Abstract
Gene therapy is a potentially powerful treatment approach that targets molecular remedies for disease. Among other challenges it remains difficult to monitor gene delivery and its downstream metabolic consequences. Approaches to MRI gene reporters have been reported but few have the potential for translation beyond isolated cell systems. Herein, we report a polycationic polymer MRI contrast agent that binds to DNA in a ratio of one monomer unit per phosphate group of DNA. Significantly, this binding event diminishes the MR contrast signal from the agent itself potentially providing a platform for imaging delivery and release of a gene into cells and tissues. Importantly, we demonstrate here the proof of concept that a positively charged polymeric contrast agent can also act as a transfection agent, delivering the gene for encoding green fluorescent protein into cells. These observations provide support for the radical, new idea of creating a combined transfection/imaging agent for monitoring gene delivery in real time by MRI.
Introduction
Gene therapy, selectively correcting the genetic code of diseased cells, offers the prospect of treating many ailments at the molecular level.1 However, introducing new genetic code into cells is not a trivial exercise. The polynucleic acids in which genes are contained consist of long, negatively charged polymers and strong Coulombic repulsions with highly negatively charged cell membranes provides a natural barrier for gene delivery. To overcome this barrier transfection agents are required to deliver genetic code across these membranes.1 Successful gene therapy also requires the subsequent delivery of the new genetic code into the cell nucleus where genetic expression takes place. Viruses, which possess an innate ability to introduce foreign genetic code into cells, have been adapted for gene therapy with some success.1–3 Viral vectors have limitations however, particularly with respect to their propensity to stimulate an immune response.4 An alternative approach frequently considered for gene therapy is to bundle the anionic nucleic acids with cationic polymers, thereby neutralizing the charge and allowing passage of the polynucleic acid into cells.1
Many cationic polymers have been investigated for their potential utility to reliably transfect cells with non-native DNA.1 Although the development of gene therapy in clinical practice has stalled at the clinical trail stage, recent developments have shown extremely promising results: for example curing adenosine deaminase (ADA) deficiency,2,3 or restoring the sight of those with Leber's congenital amaurosis.5 However, one limitation of current gene therapy technology, whether viral or polymer based, is that it is difficult to track its progress in vivo. There is considerable interest in being able to track gene therapy on two levels: first, by monitoring the distribution of the new genetic code throughout all tissue and second, assessing the outcome of gene transfection. This has stimulated modification of gene therapy agents to include reporters for detection by imaging modalities such as MRI.6–8 For practical applications it is often only possible to measure the success of gene therapy by waiting to see if the new genetic code is expressed and to measure the biological effects of that expression. Clearly, a method by which the success of gene therapy can be measured immediately and non-invasively would be a valuable addition to the gene therapy tool kit.
MRI is a technique by which images of soft tissue can be generated in a safe and non-invasive manner. As such, it is the ideal technique for consideration as a method by which gene therapy might be monitored. Recent reports have suggested that the cationic polymers used to bundle nucleic acids,9,10 and even the nucleic acids11 themselves could function as markers for gene therapy in MRI scans. These reports were based upon the idea that it is possible to apply a frequency selective, low energy pulse to protons of the polymer or nucleic acid that are in exchange with those of the bulk water of tissue. It is these bulk water protons that are monitored by MRI and by applying a frequency selective pulse we ‘saturate’, or wipe out the signal, from these exchangeable protons. When these protons move into the bulk water, the signal intensity of bulk water is reduced, generating image contrast in an MRI through a chemical exchange saturation transfer (CEST) mechanism.12,13 Although this approach allows image contrast to be generated specifically from the nucleic acid or polymer with which it was bundled, it cannot provide a means of tracking the effectiveness of gene therapy. The exchangeable protons used to probe the nucleic acid and polymer are ubiquitous in vivo and no means exist to discriminate contrast arising from the gene therapy agent and that arising from the background biological medium. Furthermore, this approach would not afford any change in contrast intensity when transfection has occurred successfully.
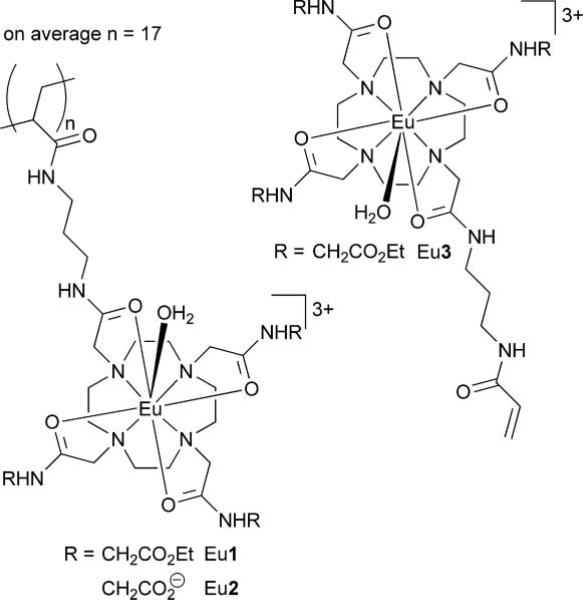
These reports suggesting that gene therapy could be imaged by MRI did, however, prompt us to consider a polymeric MRI contrast agents (Chart 1) developed recently in our lab.14 Although these polymeric contrast agents were originally conceived as a method of increasing agent delivery in targeted imaging applications, one of these polymeric agents is highly positively charged—with an average of 51 positive charges spread over an average of 17 monomer units per polymer—and thus, to our mind, potentially had the necessary attributes to facilitate gene therapy tracking by MRI. We had previously demonstrated14 that these polymeric agents could be used to generate image contrast by MRI using the same CEST mechanism used by van Zijl and coworkers.9–11 However, in this case the active component of the agent, from an MRI perspective, was a chelate of the paramagnetic ion Eu3+. Among other advantages,12 the paramagnetic CEST (PARACEST) approach allows the specific detection of the cationic polymer in biological media because activation of the agent requires that the frequency selective pre-saturation pulse be applied at ~50 ppm, far away from the resonance frequency of any exchangeable endogenous protons.
Chart 1.
Results
Cationic PARACEST polymers bind effectively to DNA
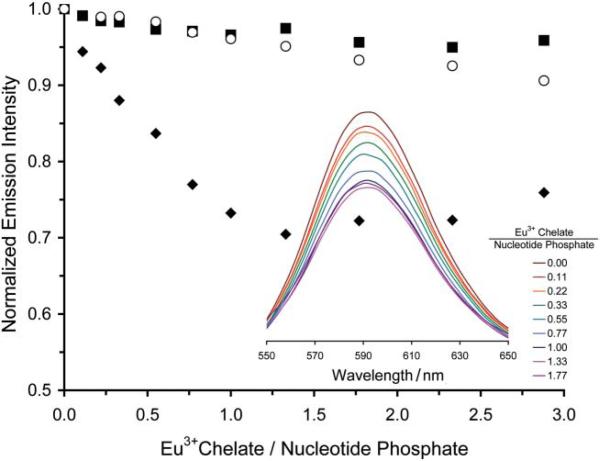
The first stage in the evaluation of the positively charged PARACEST polymer was to assess its ability to bind to DNA, without which it would most certainly be unable to mediate the transfection of cells. The bundling of DNA by cationic polymers is commonly investigated by monitoring the displacement of ethidium bromide, a DNA interchelator, that occurs when bundling polymers bind to DNA.15 This convenient method can be followed by examining the fluorescent emission of the ethidium bromide, which is strongly emissive when interchelated in DNA but only weakly emissive once displaced by binding of DNA with a cationic polymer.15 The cationic polymer Eu1, when titrated into a sample of salmon testes DNA interchelated with ethidium bromide, clearly displaced ethidium bromide as evidenced by the quenching of ethidium bromide emission at 590 nm (Fig. 1). Control experiments using the neutral polymer, Eu2 (obtained by hydrolysis of the ethyl esters of Eu1), or the monomer, Eu33+, did not displace ethidium bromide significantly. This indicates that, as expected, only the poly-cationic polymeric agent, Eu1, is capable of DNA binding. The stoichiometry of the binding event between DNA and Eu1 was determined by measuring the absorbance of a sample of DNA from salmon testes at 260 nm, which enabled the nucleotide-phosphate concentration to be calculated from previously described methods.16 This procedure demonstrated that DNA bundles with the cationic polymer Eu1 such that there is one Eu3+ monomer residue of polymer present per nucleotide phosphate group of DNA (Fig. 1).
Fig. 1.
The addition of Eu1 to a solution of DNA from salmon testes (0.6 μM) interchelated with ethidium bromide results in a decrease in emission intensity of ethidium bromide. This decrease in emission intensity indicates binding of Eu1 to DNA (◆), neither Eu2 (■) nor Eu3 (○)were found to bind to DNA. The inset shows the fluorescence spectrum (λex = 525 nm) of ethidium bromide as Eu1 is added to the same solution.
Binding to DNA suppresses CEST contrast from polymeric contrast agents
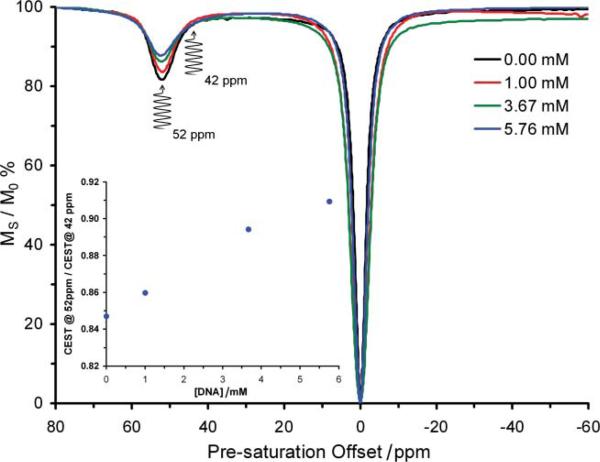
The contrast enhancing properties of Eu3+-based PARACEST agents are extremely sensitive to the local environment of the Eu3+ ion.12,13,17 We reasoned that contrast enhancing properties of Eu1 would be altered upon binding to DNA since this would almost certainly lead to changes in the local environment of Eu3+. To test this hypothesis, DNA from salmon testes was titrated into a 5 mM solution of Eu1 in PBS at pH 6.96. After each addition, the contrast agent was assessed by recording its CEST spectrum (Fig. 2). In this spectrum, the signal intensity of the bulk water signal was measured as the pre-saturation frequency was varied across the spectrum. In this way, it is possible to establish at which pre-saturation frequencies contrast enhancement occurs and the extent of that contrast enhancement. The pre-saturation frequency at which contrast enhancement is obtained from Eu1 was found to be unaffected by binding to DNA (Fig. 2); pre-saturation at +52 ppm continues to afford the greatest contrast enhancing effect from the polymer. Perhaps more significantly however, DNA binding markedly diminishes the extent to which this polymer is able to generate contrast; the contrast enhancing ability of Eu1 is found to diminish with each successive addition of DNA to the solution. As anticipated, the contrast enhancing abilities of the neutral polymer Eu2 and the monomer Eu3, which do not bind to DNA, were unaffected by the addition of DNA (ESI Fig. S1†). Even when the concentrations of Eu3+ in the polymer and the nucleotide-phosphate of DNA are almost equal (5 mM Eu3+ and 5.76 mM phosphate), the polymer Eu1 is still able to generate image contrast after pre-saturation at +52 ppm. Thus, the distribution of a gene therapy agent based on Eu1, post in vivo administration, could potentially be monitored using MRI and pre-saturation at +52 ppm. One further aspect of gene therapy could potentially be monitored by MRI using polymeric contrast agents, such as Eu1, as transfection agents. On the basis that the genetic machinery of a cell cannot read genetic code stored within a polynucleic acid introduced by gene therapy until the nucleotide has been released by the transfection agent, the change in CEST from Eu1 observed upon DNA binding may provide an entry into methods by which the expression of the introduced genetic code itself may be monitored. As the genetic machinery sets to work expressing the introduced genetic code Eu1 must come unbound from the nucleic acid, altering the magnitude of the CEST effect arising from Eu1 (Fig. 2). Unlike conventional contrast enhancing systems that would require knowledge of the agent concentration to report information such as changes in agent binding, it should be possible to relate these changes in CEST magnitude to changes in polymer-nucleic acid binding by employing a previously proposed ratiometric detection method.13,18
Fig. 2.
The CEST spectra of a 5 mM solution of Eu1 in 10 mM PBS (pH 6.96) recorded at 500 MHz and 298 K using a 600 Hz, 4 s pre-saturation pulse. The spectra are recorded at differing DNA concentrations which clearly show the CEST suppressing effect of DNA binding. The inset shows the ratio of CEST obtained by pre-saturation at +52 ppm over CEST obtained by pre-saturation at +42 ppm as a function of added DNA.
Careful examination of the CEST spectra in Fig. 2 reveals that although there is a sizable change in CEST as Eu1 binds to DNA when the pre-saturation pulse is applied at +52 ppm, there is little to no change in CEST upon binding to DNA when the pre-saturation pulse is applied at +42 ppm. That means that by taking the ratio of the CEST (contrast) generated after pre-saturation at +52 ppm and pre-saturation at +42 ppm in an MR image it would be possible to determine the amount of Eu1 that had become unbound from DNA (Fig. 2 inset). The expression of the new genetic code can only occur if the nucleic acid is no longer bundled with the polymer. Even though for successful gene therapy several factors are important, such as uptake of plasmid to the nucleus, an MR scan that indicates release of the polymer may provide an ouvre into an effective means of monitoring the success of gene therapy by imaging.
Polymeric contrast agents can mediate transfection
Binding to DNA and generating image contrast in MR images are just initial prerequisites for monitoring the progress of gene therapy by MRI. Of critical importance is that the polymeric contrast agents are themselves able to transfect cells with new genetic code. Transfecting cells with a plasmid DNA that encodes for green fluorescent protein (GFP) is a convenient method of assessing the viability of transfection agents for gene therapy. In this experiment cells that are successfully transfected with a GFP plasmid under the constitutive EF1-α promoter will begin to produce GFP and become fluorescence positive, whereas those that have not been transfected will remain negative. Samples of plasmid DNA encoding for GFP were mixed with varying amounts of Eu1 for 20 min before being introduced to HEK293 cells. Initial experiments were performed using 1.4 μg of the plasmid DNA, conditions which are known to induce a high level of transfection when lipofectamine is employed as the transfection agent (ESI Fig. S2†), varying the amount of potential transfection agent employed. Cells were then examined by fluorescence microscopy and flow cytometry to assess the extent to which the cells underwent transfection. Under these conditions it was found that cells could be transfected with foreign genetic code using Eu1 (ESI Fig. S2†). Although the levels of transfection were low, relative to the levels achieved using lipofectamine (a maximum transfection efficiency of 0.83% was obtained versus 100% for lipofectamine), the results were significantly better than those obtained with either the neutral polymer Eu2 or monomer Eu33+ for which no transfection was observed (Table 1). Varying the amount of plasmid DNA employed while holding the amount of Eu1 constant afforded comparable results (Fig. 3 and Table 2). The success, or otherwise, of transfection attempts using Eu1 was found to be highly dependent upon ratio of polymer to DNA; transfection was only observed when the Eu3+:nucleotide-phosphate ratio was between 20 and 150 (Tables 1 and 2, ESI Fig. S3†). It has been reported that chloroquine can assist transfection by promoting the release of material from endosomes.19,20 Accordingly, transfection studies were repeated in the presence of increasing concentrations of chloroquine. However, flow-cytometry data showed that neither the extent of transfection nor the level of gene expression per cell was improved by the presence of chloroquine (ESI Figs. S4 and S5†). This may indicate that bundles of plasmid DNA with Eu1 do not become entrapped in endosomes. Furthermore, the introduction of chloroquine did not result in transfection when using Eu2 or Eu33+.
Table 1.
The number of transfected HEK293 cells in each field of view of the fluorescence microscope (at 10× magnification) observed when exposed to 1.4 μg of DNA in the presence of differing amounts of each potential transfection agent
| μmol of Eu3+ | Eu1 | Eu2 | Eu33+ |
|---|---|---|---|
| 0.03 | Negative | Negative | Negative |
| 0.15 | Negative | Negative | Negative |
| 0.30 | 1–2 cells | Negative | Negative |
| 0.60 | 5–10 cells | Negative | Negative |
| 1.50 | Negative | Negative | Negative |
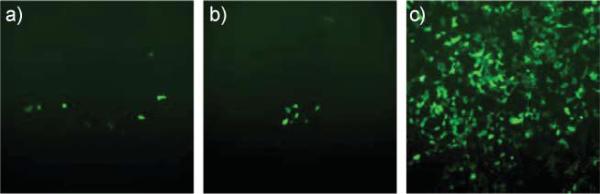
Fig. 3.
Images demonstrating the successful transfection of HEK293 cells with GFP using 0.6 mmol of Eu1 and a) 5 μg and b) 10 μg of the plasmid DNA under a fluorescence microscope at 10× magnification; c) shows the result of a control experiment using lipofectamine and 4.0 μg of DNA.
Table 2.
The number of transfected HEK293 cells in each field of view of the fluorescence microscope (at 10× magnification) observed when exposed polymer (0.6 μmol of Eu3+) in the presence of differing amounts of DNA
| μg of DNA | Eu1 | Eu2 |
|---|---|---|
| 0.0 | Negative | Negative |
| 0.5 | Negative | Negative |
| 1.0 | Very few | Negative |
| 5.0 | 10 cells | Negative |
| 10.0 | 15 cells | Negative |
One further design consideration for a gene therapy agent is whether and how fast the transfection agent is eliminated from intracellular space. Accordingly, cells were incubated with Eu1 and plasmid DNA for 4 h using the most successful ratios determined herein. After 4 h fresh cell growth medium was introduced and the cells incubated further. After washing with PBS to eliminate additional extracellular Eu1 the cells and their nuclei were lysed and the intracellular Eu3+ content determined by ICP-MS. These experiments showed that Eu1 leaves the cells fairly rapidly post-transfection (ESI Fig. S5†). Elimination from the cell interior is not a simple exponential clearance function and it appears that a certain component of Eu1, possibly the intra-nuclear component, is retained much longer.
Discussion
The polymeric cationic contrast agent Eu1 demonstrates the proof of principle that it should be possible to design MRI contrast agents that can bind to and transfect cells with nucleic acids, reporting both the location and expression of this new genetic code in MR images. Although the cationic polymer Eu1 is not an effective enough transfection agent itself to be taken forward as a potential gene therapy agent, it does—unlike the discrete monomeric agent Eu33+ and the neutral polymer Eu2—bind DNA such that ethidium bromide is completely displaced from the double helix at a binding ratio of one Eu3+ chelate per nucleotide phosphonate. However, considerably higher ratios of Eu3+ to nucleotide phosphate (20–150) are required to effect the transfection of HEK293 cells suggesting that, in this case at least, it is necessary to have many more than four positive charges per base pair before transfection can occur. It is worthy of note that if the Eu3+:nucleotide-phosphate ratio becomes too high then transfection ceases to occur, presumably this is the result of the bundle breaching the size and charge limitations that are known to determine endocytosis of the DNA-polymer bundle in these types of systems.21
The polymeric contrast agent Eu1 is capable of generating image contrast even when bound to DNA, thereby potential providing a means to track the physical location of a gene therapy agent. However, DNA binding alters the magnitude of image contrast generated by Eu1 and applying a suitable ratiometric detection method, vide infra, may provide an MRI method of detecting the unbinding of Eu1 from the delivered nucleic acid. Since unbinding is a prerequisite for gene expression, this may provide an entry into imaging the expression of genetic code delivered by gene therapy. This report demonstrates only a possible future direction of research into both gene therapy and MRI contrast agents. Clearly, for such the approach proposed herein to be viable then both the CEST and transfection efficiency of the cationic polymeric agent would have to be improved such that lower Eu3+ to nucleotide phosphate effective ratios and detection limits were achieved. At current efficiency levels the quantities of Eu3+ present in cells post-transfection with Eu1 are too low to be detectable by CEST. A further improvement in the ability of Eu1 to mediate transfection would also be required to ensure sufficient genetic delivery.
Despite these obvious limitations of Eu1 it amply proves the principle that a polymeric contrast agent itself could be used to mediate and report on transfection. Such an approach may eliminate the need to modify a successful structure to incorporate an imaging agent, with the potential risk of transfection capability loss that is associated with established labeling techniques. The structure of Eu1 may be varied almost infinitely; for example, the three amide substituents may be independently varied allowing control over the hydrophilicity and charge of each monomer residue, co-polymers can be introduced to alter the spacing of each chelate monomer,22 and the length of the polymer can be altered by changing the amount of initiator employed during synthesis.14 With such variability on offer, it is reasonable to believe that a polymer, similar to Eu1, could be developed that will bundle nucleic acids and effectively transfect cells with the genetic code bundled therein. The CEST that arises from this nucleic acid/contrast agent bundle can be used to track the location of the gene therapy agent once it has been administered. Finally the increase in CEST that is observed as the polymer is forced to unbind from the nucleic acid during gene transcription could be used to monitor the progress and overall success of the gene therapy. From this work one could envision that polymeric PARACEST contrast agents may offer an all-in-one solution to the problem of tracking gene therapy in vivo.
Experimental
Binding studies of polymeric contrast agents with DNA
Stock solutions of DNA (nucleotide phosphate concentration, 1.24 mM) (from salmon testes Sigma-Aldrich), ethidium bromide (0.76 mM) and polymer contrast agent (0.60 mM) where prepared in 10 mM PBS (pH 7.03). The nucleotide phosphate concentration of the DNA stock solution was determined from absorption measurements at 260 nm, using an extinction coefficient of 6600 M-1cm-1,23 as previously described.16 11 samples were then prepared in which 9 μL of the DNA solution and 3 μL of ethidium bromide and 1.8 mL of PBS buffer were incubated at 298 K for 30 min. 0, 2, 4, 6 10, 14, 20, 26, 34, 44 or 54 μL of the polymer stock solution was added to each sample and the volume made up to 1.866 mL by addition of water. The emission spectrum of each sample was the recorded on a Perkin Elmer LS50B flourimeter, exciting at 525 nm and using 15 nm excitation and emission slits.
Determining the effect of DNA-binding upon CEST
A stock solution of DNA (nucleotide phosphate concentration = 20 mM), which also contained polymer at a concentration of 5 mM, was prepared. 400 μL of a 5 mM polymer solution in 10 mM PBS (pH 6.96) was placed in an NMR tube and the CEST spectrum recorded at 500 MHz and 298 K by measuring the intensity of the solvent water peak after the application of a 600 Hz pre-saturation pulse for 4 s. The spectrum was generated by altering the frequency of the pre-saturation pulse from +80 to -60 ppm in 1 ppm intervals. After the acquisition of each CEST spectrum 35 μL of the DNA stock solution was added to the NMR sample and another CEST spectrum acquired.
Transfection studies
Plasmid DNA
The plasmid vector pQ100 containing an EF-1α/eGFP expression cassette was a gift from Dr J. Vieira, University of Washington. pQ100 was prepared from E. coli using the Qiagen Maxi-prep kit, as per manufacturer's instructions, and the plasmid DNA was quantified using a Nanodrop spectrophotometer.
Transfection
For transfection studies, HEK293 cells maintained in Dulbecco's modified Eagle's Medium (DMEM) supplemented with 10% fetal calf serum (FCS) and penicillin-streptomycin-l-glutamine (PSG) were split one day before transfection and plated into 6-well plates (Costar) at a density of 5 × 105 cells per well. Prior to transfection, the culture medium was removed and replaced with transfection medium. The transfection medium was produced by diluting DNA in 250 μL of OPTI-MEM and then added to a varied amount of the agent under investigation: Eu1, Eu2 or Eu33+. The solutions were vortexed and allowed to incubate for 20 min at room temperature. Transfection using Lipofectamine 2000 was included as a positive control in each case. As per manufacturers suggestion 5 μL of Lipofectamine 2000 was diluted in 250 μL of OPTI-MEM plus 1.4 or 4 μg of pQ100. Incubation of 10 μg of naked DNA diluted in Opti-MEM was included as a negative control. The whole portion of each transfection mix was added to individual wells of cells for 4 hours and then complete DMEM medium was added and the cultures incubated for an additional 48 h. For experiments involving chloroquine, DNA complexes were formed in the same manner as described above, but mixed with chloroquine (0.5, 1, or 5 μM) prior to transfer to the cell medium. The cells expressing green fluorescence protein (GFP) were visualized using a Zeiss Axiovert 40 L inverted fluorescence microscope and pictures were captured using a digital camera.
Flow Cytometry
HEK293 cells were removed from the 6-well dishes and spun at 2,000 RPM for 5 min to pellet. The pelleted cells were washed in PBS, resuspended in 500 μL PBS and analyzed for GFP on a FACS Calibur (Becton Dickinson) utilizing Cell Quest software. Final data analysis was performed utilizing FlowJo software (Tree Star, Inc.).
Polymer elimination experiments
HEK 293 cells were incubated with Eu1 (0.6 μM of Eu3+) and plasmid DNA (1.4 μg) for 4 hours after which the cell growth media was replaced with fresh DMEM media. Cells were incubated for a further period of time (0, 0.5, 1, 2, 4, 8, 24 and 75 h) before washingtwice with PBS. The cells were then lysed in 0.5mL/well of lysis buffer containing: 50 mM Tris pH 7.4, 150 mM NaCl, 5 mM MgCl2 0.5% NP-40, 1% SDS and 1 mM PMSF. The resulting samples were made up to 3 mL in 20% HNO3 and then analyzed for Eu3+ content by ICP-MS.
Supplementary Material
Acknowledgements
The authors thank the National Institutes of Health EB-11687 (MW); RR-02584 and CA-115531 (ADS); HL083194 (DNS), Oregon Nanoscience and Microtechnologies Institute, Portland State University, the Oregon Opportunity partnership for advancing biomedical research, the M.J. Murdock Charitable Trust and the Robert A Welch Foundation (AT-584) for financial support. Also, Dr Damon Sutton (UT Dallas) for helpful discussions.
Footnotes
Electronic supplementary information (ESI) available: Supplementary figures.
Notes and References
- 1.Templeton NS. Gene and Cell Therapy; Therapeutic Mechanisms and Strategies. Third Ed. CRC Press; 2009. [Google Scholar]
- 2.Blaese RM, Culver KW, Miller AD, Carter CS, Fleisher T, Clerici M, Shearer G, Chang L, Chiang Y, Tolstoshev P, Greenblatt JJ, Rosenberg SA, Klein H, Berger M, Mullen CA, Ramsey WJ, Muul L, Morgan RA, Anderson WF. Science. 1995;270:475. doi: 10.1126/science.270.5235.475. [DOI] [PubMed] [Google Scholar]
- 3.Kotani H, Newton PB, 3rd, Zhang S, Chiang YL, Otto E, Weaver L, Blaese RM, Anderson WF, McGarrity GJ. Hum. Gene Ther. 1994;5:19. doi: 10.1089/hum.1994.5.1-19. [DOI] [PubMed] [Google Scholar]
- 4.Stolberg Sheryl G. N Y Times Mag. 1999:136. [PubMed] [Google Scholar]
- 5.Bainbridge JWB, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, Viswanathan A, Holder GE, Stockman A, Tyler N, Petersen-Jones S, Bhattacharya SS, Thrasher AJ, Fitzke FW, Carter BJ, Rubin GS, Moore AT, Ali R. N. Engl. J. Med. 2008;358:2231. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 6.Chen G, Chen W, Wu Z, Yuan R, Li H, Gao J, Shuai X. Biomaterials. 2009;30:1962. doi: 10.1016/j.biomaterials.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 7.Raety JK, Liimatainen T, Wirth T, Airenne KJ, Ihalainen TO, Huhtala T, Hamerlynck E, Vihinen-Ranta M, Naervaenen A, Ylae-Herttuala S, Hakumaeki JM. Gene Ther. 2006;13:1440. doi: 10.1038/sj.gt.3302828. [DOI] [PubMed] [Google Scholar]
- 8.Kayyem JF, Kumar RM, Fraser SE, Meade TJ. Chem. Biol. 1995;2:615. doi: 10.1016/1074-5521(95)90126-4. [DOI] [PubMed] [Google Scholar]
- 9.Goffeney N, Bulte JWM, Duyn J, Bryant LH, Jr., van Zijl PCM. J. Am. Chem. Soc. 2001;123:8628. doi: 10.1021/ja0158455. [DOI] [PubMed] [Google Scholar]
- 10.McMahon MT, Gilad AA, Zhou J, Sun PZ, Bulte JWM, van Zijl PCM. Magn. Reson. Med. 2006;55:836. doi: 10.1002/mrm.20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Snoussi K, Bulte JWM, Gueron M, van Zijl PCM. Magn. Reson. Med. 2003;49:998. doi: 10.1002/mrm.10463. [DOI] [PubMed] [Google Scholar]
- 12.Woods M, Woessner DE, Sherry AD. Chem. Soc. Rev. 2006;35:500. doi: 10.1039/b509907m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sherry AD, Woods M. Ann. Rev. Biomed. Eng. 2008;10:391. doi: 10.1146/annurev.bioeng.9.060906.151929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y, Zhou Y, Ouari O, Woods M, Zhao P, Soesbe TC, Kiefer GE, Sherry AD. J. Am. Chem. Soc. 2008;130:13854. doi: 10.1021/ja805775u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kononov AI, Moroshkina EB, Tkachenko NV, Lemmetyinen H. J. Phys. Chem. B. 2001;105:535. [Google Scholar]
- 16.Aslanoglu M, Ayne G. Anal. Bioanal. Chem. 2004;380:658. doi: 10.1007/s00216-004-2797-5. [DOI] [PubMed] [Google Scholar]
- 17.Viswanathan S, Ratnakar SJ, Green KN, Kovacs Z, De Leon-Rodriguez LM, Sherry AD. Angew. Chem., Int. Ed. 2009;48:9330. doi: 10.1002/anie.200904649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wegh RT, Pikkemaat JA, Willard NP. Method for using CEST contrast agents in MRI. 2006 WO/2006/114739.
- 19.Felgner PL. Adv. Drug Delivery Rev. 1990;5:163. doi: 10.1016/s0169-409x(97)00101-4. [DOI] [PubMed] [Google Scholar]
- 20.Ahmad A, Evans HM, Ewert K, George CX, Samuel CE, Safinya CR. J. Gene Med. 2005;7:739. doi: 10.1002/jgm.717. [DOI] [PubMed] [Google Scholar]
- 21.Merdan T, Kopecek J, Kissel T. Adv. Drug Delivery Rev. 2002;54:715. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, Zhao P, Keifer GE, Sherry AD. Macromolecules. 2010;43:6616. doi: 10.1021/ma100776d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reichmann ME, Rice CA, Thomas CA, Doty P. J. Am. Chem. Soc. 1954;76:3047. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.