cloned into pGEMT Easy vector |
| pMS2-2 3′ 6807-7184 HEV | Fragment of 377 bp PCR amplified from pSG-ORF2 and cloned into SmaI site of vector |
| pSG-ORF3 | Described in reference 8 |
| pACT ORF3 | pSG-ORF3 digested with SmaI and XhoI, fragment religated |
Numbers in italics represent the RNA sequence of the HEV genome.
Intermediate vector constructs.
Yeast three-hybrid techniques.
The yeast host strain L40-coat constitutively expresses the LexA-MS2 hybrid coat protein in which the LexA DNA binding domain is at the N terminal and the MS2 coat protein is at the C terminal. Both the RNA plasmid (pIIIMS2-2) and its fusion constructs and the AD vector (pACTII) and its hybrid protein constructs were transformed in corresponding pairs to study RNA-protein interactions (Fig. 1D). HIS3 and lacZ serve as reporter genes for the three-hybrid assay. β-Galactosidase expression levels were determined either by a 5-bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) filter assay or by a liquid assay using o-nitrophenyl-d-galactoside (5, 35, 36). Constructs MS2-IRE RNA, in which the MS2 protein is 5′ of the iron response element (IRE) and construct AD-IRP, in which the iron regulatory protein (IRP) is cloned in frame downstream of the activation domain (AD), have been used as a positive control for the yeast three-hybrid experiments (26).
Gel shift assay (EMSA).
EMSA was performed as described by Martin et al. (17). Uniformly 32P-labeled RNA was produced by T7 RNA polymerase transcription from inserts cloned into the pGEMT Easy plasmid (Promega) and purified using the RN Easy kit (Qiagen). Purified probes were checked on a 6% urea-acrylamide gel to verify the integrity of RNA transcripts. Full-length ORF2 and ORF3 proteins and deletions of ORF2 were produced using an in vitro-coupled transcription-translation rabbit reticulocyte system (Promega) and verified by immunoprecipitation with anti-ORF2 and anti-ORF3 antibodies.
To detect RNA-protein interactions by EMSA, 8 μg of total protein was mixed with 50,000 cpm of uniformly 32P-labeled RNA and 20 μg of yeast tRNA and incubated for 20 min in 10 mM Tris-HCl (pH 7.5)-50 mM KCl-1 mM dithiothreitol-10% glycerol in 20 μl on ice. Reaction products were analyzed by electrophoresis on native 5 or 4% polyacrylamide gels using 50 mM Tris-glycine as a buffer and visualized by autoradiography. Mock lysates were incubated along with the labeled probe in a similar manner to rule out the possible binding of endogenous protein from the rabbit reticulocyte lysate (data not shown). For competitor binding assay, a 100-fold excess of unlabeled RNAs was incubated along with the regular reaction mixture described above.
RNA secondary-structure analysis.
RNA secondary structure was analyzed using the mfold program (http://bioinfo.rpi.edu/applications/mfold/old/RNA), based on minimum free energy calculations at 25°C.
RESULTS
The 5′ end of the HEV genomic RNA interacts with the ORF2 protein.
The different regions of the HEV genome were cloned into yeast three-hybrid vectors so as to express fusion RNA transcripts in yeast cells. The 5′ end (bases 1 to 910) of the HEV genome was cloned into the pIIIMS2-2 yeast three-hybrid vector (Table 1). Similarly, the 3′ end (bases 6807 to 7184) and the 572 nt from the middle region (bases 5108 to 5680) of the HEV genome were cloned into the pIIIMS2-2 yeast three-hybrid vector (Table 1). All the constructs were transformed into the L40-coat yeast three-hybrid host strain and grown on auxotrophic media. Independent transformants were picked and their total RNA was isolated. Transcription of the fusion RNA was checked by reverse transcription-PCR analysis. Further verification of the fusion RNA was performed by cloning each insert into the SmaI site of the IRE sequence in the pIIIMS2-2 vector (26) and checking for an interaction between IRE and IRP in the presence of 25 mM 3-aminotriazole (3-AT) on SD-His growth medium (see the legend to Fig. 2) by using the yeast three hybrid system (data not shown). 3-AT was used to measure the strength of the RNA-protein interaction. Similarly, full-length ORF2 and ORF3 genes were cloned in frame with the Gal4 activation domain to express hybrid proteins in the yeast cells as reported previously (32, 33, 34).
FIG. 2.
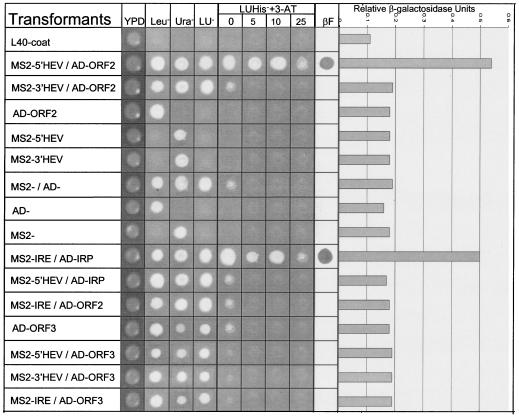
Results from the three-hybrid analysis showing 5′ HEV genomic RNA interacting with the ORF2 protein. YPD, yeast extract peptone dextrose media (nonselective); Leu−, Ura−, LU− represent SD-Leu− (synthetic dextrose complete medium lacking leucine), SD-Ura− (synthetic dextrose complete media lacking uracil), and SD-Leu− Ura− synthetic growth media. LUHis−+3-AT (synthetic dextrose complete media lacking histidine, leucine, and uracil with 3-aminotrizole) represents SD-Leu− Ura− His− synthetic medium with 0, 5, 10, and 25 mM 3-aminotrizole (3-AT) added. βF represents results from the β-galactosidase filter assay, and the bar graph represents relative β-galactosidase units from the liquid β-galactosidase assay. L40-coat is the untransformed yeast host strain. MS2-IRE/AD-IRP is the postive control used in the assay (26).
Single transformants and cotransformants were obtained in different combinations and tested for RNA-protein interactions (Fig. 2). The host yeast strain (L40-coat) showed negligible background His+ phenotype. When singly transformed host cells were analyzed, low background reporter gene activity was detected on His− plus 5 mM 3-AT medium and in β-galactosidase assays. The MS2-5′HEV/AD-ORF2 cotransformants clearly showed strong His+ prototrophy up to 15 mM 3-AT. This clone was strongly positive when tested for β-galactosidase activity, in both filter and liquid assays. However, the MS2-3′HEV/AD-ORF2 cotransformants showed no reporter gene activity. The HEV genomic RNA (nt 5108 to 5680) was also tested for interaction with AD-ORF2 and AD-ORF3. Both assays gave negative results (data not shown). Similarly, AD-ORF3, when cotransformed separately with MS2-5′ HEV and MS2-3′HEV, failed to show increased reporter gene activity.
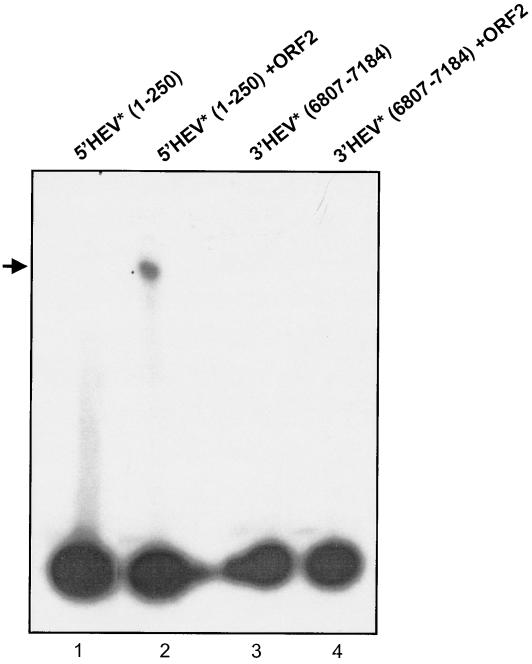
The protein binding domain was subsequently shortened to nt 1 to 250nt from nt 1 to 910, subcloned into the pIIIMS2-2 vector (Table 1), and checked for interaction with ORF2. Since this shorter RNA region showed positive interaction with the ORF2 protein, albeit of equal strength, subsequent EMSA experiments were performed using this nt 1 to 250 5′ HEV RNA. The 5′ and 3′ regions of the HEV genome were transcribed as 32P-labeled transcripts and, after purification, were incubated with unlabeled ORF2 protein in separate tubes. As negative controls, the 32P-labeled transcripts from the 5′ and 3′ genomic regions of HEV were analyzed separately on a nondenaturing 6% polyacrylamide gel. Clearly, the 5′ HEV genomic transcript containing nt 1 to 250 showed a mobility shift, indicating that the ORF2 protein was interacting with it (Fig. 3, lanes 2 and 3), in contrast to lane 1, where no protein was present. On the other hand, the 3′ genomic region and the nt 5108 to 5680 midgenomic region of HEV showed no binding to the ORF2 protein (lane 4). Similar experiments were repeated with the ORF3 protein, which showed a negative interaction with both the 5′ and 3′ genomic regions of HEV (data not shown).
FIG. 3.
In vitro gel shift assay confirming the results of the yeast three-hybrid system. 5′ HEV RNA (nt 1 to 250) and 3′HEV RNA (nt 6807 to 7184) were 32P radiolabeled. ORF2 protein was produced using a coupled transcription-translation system. The arrow shows ORF2 protein bound to 32P-labeled RNA (lanes 2 and 3). Lanes 1 and 4 are negative controls. Asterisks refer to 32P labeled transcripts.
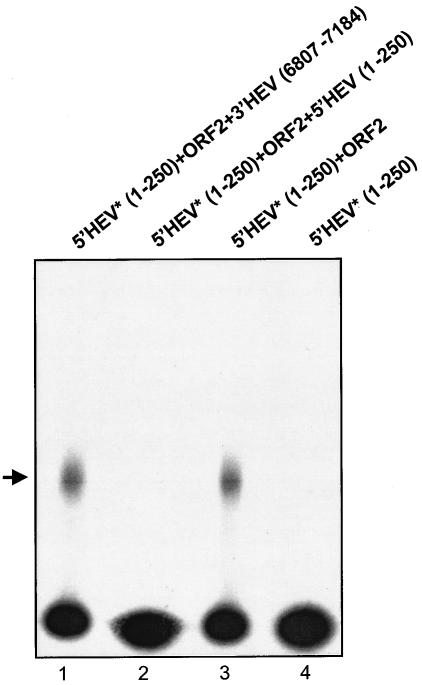
A competitor binding assay was performed to study the specificity of the interaction of the ORF2 protein with 5′ HEV RNA (nt 1 to 250). In this experiment, unlabeled 3′ HEV RNA (nt 6807 to 7184) was used as a nonspecific competitor at a 100-fold higher molar concentration (Fig. 4, lane 1). The 3′ HEV RNA (nt 6807 to 7184) did not compete for binding with the labeled 5′ HEV RNA (nt 1 to 250)-ORF2 complex, which was visible on the autoradiogram. When unlabeled 5′ HEV RNA (nt 1 to 250) at 100-fold higher molar concentrations was incubated with labeled 5′ HEV RNA (nt 1 to 250) and unlabeled ORF2 protein, it competed with the labeled 5′ HEV RNA (nt 1 to 250). This was evident by the complete disappearance of signal (lane 2). Appropriate positive and negative controls are shown in lanes 3 and 4, respectively. This experiment shows clearly that the RNA-protein interaction was specific to the 5′ HEV RNA (nt 1 to 250) transcript and full-length ORF2 protein. We further designed deletions to identify the interaction domain of ORF2 responsible for RNA binding.
FIG. 4.
Competitor binding assay. The ORF2 protein used in this experiment is unlabeled. Lane 1 contains a nonspecific competitior, 3′ HEV RNA (nt 6807 to 7184). Lane 2 contains a 100-fold excess of unlabeled 5′HEV RNA (nt 1 to 250) transcript. Lanes 3 and 4 are positive and negative controls, respectively. Asterisks refer to 32P-labeled transcripts. Nonradioactive transcripts 3′ HEV RNA (nt 6807 to 7184) and 5′ HEV RNA (nt 1 to 250) were used at 100-fold higher molar concentrations.
The N-terminal 111-amino-acid-deleted ORF2 protein interacts with 5′ HEV genomic RNA.
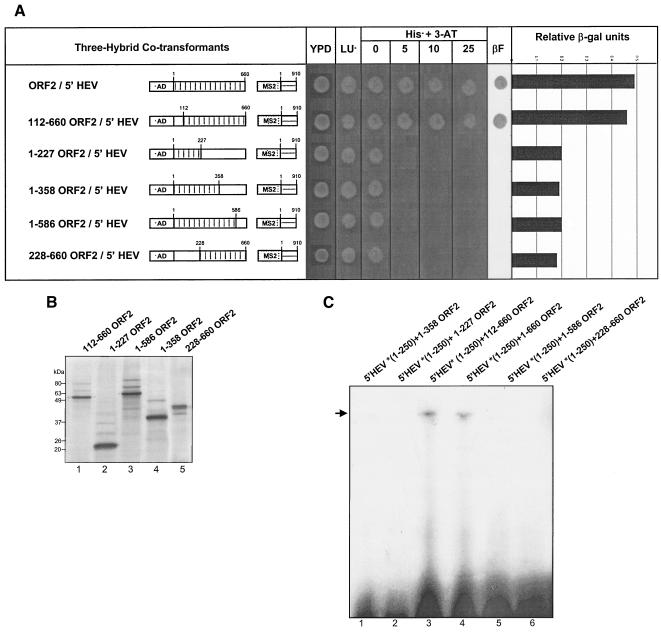
The full-length ORF2 protein was subject to a series of deletions. These deletion proteins were subsequently cloned in-frame with the Gal4 activation domain using the vector pACT2, as described in Table 1. When tested for yeast three-hybrid interactions with 5′ HEV RNA (nt 1 to 910), the constructs, AD-ORF2(1-227), AD-ORF2(1-358), AD-ORF2(1-586), and AD-ORF2(228-660) were negative. Only AD-ORF2(112-660), when tested with 5′ HEV RNA (nt 1 to 910) showed positive on the three-hybrid analysis. All three hybrid cotransformants were tested with increasing levels of 3-AT on His− media and in β-galactosidase filter and liquid assays (Fig. 5A). Although the presence of 5′ HEV RNA (nt 1 to 910) interactions with the AD-ORF2(1-227), AD-ORF2(1-358), and AD-ORF2(1-586) points toward the importance of the C-terminal region of the protein, AD-ORF2(228-660) did not show a positive interaction. This result points to an observation we have made in the past (33, 34) regarding the ORF2 protein, i.e., that its properties are lost when it is truncated beyond amino acid 111 from the N-terminal end.
FIG. 5.
Mapping the interaction domain for the ORF2 protein. (A) Amino acids 112 to 660 from the ORF2 protein are required for interaction with the 5′ HEV RNA (nt 1 to 910) region. Dotted boxes represent the AD regions which were fused in frame with the ORF2 protein (full length or deletions) shown in boxed regions with vertical lines. Checkered boxes show the MS2 regions fused with the 5′ HEV RNA (nt 1 to 910), shown as horizontal lines. Open boxes represent regions that were deleted from ORF2. The numbers above the boxed regions with vertical lines represent the first and last nucleotides of the regions included in the ORF2 deletion constructs. YPD, yeast extract peptone dextrose media (nonselective); LU−, SD-Leu− Ura− synthetic growth medium; LUHis−+3-AT, SD-Leu− Ura− His− synthetic medium with 0, 5, 10, and 25 mM 3-AT added. βF represents results from the β-galactosidase filter assay, and the bar graph represents relative β-galactosidase units from the liquid β-galactose assay. (B) Control gel showing ORF2 deletions expressed using a coupled transcription-translation expression system. Major bands show the expressed protein of interest and correspond to their calculated molecular masses. Weaker bands in each lane show nonspecific translation of rabbit reticulocyte proteins. (C) EMSA showing ORF2(112-660) interacting with the 5′ genomic region of HEV. Asterisks refer to 32P-labeled transcript.
Results obtained from the yeast three-hybrid system were verified by conventional in vitro techniques. All ORF2 deletions were expressed in a coupled transcription-translation system, checked for expressed protein (as shown in Fig. 5B), and used for EMSA with the 5′ HEV RNA transcript (Fig. 5c). Results obtained from EMSA exactly matched our observations from the yeast three-hybrid system, proving that only the full-length protein and the ORF2(112-660) deletion were capable of interacting with the 5′ genomic RNA. Due to the inherent property of the ORF2 protein losing its RNA binding activity when truncated beyond the N-terminal amino acid 111, we were unable to perform a finer mapping of the interaction domain for the ORF2 protein.
A 76-nt conserved domain from the 5′ HEV genomic RNA interacts with the ORF2 protein.
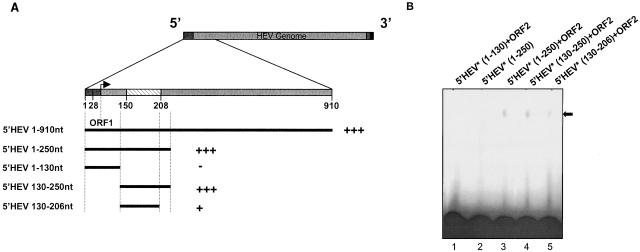
The 5′ HEV RNA (nt 1 to 910) was subcloned into smaller fragments. 5′ HEV RNA (nt 1 to 250), 5′ HEV RNA (nt 1 to 130), 5′ HEV RNA (nt 130 to 250), and 5′ HEV RNA (nt 130 to 206) were deletions of the full-length 5′ HEV RNA (nt 1 to 910) as described in Table 1. Each of these deletions was individually tested for interaction with the AD-ORF2 protein in the yeast three-hybrid assay plus EMSA. Interestingly, the 5′ HEV RNA (nt 1 to 250) transcript showed a positive interaction, as had been observed with our EMSA results described in the previous experiments. Subsequently, the two deletion transcripts, 5′ HEV RNA (nt 1 to 130) and 5′ HEV RNA (nt 130 to 250), which split the region from nt 1 to 250, were tested for interaction with ORF2. From this pair, the 5′ HEV RNA (nt 130 to 250) transcript interacted with ORF2 (Fig. 6A). The 5′ HEV RNA (nt 130 to 250) region, consisting of 120 bases, showed a considerably stronger interaction with the ORF2 protein compared with that of the 5′ HEV RNA (nt 130 to 206) region, consisting of only 76 nucleotides (Fig. 6B). Hence, the nt 130 to 206 (76 nt) of the HEV genome may contain the major interaction domain required for binding to ORF2; however, genomic sequences 44 nt downstream of this region contribute significantly to an increase in the strength of this RNA-protein interaction significantly.
FIG. 6.
Mapping the interaction domain for the 5′ HEV RNA. (A) Mapping of the interaction domain of the 5′ HEV RNA (nt 1 to 910) region. The hatched box represents the alphavirus consensus sequence. Plus signs shows a summarized result of the yeast three-hybrid interactions. (B) EMSA for the different RNA deletions from the 5′ HEV genome. Asterisks refer to 32P-labeled transcript.
DISCUSSION
We have used the yeast three-hybrid system to show that the ORF2 protein of HEV is capable of binding its viral genome at the 5′ end. The ORF3 protein does not possess this activity. Also, the ORF2-RNA interaction is specific to the 5′ end of the HEV genome and not to the 3′ end or any other region tested. Finally, we have mapped the ORF2 protein binding region on the RNA genome to a 76-nt region [5′ HEV RNA (nt 130 to 206)]. This 76-nt region of the viral genome is capable of binding the full-length ORF2 protein and its N-terminal deletion from nt 1 to 111 as well. Our previous observations of the dimerization of ORF2 (33) and its heterotypic interaction with ORF3 (34) have shown similar results, suggesting that the ORF2 protein loses its activity when truncated beyond amino acid 111 from the N terminus.
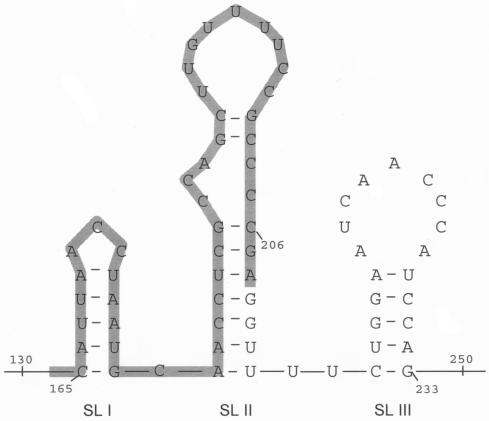
We used the mfold program to predict the RNA secondary structure for the HEV genomic region from nt 130 to 250 (which includes the 76-nt region plus the downstream 44 nt), based on minimum free energy calculations (Fig. 7). Interestingly enough, the 76-nt region, which we have shown to be responsible for binding the ORF2 protein, completely encompasses the HEV homologue of the alphavirus consensus 51-nt sequence (bases 150 to 208) (18, 20). This 51-nt conserved region is highlighted and is part of two stem-loop structures (SLI and SLII), similar to other alphaviruses such as Sindbis virus (18), Highlands J virus (28), and Semliki Forest virus (28). Looking at the secondary structures, it becomes obvious that sequences downstream of base 208 contribute to the strength of SLII. Our experimental data on the HEV RNA-ORF2 protein interaction fall in line with the in silico secondary-structure prediction, suggesting that sequences downstream of base 208 contribute to increased strength of SLII, thus strengthening the RNA-protein interaction. The 4-nt region (nt 209 to 212 in SLII) may not be essential for the RNA-protein interaction but may contribute to increasing the binding strength of the RNA-protein interaction in question. The 5′ HEV region from nt 130 to 250 also forms a third stem-loop structure (SLIII). Although not essential, this stem-loop structure may contribute significantly toward increasing the overall strength of the HEV genomic RNA-ORF2 protein interaction.
FIG. 7.
Secondary-structure prediction of the 5′ HEV genomic region from nt 130 to 250. Numbers correspond to the numbers on the HEV genome. SL I, SL II, and SL III represent the three stem-loop structures shown in the figure. The highlighted region represents the 51-nt conserved region from alphaviruses.
Since HEV is a plus-strand RNA virus, one of its structural proteins is required to show RNA binding activity for two essential viral functions - viral replication and packaging of its genome into the capsid during viral assembly. Since ORF2 is the major capsid protein, it would be the most likely candidate to bind the genomic RNA for viral packaging. We have experimentally shown that the ORF2 protein binds to the 5′-terminal region of the viral genome, thus becoming the most likely candidate to perform this biological function.
HEV is postulated to form subgenomic RNA transcripts (∼3.7 and ∼2 kb) from the 3′ (structural) region of the genome (16). Hence, it seems like a good strategic option for the virus to have its RNA encapsidation signal at the 5′ end of the genome. This will result in only the full-length genomic RNA (∼7.2 kb) being differentially recognized by the capsid protein ORF2 for headfull packaging during viral assembly in the hepatocyte.
Although data presented in this publication point to a fundamental viral process, i.e., genome encapsidation, and strongly indicates the possibility that the ORF2 protein may be responsible for bringing the genomic RNA into the capsid during assembly, direct biological evidence is difficult to obtain due to the absence of an in vitro culture system for HEV. Indirect approaches using mutational knockout of the identified interaction domain and restoration by complementary mutations are under investigation.
Acknowledgments
Technical help from Shweta Tyagi is gratefully acknowledged.
M.S. is a CSIR Junior Research Fellow and S.J. is an International Senior Research Fellow in Biomedical Sciences of the Wellcome Trust. Support for this study was provided through internal funds from the ICGEB and partially through a grant to S.J. from the Welcome Trust.
REFERENCES
- 1.Aye, T. T., T. Uchida, X. Z. Ma, F. Iida, T. Shikata, H. Zhuang, and K. M. Win. 1992. Complete nucleotide sequence of a hepatitis E virus isolated from the Xinjiang epidemic (1986-1988) of China. Nucleic Acids Res. 20:3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berke, T., and D. O. Matson. 2000. Reclassification of the Calciviridae into distinct genera and exclusion of hepatitis E virus from the family on the basis of comparative phylogenetic analysis. Arch. Virol. 145:1421-1436. [DOI] [PubMed] [Google Scholar]
- 3.Bradley, D. W. 1990. Enterically-transmitted non-A, non-B hepatitis. Br. Med. Bull. 46:442-461. [DOI] [PubMed] [Google Scholar]
- 4.Bradley, D. W., and M. A. Purdy. 1994. Molecular and serological characteristics of hepatitis E virus, p 42-45. In K. Nishioka, H. Suzuki, S. Mishiro, et al. (ed.) Viral hepatitis and liver disease. Springer-Verlag, Tokyo, Japan.
- 5.Guarente, L. 1983. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101:181-191. [DOI] [PubMed] [Google Scholar]
- 6.He, J., A. W. Tam, P. O. Yarbough, G. R. Reyes, and M. Carl. 1993. Expression and diagnostic utility of hepatitis E virus putative structural proteins expressed in insect cells. J. Clin. Microbiol. 31:2167-2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang, C., D. Nguyen, J. Fernandez, K. Y. Yun, K. E. Fry, D. W. Bradley, A. W. Tam, and G. R. Reyes. 1992. Molecular cloning and sequencing of the Mexico isolate of hepatitis E virus (HEV). Virology 191:550-558. [DOI] [PubMed] [Google Scholar]
- 8.Jameel, S., M. Zafrullah, M. H. Ozdener, and S. K. Panda. 1996. Expression in animal cells and characterization of the hepatitis E virus structural proteins. J. Virol. 70:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khudyakov, Y. E., M. O. Favorov, D. L. Jue, T. K. Hine, and H. A. Fields. 1994. Immunodominant antigenic regions in a structural protein of the hepatitis E virus. Virology 198:390-393. [DOI] [PubMed] [Google Scholar]
- 10.Khudyakov, Y. E, N. S. Khudyakov, H. A. Fields, D. Jue, C. Starling, M. O. Favorov, K. Krawczynski, L. Polish, E. Mast, and H. Margolish. 1993. Epitope mapping in proteins of hepatitis E virus. Virology 194:89-96. [DOI] [PubMed] [Google Scholar]
- 11.Khuroo, M. S. 1980. Study of an epidemic of non-A, non-B hepatitis: possibility of another human hepatitis virus distinct from post-transfusion non-A, non-B type. Am. J. Med. 68:818-823. [DOI] [PubMed] [Google Scholar]
- 12.Khuroo, M. S., M. R. Teli, S. Skidmore, M. A. Sofi, and M. Khuroo. 1981. Incidence and severity of viral hepatitis in pregnancy. Am. J. Med. 70:252-255. [DOI] [PubMed] [Google Scholar]
- 13.Koonin, E. V., A. E. Gorbalenya, M. A. Purdy, M. N. Rozanov, G. R. Reyes, and D. W. Bradley. 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-stranded RNA plant and animal viruses. Proc. Natl. Acad. Sci. USA 89:8259-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korkaya, H., S. Jameel, D. Gupta, S. Tyagi, R. Kumar, M. Zafrullah, M. Mazumdar, S. K. Lal, L. Xiaofang, D. Sehgal, S. R. Das, and D. Sahal. 2001. The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J. Biol. Chem. 276:42389-42400. [DOI] [PubMed] [Google Scholar]
- 15.Lal, S. K., P. Tulasiram, and S. Jameel. 1997. Expression and characterization of the hepatitis E virus ORF3 protein in the methylotrophic yeast, Pichia pastoris. Gene 190:63-67. [DOI] [PubMed] [Google Scholar]
- 16.Li, F., H. Zhuang, S. Kolivas, S. A. Locarnini, and D. A. Anderson. 1994. Persistent and transient antibody responses to hepatitis E virus detected by Western immunoblot using open reading frame 2 and 3 and glutathione S-transferase fusion proteins. J. Clin. Microbiol. 32:2060-2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin, F., A. Schaller, S. Eglite, D. Schumperli, and B. Muller. 1997. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 16:769-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niesters, H. G. M., and J. H. Strauss. 1990. Mutagenesis of the conserved 51-nucleotide region of Sindbis virus. J. Virol. 64:1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panda, S. K., I. H. Ansari, H. Durgapal, S. Agrawal, and S. Jameel. 2000. The in vitro-synthesized RNA from a cDNA clone of hepatitis E virus is infectious. J. Virol. 74:2430-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panda, S. K., and S. Jameel. 1997. Hepatitis E virus: from epidemiology to molecular biology. Viral Hepatitis Rev. 3:227-251. [Google Scholar]
- 21.Purcell, R. H. 2001. Hepatitis E virus, p. 3051-3061. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippinscott-Raven, Philadelphia, Pa.
- 22.Purcell, R. H., and J. R. Ticehurst. 1988. Enterically transmitted non-A, non-B hepatitis: epidemiology and clinical characteristics, p. 131-137. In A. J. Zuckerman (ed.), Viral hepatitis and liver disease. Alan R. Liss, Inc., New York, N.Y.
- 23.Purdy, M. A., A. W. Tam, C. C. Huang, P. O. Yarbough, and G. R. Reyes. 1993. Hepatitis E virus: a non-enveloped member of the “alpha-like” RNA virus supergroup? Semin. Virol. 4:319-326. [Google Scholar]
- 24.Reyes, G. R., C. C. Huang, A. W. Tam, and M. A. Purdy. 1993. Molecular organization and replication of hepatitis E virus (HEV). Arch. Virol. 7:15-25. [DOI] [PubMed] [Google Scholar]
- 25.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Sengupta, D. J., B. Zhang, B. Kraemer, P. Pochart, S. Fields, and M. Wickens. 1996. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl. Acad. Sci. USA 93:8496-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava, R., and S. K. Lal. 2002. A liquid synchronized-growth culture assay for the identification of true positive and negative yeast three-hybrid transformants. Lett. Appl. Microbiol. 34:300-303. [DOI] [PubMed] [Google Scholar]
- 28.Strauss E. G., and J. H. Strauss. 1986. Structure and replication of the alphavirus genome, p. 35-90, In S. Schlesinger and M. J. Schlesinger (ed.), The Togaviridae and Flaviviridae. Plenum Publishing Corp., New York, N.Y.
- 29.Tam, A. W., M. M. Smith, M. E. Guerra, C. C. Huang, D. W. Bradley, K. E. Fry, and G. R. Reyes. 1991. Hepatits E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185:120-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torresi, J., F. Li, S. A. Locamini, and D. A. Anderson. 1999. Only the non-glycosylated fraction of hepatitis E virus capsid (open reading frame 2) protein is stable in mammalian cells. J. Gen. Virol. 80:1185-1188. [DOI] [PubMed] [Google Scholar]
- 31.Tsarev, S. A., S. U. Emerson, G. R. Reyes, T. S. Tsareva, L. J. Legters, I. A. Malik, M. Iqbal, and R. H. Purcell. 1992. Characterization of a prototype strain of hepatitis E virus. Proc. Natl. Acad. Sci. USA 89:559-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tyagi, S., S. Jameel, and S. K. Lal. 2001. Self-association and mapping of the interaction domain of hepatitis E virus ORF3 protein. J. Virol. 75:2493-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tyagi, S., S. Jameel, and S. K. Lal. 2001. A yeast two-hybrid study on self-association of the ORF2 protein of hepatitis E virus. Biochem. Biophys. Res. Commun. 284:614-621. [DOI] [PubMed] [Google Scholar]
- 34.Tyagi, S., H. Korkaya, M. Zafrullah, S. Jameel, and S. K. Lal. 2002. The phosphorylated form of the ORF3 protein of hepatitis E virus interacts with its non-glycosylated form of the major capsid protein, ORF2. J. Biol. Chem. 277:22759-22767. [DOI] [PubMed] [Google Scholar]
- 35.Tyagi, S., and S. K. Lal. 2000. Combined transformation and genetic technique verification of protein-protein interactions in the yeast two-hybrid system. Biochem. Biophys. Res. Commun. 277:589-593. [DOI] [PubMed] [Google Scholar]
- 36.Tyagi, S., J. P. Salier and S. K. Lal. 2002. The liver-specific human alpha(1)-microglobulin/bikunin precursor (AMBP) is capable of self-association. Arch. Biochem. Biophys. 399:66-72. [DOI] [PubMed] [Google Scholar]
- 37.Zafrullah, M., M. H. Ozdener, S. K. Panda, and S. Jameel. 1997. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J. Virol. 71:9045-9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, B., M. Gallegos, A. Puoti, E. Durkin, S. Fields, J. Kimble, and M. P. Wickens. 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390:477-484. [DOI] [PubMed] [Google Scholar]