Abstract
Background
Infection with H. pylori is important in the etiology of gastric cancer. Gastric cancer is infrequent in Africa, despite high frequencies of H. pylori infection, referred to as the African enigma. Variation in environmental and host factors influencing gastric cancer risk between different populations have been reported but little is known about the biological differences between gastric cancers from different geographic locations. We aim to study genomic instability patterns of gastric cancers obtained from patients from United Kingdom (UK) and South Africa (SA), in an attempt to support the African enigma hypothesis at the biological level.
Methods
DNA was isolated from 67 gastric adenocarcinomas, 33 UK patients, 9 Caucasian SA patients and 25 native SA patients. Microsatellite instability and chromosomal instability were analyzed by PCR and microarray comparative genomic hybridization, respectively. Data was analyzed by supervised univariate and multivariate analyses as well as unsupervised hierarchical cluster analysis.
Results
Tumors from Caucasian and native SA patients showed significantly more microsatellite instable tumors (p < 0.05). For the microsatellite stable tumors, geographical origin of the patients correlated with cluster membership, derived from unsupervised hierarchical cluster analysis (p = 0.001). Several chromosomal alterations showed significantly different frequencies in tumors from UK patients and native SA patients, but not between UK and Caucasian SA patients and between native and Caucasian SA patients.
Conclusions
Gastric cancers from SA and UK patients show differences in genetic instability patterns, indicating possible different biological mechanisms in patients from different geographical origin. This is of future clinical relevance for stratification of gastric cancer therapy.
Background
Gastric cancer is the second most common cause of cancer death worldwide, but incidence and mortality rates show large variations across different countries. Japan and China show the highest incidence rates of gastric cancer of 80-115 cancers/100,000 population and 32-59 cancers/100,000 population respectively, while in other Asian counties, such as India, Bangladesh, and Thailand, the incidence rates are much lower (10.6, 1.3, and 7.1 per 100,000 populations, respectively). Also within Europe, incidence and mortality rates differ between countries. Portugal has the highest incidence rates (33.2/100,000) whereas other countries in Western Europe show incidence rates of 19.4 per 100,000 populations. In the Netherlands it ranks fifth as a cause of cancer death with incidence rates of 14.6/100,000. In Africa, gastric cancer is infrequent, with incidence rates varying between 6.9/100,000 in Northern Africa, 12.9/100,000 in Eastern Africa, 11.9/100,000 in Southern Africa and 7.0/100,000 in Western Africa (Table 1) [1-3].
Table 1.
Incidence rates of gastric cancers per 100,000 populations.
| Incidence rates | Incidence rates | ||
|---|---|---|---|
| Japan | 80-115 | Netherlands | 14.6 |
| China | 32-59 | Western Europe | 19.4 |
| India | 10.6 | Northern Africa | 6.9 |
| Bangladesh | 1.3 | Eastern Africa | 12.9 |
| Thailand | 7.1 | Southern Africa | 11.9 |
| Portugal | 33.2 | Western Africa | 7.0 |
According to the Correa model, intestinal type gastric cancers arise through a sequence of events, starting with chronic active gastritis due to infection with Helicobacter pylori (H. pylori). This chronic inflammatory process may lead to atrophy, intestinal metaplasia followed by dysplasia and eventually may lead to invasive adenocarcinoma [4].
The mechanism by which H. pylori contributes to gastric carcinogenesis is still largely unknown. However, we do know that gastric cancer is the result of accumulation of (epi)genetic changes. In gastric cancer, at least two types of genetic instability play a role. Microsatellite instability (MSI) occurs in cancers associated with Lynch syndrome or hereditary non-polyposis colorectal cancer (HNPCC), and in 10-15% of sporadic gastric cancers due to hMLH1 promoter hypermethylation [5,6]. However, the majority of gastric cancers show chromosomal instability, resulting in DNA copy number aberrations that can be analyzed in detail by high resolution array comparative genomic hybridization (array CGH). In a previous study using chromosome based comparative genomic hybridization (CGH), we were unable to demonstrate that there are specific chromosomal alterations which are associated with H. pylori infection [7].
Infection with H. pylori is important in the etiology of gastric cancer, consequently high incidences of gastric cancer are observed in areas with high prevalence of H. pylori infection, like Asia. However, despite high frequencies of H. pylori infection in Africa, gastric cancer is infrequent in Africa, a phenomenon often referred to as the 'African enigma' [8,9]. We hypothesize that geographical differences in environmental factors, including infection with H. pylori, and host factors are reflected by different biological characteristics of the tumors from those areas. Therefore, we compared MSI status and DNA copy number profiles in gastric cancer patients from United Kingdom (UK) and South Africa (SA).
Methods
Material
A total of 67 gastric adenocarcinomas were included in this study. Of these, 33 gastric adenocarcinomas were obtained from Leeds (Leeds, General Infirmary, UK) and 34 gastric adenocarcinomas were obtained from Pretoria (Prinshof Campus, Pretoria, South Africa), of which 25 were obtained from native South African patients (native SA) and 9 from Caucasian South African patients (Caucasian SA), respectively. All tumors were randomly selected after testing for proper DNA quality as previously described [10]. All gastric adenocarcinomas were staged according to the TNM classification (5th edition) for the grading and to the Laurén's classification for morphology [11]. The study was approved by the Institutional Review Board and was in accordance with local medical ethical regulations.
DNA isolation procedure
DNA was isolated from formalin-fixed and paraffin embedded gastric cancer material as described previously,[12,13] using the QIAamp microkit (Qiagen, Hilden, Germany). DNA concentrations were measured using a Nanodrop ND-1000 spectrophotometer (Isogen, IJsselstein, The Netherlands) and DNA quality was assessed by isothermal amplification [10]. Genomic DNA isolated from peripheral blood obtained from eighteen healthy females or males was pooled to use as normal reference.
Microsatellite instability (MSI) analysis
MSI analysis was performed using the MSI Analysis System (MSI Multiplex System Version 1.1, Promega) consisting of five nearly monomorphic mononucleotide markers (BAT-25, BAT-26, NR-21, NR-24, MONO-27) according to the manufacturer's instructions. PCR products were separated by capillary electrophoresis using an ABI 3130 DNA sequencer (Applied Biosystems, Foster City, CA, USA), and analyzed using GeneScan 3100 (Applied Biosystems, Foster City, CA, USA). An internal lane size standard was added to the PCR samples for accurate sizing of alleles and to adjust for run-to run variations. When all markers were stable, the tumor was interpreted as microsatellite stable (MSS). The tumor was interpreted as MSI-low (MSI-L) if one marker was instable and MSI-high (MSI-H) if two or more markers showed instability. MSI-L tumors were included in the MSS category in further analysis. Due to polymorphisms[14] in the South African population, native South African tumors were classified as MSI when three or more markers were instable.
Array CGH
Array CGH was performed as described before [12,15]. Briefly, 600 ng tumor and normal reference DNAs were labeled by random priming (Bioprime DNA Labeling System, Invitrogen, Breda, The Netherlands) and hybridized onto a BAC array containing approximately 6000 clones, consisting of the Sanger BAC clone set with an average resolution along the whole genome of 1.0 Mb, the OncoBac set, containing approximately 600 clones corresponding to 200 cancer-related genes, and selected clones of interest obtained from the Children's Hospital Oakland Research Institute (CHORI) to fill gaps larger than 1 Mb on chromosome 6 and to have full coverage contigs of regions on chromosome 8, 13 and 20. All clones were printed in triplicate on Nexterion slides (Schott Nexterion, Jena, Germany). Subsequent analysis was performed according to the clone position from the UCSC May 2004 freeze of the Human Genome Golden Path http://genome.ucsc.edu.
Image acquisition and data analysis
Images of the arrays were acquired by scanning (Agilent DNA Microarray scanner, Agilent Technologies, Palo Alto, USA) and Bluefuse software version 3.4 (BlueGnome, Cambridge, UK) was used for automatic feature extraction. Spots were excluded when the quality flag was below 1 or the confidence value was below 0.1. Log2 tumor to normal ratio was calculated for each clone and median block normalization was used to normalize the data. Quality of array CGH profiles was measured by calculating a median absolute deviation value of chromosome 2 (MAD2) [10]. Array CGH profiles with MAD2 values >0.18 were excluded from further analysis. For determining copy number gains and losses, the R package CGH call was used [16]. Output of the CGH call analysis was used for CGH region analysis to compress the data, using a threshold for average error rate of 0.001 [17]. Hierarchical cluster analysis was performed using the WECCA program, with the parameter total linkage [18].
Array data can be accessed using the Gene Expression Omnibus (GEO) http://www.ncbi.nlm.nih.gov/geo/, under accession number GSE22789.
Statistical analysis
Significance of differences for categorical variables between different categories was tested using a chi-square test. One-way ANOVA with Bonferroni correction was used to calculate significant differences for continuous variables between Caucasian SA, native SA, and UK patients (SPSS 12.0.1 for Windows, SPSS Inc, Chicago, IL, USA). P values less than 0.05 were considered to be significant.
Supervised analysis was performed using the non-parametric Mann-Whitney two-sample test (CGH test [19]). Alterations in patterns between different tumor groups were compared using a binomial differential proportion test. The test procedure included a permutation-based false discovery rate correction for multiple testing [20]. Two-sided p values less than 0.05 and false discovery rates below 0.15 were considered to be significant.
Results
Clinicopathological data
The mean age of the UK gastric cancer patients was 73.3 years (range 51-96), mean age of the Caucasian SA patients was 68.0 years (range 56-84) and the mean age of the native SA patients was 56.5 years (range 29-79). One-way ANOVA with Bonferroni correction yielded a significant difference between the mean age of the patients between native and Caucasian SA patients (p = 0.03) and between native SA and UK patients (p < 0.001), but not between Caucasian SA and UK patients (n.s).
There was no significant difference between patients of different geographical location and gender, tumor stage (T-category) and lymph node stage (N-category). UK gastric cancers showed significantly more diffuse type morphology compared to South African gastric cancers (p = 0.002). Overview of patient and tumor characteristics is given in Table 2.
Table 2.
Tumor and patient characteristics of the 67 tumors used for MSI and array CGH analysis.
| ID | gender | age | Tumor type | T | N | origin | MSI status | Cluster number | Cluster order | % events | % gains | % losses |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | F | 62 | intestinal | T2 | N1 | Cauc SA | MSS | 5 | 34 | 3.2 | 3.2 | 0 |
| 2 | M | 73 | intestinal | T2 | N2 | Cauc SA | MSS | 5 | 28 | 16.9 | 13.4 | 3.4 |
| 3 | F | 59 | intestinal | T2 | N1 | Cauc SA | MSS | 5 | 26 | 26.5 | 15.1 | 11.4 |
| 4 | F | 74 | intestinal | T2 | N1 | Cauc SA | MSS | 5 | 27 | 15.7 | 9.3 | 6.5 |
| 5 | M | 56 | intestinal | T3 | N1 | Cauc SA | MSI | - | - | 15.6 | 10.3 | 5.3 |
| 6 | M | 57 | intestinal | T2 | N1 | Cauc SA | MSS | 6 | 48 | 3.4 | 2.4 | 1.0 |
| 7 | F | 84 | intestinal | T1 | N0 | Cauc SA | MSS | 6 | 43 | 10.9 | 7.9 | 3.1 |
| 8 | F | 79 | intestinal | T3 | N0 | Cauc SA | MSI | - | - | 17.6 | 16.8 | 0.8 |
| 9 | M | 68 | intestinal | T3 | N2 | Cauc SA | MSS | 5 | 25 | 32.1 | 24.3 | 7.7 |
| 10 | M | 65 | intestinal | T2 | N0 | native SA | MSI | - | - | 1.7 | 1.7 | 0 |
| 11 | M | 57 | intestinal | T1 | N0 | native SA | MSI | - | - | 0 | 0 | 0 |
| 12 | F | 29 | intestinal | T4 | N0 | native SA | MSS | 6 | 37 | 28.3 | 26.2 | 2.1 |
| 13 | F | 59 | intestinal | T3 | N1 | native SA | MSS | 6 | 42 | 10.6 | 5.3 | 5.3 |
| 14 | M | 66 | intestinal | T2 | N1 | native SA | MSS | 5 | 31 | 6.4 | 6.4 | 0 |
| 15 | M | 46 | intestinal | T3 | N2 | native SA | MSS | 5 | 35 | 3.2 | 3.2 | 0 |
| 16 | F | - | diffuse | T4 | N2 | native SA | MSS | 6 | 44 | 13.2 | 12.4 | 0.7 |
| 17 | M | 51 | intestinal | T3 | N1 | native SA | MSS | 5 | 32 | 6.1 | 6.1 | 0 |
| 18 | F | 49 | intestinal | T3 | N1 | native SA | MSS | 4 | 20 | 29.1 | 18.2 | 10.9 |
| 19 | M | 56 | intestinal | T3 | N1 | native SA | MSS | 4 | 19 | 26.0 | 10.3 | 15.7 |
| 20 | M | 48 | intestinal | T3 | N2 | native SA | MSS | 5 | 36 | 5.4 | 5.4 | 0 |
| 21 | M | 65 | mixed | T3 | N1 | native SA | MSI | - | - | 18.2 | 16.3 | 1.9 |
| 22 | M | 60 | intestinal | T2 | - | native SA | MSS | 6 | 41 | 14.0 | 11.3 | 2.7 |
| 23 | F | 63 | intestinal | - | - | native SA | MSS | 6 | 38 | 17.1 | 10.7 | 6.4 |
| 24 | F | 54 | papillary | T2 | N0 | native SA | MSS | 3 | 14 | 44.8 | 20.6 | 24.2 |
| 25 | M | 67 | intestinal | T3 | N1 | native SA | MSS | - | - | - | - | - |
| 26 | M | 31 | intestinal | T3 | N1 | native SA | MSS | 5 | 24 | 41.1 | 28.4 | 12.7 |
| 27 | M | 43 | intestinal | T3 | N1 | native SA | MSI | - | - | 9.9 | 9.9 | 0 |
| 28 | F | 71 | intestinal | T3 | - | native SA | MSS | 5 | 33 | 3.7 | 2.7 | 1.0 |
| 29 | F | 77 | intestinal | T3 | N1 | native SA | MSS | 5 | 30 | 12.7 | 7.1 | 5.5 |
| 30 | M | 57 | intestinal | T2 | N0 | native SA | MSS | - | - | - | - | - |
| 31 | M | 79 | intestinal | T3 | N2 | native SA | MSI | - | - | 2.5 | 2.5 | 0 |
| 32 | M | 57 | intestinal | T4 | N0 | native SA | MSI | - | - | 17.8 | 16.8 | 1.0 |
| 33 | M | 56 | mixed | T3 | N1 | native SA | MSS | 4 | 17 | 42.2 | 21.3 | 20.9 |
| 34 | F | 49 | mixed | T3 | N3 | native SA | MSS | 5 | 29 | 22.3 | 19.2 | 3.2 |
| 35 | F | 82 | diffuse | T1 | N0 | UK | MSS | 6 | 46 | 11.2 | 10.3 | 1.0 |
| 36 | M | 81 | diffuse | T3 | N2 | UK | MSS | 4 | 22 | 15.7 | 7.2 | 8.5 |
| 37 | M | 71 | diffuse | T2 | N1 | UK | MSS | 7 | 53 | 18.4 | 14.7 | 3.7 |
| 38 | M | 73 | intestinal | T2 | N0 | UK | MSS | 2 | 11 | 26.0 | 17.2 | 8.8 |
| 39 | F | 65 | diffuse | T2 | N3 | UK | MSS | 6 | 45 | 15.0 | 12.0 | 3.1 |
| 40 | F | 58 | diffuse | T3 | N3 | UK | MSS | 1 | 6 | 25.7 | 13.7 | 12.0 |
| 41 | F | 51 | diffuse | T3 | N3 | UK | MSS | 1 | 7 | 22.4 | 16.5 | 6.0 |
| 42 | M | 91 | intestinal | T1 | N0 | UK | MSS | 7 | 56 | 49.2 | 27.6 | 21.6 |
| 43 | M | 71 | diffuse | T3 | N2 | UK | MSS | 7 | 49 | 40.6 | 19.8 | 20.8 |
| 44 | M | 73 | intestinal | T2 | N1 | UK | MSS | 1 | 5 | 21.0 | 8.2 | 12.9 |
| 45 | M | 64 | diffuse | T2 | N0 | UK | MSS | 7 | 50 | 14.9 | 10.3 | 4.6 |
| 46 | F | 71 | intestinal | T1 | N0 | UK | MSS | 1 | 4 | 40.8 | 13.4 | 27.4 |
| 47 | F | 60 | intestinal | T4 | N1 | UK | MSS | 3 | 16 | 30.1 | 15.6 | 14.5 |
| 48 | F | 96 | diffuse | T3 | N0 | UK | MSS | 2 | 12 | 41.0 | 25.0 | 16.0 |
| 49 | M | 91 | mixed | T3 | N0 | UK | MSS | 6 | 40 | 16.0 | 11.4 | 4.6 |
| 50 | M | 81 | diffuse | T2 | N0 | UK | MSS | 2 | 10 | 37.7 | 17.2 | 20.4 |
| 51 | F | 83 | intestinal | T2 | N0 | UK | MSS | 7 | 51 | 14.5 | 10.5 | 4.0 |
| 52 | F | 82 | intestinal | T3 | N1 | UK | MSS | 6 | 47 | 2.2 | 2.2 | 0 |
| 53 | F | 77 | intestinal | T3 | N1 | UK | MSS | 6 | 39 | 19.5 | 11.1 | 8.5 |
| 54 | M | 74 | mixed | T2 | N1 | UK | MSS | 2 | 8 | 44.8 | 22.4 | 22.3 |
| 55 | F | 59 | diffuse | T3 | N1 | UK | MSS | 3 | 15 | 26.4 | 18.1 | 8.2 |
| 56 | M | 77 | mixed | T3 | N2 | UK | MSS | 1 | 3 | 24.1 | 13.7 | 10.3 |
| 57 | M | 75 | intestinal | T2 | N0 | UK | MSS | 2 | 9 | 36.9 | 18.2 | 18.7 |
| 58 | M | 64 | diffuse | T3 | N3 | UK | MSS | 2 | 13 | 35.3 | 20.2 | 15.1 |
| 59 | M | 71 | intestinal | T3 | N1 | UK | MSS | 1 | 2 | 31.0 | 11.7 | 19.3 |
| 60 | F | 74 | diffuse | T3 | N2 | UK | MSS | 4 | 23 | 23.2 | 14.7 | 8.4 |
| 61 | M | 81 | intestinal | T2 | N0 | UK | MSI | - | - | 10.7 | 8.7 | 2.0 |
| 62 | F | 74 | mixed | T3 | N2 | UK | MSS | 4 | 18 | 30.4 | 17.9 | 12.5 |
| 63 | M | 67 | mixed | T3 | N1 | UK | MSS | 7 | 52 | 12.3 | 6.4 | 5.9 |
| 64 | M | 73 | intestinal | T3 | N1 | UK | MSS | 4 | 21 | 15.2 | 7.3 | 7.9 |
| 65 | F | 66 | mixed | T3 | N2 | UK | MSS | 7 | 55 | 34.3 | 18.7 | 15.6 |
| 66 | M | 82 | intestinal | T1 | N1 | UK | MSS | 7 | 54 | 46.7 | 27.4 | 19.3 |
| 67 | F | 62 | mixed | T3 | N0 | UK | MSS | 1 | 1 | 44.0 | 20.7 | 23.3 |
Percentages of events, gains and losses are given for all tumors of sufficient array CGH quality. Cluster number and order are listed for the 56 cases included in the cluster analysis.
F: female, M: male, T: T-stage, N: N-stage MSS: microsatellite stable, MSI: microsatellite instable, Cauc SA: Caucasian South African patients, native SA: native South African patients, UK: patients from United Kingdom, -: unknown.
Microsatellite instability (MSI) analysis
Two out of nine (22%) Caucasian SA gastric cancers, six out of 25 (24%) native SA gastric cancers, and one out of 33 (3%) UK gastric cancers showed MSI. All other gastric cancers were MSS (Table 2). Pearson chi-square yielded a significant difference between the three different tumor groups and MSI status (p < 0.05).
Hierarchical cluster analysis
We analyzed DNA from all gastric cancers by genome-wide array CGH analysis to unravel DNA copy number changes in tumors from different geographical location. MSI positive gastric cancers and gastric cancers with array CGH profiles with a MAD2 value above 0.18 were excluded for cluster analysis leaving 56 tumors (from 32 UK, 17 native SA and 7 Caucasian SA patients) for further analysis.
Hierarchical cluster analysis yielded seven clusters which were significantly correlated with gastric cancers of different geographical origin (p < 0.001) (Figure 1). Clusters 1, 2 and 7 obtained only gastric cancers from UK patients. Clusters 3 and 4 comprised of gastric cancers from UK and native SA patients. Cluster 5 contained only gastric cancers from SA patients, and cluster 6 contained a mixture of tumors of all three groups (Table 2).
Figure 1.
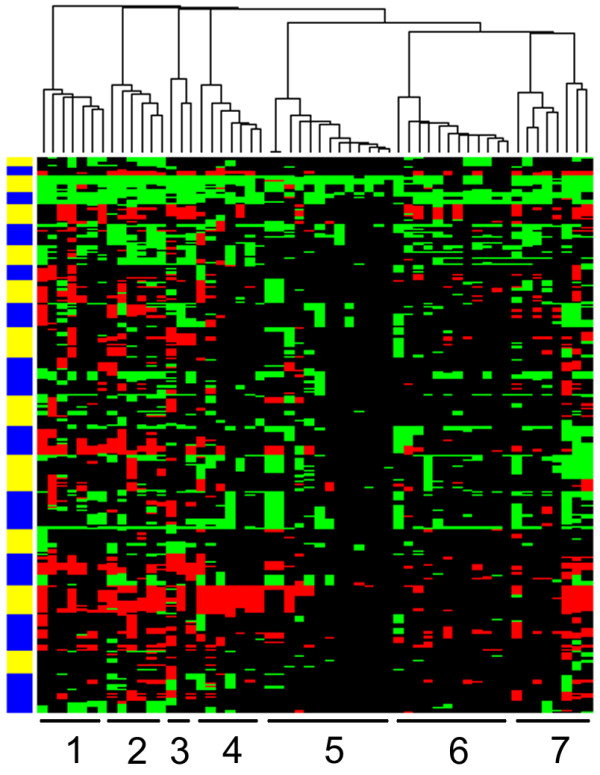
Cluster analysis of 56 gastric adenocarcinomas of which 32, 17 and 7 were obtained from UK, native SA and Caucasian SA patients, respectively. Hierarchical cluster analysis yielded 7 clusters significantly correlated with geographical origin of the tumors (p < 0.001). Columns represent the different tumors and rows represent the different chromosomal regions, with chromosome 1 at the bottom and chromosome 22 at the top of the heatmap. DNA copy number gains and losses are indicated in green and red, respectively. The yellow and blue bar next to the cluster represents the chromosome separation.
UK patients showed significantly more gastric adenocarcinomas of the diffuse type according to the Laurén classification[11] compared to SA patients (p = 0.002). We therefore repeated the cluster analysis including only intestinal type gastric carcinomas. Cluster membership of the remaining 12 tumors from UK patients and 12 and 7 tumors from native SA and Caucasian SA patients, respectively, was again significantly correlated to geographical origin of the patient (p < 0.001). Moreover, when analyzing only UK gastric cancers, hierarchical cluster analysis did not separate intestinal and diffuse type gastric cancers, nor were any significant differences observed between these two morphological tumor types with supervised analysis using CGH test.
Cluster membership was independent of gender, tumor stage, lymph node stage and of age of the patients (categorized as < 50 years of age versus ≥ 50 years of age).
DNA copy number changes
We first compared the number of events, which was defined as percentage of clones showing a gain or loss. Gastric cancers from UK patients showed a higher number of events (27% (range 2-49%)) compared to cancers from Caucasian SA (16% (range 3-32%)) and native SA patients (16% (range 0-45%)) (p = 0.005). Cancers from UK, Causasian SA and native SA patients showed 15% (range 2-28%), 11% (range 2-24%) and 11% (range 0-28%) of gained clones respectively, and 12% (range 0-27%), 4% (range 0-11%) and 5% (0-24%) clones showing a loss, respectively. A significant difference in the percentage of clones showing a loss was observed between UK patients and Caucasian SA patients (p = 0.002) and between UK patients and native SA patients (p = 0.02).
Also, when looking only at microsatellite stable gastric cancers UK patients showed a higher number of events (27% (range 2-49%)) compared to microsatellite stable cancers from Caucasian SA and native SA patients (16% (range 3-32%) and 19% (range 3-45%), respectively; p = 0.04). Microsatellite stable cancers from UK, Caucasian SA and native SA patients showed 15% (range 2-28%), 12% (range 2-24%) and 13% (range 3-28%) of gained clones, respectively. There was again a significant difference in percentages of clones showing a loss between cancers from UK and Caucasian SA patients (12% (range 0-27%) and 4% (range 0-11%), respectively; p = 0.04) and between cancers from UK and native SA patients (12% (range 0-27%) and 7% (range 0-24%) respectively; p = 0.04).
An overview of frequently altered (>30%) chromosomal regions with gains and losses per tumor group is given in Tables 3, 4 and 5. Most frequently altered (>30%) chromosomal regions observed in the UK tumors were gains on chromosomes 1p, 1q, 5p, 6p, 7p, 7q, 8q, 9q, 10p, 10q, 11p, 11q, 13q, 14q, 16p, 16q, 17p, 17q, 19p, 19q, 20p, 20q, 21q and 22q and losses on chromosomes 1p, 1q, 3p, 4p, 4q, 5q, 9p, 12q, 13q, 14q, 15q, 17q, 18q and 21q (Table 3). Most frequent DNA copy number aberrations in the native SA patients were gains on the chromosomal regions 7p, 7q, 8q, 9q, 17q, 19p, 19q, 20p and 20q, and losses on 3p and 4q (Table 4). Most frequently altered chromosomal regions in Caucasian SA patients were gains on 3q, 5p, 7p, 7q, 8p, 8q, 9q, 11q, 16p, 17q, 19p, 19q, 20p and 20q and losses on 3p, 4q and 9p (Table 5). A summary of frequencies of gains and losses of all gastric cancers per tumor group is presented in Figures 2 (UK), 3 (native SA) and 4 (Caucasian SA).
Table 3.
Detailed overview of frequent DNA copy number aberrations (>30%) of tumors from UK patients.
| chromosomal aberrations | flanking clones | position (bp) | segment size | |||
|---|---|---|---|---|---|---|
| gains | losses | start | end | start | end | (Mb) |
| 1p36.33-p36.21 | RP11-206L10 | RP4-636F13 | 672780 | 12417597 | 11.74 | |
| 1p31.1 | RP5-944F13 | RP11-246O4 | 69815162 | 83112098 | 13.30 | |
| 1p21.3-p13.3 | RP11-146P11 | RP5-1077K16 | 95695632 | 107389118 | 11.69 | |
| 1q21.2-q23.1 | RP4-790G17 | RP11-214H6 | 146971278 | 153444622 | 6.47 | |
| 1q31.1-q31.3 | RP11-134C1 | RP11-75C23 | 184717073 | 194242465 | 9.53 | |
| 1q32.1-q32.2 | RP11-150l7 | RP11-564A8 | 197877387 | 203602276 | 5.72 | |
| 3p26.3 | RP11-385A18 | RP11-129K1 | 46140 | 2377366 | 2.33 | |
| 3p25.1-p24.1 | RP11-255O19 | RP11-99M10 | 15780361 | 30799547 | 15.02 | |
| 3p14.2 | RP11-170K19 | RP11-114P15 | 59701329 | 62639806 | 2.94 | |
| 3p12.3-p11.2 | RP11-103P13 | RP11-91M15 | 75146113 | 96627928 | 21.48 | |
| 4p16.1-q35.2 | RP11-61G19 | CTC-963K6 | 10275012 | 191158370 | 180.88 | |
| 5p15.33 | RP11-811I15 | CTD-2265D9 | 70262 | 2671745 | 2.60 | |
| 5q11.1-q23.3 | RP11-269M20 | RP11-114H7 | 49913067 | 130460728 | 80.55 | |
| 6p21.32-p21.1 | RP11-79I1 | RP11-121G20 | 33123932 | 44385866 | 11.26 | |
| 7p22.3-p22.1 | RP11-713A20 | RP11-161C7 | 106471 | 6396697 | 6.29 | |
| 7p11.2 | RP11-449G3 | RP11-34J24 | 54413814 | 55403627 | 0.99 | |
| 7q22.1 | RP11-10D8 | RP11-163M5 | 98067793 | 101528379 | 3.46 | |
| 7q36.1 | RP11-89P11 | RP11-43l19 | 147485335 | 151131938 | 3.65 | |
| 7q36.3 | RP11-58F7 | RP11-120H14 | 157072238 | 158524109 | 1.45 | |
| 8q24.12-q24.3 | RP11-22A24 | RP5-1109M23 | 120711365 | 146238749 | 25.53 | |
| 9p24.3-p21.1 | RP11-48M17 | RP11-141J7 | 2136329 | 32469400 | 30.33 | |
| 9q33.3-q34.3 | RP11-205K6 | RP11-424E7 | 126296075 | 138363252 | 12.07 | |
| 10p15.3 | RP11-631M21 | RP11-74N14 | 50000 | 1789100 | 1.74 | |
| 10p15.2 | RP11-195B3 | 3293007 | 3338470 | 0.05 | ||
| 10q22.1 | RP11-91A1 | RP11-28E3 | 72033907 | 73573433 | 1.54 | |
| 11p15.5-p15.4 | CTC-908H22 | RP11-304P12 | 178227 | 3140168 | 2.96 | |
| 11q12.2-q13.5 | RP11-286N22 | RP11-30J7 | 60851860 | 76232373 | 15.38 | |
| 12q21.2-q22 | RP1-97G4 | RP11-2K12 | 76228586 | 91346299 | 15.12 | |
| 13q11-q12.11 | RP11-94A1 | RP11-61K9 | 18360157 | 19386914 | 1.03 | |
| 13q21.2-q21.33 | RP11-310K10 | RP11-451E2A | 60721181 | 71574154 | 10.85 | |
| 13q32.3 | RP11-19J14 | RP11-113F15 | 97851594 | 99328275 | 1.48 | |
| 13q33.3-q34 | RP11-61I17 | RP11-569D9 | 108847725 | 114103243 | 5.26 | |
| 14q12 | RP11-330O19 | RP11-109D12 | 25538832 | 26345513 | 0.81 | |
| 14q21.1-q21.3 | RP11-88D14 | RP11-94K16 | 36949212 | 48298851 | 11.35 | |
| 14q31.1-q31.3 | RP11-46l17 | RP11-88N20 | 78630860 | 86772926 | 8.14 | |
| 14q32.31-q32.33 | RP11-367F11 | RP11-815P21 | 101467534 | 105159201 | 3.69 | |
| 15q14 | RP11-294M6 | RP11-79A5 | 33841939 | 35953471 | 2.11 | |
| 16p13.3 | CTD-2148K8 | RP11-89M4 | 87754 | 4697230 | 4.61 | |
| 16p13.2-p13.11 | RP11-475D10 | RP11-489O1 | 8598165 | 15572359 | 6.97 | |
| 16p12.1-p11.2 | RP11-142A12 | RP11-18H23 | 26595069 | 31443695 | 4.85 | |
| 16q21-q22.1 | RP11-52B24 | RP11-394B2 | 63708677 | 69365102 | 5.66 | |
| 16q23.3-q24.3 | RP11-483P21 | RP11-566K11 | 82361609 | 88613383 | 6.25 | |
| 17p13.3-p13.1 | RP11-411G7 | RP11-89A15 | 427024 | 8365794 | 7.94 | |
| 17p11.2 | RP11-524F11 | RP1-162E17 | 17343389 | 19251691 | 1.91 | |
| 17q11.2-q21.31 | RP11-138P22 | RP11-374N3 | 23133763 | 41096064 | 17.96 | |
| 17q21.32-q21.33 | RP11-234J24 | RP11-506D12 | 42655422 | 46333070 | 3.68 | |
| 17q22-q23.2 | RP11-143M4 | RP11-139B3 | 47607556 | 51363278 | 3.76 | |
| 17q24.3-q25.3 | RP11-65C22 | RP11-258N23 | 68165339 | 78308832 | 10.14 | |
| 18q11.2-q23 | RP11-5G23 | RP11-396D4 | 21431314 | 71337306 | 49.91 | |
| 19p13.3-q13.43 | RP11-110A24 | GS1-1129C9 | 134914 | 63771717 | 63.64 | |
| 20p13-q13.33 | RP11-530N10 | CTB-81F12 | 9943 | 62393015 | 62.38 | |
| 21q11.2-q22.11 | RP11-72P4 | RP11-41N19 | 13857799 | 30673984 | 16.82 | |
| 22q11.1-q11.21 | RP11-81H21 | RP11-586I18 | 14754982 | 18976359 | 4.22 | |
| 22q12.3-q13.33 | RP11-90I17 | CTB-99K24 | 35686144 | 49397088 | 13.71 | |
Table 4.
Detailed overview of frequent DNA copy number aberrations (>30%) of tumors from native SA patients.
| chromosomal aberrations | flanking clones | position (bp) | segment size | |||
|---|---|---|---|---|---|---|
| gains | losses | start | end | start | end | (Mb) |
| 3p14.2 | RP11-734E15 | RP11-137N22 | 59105371 | 61252524 | 2.15 | |
| 4q35.2 | RP11-354H17 | CTC-963K6 | 190095484 | 191158370 | 1.06 | |
| 7p22.3-p11.2 | RP11-713A20 | RP11-80l24 | 106471 | 55784518 | 55.68 | |
| 7q22.1 | RP11-10D8 | RP11-163M5 | 98067793 | 101528379 | 3.46 | |
| 8q24.3 | RP11-472K18 | RP5-1109M23 | 144481535 | 146238749 | 1.76 | |
| 9q33.3-q34.3 | RP11-91G7 | RP11-424E7 | 124316484 | 138363252 | 14.05 | |
| 17q12-q21.31 | RP11-893G17 | RP11-392O1 | 31506328 | 39091575 | 7.59 | |
| 17q21.32-q21.33 | RP1-62O9 | RP11-506D12 | 44647598 | 46333070 | 1.69 | |
| 17q23.2-q25.3 | RP11-579A4 | RP11-258N23 | 54149948 | 78451750 | 24.30 | |
| 19p13.3 | RP11-110A24 | CTC-1482H14 | 134914 | 5154803 | 5.02 | |
| 19p13.2-p13.11 | RP11-197O4 | RP11-88I12 | 10248852 | 19023254 | 8.77 | |
| 19q12-q13.34 | CTC-1459F4 | GS1-1129C9 | 32889410 | 63771717 | 30.88 | |
| 20p13-q13.33 | RP11-530N10 | CTB-81F12 | 9943 | 62393015 | 62.38 | |
Table 5.
Detailed overview of frequent DNA copy number aberrations (>30%) of tumors from Caucasian SA patients.
| chromosomal aberrations | flanking clones | position (bp) | segment size | |||
|---|---|---|---|---|---|---|
| gains | losses | start | end | start | end | (Mb) |
| 3p14.2 | RP11-48E21 | RP11-641C17 | 60380670 | 60705094 | 0.32 | |
| 3q26.2-q26.31 | RP11-669J9 | RP11-44A1 | 172392313 | 173855790 | 1.46 | |
| 4q32.1-q35.2 | RP11-192D11 | CTC-963K6 | 159886665 | 191158370 | 31.27 | |
| 5p13.1-p12 | RP11-17J3 | RP11-55O15 | 40113135 | 44396362 | 4.28 | |
| 7p22.3-p21.3 | RP11-713A20 | RP11-505D17 | 106471 | 7932634 | 7.83 | |
| 7q22.1 | RP4-550A13 | RP11-333G13 | 98512376 | 101153193 | 2.64 | |
| 8p23.1 | RP11-241P12 | RP11-589N15 | 9788949 | 11803111 | 2.01 | |
| 8q22.1-q22.3 | RP11-664H21 | RP11-132E3 | 98618965 | 105402542 | 6.78 | |
| 8q24.21 | RP11-28I2 | RP11-1142f3 | 127563658 | 129620230 | 2.06 | |
| 9p24.1-p23 | RP11-165O14 | RP11-91E3 | 5873408 | 9689968 | 3.82 | |
| 9q33.3-q34.3 | RP11-62A6 | RP11-424E7 | 124479347 | 138363252 | 13.88 | |
| 11q13.3-q13.5 | RP11-554A11 | RP11-98G24 | 68509550 | 77008323 | 8.50 | |
| 16p11.2 | RP11-110P16 | RP11-388M20 | 28675396 | 31163676 | 2.49 | |
| 17q12-q21.1 | RP5-986F12 | RP11-94L15 | 33099924 | 35227135 | 2.13 | |
| 17q25.1-q25.3 | RP11-41E12 | RP11-258N23 | 68729134 | 78451750 | 9.72 | |
| 19p13.3-p13.11 | RP11-110A24 | RP11-88I12 | 134914 | 19023254 | 18.89 | |
| 19q13.11-q13.43 | CTC-1325L16 | GS1-1129C9 | 37623641 | 63771717 | 26.15 | |
| 20p13-q13.33 | RP11-48M7 | CTB-81F12 | 3728265 | 62393015 | 58.66 | |
Figure 2.

Frequencies of gains and losses throughout the genome in all gastric adenocarcinomas from UK patients. Clones are sorted by position per chromosome (1-22). Vertical lines indicate transition between chromosomes; dashed vertical lines indicate centromere position.
Figure 3.
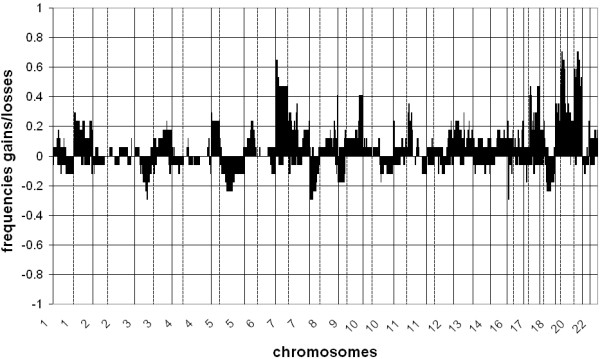
Frequencies of gains and losses throughout the genome in all gastric adenocarcinomas from native SA patients. Clones are sorted by position per chromosome (1-22). Vertical lines indicate transition between chromosomes; dashed vertical lines indicate centromere position.
Figure 4.
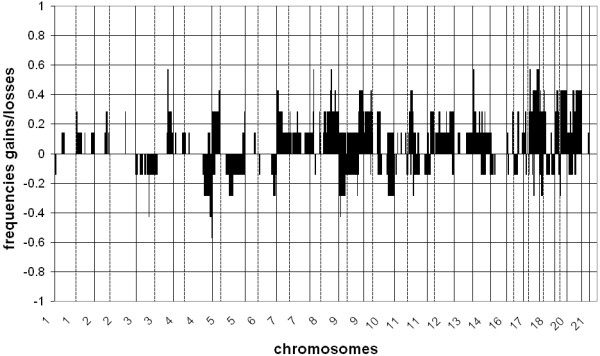
Frequencies of gains and losses throughout the genome in all gastric adenocarcinomas from Caucasian SA patients. Clones are sorted by position per chromosome (1-22). Vertical lines indicate transition between chromosomes; dashed vertical lines indicate centromere position.
Supervised analysis
To identify biological differences between gastric cancers from different geographical origin, native SA tumors were compared with UK tumors using CGH test. Only MSS tumors were included in the supervised analysis. In total, 133 regions, located on different chromosomes, were significantly different (p < 0.05 and fdr≤0.15) between these two patient groups. An overview of the significant chromosomal regions, including the fdr rates, is given in Table 6. No significant differences were found between gastric cancers from UK and Caucasian SA patients or between gastric cancers from native and Caucasian SA patients.
Table 6.
Detailed overview of the supervised analysis using CGH test.
| cytoband | region start (bp) | region end (bp) | p-value | fdr | cytoband | region start (bp) | region end (bp) | p-value | fdr |
|---|---|---|---|---|---|---|---|---|---|
| 1p36.33 | 672780 | 1359795 | 0.04 | 0.15 | 11p14.2-p14.1 | 27033269 | 27371257 | 0.05 | 0.15 |
| 1p36.32-p36.31 | 3386389 | 6294064 | 0.03 | 0.12 | 11q13.3 | 68509550 | 69323966 | 0.04 | 0.14 |
| 1p36.21-p36.13 | 12798944 | 15683816 | 0.03 | 0.12 | 11q13.3-q13.4 | 69314721 | 70472869 | 0.04 | 0.14 |
| 1p31.2 | 67178936 | 69303906 | 0.04 | 0.14 | 11q22.1-q22.2 | 98930611 | 101405228 | 0.03 | 0.12 |
| 1p31.1 | 69815162 | 76679895 | 0.01 | 0.06 | 11q22.2-q22.3 | 102010610 | 102424014 | 0.03 | 0.12 |
| 1p31.1 | 77428804 | 77820126 | <0.01 | 0.04 | 12q21.2 | 76570565 | 78724263 | 0.04 | 0.14 |
| 1p21.2-p21.1 | 101684496 | 104502748 | 0.03 | 0.12 | 13q21.31-q21.33 | 61335626 | 69275204 | 0.04 | 0.14 |
| 1q31.1-q31.2 | 184717073 | 188976520 | 0.01 | 0.09 | 14q21.1 | 39694531 | 42171623 | 0.02 | 0.10 |
| 1q31.2-q31.3 | 189822405 | 193082884 | 0.02 | 0.10 | 14q21.2 | 42965408 | 44043547 | 0.01 | 0.09 |
| 1q31.3 | 193336091 | 194242465 | 0.01 | 0.08 | 14q21.2-q21.3 | 45258184 | 48298851 | 0.03 | 0.12 |
| 1q31.3 | 195068870 | 195629725 | 0.03 | 0.12 | 15q14 | 33841939 | 35953471 | 0.04 | 0.14 |
| 3p26.3 | 46140 | 2377366 | <0.01 | 0.03 | 15q22.2 | 57373165 | 61214280 | 0.02 | 0.09 |
| 3p24.3 | 17181327 | 18148477 | <0.01 | 0.03 | 15q23 | 65816865 | 68768615 | 0.02 | 0.09 |
| 3p24.3 | 19033520 | 21742232 | <0.01 | 0.03 | 15q23-q24.2 | 69188639 | 73645336 | 0.05 | 0.15 |
| 3p24.3-p24.1 | 22747912 | 27531283 | 0.02 | 0.10 | 15q24.2-q25.2 | 74334873 | 81024206 | 0.03 | 0.12 |
| 3p21.31 | 46545403 | 47371983 | 0.05 | 0.15 | 16p13.2-p13.12 | 8777494 | 12522798 | 0.05 | 0.15 |
| 3p21.31 | 47384745 | 50114898 | 0.04 | 0.14 | 16p11.2 | 28675396 | 31163676 | <0.01 | 0.04 |
| 3p21.31-p21.2 | 50533656 | 51418837 | 0.01 | 0.09 | 16q24.1 | 82993991 | 83622386 | 0.05 | 0.15 |
| 3p21.31-p21.2 | 51390596 | 52007218 | 0.02 | 0.09 | 16q24.1 | 84123856 | 84922042 | 0.02 | 0.09 |
| 3p21.1 | 52658450 | 53621497 | 0.01 | 0.09 | 16q24.2-q24.3 | 86986672 | 87452904 | 0.03 | 0.12 |
| 3p12.3 | 76450939 | 79097142 | 0.02 | 0.09 | 16q24.3 | 87848795 | 88465228 | 0.02 | 0.09 |
| 3p12.3-p12.2 | 79197544 | 82066233 | <0.01 | 0.04 | 16q24.3 | 88398231 | 88613383 | 0.01 | 0.07 |
| 3p12.2-p12.1 | 82657794 | 84801874 | <0.01 | 0.03 | 17p13.3 | 427024 | 1071560 | <0.01 | 0.03 |
| 3p12.1-p11.2 | 85234579 | 87730259 | <0.01 | 0.03 | 17p13.3-p13.2 | 2026966 | 4169913 | 0.04 | 0.14 |
| 3p11.1 | 88915283 | 89771786 | <0.01 | 0.04 | 17p13.2 | 4810523 | 5166678 | 0.01 | 0.09 |
| 3q11.2 | 95569618 | 96250439 | 0.01 | 0.05 | 17p13.1 | 6780962 | 7477414 | <0.01 | 0.03 |
| 3q25.1-q25.33 | 151508469 | 161536133 | 0.03 | 0.12 | 17p13.1 | 7436435 | 7682367 | <0.01 | 0.03 |
| 3q25.33-q26.1 | 162044657 | 164516640 | 0.03 | 0.12 | 17p11.2 | 17343389 | 19251691 | 0.04 | 0.14 |
| 3q26.1 | 165289729 | 167697600 | 0.05 | 0.15 | 17q11.2 | 23133763 | 28423820 | 0.02 | 0.09 |
| 3q26.31-q26.32 | 174848631 | 180449332 | 0.05 | 0.15 | 17q11.2-q12 | 28664889 | 29147086 | 0.02 | 0.09 |
| 4p15.32-p14 | 17799729 | 36260048 | 0.02 | 0.11 | 17q25.1 | 69709369 | 71238958 | 0.01 | 0.09 |
| 4p14 | 36998587 | 37497326 | 0.01 | 0.09 | 17q25.1-q25.3 | 71406319 | 77588058 | 0.03 | 0.12 |
| 4p14 | 37851044 | 38103870 | 0.02 | 0.11 | 18q11.2 | 21431314 | 21774952 | 0.01 | 0.05 |
| 4p14-p12 | 38087293 | 46781512 | 0.01 | 0.09 | 18q11.2 | 22104619 | 22931012 | <0.01 | 0.04 |
| 4q12 | 58332155 | 59148507 | 0.04 | 0.14 | 18q12.1 | 23970882 | 24526449 | 0.02 | 0.10 |
| 4q12-q13.1 | 59679465 | 62653308 | 0.02 | 0.10 | 18q12.1 | 25034674 | 26878315 | 0.01 | 0.08 |
| 4q13.3 | 74322497 | 76429795 | 0.03 | 0.12 | 18q12.1 | 27447009 | 28415095 | 0.01 | 0.09 |
| 4q13.3-q21.21 | 76495364 | 79353372 | 0.02 | 0.09 | 18q12.1 | 29409483 | 30219417 | 0.01 | 0.09 |
| 4q21.21 | 79222718 | 80683287 | 0.03 | 0.12 | 18q12.1-q12.2 | 30773824 | 31588529 | 0.01 | 0.06 |
| 4q33-q34.3 | 172094999 | 178721484 | 0.02 | 0.10 | 18q22.1 | 60340433 | 61310295 | 0.03 | 0.12 |
| 4q34.3 | 178969859 | 179599683 | 0.01 | 0.09 | 18q22.1 | 61623805 | 62645202 | 0.03 | 0.12 |
| 4q34.3 | 180819690 | 183096645 | 0.02 | 0.10 | 18q22.1-q22.3 | 63092764 | 68041625 | 0.03 | 0.12 |
| 4q35.1 | 184503994 | 186178296 | 0.03 | 0.12 | 19p13.3 | 134914 | 913289 | 0.01 | 0.08 |
| 5q11.2 | 50971745 | 52055659 | 0.04 | 0.14 | 19p13.3 | 902641 | 5009969 | <0.01 | 0.04 |
| 5q11.2 | 52909242 | 56437163 | 0.03 | 0.12 | 19p13.3 | 5663923 | 6519297 | <0.01 | 0.01 |
| 5q11.2-q12.1 | 56921490 | 58947885 | 0.02 | 0.09 | 19p13.3-p13.2 | 6523443 | 9826740 | <0.01 | <0.01 |
| 5q14.3 | 82802677 | 86118543 | 0.05 | 0.15 | 19p13.2-p13.12 | 10248852 | 15116365 | <0.01 | 0.03 |
| 5q23.2 | 122463056 | 123527915 | 0.05 | 0.15 | 19p13.12-p13.11 | 15415833 | 17777501 | <0.01 | 0.01 |
| 7p22.1-p21.3 | 6983150 | 7932634 | <0.01 | 0.02 | 19p13.11 | 18202507 | 19023254 | <0.01 | 0.03 |
| 7p21.3-p21.2 | 9256088 | 14085902 | <0.01 | <0.01 | 19p12 | 19877150 | 21504328 | 0.01 | 0.05 |
| 7p21.2-p21.1 | 14342152 | 19957111 | <0.01 | 0.01 | 19p12 | 22133662 | 22949959 | 0.01 | 0.05 |
| 7q22.1 | 98651132 | 100929260 | <0.01 | 0.03 | 19q12 | 33315121 | 33507712 | 0.02 | 0.10 |
| 8q24.21 | 130562467 | 131126185 | 0.05 | 0.15 | 19q12 | 34159370 | 34664148 | 0.02 | 0.11 |
| 8q24.22 | 131641447 | 133561490 | 0.01 | 0.09 | 19q12 | 34960906 | 35766560 | 0.02 | 0.11 |
| 8q24.22 | 134537164 | 136154996 | 0.02 | 0.11 | 19q12-q13.11 | 36958851 | 37518517 | 0.05 | 0.15 |
| 8q24.22-q24.23 | 136472354 | 138543270 | 0.04 | 0.14 | 19q13.12-q13.13 | 40883472 | 43004503 | 0.03 | 0.13 |
| 8q24.3 | 141395868 | 142790557 | 0.01 | 0.05 | 19q13.2-q13.32 | 46498596 | 50725496 | 0.03 | 0.12 |
| 8q24.3 | 144790054 | 145357620 | 0.01 | 0.09 | 19q13.32-q13.33 | 52665374 | 54718281 | 0.03 | 0.12 |
| 8q24.3 | 145585590 | 145953950 | 0.02 | 0.11 | 19q13.33-q13.43 | 55461670 | 63771717 | <0.01 | 0.01 |
| 8q24.3 | 145893230 | 146238749 | 0.04 | 0.14 | 21q11.2-q21.1 | 13857799 | 18774434 | <0.01 | 0.03 |
| 9p24.1-p23 | 8398601 | 9689968 | 0.04 | 0.14 | 21q21.1 | 20982315 | 21411332 | <0.01 | 0.03 |
| 9p23 | 9684353 | 10554235 | 0.04 | 0.14 | 21q21.1-q21.2 | 22151920 | 22943367 | <0.01 | 0.03 |
| 10q22.1 | 70824040 | 71674097 | 0.03 | 0.12 | 21q21.2 | 23491399 | 25568510 | <0.01 | 0.01 |
| 10q22.1 | 72033907 | 73573433 | 0.02 | 0.09 | 21q21.3 | 26174732 | 26923374 | 0.02 | 0.09 |
| 10q26.3 | 135110821 | 135301208 | 0.03 | 0.12 | 21q21.3-q22.11 | 28596969 | 30855176 | 0.03 | 0.12 |
| 11p15.5 | 178227 | 626401 | 0.01 | 0.05 | 22q13.33 | 48473404 | 49397088 | 0.04 | 0.15 |
| 11p15.5 | 1299306 | 1785278 | <0.01 | 0.03 |
In total, 133 regions were significantly different (p < 0.05 and fdr≤0.15) between UK and native SA tumors. The chromosomal regions, including the start and end positions, and the fdr rates are listed.
Discussion
One of the main risk factors contributing to gastric cancer is infection with H. pylori, which causes a chronic active gastritis [4,21]. In South Africa, gastric cancer is infrequent, while the prevalence of H. pylori infection is very high. Although differences in genotypes of H. pylori exist in different geographic areas, this African enigma can not only be explained by differences in virulent strains of H. pylori [22-24]. High prevalence of vacA s1b strain is observed in South Africa as well as in Brazil and Portugal, countries with high incidences of gastric cancer,[25-27] and frequencies of CagA antibodies were similar between patients with gastric neoplasia compared to healthy controls [28]. The prevalence of the different virulent strains in the present study is unknown. Since H. pylori is thought to play a major role in the initiation phase of gastric cancer development and most often already disappeared at time of gastric cancer diagnosis, it is impossible to accurately retrieve this information.
Besides the virulence of the infecting H. pylori strain, other factors influence gastric cancer risk, including environmental factors such as diet and socioeconomic status, and host factors, such as polymorphisms, which are involved in the inflammatory response to the infection [29,30]. Knowing that the prevalence of H. pylori infection and incidences of gastric cancer are different in South Africa and Western Europe, we aimed to study if this would reflect in different patterns of gastric carcinogenesis.
The concept of the African enigma has been challenged since it has been suggested that the enigma could be explained due to lack of infrastructure and access to hospitals and care in African countries resulting in incomplete reporting of gastric cancer. However the incidence of gastric cancer would have been so dramatically underestimated that it has been stated that under-reporting by itself could not explain the lower frequencies of gastric cancer in African countries [31]. Also, when using the proportional frequency of gastric cancer compared to other cancer types in Africa, gastric cancer incidence remains very low [8]. Another criticism on the African enigma has been the high prevalence of HIV infection. A relatively large part of the African population would die of HIV before the age in which gastric cancer becomes more frequent. However, the low gastric cancer incidence in Africa was described before the HIV epidemic.
South African patients showed significantly more microsatellite instable gastric cancers compared to Western European patients. Also at the level of chromosomal instability clear differences were found, reflected by a significant correlation between cluster membership and geographical tumor origin, i.e. UK, native SA and Caucasian SA. Microsatellite instable gastric cancers are described to have fewer chromosomal aberrations compared to microsatellite stable gastric cancers [32,33]. To rule out that tumors from South African patients cluster together by hierarchical cluster analysis due to the fact that these tumors show higher frequencies of microsatellite instability, only microsatellite stable gastric cancers were included in the hierarchical cluster analysis.
Not much has been reported about microsatellite status in gastric cancers from African patients. One study reported infrequent microsatellite instable gastric cancers in South African patients[34] which is in contrast with our findings which show a higher frequency of microsatellite instability in gastric cancers from SA patients compared to UK patients. Based on the present data, MSI does play an important role in gastric carcinogenesis in South Africa.
Several chromosomal aberrations are common in the three different tumor groups analyzed, including gains of chromosomes 7, 8q, 9q, 17q, 19 and 20 and losses of 3p and 4q, while other chromosomal changes are specific for each tumor group. In addition, gastric cancers from UK patients showed a significantly higher number of clones showing a loss compared to gastric cancers form South African patients. These results indicate different patterns of chromosomal instabilities in gastric cancers correlating to geographical origin of the patient.
The chromosomal aberrations of the UK tumors are comparable to other array CGH studies analyzing Western European tumors [12,35-37]. To the best of our knowledge, this is the first array CGH study on gastric cancers from South African patients. Since several chromosomal regions are significantly different between gastric cancers from different geographical origin, and each region comprises multiple genes, further studies are needed to pinpoint candidate genes contributing to the differences in genomic profiles.
The higher frequency of diffuse gastric cancers from UK patients compared to the SA patients in the present study could be considered as a confounding factor. Contradicting results have been published either describing different or similar patterns of DNA copy number aberrations between intestinal and diffuse type gastric cancers [7,33,37-39]. In the context of the present study we believe that the differences in DNA copy number aberrations between UK and SA gastric cancers are independent of the histological tumor type. When repeating the cluster analysis with intestinal type carcinomas only, cluster membership again was significantly correlated with geographical origin of the tumors. Also supervised data analysis, i.e. testing copy number status of all genomic loci, did not reveal any significant differences in DNA copy number changes between intestinal and diffuse gastric cancers from UK patients. Furthermore, hierarchical cluster analysis including UK gastric cancers only did not separate intestinal and diffuse type gastric cancers. We therefore do not believe our findings to be influenced by distribution of histological types in this series. Question remains why diffuse type gastric cancers were more frequently observed in gastric cancers from UK patients compared to SA patients. Besides being a confounding factor, we can hypothesize that mutation of E-cadherin (CDH1), or other mechanisms disrupting the CDH1 gene function such as epigenetic mechanisms or miRNAs, playing an important role in diffuse type gastric cancer, might play a minor role in SA gastric cancer patients due to different pathways of carcinogenesis, as shown in the present study by differences in patterns of DNA copy number aberrations. Also, the prevalence of H. pylori infection is very high in South Africa, and H. pylori infection mainly plays a role in intestinal type gastric cancers. This could also explain the higher number of intestinal type gastric cancers in SA patients.
Further, with respect to copy number changes in relation to histological types, chromosomal gains of 8q and 17q and losses of 3p have been described to be associated with intestinal type gastric cancers [33]. On the other hand, gains of 8q and 17q have been reported to be altered predominantly in diffuse type gastric cancers [38]. In the present study gains on chromosomes 8q and 17q and losses on 3p were common to both intestinal and diffuse type gastric cancers. In addition, these aberrations also were common in tumors from both UK and SA patients. Gains on chromosomes 13q and 19q have been found more frequently in diffuse type gastric cancers [33,36,38]. Again, in the present study, gains of these chromosomes were observed equally in intestinal and diffuse type gastric cancers. Gain of 19q was frequently observed in tumors from both geographical origins. Although gain of 13q was observed less frequently in tumors from SA patients compared to tumors from UK patients, still around 20-25% of the tumors of native SA patients show a gain of chromosome 13q, making it unlikely that tumor type has influenced cluster membership.
A limitation of the present study is the fact that native SA gastric cancer patients were significantly younger compared to Caucasian SA and UK gastric cancer patients. We previously showed that gastric cancers of young and elderly patients have different patterns of chromosomal aberrations [12]. We cannot rule out that also in these series, age might contribute to differences in DNA copy number profiles, however cluster analysis showed that gastric cancers from native SA patients were more similar to cancers from Caucasian SA patients, who have similar age as UK patients, indicating that cluster membership is independent of age in this respect. Overall, most differences were observed between UK and native SA tumors.
We realize that the present study is based on a heterogeneous group of gastric cancer patients, with different genetic background and different environmental factors, including H. pylori, diet and socioeconomic status, influencing gastric cancer risk. Statements on genotype influencing gastric cancer are very difficult to make since the degree of heterogeneity within each different patient group, i.e. UK, native SA and Caucasian SA, is unknown.
The patterns of genomic alterations in gastric cancers from UK and SA patients could gain clinical relevance in the future. In addition to surgery, gastric cancer treatment increasingly includes (neo)adjuvant chemotherapy and/or radiotherapy, however still without patients being stratified based on biological characteristics of their tumors. Clinical trials are underway in which also the value of genetic markers for predicting response to therapy are studied. In the end, stratification for therapy may include genomic alterations observed in tumors of patients from different geographical origin.
Conclusions
We showed that gastric cancers of UK and SA patients are different in their patterns of genomic instability. Gastric cancers from SA patients show higher frequencies of microsatellite instability and different patterns of chromosomal aberrations compared to gastric cancers from UK patients. These results may suggest different molecular pathways of gastric carcinogenesis, consistent with the African enigma hypothesis. Further studies are needed to explore the link between H. pylori and other environmental factors, as well as host factors, such as polymorphisms influencing gastric cancer susceptibility, in relation to the patterns of genomic instability in gastric cancers from these different geographic areas.
Competing interests
The authors declare that there are no competing interests.
Authors' contributions
TB performed part of the DNA isolations, array CGH experiments and MSI analysis, performed data analysis and wrote the manuscript, ML collected the material from South African patients, performed part of the DNA isolations and array CGH experiments and helped in conceiving the study. NG revised the carcinomas included in the study and helped in coordinating the study and writing of the manuscript, MT helped in performing array CGH experiments and MSI analysis, BC helped in coordinating the study, BY provided the facilities for the microarray experiments, HG provided the material from the United Kingdom, CM, CV, and SM were involved in conceiving and coordinating the study, GM was involved in coordinating the study and writing of the manuscript. All authors read and approved the final version of the manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Tineke E Buffart, Email: t.buffart@vumc.nl.
Melanie Louw, Email: melanie.louw@up.ac.za.
Nicole CT van Grieken, Email: nct.vangrieken@vumc.nl.
Marianne Tijssen, Email: m.tijssen@vumc.nl.
Beatriz Carvalho, Email: b.carvalho@vumc.nl.
Bauke Ylstra, Email: b.ylstra@vumc.nl.
Heike Grabsch, Email: H.I.Grabsch@leeds.ac.uk.
Chris JJ Mulder, Email: cjmulder@vumc.nl.
Cornelis JH van de Velde, Email: C.J.H.van_de_Velde@lumc.nl.
Schalk W van der Merwe, Email: svdmerwe1@mweb.co.za.
Gerrit A Meijer, Email: ga.meijer@vumc.nl.
Acknowledgements
We thank the Mapping Core and Map Finishing groups of the Wellcome Trust Sanger Institute for initial clone supply and verification. We thank Peter van der Vlies and Klaas Kok for providing the BAC arrays. This work was financially supported by the Dutch Cancer Society grant-KWF 2004-3051 and the Pathological Society Pilot Study Grant - August 2007.
References
- Ferlay J, Autier P, Boniol M, Heanue M, Colombet M, Boyle P. Estimates of the cancer incidence and mortality in Europe in 2006. Ann Oncol. 2007;18:581–592. doi: 10.1093/annonc/mdl498. [DOI] [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Singh K, Ghoshal UC. Causal role of Helicobacter pylori infection in gastric cancer: an Asian enigma. World J Gastroenterol. 2006;12:1346–1351. doi: 10.3748/wjg.v12.i9.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa P, Haenszel W, Cuello C, Tannenbaum S, Archer M. A model for gastric cancer epidemiology. Lancet. 1975;2:58–60. doi: 10.1016/S0140-6736(75)90498-5. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Perucho M. Cancer of the microsatellite mutator phenotype. Biol Chem. 1996;377:675–684. [PubMed] [Google Scholar]
- van Grieken NC, Weiss MM, Meijer GA, Hermsen MA, Scholte GH, Lindeman J. et al. Helicobacter pylori-related and -non-related gastric cancers do not differ with respect to chromosomal aberrations. J Pathol. 2000;192:301–306. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH697>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Holcombe C. Helicobacter pylori: the African enigma. Gut. 1992;33:429–431. doi: 10.1136/gut.33.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunet N, Barros H. Helicobacter pylori infection and gastric cancer: facing the enigmas. Int J Cancer. 2003;106:953–960. doi: 10.1002/ijc.11306. [DOI] [PubMed] [Google Scholar]
- Buffart TE, Tijssen M, Krugers T, Carvalho B, Smeets SJ, Brakenhoff RH. et al. DNA quality assessment for array CGH by isothermal whole genome amplification. Cell Oncol. 2007;29:351–359. doi: 10.1155/2007/709290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurén P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attemt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- Buffart T, Carvalho B, Hopmans E, Brehm V, Klein-Kranenbarg E, Schaaij-Visser T. et al. Gastric cancers in young and elderly patients show different genomic profiles. J Pathol. 2007;211:45–51. doi: 10.1002/path.2085. [DOI] [PubMed] [Google Scholar]
- Weiss MM, Hermsen MA, Meijer GA, van Grieken NC, Baak JP, Kuipers EJ. et al. Comparative genomic hybridisation. Mol Pathol. 1999;52:243–251. doi: 10.1136/mp.52.5.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhard O, Cattaneo F, Wong YF, Yim SF, Friedman E, Flejou JF. et al. Multipopulation analysis of polymorphisms in five mononucleotide repeats used to determine the microsatellite instability status of human tumors. J Clin Oncol. 2006;24:241–251. doi: 10.1200/JCO.2005.02.7227. [DOI] [PubMed] [Google Scholar]
- Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J. et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- van de Wiel MA, Kim KI, Vosse SJ, van Wieringen WN, Wilting SM, Ylstra B. CGHcall: calling aberrations for array CGH tumor profiles. Bioinformatics. 2007;23:892–894. doi: 10.1093/bioinformatics/btm030. [DOI] [PubMed] [Google Scholar]
- van de Wiel MA, van Wieringen WN. CGH regions:dimension reduction for array CGH data with minimal information loss. Cancer Informatics. 2007;2:55–63. [PMC free article] [PubMed] [Google Scholar]
- van Wieringen WN, van de Wiel MA, Ylstra B. Weighted clustering of called array CGH data. Biostatistics. 2008;9:484–500. doi: 10.1093/biostatistics/kxm048. [DOI] [PubMed] [Google Scholar]
- van de Wiel MA, Smeets SJ, Brakenhoff RH, Ylstra B. CGHMultiArray: exact P-values for multi-array comparative genomic hybridization data. Bioinformatics. 2005;21:3193–3194. doi: 10.1093/bioinformatics/bti489. [DOI] [PubMed] [Google Scholar]
- Manduchi E, Grant GR, McKenzie SE, Overton GC, Surrey S, Stoeckert CJ Jr. Generation of patterns from gene expression data by assigning confidence to differentially expressed genes. Bioinformatics. 2000;16:685–698. doi: 10.1093/bioinformatics/16.8.685. [DOI] [PubMed] [Google Scholar]
- Graham DY. Helicobacter pylori infection is the primary cause of gastric cancer. J Gastroenterol. 2000;35(Suppl 12):90–97. [PubMed] [Google Scholar]
- Bravo LE, van Doom LJ, Realpe JL, Correa P. Virulence-associated genotypes of Helicobacter pylori: do they explain the African enigma? Am J Gastroenterol. 2002;97:2839–2842. doi: 10.1111/j.1572-0241.2002.07031.x. [DOI] [PubMed] [Google Scholar]
- Correa P, Piazuelo MB. Natural history of Helicobacter pylori infection. Dig Liver Dis. 2008. [DOI] [PMC free article] [PubMed]
- Yamaoka Y, Kato M, Asaka M. Geographic differences in gastric cancer incidence can be explained by differences between Helicobacter pylori strains. Intern Med. 2008;47:1077–1083. doi: 10.2169/internalmedicine.47.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG, Queiroz DM, Mendes EN, Evans DJ Jr. Helicobacter pylori cagA status and s and m alleles of vacA in isolates from individuals with a variety of H. pylori-associated gastric diseases. J Clin Microbiol. 1998;36:3435–3437. doi: 10.1128/jcm.36.11.3435-3437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd M, Lastovica AJ, Atherton JC, Louw JA. Heterogeneity in the Helicobacter pylori vacA and cagA genes: association with gastroduodenal disease in South Africa? Gut. 1999;45:499–502. doi: 10.1136/gut.45.4.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doorn LJ, Figueiredo C, Rossau R, Jannes G, van AM, Sousa JC. et al. Typing of Helicobacter pylori vacA gene and detection of cagA gene by PCR and reverse hybridization. J Clin Microbiol. 1998;36:1271–1276. doi: 10.1128/jcm.36.5.1271-1276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal UC, Tiwari S, Dhingra S, Pandey R, Ghoshal U, Tripathi S. et al. Frequency of Helicobacter pylori and CagA antibody in patients with gastric neoplasms and controls: the Indian enigma. Dig Dis Sci. 2008;53:1215–1222. doi: 10.1007/s10620-008-0229-7. [DOI] [PubMed] [Google Scholar]
- El-Omar EM, Carrington M, Chow WH, McColl KE, Bream JH, Young HA. et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- Kikuchi S. Epidemiology of Helicobacter pylori and gastric cancer. Gastric Cancer. 2002;5:6–15. doi: 10.1007/s101200200001. [DOI] [PubMed] [Google Scholar]
- Muñoz N, Pisani P. Helicobacter pylori and gastric cancer. Eur J Gastroenterol Hepatol. 1994;6:1097–1103. [Google Scholar]
- Vauhkonen H, Vauhkonen M, Sajantila A, Sipponen P, Knuutila S. Characterizing genetically stable and unstable gastric cancers by microsatellites and array comparative genomic hybridization. Cancer Genet Cytogenet. 2006;170:133–139. doi: 10.1016/j.cancergencyto.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Wu MS, Chang MC, Huang SP, Tseng CC, Sheu JC, Lin YW. et al. Correlation of histologic subtypes and replication error phenotype with comparative genomic hybridization in gastric cancer. Genes Chromosomes Cancer. 2001;30:80–86. doi: 10.1002/1098-2264(2000)9999:9999<::AID-GCC1062>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Chetty R, Naidoo R, Tarin M, Sitti C. Chromosome 2p, 3p, 5q and 18q status in sporadic gastric cancer. Pathology. 2002;34:275–281. doi: 10.1080/00313020220131354. [DOI] [PubMed] [Google Scholar]
- Carvalho B, Buffart TE, Reis RM, Mons T, Moutinho C, Silva P. et al. Mixed gastric carcinomas show similar chromosomal aberrations in both their diffuse and glandular components. Cell Oncol. 2006;28:283–294. doi: 10.1155/2006/650620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllykangas S, Junnila S, Kokkola A, Autio R, Scheinin I, Kiviluoto T. et al. Integrated gene copy number and expression microarray analysis of gastric cancer highlights potential target genes. Int J Cancer. 2008;123:817–825. doi: 10.1002/ijc.23574. [DOI] [PubMed] [Google Scholar]
- Vauhkonen H, Vauhkonen M, Sajantila A, Sipponen P, Knuutila S. DNA copy number aberrations in intestinal-type gastric cancer revealed by array-based comparative genomic hybridization. Cancer Genet Cytogenet. 2006;167:150–154. doi: 10.1016/j.cancergencyto.2005.11.014. [DOI] [PubMed] [Google Scholar]
- Koo SH, Kwon KC, Shin SY, Jeon YM, Park JW, Kim SH. et al. Genetic alterations of gastric cancer: comparative genomic hybridization and fluorescence In situ hybridization studies. Cancer Genet Cytogenet. 2000;117:97–103. doi: 10.1016/S0165-4608(99)00152-1. [DOI] [PubMed] [Google Scholar]
- Noguchi T, Wirtz HC, Michaelis S, Gabbert HE, Mueller W. Chromosomal imbalances in gastric cancer. Correlation with histologic subtypes and tumor progression. Am J Clin Pathol. 2001;115:828–834. doi: 10.1309/2Q9E-3EP5-KYPK-VFGQ. [DOI] [PubMed] [Google Scholar]


