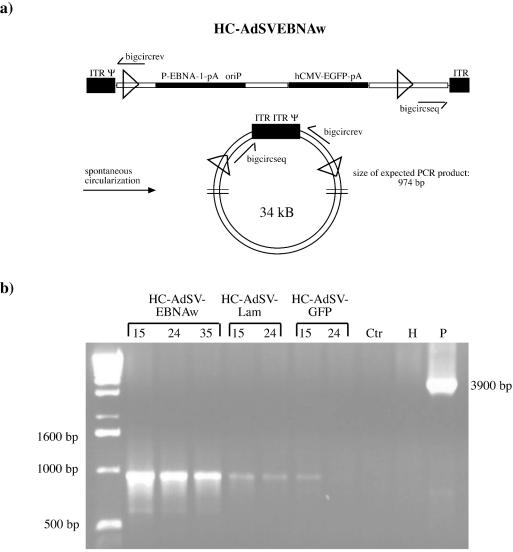
Abstract
High-capacity “gutless” adenovirus vectors (HC-AdV) mediate long-term transgene expression in resting cells in vitro and in vivo because of low toxicity and immunogenicity. However, in proliferating cells, expression is transient since HC-AdV genomes do not possess elements that allow for replication and segregation of the replicated genomes to daughter cells. We developed a binary HC-AdV system that, under certain conditions, allows for significantly prolonged episomal maintenance of HC-AdV genomes in proliferating tissue culture cells, resulting in sustained transgene expression. After transduction of target cells the linear HC-AdV genomes were circularized by the DNA recombinase FLPe, which was expressed from the second HC-AdV. The oriP/EBNA-1 replication system derived from Epstein-Barr virus, as well as the human replication origin from the lamin B2 locus, were used as cis elements to test for replication of the 28-kb circular vector genomes with or without selective pressure. Depending on the system, up to 98% of the circularized genomes were replicated and segregated to daughter cells, as demonstrated by Southern assays and as confirmed by monitoring EGFP transgene expression. Surprisingly, in the absence of FLPe recombinase, a small but significant number of HC-AdV genomes spontaneously circularized after transduction of target cells. These circles, found to contain end-to-end joined adenovirus termini, replicated with increased efficiency compared to vectors circularized by FLPe. After further improvements, this HC-AdV system might be suitable for gene therapy applications requiring long-term transgene expression.
Gene transfer with adenovirus vectors has been extensively used for functional studies in vitro and in vivo and in clinical studies. Adenovirus (Ad) vectors are nonintegrating vectors and, therefore, their genome is lost in proliferating cells. This even holds true for high-capacity “gutless” Ad vectors (HC-AdVs) that are devoid of viral coding sequences and allow for long-term in vivo gene expression in resting cell types (8, 10, 16, 21). In principle, there could be two ways to achieve vector genome maintenance in proliferating cells after Ad vector-mediated gene transfer: chromosomal integration or episomal replication. Chromosomal integration by integrases or transposable elements is random and bears the risk of activation or inactivation of genes at or next to the integration site. In addition, the integrated transgene may be subject to locus-dependent influences on expression levels. In contrast, an episomal HC-AdV system tuned to use the cellular replication machinery for episomal replication of the vector genomes could have significant advantages. However, several obstacles must be overcome to develop an episomal HC-AdV system. First, it must provide mechanisms that ensure correct full-length replication of both strands. In principle, this could be achieved by converting the linear vector genomes into circular episomes after transduction of the target cells. Second, these circular episomes must be designed to allow both for replication in a cell cycle-controlled manner and for proper segregation of the replicated genomes.
We developed a binary HC-AdV system that is based on two different HC-AdVs: one expressing the site-specific recombinase FLPe (4) and the other carrying two Frt recognition sites for the recombinase in a direct orientation. After cotransduction of target cells with both vectors, the FLPe recombinase excises and circularizes the DNA segment located in between the Frt sites, thereby generating up to 28-kb circular episomes. To allow for cell cycle-controlled replication of the FLPe-generated circular episomes, we used two independent strategies. The first relies on the oriP/EBNA-1 replication system derived from Epstein-Barr virus (EBV) (24, 25). oriP is the latent origin of replication of EBV and consists of two elements: the family of repeats FR and the dyad symmetry region DS. Both are necessary for efficient replication of circular episomes and, together with EBNA-1, allow for segregation of the replicated genomes to daughter cells. The major advantage of oriP/EBNA-1 episomes is that their replication is cell cycle controlled and occurs only once per cell cycle. As far as known up to now, EBNA-1 is believed not to be oncogenic and does not seem to be immunogenic (3). However, EBNA-1 is a viral protein, and its synthesis could lead to adverse side effects in the context of therapeutic approaches. Therefore, we analyzed a second strategy for the replication of FLPe-generated circular episomes that is based on the human origin of replication derived from the lamin B2 locus (1, 2, 13) and does not rely on the expression of a viral protein. Currently, this origin of replication is considered to be the best-characterized human replication origin since initiation of replication from this origin has been demonstrated by different independent methods and in different cell lines. Furthermore, footprint analysis have revealed its cell cycle-dependent association with replication complex proteins.
We compared the performance of both oriP/EBNA-1 and human lamin B2 origin-based episomal HC-Ad vector systems with or without selective pressure in different human cell lines.
MATERIALS AND METHODS
Plasmids.
To generate pAdFLPe, a shuttle plasmid for the construction of the FLPe-expressing vector HC-AdFLPe, the SalI/PacI fragment of pCAGS-FLPe (a gift of Francis Stewart, Heidelberg, Germany) containing the CAGS-promoter and FLPe cDNA, was isolated. An oligodesoxyribonucleotide adapter consisting of the oligodesoxyribonucleotides adapt1 (5′-TAACGACACGG-3′) and adapt2 (5′-GATCCCGTGTCGTTAAT-3′) was ligated to the PacI end of the isolated fragment. The ligation product was digested with BamHI and cloned into the BamHI/XhoI restriction sites of pCEP4 (Invitrogen). The resulting plasmid was digested with NotI/SalI, and the 3,500-bp fragment containing the complete expression cassette for FLPe [including simian virus 40 poly(A) signals] was isolated, blunt ended with Klenow polymerase, and cloned into the EcoRV site of the HC-AdV shuttle plasmid pSTK129 (S. Kochanek, unpublished results).
pFrt#39, a plasmid containing two FLP recognition sites Frt in direct orientation, was generated by annealing the oligodesoxyribonucleotides frtseq (5′-GGAGAAGTTCCTATTCCGAAGTTCCTATTCTCTAGAAAGTATAGGAACTTCAAGC-3′) and frtrev (5′-GCTTGAAGTTCCTATACTTTCTAGAGAATAGGAACTTCGGAATAGGAACTTCTCC-3′) and cloning the products successively and in direct orientation into the SrfI and Kpn2I restriction sites of the HC-AdV shuttle plasmid pSTK129. The distance between the two Frt sites in pFrt#39 is 22 kb. This plasmid has two unique restriction sites NotI and SwaI between the Frt sites. These sites were used to generate the derivatives of pFrt#39.
pHyg22 is a derivative of pFrt#39 containing the two Frt sites in parallel orientation and an expression cassette for a hygromycin resistance-enhanced green fluorescent protein (EGFP) fusion protein. This hCMV-driven expression cassette was isolated from pHyg-EGFP (Clontech) as an ClaI/BglII fragment, blunt ended with Klenow polymerase, and placed into the NotI restriction site of pFrt#39.
pHygLam is based on pHyg22 and additionally contains the 1,200-bp core region of the origin of replication from the human lamin B2 locus. This region was amplified in 25 cycles by PCR with the primers humlam1 (5′-AGACTGATTTAAATCAGACGCCACCCAGCCCTGG-3′) and humlam2 (5-AGACTGATTTAAATCGGAGCCGAAGCCGCCCTC-3′) from genomic DNA prepared from the human N52E6 cell line (20). The PCR product was purified and sequenced. To generate compatible ends, it was digested with SwaI and finally cloned into the SwaI site of pHyg22.
pHygEBNA is based on pHyg22 as well. It contains an SRα promoter-driven expression cassette for EBNA-1 and the EBV origin of replication oriP. The EBNA-1 expression cassette was isolated together with oriP as a PvuII fragment from p2747-1 (a gift from W. Hammerschmidt, Munich, Germany), blunt ended with Klenow polymerase, and cloned into the SwaI site of pHyg22.
pSVGFP is based on pFrt#39. In addition to the Frt sites, it contains an hCMV-driven expression cassette for EGFP. This expression cassette was isolated as an AflII/AflIII fragment from pEGFP-N1 (Clontech), blunt ended with Klenow polymerase, and cloned into the NotI site of pFrt#39.
pSVLam is based on pSVGFP and contains the same 1,200-bp human lamin B2 locus fragment as pHygLam. This fragment was amplified as described before, digested with SwaI, and cloned into the SwaI site of pSVGFP.
pSVEBNAs is based on pSVGFP. It contains the same expression cassette for EBNA-1 and oriP as pHygEBNA and was generated the same way.
pSVEBNAsΔFR is also based on pSVGFP. It contains the same expression cassette for EBNA-1 as pSVEBNAs but has a truncated form of oriP lacking the family of repeats element. This element was removed from p2747-1 by digestion with HindIII and MluI, creating blunt ends with Klenow polymerase, and religation. The expression cassette for EBNA-1 and the truncated oriP were isolated as a PvuII fragment, blunt ended with Klenow polymerase, and cloned into the SwaI site of pSVGFP.
pSVEBNAw contains the EBNA-1 gene and oriP from the EBV strain B95-8. The expression cassette for EBNA-1 and oriP were isolated from pCEP4 (Invitrogen) by digestion with SalI/ClaI, generating blunt ends with Klenow polymerase, and cloning the fragment into the SwaI site of pSVGFP.
HC-AdVs.
The HC-AdVs were generated as described elsewhere (17, 21). In brief, the bacterial backbone was released from the shuttle plasmids by digestion with PmeI. The linearized shuttle plasmids were transfected into 293-cre66 cells (G. Schiedner, unpublished results) by calcium phosphate coprecipitation. At 4 to 6 h before transfection, the 293-cre66 cells had been infected with helper virus AdLC8cluc (at a multiplicity of infection [MOI] of 5). Vectors were serially amplified in 293-cre66 cells, purified by CsCl equilibrium density centrifugation, and desalted by using PD-10 columns (Amersham). The infectious and particle titers were determined by the slot blot procedure (9), and the integrity of the vector genomes was confirmed by restriction digestion. In addition, the helper virus contamination was determined by the slot blot procedure and found to be between 0.1 and 0.4%.
Cell lines.
HeLa cells (ATCC CCL-2) and A549 cells (ATCC CCL-185) were grown in monolayers in minimal essential medium (Gibco) supplemented with 10% fetal calf serum (FCS) and penicillin-streptomycin (Gibco). The HeLaEBNA1 cell line was a gift of W. Hammerschmidt and was grown in monolayers in α-minimal essential medium (Gibco) supplemented with 10% FCS and penicillin-streptomycin.
Replication assays under selective pressure.
A total of 2 × 106 A549 and HeLa cells was cotransduced with HC-AdFLPe and HC-AdHyg22, HC-AdHygLam, or HC-AdHygEBNA (20 MOI each), respectively. At 36 h posttransduction the medium was supplemented with 200 μg of hygromycin B/ml. At day 4 posttransduction, cells were passaged 1:5. In order to have stringent selective conditions, the medium was exchanged every 2 days. During the following weeks, the cells were passaged 1:5 twice per week. Genomic DNA of cell aliquots from each passage was prepared.
Replication assays without selective pressure.
A total of 2 × 106 A549, HeLa, or HeLaEBNA1 cells was cotransduced with HC-AdFLPe (20 or 100 MOI) and the different substrate vectors (20 or 100 MOI each). Control experiments were performed without cotransduction with HC-AdFLPe. After transduction, cells were grown to confluency and split twice per week in a way that allowed us to reach >90% confluency between two passages. Aliquots from cells of every passage were subjected to flow cytometry analysis for EGFP expression, and from every aliquot genomic DNA was prepared.
Isolation of genomic DNA.
Genomic DNA was prepared by QiaAmp DNA MiniKits (Qiagen). After elution from the columns, the DNA was ethanol precipitated, washed, and resuspended in 10 mM Tris-Cl-1 mM EDTA (pH 8.5). The concentration was then determined by measuring the optical density at 260 nm.
Southern transfer assays.
Portions (8 to 15 μg) of genomic DNA were digested with HindIII, and fragments were separated on a 0.8% agarose gel and subjected to standard Southern transfer onto positively charged nylon membranes (Pall). Hybridization was performed with a radiolabeled 600-bp probe derived from the intronic HPRT (hypoxanthine phosphoribosyltransferase) intronic stuffer DNA contained in all vectors and cell lines used.
Preparation of radiolabeled DNA probe.
A 600-bp DNA fragment from the intronic HPRT stuffer region of all vectors was amplified from pFrt#39 by PCR with the primers probeseq (5′-TGTGATGCCTGCCCCAGTAT-3′) and proberev (5-TTCTTAGCTCAAGTGCGTCCA-3) in 25 cycles. The PCR product was purified by the QiaQuick PCR purification kit (Qiagen). Subsequently, DNA was labeled with [32P]dCTP by using the DNA labeling kit RediPrimeII (Amersham Pharmacia).
Signal quantification and copy number calculation.
A PhosphorImager system and the software ImageQuaNT (Molecular Dynamics) were used to quantify the radioactive signals on nylon membranes after DNA transfer and hybridization. Copy numbers were determined by comparing the signal intensities of the vector fragments with DNA fragments from untransduced cells.
Flow cytometry.
Flow cytometry was performed with a Becton Dickinson flow cytometer to quantify the number of cells showing EGFP fluorescence after transduction with HC-AdVs. In brief, cells were washed twice with phosphate-buffered saline (PBS), trypsinated, and resuspended in PBS-2% FCS-20 mM EDTA. Analysis was performed in a Becton Dickinson flow cytometer with appropriate filter settings. The data were quantified by using the CellQuest software package.
PCR for detection of full-length ciruclar vector genomes.
To detect circular vector genomes containing the Ad5 inverted terminal repeats (ITRs), a 25-cycle PCR with the primers bigcircseq (5′-AGATACGAGCATGGATTCTTGG-3′) and bigcircrev (5′-CCACCTTCTCTTCCCACACG-3′) was performed (Tm = 55°C), and 1/5 of each reaction was analyzed on a 1.5% agarose gel.
Calculation of maintenance values.
To calculate maintenance values from the flow cytometry data, we converted the number of days posttransduction into the number of population doublings posttransduction as follows: d(t) = n(t)×(ln x/ln 2), where d(t) is the number of population doublings at day t posttransduction, n(t) is the number of passages at day t posttransduction, and x is the reciprocal of the used split rate. This conversion allowed for normalization of individual population doubling rates of different cell lines. The maintenance value M was then calculated as follows: M = −(100/m), where m is the slope of the linear range of the decreasing number of EGFP-positive cells posttransduction. The coefficient of correlation was determined for the linear ranges and had to be >0.98.
RESULTS
Formation of replicons under selective pressure.
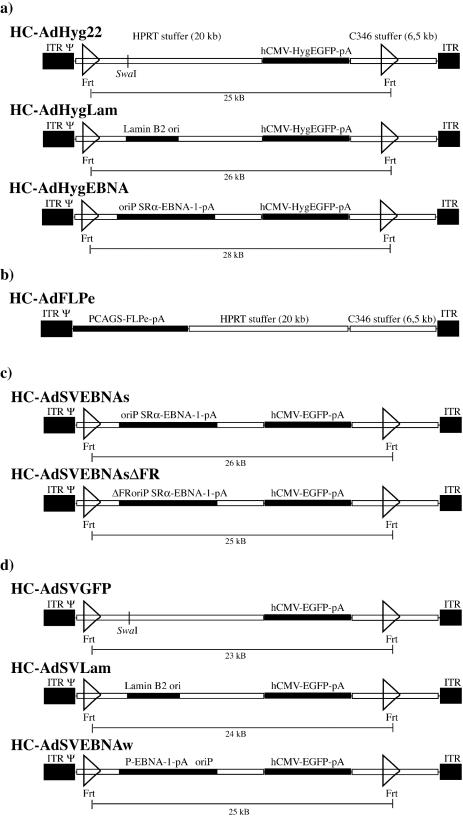
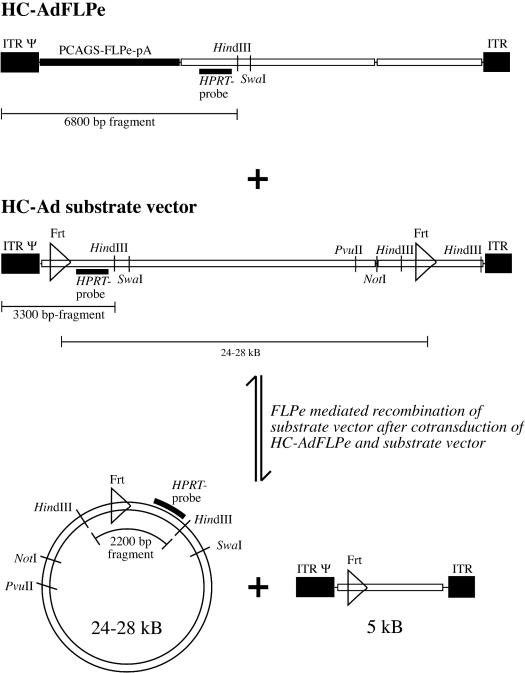
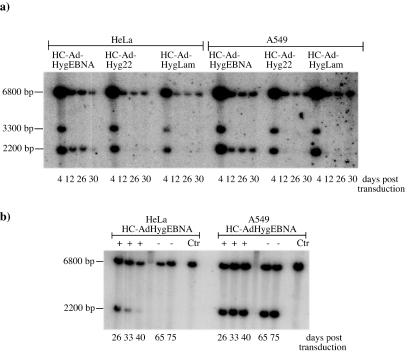
In order to test the ability of the oriP/EBNA-1 system and the human lamin B2 origin of replication to mediate the formation of replicons in an HC-AdV system, we constructed the vectors HC-AdFLPe, HC-AdHyg22, HC-AdHygEBNA, and HC-AdHygLam (Fig. 1a and b). HC-AdFLPe is an expression vector for the site-specific recombinase FLPe. The vectors HC-AdHyg22, HC-AdHygEBNA, and HC-AdHygLam are referred to as substrate vectors below because they carry two Frt sites in parallel orientation and are substrates for the FLPe recombinase to be circularized upon FLPe expression. Figure 2 illustrates the recombination of a substrate vector and the specific DNA fragments of linear and circularized substrate vectors that can be detected after digestion with HindIII in a Southern transfer assay. To test the ability of these HC-AdVs to form stable replicons under selective pressure in fast-proliferating human tumor cell lines, we cotransduced 2 × 106 A549 or HeLa cells with HC-AdFLPe (20 MOI) and HC-AdHyg22, HC-AdHygLam, or HC-AdHygEBNA (20 MOI each), respectively. Genomic DNA prepared at different time points posttransduction was subjected to a Southern transfer assay described previously (Fig. 3a). Comparison of the signal intensities at 2,200 bp (recombined, circular substrate vectors) and 3,300 bp (unrecombined, linear substrate vectors) revealed that 70 to 80% of all substrate vectors were recombined and circularized by FLPe in HeLa and A549 cells. The circularized genome of HC-AdHygEBNA carrying the oriP/EBNA-1 replication system was maintained at one to three copies/cell in both cell lines up to day 30 (the weak signal for the circular genome of HC-AdHygEBNA at day 30 posttransduction was due to a smaller amount of DNA loaded; for the copy number calculation, this was corrected). In contrast, the linear genome of this vector, as well as the circular and linear genomes of HC-AdHyg22 and HC-AdHygLam, was lost rapidly from either cell line between days 4 and 12 posttransduction. To analyze whether the replicons formed in both cell lines would be further maintained without selective pressure, we terminated selection at day 50 posttransduction. Figure 3b shows the results of the Southern assay at late time points posttransduction and after the removal of selective pressure. In HeLa cells a decreasing copy number of circular HC-AdHygEBNA genomes was detected until day 40. In A549 cells two or three copies of circular HC-AdHygEBNA per cell were detected. Apparently, this copy number was stable between days 33 and 75 posttransduction. Removal of the selective pressure on day 50 posttransduction had no influence on the maintenance of the replicons.
FIG. 1.
(a, c, and d) HC-Ad vectors used for establishing replicons with (a) or without (c and d) selective pressure. All vectors are based on the same backbone, containing 20-kb intronic stuffer DNA derived from the human HPRT locus (HUMHPRTB, gene map positions 1777 to 21729) and a 6.5-kb intronic stuffer DNA fragment from C346 (HUMDXS455A, cosmid map positions 10205 to 16750). They contain the left terminus of Ad5 (nucleotides 1 to 440; ITR), the right terminus (nucleotides 35818 to 35935; ITR), and the packaging signal from Ad5 (Ψ). Open triangles represent the FLP recognition sites Frt. (b) Vector for expression of the FLPe recombinase.
FIG. 2.
The FLPe-mediated circularization of substrate vectors and detection by restriction and Southern transfer. HC-AdFLPe is used to express the site-specific recombinase FLPe that in turn recognizes the Frt sites in the substrate vector and excises and circularizes the DNA fragment between the Frt recognition sites. The circular genomes are 24 to 28 kb in size, depending on the type of substrate vector. HC-AdFLPe remains unchanged. After restriction with HindIII, a 2,200-bp fragment represents the recombined circular genomes of the substrate vector. A 3,300-bp fragment represents unrecombined linear substrate vector genomes, and a 6,800-bp fragment represents the FLPe expression vector HC-AdFLPe. The HPRT probe used here detects an additional cellular DNA fragment of 6,800 bp (not shown).
FIG. 3.
Southern blot analysis of genomic DNA from HeLa and A549 cells after cotransduction with HC-AdFLPe (20 MOI) and the indicated substrate vectors (20 MOI each) at early (a) and late (b) time points posttransduction. Signals at 2,200 bp indicate recombined, circular substrate vectors; signals at 3,300 bp indicate unrecombined, linear substrate vectors. HC-AdFLPe and the cellular HPRT fragment are represented by the signals at 6,800 bp. +, selection with hygromycin. Selection was terminated at day 50 posttransduction (−). Ctr, untransduced control cells.
The Southern transfer in Fig. 3 demonstrates that integration of unrecombined, linear vector genomes or concatemers over the ITR ends did not take place, since additional signals larger than 3,300 bp would have occurred. To ensure that the signals at 2,200 bp originated from episomal replicons rather than integrated vector genomes, we performed an additional Southern transfer with genomic DNA from A549 cells at 75 days posttransduction. The DNA was restricted with the 6-bp cutter PvuII (which has a unique site in the circular episome), transferred onto a nylon membrane, and hybridized with the HPRT probe used previously. This Southern transfer yielded one signal for the episome in the expected size of 28 kb and another signal of the same intensity at 20 kb representing the cellular HPRT locus (data not shown). Since there were no additional bands observed and the ratio of signal intensities for the episome and the cellular HPRT locus remained the same as in Fig. 3b, we conclude that the episomes did not integrate into the host cell's genome. Furthermore, this finding demonstrates that the circular episomes were stable and did not rearrange.
In flow cytometry analyses we found only very low EGFP-specific fluorescence in A549 and HeLa cells at 48 h posttransduction, with up to 100 MOI of the different substrate vectors expressing the hygromycin resistance-EGFP fusion protein (data not shown). The EGFP-specific fluorescence was too weak to reliably allow monitoring transgene expression over extended time periods. Therefore, we generated a new set of substrate vectors, analogous to those shown in Fig. 1a, which lack the hygromycin resistance gene but share an hCMV-driven expression cassette for strong expression of EGFP (Fig. 1c).
Formation of autonomous oriP replicons without selective pressure.
In order to analyze the formation of oriP/EBNA-1 replicons without selective pressure and to monitor the number of EGFP-positive cells by flow cytometry, we cotransduced 2 × 106 HeLa cells each with HC-AdFLPe and HC-AdSVEBNAs or HC-AdSVEBNsΔFR (see Fig. 1b and c) (100 MOI each), respectively. HC-AdSVEBNAs contained the same sequences for expression of EBNA-1 and the same replication origin oriP as HC-AdHygEBNA but lacked the option for selection. HC-AdSVEBNAsΔFR harbored a truncated form of oriP, which lacked the family of repeats element FR. This vector was expected to be impaired in proper segregation of the replicated genomes to daughter cells. Figure 4a shows the time course of EGFP expression in HeLa cells postcotransduction; Fig. 4b shows the corresponding Southern blot membrane. The number of EGFP-positive HeLa cells rapidly decreased from 100 to 4% between days 4 and 20 posttransduction with HC-AdSVEBNAs. After transduction with the same amount of HC-AdSVEBNAsΔFR, the number of EGFP-expressing cells dropped more rapidly to 4% at day 14 posttransduction. From the corresponding Southern transfer experiment, it was apparent that the circular genomes of HC-AdSVEBNAs were maintained slightly better than those of HC-AdSVEBNAsΔFR (Fig. 4b). We repeated this set of experiments with 20 MOI of each vector in HeLa cells and 20 and 100 MOI in A549 cells without finding any significant differences in vector maintenance either at the levels of transgene expression or in copy number (data not shown). Interestingly, we observed ∼2-fold-reduced cell population doubling rates after transduction with HC-AdSVEBNAs, HC-AdSVEBNAsΔFR, or HC-AdHygEBNA (>20 MOI each). Reduced population doubling rates of morphologically intact cells were seen both in HeLa as well as in A549 cells without selection and were only observed after transduction with vectors that expressed EBNA-1. This effect was independent of cotransduction with HC-AdFLPe. If we assume that the strong overexpression of EBNA-1 from the SRα promoter could influence cell growth kinetics and thereby create selective disadvantages for cells carrying oriP/EBNA-1 replicons, we generated HC-AdSVEBNAw, which contained the oriP and the EBNA-1 gene from EBV strain B95-8 (Fig. 1d). Figure 5 shows the amount of EBNA-1 protein expressed by the different HC-AdVs in a Western transfer experiment. HC-AdSVEBNAw expressed less than 1/10 of EBNA-1 compared to HCAdSVEBNAs, HC-AdSVEBNAsΔFR, and HC-AdHygEBNA.
FIG. 4.
(a) Time course of EGFP expression in HeLa cells after cotransduction with HC-AdFLPe (100 MOI) and HC-AdSVEBNAs or HC-AdSVEBNAsΔFR (100 MOI each), respectively. Cells were analyzed for EGFP-specific fluorescence by flow cytometry. (b) Southern blot analysis of genomic DNA from the same HeLa cells. The circular substrate vector genomes are indicated by signals at 2,200 bp; the linear substrate vector genomes are indicated by signals at 3,300 bp. Ctr, untransduced control cells.
FIG. 5.
Western transfer membrane for the detection of EBNA-1 protein. A total of 2 × 106 293 cells were transduced with the indicated amounts of the indicated vectors. At 48 h posttransduction, total protein lysates were prepared, and equal amounts of lysates were subjected to Western transfer and detection of EBNA-1. Cells transduced with vectors containing the SRα promoter-driven expression cassette for EBNA-1 synthesized ∼10-fold more EBNA-1 than cells transduced with HC-AdSVEBNAw. Ctr, lysate from untransduced control cells.
Formation of oriP replicons without selective pressure and comparison to lamin origin-based replicons.
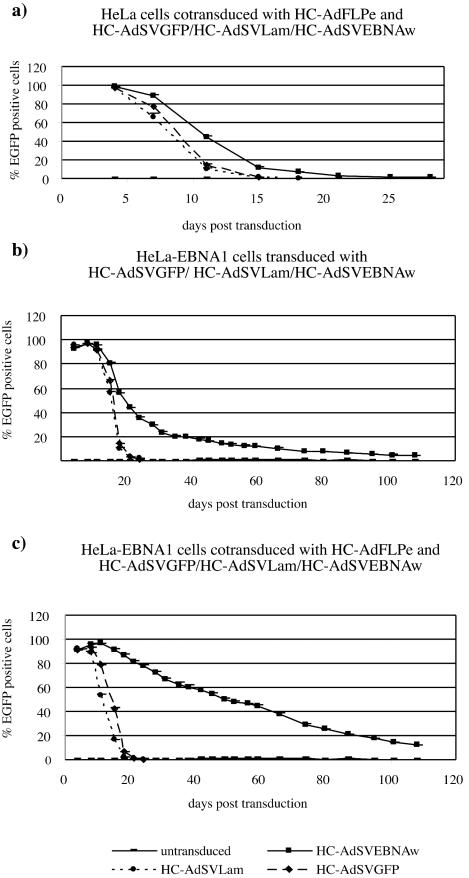
We cotransduced HeLa and A549 cells with HC-AdFLPe (100 MOI) and HC-AdSVGFP, HC-AdSVLam, or HC-AdSVEBNAw (100 MOI each), respectively. Figure 6a shows the time course of EGFP-expressing HeLa cells posttransduction. HC-AdSVEBNAw was maintained slightly more efficiently (4% EGFP-positive cells at day 21) than HC-AdSVLam or HC-AdSVGFP (4% EGFP-positive cells at day 14), but stable replicons were not established. This observation was independent of the amount of vector used for initial transduction (20 or 100 MOI) and independent of the transduced cell type (HeLa or A549 cells [data not shown]). In addition, we did not observe reduced population doubling rates after transduction with an MOI of HC-AdSVEBNAw of >20, suggesting no selective disadvantage for cells carrying circular HC-AdSVEBNAw genomes.
FIG. 6.
(a) Time course of EGFP-expressing HeLa cells after cotransduction with HC-AdFLPe (100 MOI) and the indicated substrate vectors (100 MOI each). (b) Time course of EGFP-expressing HeLaEBNA1 cells after transduction with the indicated substrate vectors but without cotransduction with HC-AdFLPe. (c) Time course after cotransduction with HC-AdFLPe (100 MOI).
To confirm that EBNA-1 expression from HCAdSVEBNAw was not too weak to allow for establishing stable oriP replicons, we repeated this set of experiments with HeLa-EBNA1 cells that constitutively expressed EBNA-1. Figure 6b shows the time course of EGFP expression after transduction with HC-AdSVGFP, HC-AdSVLam, and HC-AdSVEBNAw (100 MOI each), respectively. Figure 6c illustrates the time course after cotransduction with HC-AdFLPe (100 MOI). In both cases, the vectors HC-AdSVGFP and HC-AdSVLam were rapidly lost from dividing cells. At day 20 posttransduction fewer than 1% of the cells remained EGFP positive, a result independent of cotransduction with HC-AdFLPe. In contrast, after cotransduction with HC-AdSVEBNAw and HC-AdFLPe, the number of EGFP-positive cells only slowly decreased, with 50% positive cells remaining at day 50 and 15% EGFP-positive cells at day 110 posttransduction. Interestingly, after transduction with HC-AdSVEBNAw alone, the number of EGFP-expressing cells dropped rapidly between day 4 and 30 posttransduction. Starting at day 30 posttransduction, the loss rate was slowed down, and between days 30 and 110 the number of EGFP-positive cells decreased slowly in a linear way, with >4% remaining EGFP positive at day 110 posttransduction.
Genomic DNA of cells at different time points posttransduction was prepared and subjected to Southern blot analysis (Fig. 7). The linear (3,300 bp) and circular (2,200 bp) genomes of HC-AdSVGFP and HC-AdSVLam were lost from HeLaEBNA1 cells between days 4 and 15 posttransduction. In contrast, the circular genomes of HC-AdSVEBNAw initially dropped to 12 to 15 copies per cell at day 15 posttransduction and remained quite stable at 11 copies per cell on day 24, 9 copies per cell on day 35, and 7 copies per cell on day 60 posttransduction. This revealed a maintenance of 98% of the vector genomes per cell population doubling and correlates well with the flow cytometry data (Fig. 6c). At late time points posttransduction (days 64 to 122, Fig. 7b), we could still detect circular genomes of HC-AdSVEBNAw in HeLaEBNA1 cells. At day 122 posttransduction about one copy per cell was found. Together with the flow cytometry data underlying Fig. 6b and c, these results suggest nearly symmetrical segregation of the vector genomes during cell division, because in the flow cytometry histograms we could not observe subpopulations of cells showing significantly higher or lower fluorescence during the time course of the experiment (data not shown).
FIG. 7.
Southern blot analysis of genomic DNA from HeLaEBNA1 cells transduced with the indicated substrate vectors at early time points posttransduction (a) and at late time points posttransduction (b). −FLPe, no cotransduction with HC-AdFLPe; +FLPe, cotransduction with HC-AdFLPe. Circular substrate vector genomes are represented by signals at 2,200 bp; linear substrate vector genomes are represented by signals at 3,300 bp. Ctr, DNA from untransduced control cells.
After transduction with HC-AdSVEBNAw but without cotransduction with HC-AdFLPe, we could observe a small number of cells expressing EGFP up to day 110 posttransduction (Fig. 6b). The Southern blot analysis in Fig. 7 shows that a very low copy number of this vector could be found in HeLaEBNA1 cells at least until day 60 posttransduction. We noted that the signal for this vector genome appeared to be slightly larger than the expected 3,300 bp (see the molecular analysis below).
To evaluate whether the amount of EBNA-1 protein might influence the efficiency of replicon formation or episome maintenance, we cotransduced HeLaEBNA1 cells with HC-AdFLPe (20 MOI) and HC-AdHygEBNA, HC-AdSVEBNAs, or HC-AdSVEBNAsΔFR (20 MOI each), respectively. Figure 8a shows the results of flow cytometry analysis after cotransduction with HC-AdSVEBNAs and HC-AdSVEBNAsΔFR; Fig. 8b shows the corresponding Southern blot membrane. HC-AdSVEBNAs was, although maintained slightly longer than HC-AdSVEBNAsΔFR, rapidly lost from HeLaEBNA1 cells within 28 days posttransduction. HC-AdHygEBNA was lost during the same time period. Although the vector HC-AdSVEBNAs was maintained longer in HeLaEBNA1 cells than was HC-AdSVLam or HC-AdSVGFP, no stable replicons were formed.
FIG. 8.
(a) Time course of EGFP-expressing HeLaEBNA cells after cotransduction with HC-AdFLPe (20 MOI) and the indicated substrate vectors (20 MOI each). Cells transduced with HC-AdSVEBNAs remained EGFP positive for longer time periods than did cells transduced with HC-AdSVEBNAsΔFR, which carried a truncated oriP. (b) The corresponding Southern blot membrane. The circular genomes of HC-AdSVEBNAs and HC-AdSVEBNAw are represented by signals at 2,200 bp; their linear counterparts are represented by signals at 3,300 bp. HC-AdHygEBNA was not included in the flow cytometry experiments due to weak EGFP-specific fluorescence.
Molecular analysis of episomes formed by HC-AdSVEBNAw in HeLaEBNA cells.
Surprisingly, the vector HC-AdSVEBNAw was maintained in HeLaEBNA1 cells without cotransduction with HC-AdFLPe and subsequent FLPe-mediated circularization of the vector genomes.
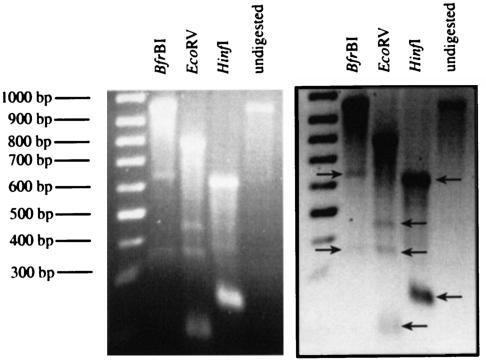
The vector signal in the Southern blot assay originated from a DNA fragment that was slightly larger than expected (Fig. 7a). One possible explanation for this larger DNA fragment was the formation of circular DNA molecules, including the Ad5 ITRs of the vector independent of cotransduction with HC-AdFLPe (see Fig. 2, in the case of large circles including both ITRs, the HindIII site next to the right ITR would lead to a fragment slightly larger than 3,300 bp in the described Southern transfer assay). To evaluate whether circular DNA molecules, including vector ITRs, had formed, we analyzed by PCR the genomic DNA prepared from HeLaEBNA1 cells transduced with HC-AdSVEBNAw. Figure 9a shows the design of the PCR: a product is only formed in the presence of circular DNA templates that had formed through joining of the termini. The results of this PCR suggested that the vectors HC-AdSVEBNAw, HC-AdSVLam, and HC-AdSVGFP were able to form circular DNA molecules without the action of FLPe in HeLaEBNA1 cells (Fig. 9b). To analyze these circles further, we tried to sequence the ITR junctions of the PCR products. At the ITR junction site the obtained signals collapsed, indicating a mixture of different sequences. Therefore, we assumed that the large circles contained end-to-end junctions with a partial and variable loss of 1 or 2 bp at the very ends of the ITRs. Since symmetric head-to-tail junctions of the ITRs should lead to the formation of new restriction sites, we analyzed the PCR product obtained from genomic DNA of HeLaEBNA1 cells prepared 15 days after transduction with HC-AdSVEBNAw by restriction digestion. Table 1 gives an overview over the enzymes selected, and the expected fragment sizes for circular molecules containing either the full-length ITRs or ITR junctions, which lack the terminal base pairs of both ITRs. Figure 10 shows the result of the restriction analysis. Digestion of the PCR product with BfrBI led to the formation of a 974-bp fragment and of additional fragments of 623 and 351 bp. This indicated that a part of the large circles formed contained the full-length ITRs and another part lacked the two terminal base pairs. This finding was confirmed by digestion with EcoRV, which resulted in fragments of 800 and 174 bp for molecules containing full-length ITRs and additional fragments of 455 and 343 bp, indicating ITR junctions that lacked the terminal base pair of each ITR.
FIG. 9.
(a) PCR assay for detection of circular vector genomes that have been generated by joining the vector ITRs. The indicated primers bigcircseq and bigcircrev can only form a product when there are appropriate joint circular vector genomes. The expected PCR product is 974 bp. (b) PCR with genomic DNA from HeLaEBNA1 cells transduced with the indicated substrate vectors (100 MOI each). Genomic DNA was prepared at different time points posttransduction (numbers above lanes). Ctr, genomic DNA from untransduced control cells; H, water control; P, pFrt#39 (which was used as a plasmid control and should result in a 3,900-bp product).
TABLE 1.
Restriction analysis of the PCR product obtained from ITR-ITR junctions
| Circle type | Size(s) (bp) of PCR product fragments obtained with:
|
||
|---|---|---|---|
| BfrBI | EcoRV | HinfI | |
| Circles containing full-length ITRs | 623, 351 | 800, 174 | 628, 274, 86, 13 |
| Circles containing ITRs that lack the terminal base pair | 972 | 455, 343, 174 | 627, 247, 85, 13 |
FIG. 10.
Restriction analysis of the PCR product obtained from genomic DNA prepared from HeLaEBNA1 cells at day 15 posttransduction with HC-AdSVEBNAw.
Calculation of maintenance values for HC-AdSVEBNAw.
Based on the observation that the decrease in the number of EGFP-positive cells posttransduction showed a linear range, we calculated maintenance values that allow for quantitative description and comparison of the maintenance of different vectors in cell lines with different proliferation rates. At first, we converted the number of days posttransduction into the number of passages posttransduction. The slope of the resulting straight line was then taken as a measurement for maintenance. Table 2 shows the maintenance values for HC-AdSVEBNAw with or without cotransduction with HC-AdFLPe in HeLaEBNA1 cells in comparison to HC-AdSVGFP and HC-AdSVLam. The values indicated the efficient maintenance of FLPe-circularized HC-AdSVEBNAw and of spontaneously circularized HC-SVEBNAw. Surprisingly, the latter was maintained even more efficiently.
TABLE 2.
Maintenance values of different substrate vectors in HeLaEBNA1 cells with (+) and without (−) cotransduction with HC-AdFLPe
| Vector |
M value
|
|
|---|---|---|
| +HC-AdFLPe | −HC-AdFLPe | |
| HC-AdSVGFP | 6 | 5 |
| HC-AdSVLam | 5 | 5 |
| HC-AdSVEBNAw | 51 | 141 |
DISCUSSION
We used the oriP/EBNA-1 replication system from EBV and a human origin of replication to analyze the establishment of episomal replicons in human cells after gene transfer with high-capacity adenovirus vectors. The vector HC-AdHygEBNA containing the oriP/EBNA-1 replication system was able to form replicons in the fast-proliferating human tumor cell lines HeLa and A549 that were stable under initial selection with hygromycin B. In A549 cells the replication and/or segregation of replicated molecules to daughter cells was even more efficient than in HeLa cells. Several cellular factors have been suggested to support efficient replication of oriP/EBNA-1- based episomes, such as the telomeric repeat binding factor 2 and its interacting partner hRap1, as well as the telomer-associated poly(ADP-ribose)-polymerase and the p32/Tat-associated proteins (5, 23). Different levels of these or additional cellular factors in A549 and HeLa cells might explain the observed differences in the efficiency of replication and/or segregation of circular HC-AdHygEBNA genomes. In addition, the presented vector system can probably be optimized with regard to the duration of selection and the antibiotics used for selection. The observation that in A549 cells the oriP replicons remained stable even after the removal of selection suggests that such a system could potentially be used for ex vivo gene therapy applications that include clonal selection and analysis of transduced cells. In contrast, the incorporation of the core region of a human origin of replication derived from the lamin B2 locus into the vector HC-AdHygLam did not provide prolonged episomal maintenance of the vector genomes. Compared to HC-AdHyg22, a vector containing 22-kb human DNA, no differences in genome maintenance were detected. With the assays applied here, we could not distinguish whether the circular genomes of HC-AdHygLam and HC-AdHyg22 did not replicate efficiently or were impaired in proper segregation of replicated genomes or both. Nevertheless, the vector system presented here is the first that permits high-titer production, efficient delivery of circular episomes free of viral coding or bacterial DNA sequences and up to 30 kb in size. These features render it favorable for maintenance studies of large episomes free of viral coding or bacterial DNA sequences. It has been shown that human DNA fragments larger than 20 kb in size have the potential to replicate autonomously in human cell lines (6, 7, 12). However, additional elements are needed to allow for segregation of the replicated molecules. The HC-Ad vector system can easily be modified to test additional elements such as, for example, matrix attachment regions on their influence on vector genome maintenance.
The prerequisites for the formation of stable replicons without selective pressure remain partially unclear. Neither the vectors based on the oriP/EBNA-1 replication system nor the vectors carrying the lamin B2 origin of replication were able to form stable replicons in A549 or HeLa cells without selective pressure. The comparison of vectors containing the lamin B2 origin with vectors harboring the oriP/EBNA-1 replication system revealed the latter to be maintained slightly longer in proliferating cells, suggesting at least low-level replication of the oriP vectors. From studies with plasmid-based oriP systems and recombinant EBV, it is known that only a small percentage of oriP episomes introduced into human cells is able to form stable replicons. The reasons for this are largely unknown, and a rare epigenetic event was suggested (15). Furthermore, Leight and Sugden could show that, after transfection of oriP plasmids into cell clones already carrying stable oriP replicons, the majority of the newly introduced plasmids were lost, and these authors assumed an influence of chromatin structure on episome maintenance (15).
Surprisingly, the vector HC-AdSVEBNAw was efficiently maintained without selection in HeLaEBNA1 cells. This finding is in sharp contrast to studies that analyzed the maintenance of oriP-based plasmids in human cell lines constitutively expressing EBNA-1 since such plasmids were rapidly lost (15). Several reasons or combinations thereof might explain the maintenance of HC-AdSVEBNAw in HeLaEBNA1 cells. First, the HeLaEBNA1 cell line could provide mechanisms that facilitate the formation of certain chromatin structures or other epigenetic events, thus allowing for efficient vector genome maintenance. Second, the HC-Ad vector itself could be advantageous compared to plasmid systems simply because of its size and as-yet-unidentified additional elements provided by the human intronic stuffer DNA. Third, the amount of EBNA-1 protein expressed by the vector might influence the maintenance of replicons without selective pressure. The vector HC-AdHygEBNA carried a strong expression cassette for EBNA-1 and could be efficiently maintained under selective pressure in HeLa and A549 cells. This observation substantiates that in principle this vector provides all elements necessary for the formation of stable oriP replicons. Nevertheless, without selection it was rapidly lost from HeLaEBNA1 cells, whereas the vector HC-AdSVEBNAw was maintained with high efficiency in this cell line. The finding that cells transduced with HC-AdVs carrying the SRα promoter-driven expression cassette for EBNA-1 exhibited reduced population doubling rates suggests that strong overexpression of EBNA-1 protein could mean a selective disadvantage. This disadvantage could probably be outcompeted by selection with hygromycin. Further analysis that includes different expression cassettes for EBNA-1 and different variants of EBNA-1 will elucidate its effect on cell growth kinetics.
Similar vector systems based on adenovirus vectors with E1/E3 or E1/E4 deleted have recently been published (11, 14, 22). These systems utilize the cre recombinase for excision and circularization of DNA fragments from first-generation vector genomes. This was done either in a binary vector system with one vector expressing the cre recombinase and the other bearing loxP recognition sites or by a single vector with inducible expression of cre (11). In all cases, the generated episomes were comparatively small (up to 8 kb), thus limiting the uptake of therapeutic transgenes in addition to the regulatory elements needed for maintenance. Furthermore, all episomes retained adenovirus coding sequences. Tan et al. observed toxic effects of their vector system ascribed to residual expression of adenovirus proteins. Another essential aspect of the design of the vector system described by Tan et al. was that EBNA-1 was only expressed after cre-mediated episome formation. This design was found to be necessary for the successful production of the Ad vectors because attempts to construct Ad recombinants containing both a functional expression cassette for EBNA-1 and oriP were unsuccessful. In contrast, with our HC-AdV production system we did not have difficulties in generating high-titer HC-Ad vectors expressing EBNA-1 and harboring oriP. By infecting the 293-cre66 production cells with helper virus 4 to 6 h before transfection or infection with vector plasmid/vector, we ensured that the replication of the vectors could take place in the absence of significant amounts of EBNA-1 protein, thereby eliminating interference between adenoviral replication and EBNA/oriP. In summary, the generation of episomes up to 28 kb in size optionally being free of any viral coding sequences, the lack of toxic side effects induced by Ad coding sequences and the ease of production are advantages of the HC-Ad vector system presented here.
Surprisingly, the vector HC-AdSVEBNAw was maintained in HeLaEBNA1 cells even without cotransduction with HC-AdFLPe. PCR analysis of the replicons revealed that this vector had formed large circular DNA molecules containing the Ad5 ITRs in a head-to-tail fashion. Sequence analysis and restriction digestion demonstrated that the maintained replicons consisted of molecules with sequence variations at the ITR-ITR junction. Our data suggest that ca. 1 to 3% of the transduced genomes were spontaneously converted into circular molecules. Spontaneous circularization of adenoviral DNA was described 20 years ago by Ruben et al. (18, 19). These authors demonstrated the formation of circular DNA molecules after infection of several rat cell lines and HeLa cells, and they postulated that the terminal protein could possess an enzymatic strand ligation activity. The spontaneous formation of circular vector genomes might be useful for establishing circular replicons in ex vivo approaches without the use of additional site-specific recombinases such as FLPe. This could further reduce the risk of recombinase-mediated adverse events. The maintenance of the HC-AdSVEBNAw circles, including the Ad5 ITRs, was the highest observed in the present study. We speculate that the presence of the ITR sequences in this vector might have somehow stabilized the genomes by forming secondary structures that facilitate the opening of chromatin. Therefore, we suggest that the incorporation of Ad5 ITRs in future vector systems might be a promising approach to further increase the stability of episomal vector genomes in proliferating cells.
In addition to its use in potential ex vivo gene therapy applications, the vector system described here might be a helpful tool for the analysis of features required for episomal replication; since large episomes can be delivered to a wide variety of cells with high efficiency and in a controlled manner, any sequences up to 28 kb in size can be incorporated, and the maintenance of genomes and transgene expression can be easily monitored.
Acknowledgments
We thank Francis Stewart for providing pCAGS-FLPe, the expression plasmid for the FLPe recombinase, and Frank Graham for the helper virus AdLC8cluc. The EBNA-1 antibody and the HeLaEBNA1 cell line were a generous gift from W. Hammerschmidt.
F.K. was sponsored by the Boehringer Ingelheim Foundation. This study was supported by a grant from the Federal Ministry of Education and Research (FKZ 01KS9502) and the Center for Molecular Medicine Cologne.
REFERENCES
- 1.Abdurashidova, G., M. Deganuto, R. Klima, S. Riva, G. Biamonti, M. Giacca, and A. Falaschi. 2000. Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287:2023-2026. [DOI] [PubMed] [Google Scholar]
- 2.Abdurashidova, G., S. Riva, G. Biamonti, M. Giacca, and A. Falaschi. 1998. Cell cycle modulation of protein-DNA interactions at a human replication origin. EMBO J. 17:2961-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, S., E. Livanos, and J. M. Vos. 1995. Therapeutic gene delivery in human B lymphoblastoid cells by engineered non-transforming infectious Epstein-Barr virus. Nat. Med. 1:1303-1308. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz, F., P. O. Angrand, and A. F. Stewart. 1998. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat. Biotechnol. 16:657-662. [DOI] [PubMed] [Google Scholar]
- 5.Deng, Z., L. Lezina, C. J. Chen, S. Shtivelband, W. So, and P. M. Lieberman. 2002. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell 9:493-503. [DOI] [PubMed] [Google Scholar]
- 6.Haase, S. B., and M. P. Calos. 1996. Replication control of autonomously replicating human sequences. Nucleic Acids Res. 19:5053-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinzel, S. S., P. J. Krysan, C. T. Tran, and M. P. Calos. 1991. Autonomous DNA replication in human cells is affected by the size and the source of the DNA. Mol. Cell. Biol. 11:2263-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kochanek, S. 1999. High-capacity adenoviral vectors for gene transfer and somatic gene therapy. Hum. Gene Ther. 10:2451-2459. [DOI] [PubMed] [Google Scholar]
- 9.Kreppel, F., V. Biermann, S. Kochanek, and G. Schiedner. 2002. A DNA-based method to assay total and infectious particle contents and helper virus contamination in high-capacity adenoviral vector preparations. Hum. Gene Ther. 13:1151-1156. [DOI] [PubMed] [Google Scholar]
- 10.Kreppel, F., T. T. Luther, I. Semkova, U. Schraermeyer, and S. Kochanek. 2002. Long-term transgene expression in the RPE after gene transfer with a high-capacity adenoviral vector. Investig. Ophthalmol. Vis. Sci. 43:1965-1970. [PubMed] [Google Scholar]
- 11.Krougliak, V. A., N. Krougliak, and R. C. Eisensmith. 2001. Stabilization of transgenes delivered by recombinant adenovirus vectors through extrachromosomal replication. J. Gene Med. 3:51-58. [DOI] [PubMed] [Google Scholar]
- 12.Krysan, P. J., S. B. Haase, and M. P. Calos. 1989. Isolation of human sequences that replicate autonomously in human cells. Mol. Cell. Biol. 9:1026-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar, S., M. Giacca, P. Norio, G. Biamonti, S. Riva, and A. Falaschi. 1996. Utilization of the same DNA replication origin by human cells of different derivation. Nucleic Acids Res. 24:3289-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leblois, H., C. Roche, N. di Falco, C. Orsini, P. Yeh, and M. Perricaudet. 2000. Stable transduction of actively dividing cells via a novel adenoviral/episomal vector. Mol. Ther. 1:314-322. [DOI] [PubMed] [Google Scholar]
- 15.Leight, E. R., and B. Sugden. 2001. Establishment of an oriP replicon is dependent upon an infrequent, epigenetic event. Mol. Cell. Biol. 21:4149-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morral, N., W. O'Neal, K. Rice, M. Leland, J. Kaplan, P. A. Piedra, H. Zhou, R. J. Parks, R. Velji, E. Aguilar-Cordova, S. Wadsworth, F. L. Graham, S. Kochanek, K. D. Carey, and A. L. Beaudet. 1999. Administration of helper-dependent adenoviral vectors and sequential delivery of different vector serotype for long-term liver-directed gene transfer in baboons. Proc. Natl. Acad. Sci. USA 96:12816-12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parks, R. J., L. Chen, M. Anton, U. Sankar, M. A. Rudnicki, and F. L. Graham. 1996. A helper-dependent adenovirus vector system: removal of helper virus by cre-mediated excision of the viral packaging signal. Proc. Natl. Acad. Sci. USA 93:13565-13570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson, A. J., H. B. Younghusband, and A. J. Bellett. 1973. A circular DNA-protein complex from adenoviruses. Virology 56:54-69. [DOI] [PubMed] [Google Scholar]
- 19.Ruben, M., S. Bacchetti, and F. Graham. 1983. Covalently closed circles of adenovirus 5 DNA. Nature 301:172-174. [DOI] [PubMed] [Google Scholar]
- 20.Schiedner, G., S. Hertel, and S. Kochanek. 2000. Efficient transformation of primary human amniocytes by E1 functions of Ad5: generation of new cell lines for adenoviral vector production. Hum. Gene Ther. 11:2105-2116. [DOI] [PubMed] [Google Scholar]
- 21.Schiedner, G., N. Morral, R. J. Parks, Y. Wu, S. C. Koopmans, C. Langston, F. L. Graham, A. L. Beaudet, and S. Kochanek. 1998. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat. Genet. 18:180-183. [DOI] [PubMed] [Google Scholar]
- 22.Tan, B. T., L. Wu, and A. J. Berk. 1999. An adenovirus-Epstein-Barr virus hybrid vector that stably transforms cultured cells with high effciency. J. Virol. 73:7582-7589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang, Y., J. E. Finan, J. M. Middeldorp, and S. D. Hayward. 1997. P32/tap, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology 236:18-29. [DOI] [PubMed] [Google Scholar]
- 24.Yates, J. L., and N. Guan. 1991. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J. Virol. 65:483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 313:812-815. [DOI] [PubMed] [Google Scholar]