Abstract
The E3 region of adenovirus codes for several membrane proteins, most of which are involved in immune evasion and prevention of host cell apoptosis. We explored the topology and targeting mechanisms of E3-6.7K, the most recently described member of this group, by using an in vitro translation system supplemented with microsomes. Here, we present evidence that E3-6.7K, one of the smallest signal-anchor proteins known, translocates across the membrane of the endoplasmic reticulum in a posttranslational, ribosome-independent, yet ATP-dependent manner, reminiscent of the translocation of tail-anchored proteins. Our analysis also demonstrated that E3-6.7K could achieve several distinct topological fates. In addition to the previously postulated type III orientation (N-luminal/C-cytoplasmic, termed NtmE3-6.7K), we detected a tail-anchored form adopting the opposite orientation (N-cytoplasmic/C-luminal, termed CtmE3-6.7K) as well as the possibility of a fully translocated form (N and C termini are both translocated, termed NCE3-6.7K). Due to the translocation of a positively charged domain, both the CtmE3-6.7K and NCE3-6.7K topologies of E3-6.7K constitute exceptions to the “positive inside” rule. The NtmE3-6.7K and NCE3-6.7K are the first examples of posttranslationally translocated proteins in higher eukaryotes that are not tail anchored. Distinct topological forms were also found in transfected cells, as both N and C termini of E3-6.7K were detected on the extracellular surface of transfected cells. The demonstration of unexpected topological forms and translocation mechanisms for E3-6.7K defies conventional thinking about membrane protein topogenesis and advises that both the mode of targeting and topology of signal-anchor proteins should be determined experimentally.
Group C adenovirus membrane proteins are encoded by the E3 region, a cassette of genes involved in immune evasion (55). More specifically, E3/10.4K, E3/14.5K (receptor internalization and degradation α [RIDα] and RIDβ, respectively) (1, 9, 43-45), E3/14.7K (16), and E3-6.7K (1, 35) prevent death receptor-induced apoptosis and release of inflammatory mediators from infected cells while E3/19K inhibits antigen presentation by retaining the major histocompatibility class I complex in the endoplasmic reticulum (ER) (23, 38).
It was recently shown that a subset of E3-6.7K is localized at the plasma membrane, where it is required by the RID complex in downregulating the surface levels of tumor necrosis factor-related apoptosis-inducing ligand receptor 1 (TRAIL-R1) and TRAIL-R2 (1). The requirement for E3-6.7K in RID-mediated downregulation of TRAIL-R1 has been disputed by others (45). Nevertheless, we showed that even in the absence of other viral proteins, E3-6.7K maintains ER calcium homeostasis, confers resistance to death receptor- and calcium ionophore-induced apoptosis, and reduces the tumor necrosis factor-induced release of arachidonic acid from transfected cells (35).
E3-6.7K was previously described as a small membrane glycoprotein localized primarily in the ER (54). It was presumed to be a type III glycoprotein because its only asparagine residues that are candidate acceptors for Asn-linked glycosylation are found in the N-terminal region of the protein (53). It contains a single hydrophobic stretch that initiates membrane insertion and also acts as a transmembrane domain, hence, acting as a signal-anchor (SA) sequence. Curiously, when E3-6.7K was fused to its cocistronic partner, E3/19K, the fusion protein was fully translocated across the membrane of the ER and directed E3/19K to the membrane of the ER (53). This indicates that the SA domain of E3-6.7K does not act as a stop-transfer sequence.
SA-containing proteins can achieve either an N-cytoplasmic/C-luminal orientation, as seen for type II proteins, or an N-luminal/C-cytoplasmic orientation, as seen for type III transmembrane proteins (reviewed by Goder and Spiess [15]). Statistically, the topology of SA-containing proteins is determined by the distribution of charged residues (49) or the net charge difference between the regions of the polypeptide flanking the transmembrane domain (17). The presence of positively charged residues in a flanking domain correlates with its retention in the cytoplasm (this observation is also called the positive inside rule) (49). One possible explanation is that positive residues found in the cytoplasmic domain of these proteins interact with the anionic phospholipids on the cytoplasmic face of the membrane (48). It has also been observed that the length and hydrophobicity of the transmembrane domain also affects the orientation of SA proteins, with long hydrophobic regions favoring the type III orientation (40, 50). Glycosylation of luminal exposed domains can also influence the topology of proteins by arresting lumenal exposed glycosylation acceptor sites in the ER (14). In the case of some SA proteins, the topology was also determined by the folding state of the regions flanking the transmembrane domain (7).
SA-containing proteins can use both co- and posttranslational modes of targeting to the ER. This is to be contrasted with the mode of targeting employed by proteins containing a cleavable signal sequence such as secreted soluble and type I (N-luminal/C-cytoplasmic oriented) proteins, where translocation occurs cotranslationally and requires the targeting mechanism of the signal receptor particle (SRP) (12, 13). Most SA domain-containing proteins employ a cotranslational, SRP-mediated targeting mechanism. This has been observed in such type II proteins as the invariant chain protein (30) and the transferrin receptor (58) and in type III proteins like synaptotagmin II (24). There are also examples, albeit very few in higher eukaryotes, of proteins that are posttranslationally translocated. In fact, the only such proteins have their SA domains in close proximity to their C termini (tail anchored [TA] proteins). TA proteins include synaptobrevin (26, 51), cytochrome b5, bcl-2 (22), and the vaccinia virus H3L protein (6). These proteins insert in the ER membrane posttranslationally, in the absence of the SRP or the Sec61 complex, and assume a type II topology.
The lack of an N-terminal cleavable signal sequence and the short length of the protein (61 amino acids) suggest that E3-6.7K may not be a good substrate for the SRP and may employ an unusual mode of targeting across the membrane of the ER. Its membrane topology and targeting are important in determining its mode of action as an antiapoptotic protein and may shed some light on the targeting mechanisms and properties of small and hydrophobic membrane proteins.
MATERIALS AND METHODS
Materials and reagents.
The RiboMax large-scale RNA production system-T7 and the nuclease-treated rabbit reticulocyte lysate and RNasin were obtained from Promega Corp. (Madison, Wis.). Tissue culture supplies such as Dulbecco minimal essential medium, penicillin, streptomycin sulfate, and l-glutamine were obtained from Invitrogen while fetal calf serum was obtained from VWR Canada. Grade VI apyrase, [γ-S]ATP, tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (TPCK-trypsin), proteinase K (ProtK), and phenylmethylsulfonyl fluoride (PMSF) were obtained from Sigma. Peptide-N-glycosidase F (PNGaseF) was obtained from Boehringer Mannheim, and puromycin was obtained from Clontech. Microsomes from Raji cells were prepared as previously described by Levy et al. (28). A polyclonal rabbit antiserum raised against a peptide corresponding to residues 47 to 61 in the sequence of Ad2 E3-6.7K, referred to here as R47-61, was obtained as a generous gift from W. S. M. Wold (St. Louis University, St. Louis, Mo.).
Production of N-terminal or C-terminal FLAG-tagged E3-6.7K.
The E3 region plasmid pBR-E3 containing the EcoRI D fragment of Ad2 was a gift from W. S. M. Wold (St. Louis University). The E3-6.7K and the E3/19K coding regions were isolated and subcloned in pGEM 3Zf(+) under the control of the T7 promoter and with the E3 polyadenylation signal. E3-6.7K cDNA was subcloned from pGEM 3Zf(+) into pcDNA3.1 (Invitrogen) in frame with sequences encoding a FLAG tag at the 5′ end to generate pFLAG-6.7K, a vector that expresses E3-6.7K with a FLAG tag at the N terminus. E3-6.7K was amplified by PCR from pFLAG-6.7K and subcloned into pIRES-hrGFP-1a (Stratagene) to generate pGFP/6.7K-FLAG, a vector that cocistronically expresses E3-6.7K with a FLAG tag at the C terminus and green fluorescent protein (GFP), allowing for the identification of transfected cells.
Immunofluorescence staining.
HeLa cells were transiently transfected with pGFP, pGFP/6.7K-FLAG, or pFLAG-6.7K by using FuGENE 6 (Roche). Transfected cells were fixed with 2% paraformaldehyde for 20 min and, where indicated, permeabilized with 0.1% saponin in 2% bovine serum albumin (BSA) in phosphate-buffered saline for 15 min. Nonpermeabilized cells were treated with 2% BSA for 15 min. Cells were then blocked with 2% BSA for 1 h and reacted with 1:1,000-diluted M2 anti-FLAG primary antibody (Sigma) in 2% BSA for 30 min. After incubation, the cells were washed extensively with 2% BSA, incubated with 1:1,000-diluted Alexa Fluor 568 goat anti-mouse secondary antibody (Molecular Probes) in 2% BSA for 30 min at room temperature in the dark, and washed five times with 2% BSA. Cells were treated with SlowFade Antifade (Molecular Probes) and analyzed by confocal microscopy using a Bio-Rad Radiance Plus on an inverted Zeiss Axiovert with DIC optics and Lasersharp software (Bio-Rad).
In vitro transcription and translation.
Transcription of E3-6.7K and E3/19K cDNA from the pGEM vector under the control of the T7 promoter was carried out according to the manufacturer's protocol by using the RiboMax large-scale RNA production system-T7 (Promega Corp.). Translation was carried out for 60 min at 30°C using a nuclease-treated rabbit reticulocyte lysate system (Promega Corp.) in the presence of 37 kBq of [l-35S]cysteine (Amersham)/μl and containing 70% reticulocyte lysate, 79 mM potassium acetate, 0.5 mM magnesium acetate, 1 U of RNasin/μl, and 0.5 mM PMSF. Unless otherwise indicated, translation was performed in the presence of 8 μl of microsomes for every 50-μl reaction mixture.
Immunoprecipitation and SDS-PAGE electrophoretic analysis.
Unless otherwise indicated, the translation reaction mixture was diluted in 10 volumes of TNE buffer (20 mM Tris [pH 7.4], 150 mM NaCl, and 5 mM EDTA) and centrifuged for 10 min at 12,000 × g. The microsomal pellet was solubilized in TNE buffer supplemented with 1% Triton X-100 and 2 mM PMSF. Samples were precleared overnight with protein A-Sepharose CL-4B and then immunoprecipitated from the supernatant with R47-61 antiserum for 2 h at 4°C followed by purification of immune complexes with protein A-Sepharose CL-4B for 2 h at 4°C. The immune complex-bead slurry was washed three times in 1% Triton X-100 and TNE buffer, and one last wash was done in TNE buffer without detergent. Protein samples or bead slurries were boiled for 10 min in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (0.6 M Tris [pH 7.0], 3% [wt/vol] SDS, 10% [vol/vol] glycerol, 70 mM dithiothreitol, 0.01% Serva blue G) followed by treatment with 5 μl of 0.5 M iodoacetamide for 15 min. The products were examined by Tricine-SDS-PAGE according to the method of Schägger and von Jagow (41) with a 16.5% T, 3% C separating gel and a 10% T, 3% C spacer gel (where percent T represents the total concentration of both monomers, acrylamide, and bisacrylamide and percent C represents the percentage of cross-linker bisacrylamide relative to the total concentration).
Posttranslational translocation assays.
To assay posttranslational translocation, translation was performed in the absence of microsomes for 1 h. Various amounts of cycloheximide (as indicated in Fig. 2) were added, and samples were incubated at room temperature for 15 min, 8 μl of microsomes was added, and the mixture was incubated for an additional hour. Alternatively, translation reaction mixtures were treated with 2 mM puromycin (final concentration) for 30 min at 30°C, and the polysomes were sedimented by centrifugation at 100,000 rpm in a Beckman tabletop ultracentrifuge with a TLA100.1 rotor. The supernatant containing ribosome-free polypeptide chains was incubated with 8 μl of microsomes for an additional 30 min at 30°C. Where indicated, the supernatant and the microsomes were pretreated for 15 min at 30°C with 1 mM [γ-S]ATP or 50 U of grade VI apyrase/ml prior to incubation with microsomes.
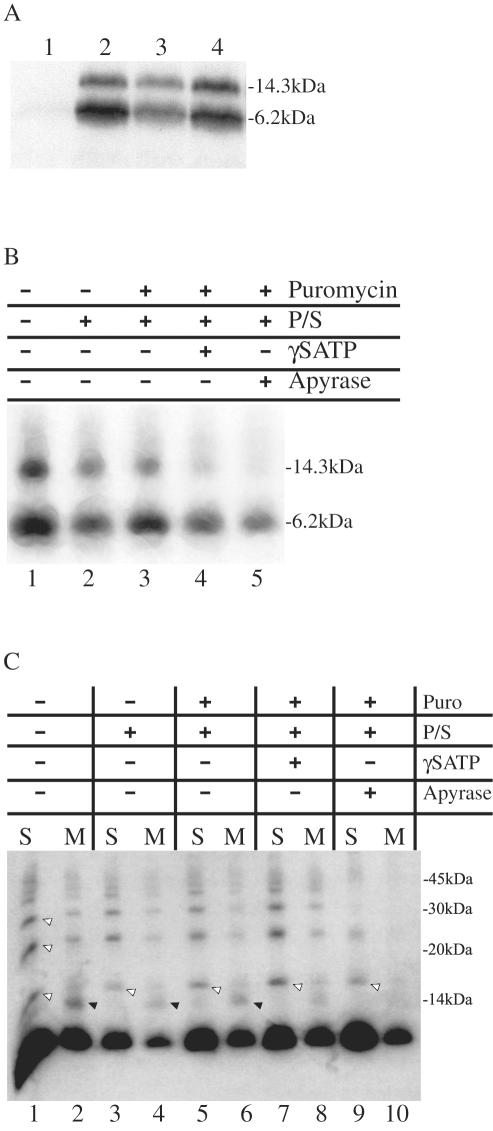
FIG. 2.
E3-6.7K translocates posttranslationally. (A) In vitro-synthesized E3-6.7K translocated across the membrane of the microsomes whether microsomes were present during translation (lane 4) or whether they were added after translation was stopped with 1 or 10 mM cycloheximide (lanes 2 and 3, respectively). Lane 1, E3-6.7K mRNA was translated for 1 h in the presence of 1 mM cycloheximide to indicate an effective translation block. All samples were immunoprecipitated with R47-61 antiserum and analyzed by SDS-PAGE. (B) Translation was carried out in the absence of microsomes, then the samples were treated with 2 mM puromycin (lanes 3, 4, and 5) to release the nascent chains, and then we sedimented the polysomes (P/S) (lanes 2 to 5). The resulting pool of proteins was then exposed tomicrosomes to assay translocation. The ribosome-free nascent chains and the microsomes were treated with [γ-S]ATP or apyrase, lanes 4 and 5, respectively, prior to coincubation. After translocation, half of the total reaction was pelleted and washed in TNE buffer and the resulting microsomes were immunoprecipitated with R47-61 antiserum and examined by SDS-PAGE and fluorography. (C) The other half of the translocation reaction mixture was extracted with 0.2 M Na2CO3 and subjected to alkaline flotation (1.8, 1.25, and 0.25 M sucrose steps in 0.1 M Na2CO3). The 1.8 M layer contained soluble material (S), and the 1.25 and 0.25 M pooled fractions contained floated membrane material (M). The M and S fractions were immunoprecipitated with R47-61 antiserum and examined by SDS-PAGE and fluorography. The black arrows (◂) indicate the 14-kDa glycosylated form of E3-6.7K, and the white arrows (◃) indicate SDS-resistant complexes of multimers of E3-6.7K. The positions of protein molecular mass markers are shown at the right side of the panels. +, compound present; −, compound absent.
Alkaline extraction and alkaline flotation of microsomes.
High-salt alkali extraction was performed by diluting the microsome suspension at the end of the translation-translocation reaction in 20 volumes of 0.1 M sodium carbonate, pH 11.5, followed by centrifugation and reextraction. Alkaline flotation experiments were performed as described by Kida et al. (24). After posttranslational translocation assays were performed, one half of the total reaction mixture was diluted in 10 volumes of TNE buffer and subjected to centrifugation for 10 min at 12,000 × g followed by resuspension in TNE buffer and recentrifugation. The washed microsomal pellet was then lysed in TNE and 1% Triton X-100, immunoprecipitated with R47-61 antiserum, and examined by Tricine-SDS-PAGE (see Fig. 2B). The other half of the posttranslational translocation reaction mixture was mixed with an equal volume of 0.2 M Na2CO3 and incubated for 30 min on ice. The samples were then mixed with 2.5 M sucrose to result in a 1.8 M sucrose final concentration, transferred to centrifugation tubes, and overlaid with an equal volume of 1.25 M sucrose-0.1 M Na2CO3 and with a 0.25 M sucrose-0.1 M Na2CO3 layer. The gradients were spun at 100,000 rpm for 90 min in a Beckman tabletop ultracentrifuge with a TLA100.1 rotor. The upper layers of alkali-treated membrane fractions (1.25 M and 0.25 M sucrose layers not including the 1.8 to 1.25 M interphase) were pooled and labeled M (membrane associated) while the bottom layer (1.8 M sucrose) was labeled S (containing extracted peripheral proteins and soluble proteins). The samples were diluted with lysis buffer, TNE, and 1% Triton X-100, precleared overnight with protein A-Sepharose CL-4B, immunoprecipitated with R47-61 antiserum, and examined by Tricine-SDS-PAGE (see Fig. 2C).
Protease protection assays.
To assay the membrane topology, translation was terminated with the addition of 100 μg of cycloheximide/ml, and then the microsomes were pelleted at 18,000 × g, washed in STKMM buffer (250 mM sucrose, 50 mM TEA-HCl [pH 7.5], 50 mM potassium acetate, 2.2 mM magnesium acetate, 0.1% β-mercaptoethanol), resuspended in STKMM buffer with 1 mM CaCl2, and allowed to stabilize for 10 min on ice. ProtK was added at a final concentration of 300 μg/ml in the presence or absence of 2% NP-40, and the samples were incubated for 30 min on ice. Preactivated TPCK-trypsin (final concentration, 750 μg/ml) was added in the presence or absence of 2% NP-40, and samples were incubated for 30 min at room temperature. PMSF was added to a final concentration of 2 mM to stop digestion, and membranes were pelleted, solubilized in TNE plus 1% Triton X-100 supplemented with 2 mM PMSF, and immunoprecipitated. Alternatively, if the microsomes were separated from the translation reaction mixture prior to protease digestion, the microsomal pellet was simply boiled in SDS-PAGE sample buffer and examined by electrophoresis (Fig. 3C).
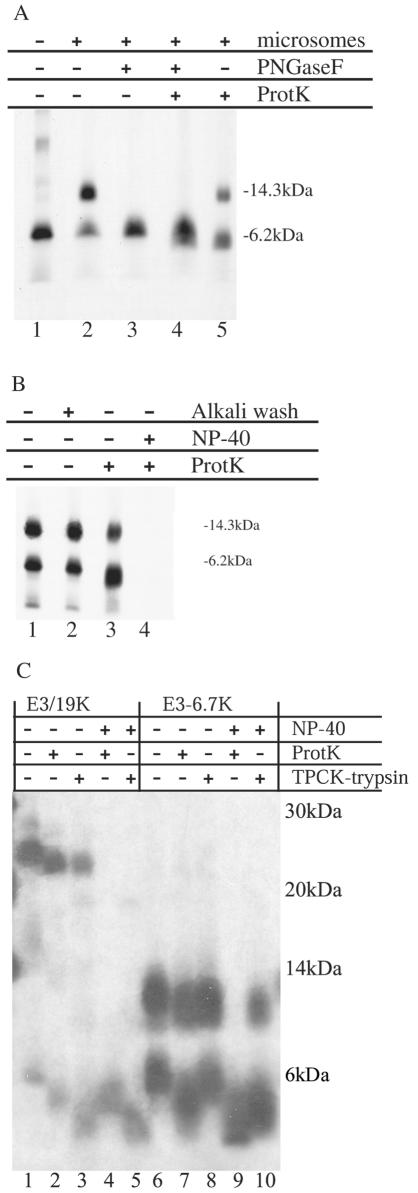
FIG. 3.
In vitro analysis of the topology adopted by E3-6.7K indicates various membrane configurations. E3-6.7K mRNA was translated in the presence of microsomes to generate a radiolabeled membrane-embedded protein, and then the microsomes were isolated, protease treated, lysed, and immunoprecipitated with R47-61 antisera (in panels A and B) or examined without further immunoprecipitation by SDS-PAGE (in panel C). The positions of protein molecular mass markers are shown at the right side of the panels. (A) Lane 1 shows the primary translation product of E3-6.7K in the absence of microsomes.The microsome-embedded protein is treated with 300 μg of ProtK/ml to identify the membrane-protected domains in lanes 4 and 5. Lanes 3 and 4 show the immunoprecipitated protein following treatment with PNGaseF. (B) Microsomes containing newly synthesized protein were subjected to either high-salt-alkali extraction at pH 11.5 (lane 2) or ProtK digestion in the absence (lane 3) or presence (lane 4) of 2% NP-40 detergent. (C) The protease digestion pattern of membrane-embedded E3-6.7K is examined in comparison with the one of E3/19K, a well-characterized type I protein. Membrane-embedded E3-6.7K or E3/19K was treated with 300 μg of ProtK/ml or 750 μg of TPCK-trypsin/ml in the presence or absence of 2% NP-40 detergent, as indicated. After proteolysis, the samples were boiled in SDS-PAGE loading buffer and examined by SDS-PAGE. +, compound present; −, compound absent.
RESULTS
E3-6.7K translocates posttranslationally across the membrane of the ER.
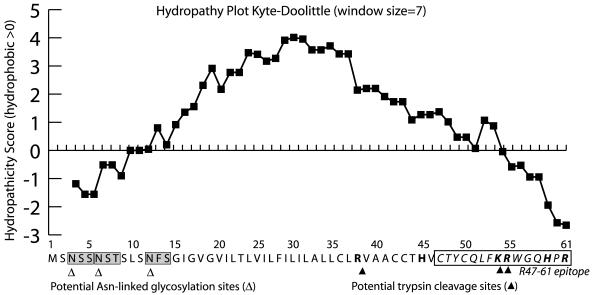
It has been previously postulated that E3-6.7K may be posttranslationally translocated or posttranslationally glycosylated (54). Sequence analysis of E3-6.7K with the Kyte and Doolittle algorithm (27) predicts a single transmembrane, SA, domain between residues 19 and 39 in the polypeptide chain (hydropathicity score greater than 2) (Fig. 1). Please note that all three possible Asn-linked glycosylation sites are N terminal to the transmembrane domain while the charged residues are all basic and C terminal (at physiologic pH) (Fig. 1). The E3-6.7K protein is present in infected cells in two forms, a high-mannose glycosylated 14-kDa form and an unglycosylated 6- to 7-kDa form (54). We noticed the appearance of both of the previously observed forms of E3-6.7K in microsomes following in vitro translation of E3-6.7K mRNA (Fig. 2 ).
FIG. 1.
Sequence analysis of E3-6.7K. A Kyte-Doolittle hydropathy plot of E3-6.7K displays the highly hydrophobic character of this protein. The three predicted Asn-linked glycosylation sites are marked with white triangles (▵). Trypsin targets the peptide bond on the carboxyl side of arginine and lysine residues, and is indicated on the figure as filled triangles (▴). Residues 47 to 61 corresponding to the peptide epitope used to raise the R47-61 rabbit polyclonal antisera are italicized and have an outside border. The positively charged residues are shown in bold.
To investigate whether E3-6.7K is posttranslationally translocated across the ER membrane, we used cycloheximide to block protein synthesis following the in vitro translation reaction. We assayed translocation in the absence of translation by adding microsomes after translation was stopped. The appearance of the higher molecular mass glycosylated E3-6.7K of 14 kDa was used to establish the success of the translocation reaction. We observed that the E3-6.7K protein can translocate across the membrane of the ER in the absence of protein synthesis (Fig. 2A, lanes 2 and 3) as efficiently as when translation is not inhibited (Fig. 2A, lane 4). As observed in Fig. 2A, the lowest concentration of cycloheximide used was sufficient to block protein synthesis when added at the beginning of the translation reaction (Fig. 2A, lane 1).
We assayed whether bound ribosomes or ATP influence E3-6.7K's translocation across microsomal membranes. As a reporter for complete translocation we relied on the acquisition of Asn-linked glycosylation, carried out by enzymes inside the ER lumen (microsomal lumen). Several studies have employed Asn-linked glycosylation in combination with apyrase treatment to establish the energy requirements for an in vitro translocation reaction (21, 47, 56). This is because microsomes contain preassembled oligosaccharyl-dolichol donor molecules and the transfer of Glc3Man9GlcNAc2 onto polypeptides entering the ER is not ATP dependent. In this study, we released the nascent chains from polysomes by using puromycin and then we sedimented the polysomes by high-speed centrifugation. The polysome-free supernatant was then tested for its ability to support translocation of E3-6.7K in the presence or absence of apyrase or nonhydrolyzable ATP. We found that newly synthesized E3-6.7K chains were in a translocation-competent state in the absence of bound ribosomes (Fig. 2B, lane 1 versus lanes 2 and 3). We also analyzed the energy requirements for the translocation of E3-6.7K. Before the translocation reaction occurred, the nascent chain and microsomes were depleted of ATP through apyrase treatment or were treated with a nonhydrolyzable ATP analogue ([γ-S]ATP). The results demonstrate that the translocation across the membrane into the lumen of the ER, accompanied by Asn-linked glycosylation and the appearance of the 14kDa form was dependent on ATP. Conversely, association of the 6.7-kDa nonglycosylated form of E3-6.7K with the membranes appeared to be ATP independent (Fig. 2B, lanes 4 and 5).
In the absence of Asn-linked glycans, it is difficult to say whether the 6.7-kDa unglycosylated form of the protein was peripherally or integrally associated with membranes following ATP depletion or whether it simply precipitated onto membranes in the absence of ATP. Therefore, it was necessary to subject the microsomes and embedded proteins to alkaline extraction and flotation gradient analysis, which is effective in removing peripheral membrane proteins (24). We observed that a large proportion of nascent chains failed to associate with microsomes (Fig. 2C, 6.7-kDa band in odd-numbered lanes). Even in the absence of ATP, some of the nascent chains were still able to associate in a stable fashion with the membranes, giving rise to an unglycosylated integral membrane protein of 6.7 kDa (Fig. 2C, lanes 8 and 10). The 14-kDa form appeared in the membrane-associated fraction (M) as an integral membrane protein (Fig. 2C, lanes 2, 4, and 6) only in the presence of hydrolyzable ATP (Fig. 2C, lanes 8 and 10 versus lane 6). We observed that depletion of ATP through apyrase treatment was far more effective in blocking the translocation step than competition with the nonhydrolyzable ATP analogue [γ-S]ATP (Fig. 2B, lane 5 versus 4, and C, lane 10 versus 8). As a result, membrane-bound E3-6.7K can be observed, albeit in decreased amounts, in Fig. 2C, lane 8. Soluble fractions also contained slower-migrating radiolabeled proteins of 16, 24, and 32 kDa, which could represent SDS-resistant multimers of E3-6.7K (Fig. 2C, lane 1). Proteins complexes of 24 and 32 kDa were also seen in the membrane fraction, possibly representing membrane-integrated, higher-order structures of E3-6.7K. Further studies will be necessary to establish the existence and significance of such structures in vivo.
The data presented here suggest that E3-6.7K is capable of translocating across the ER membrane following complete synthesis. One reason for this could be that the lack of a signal sequence and the length of the E3-6.7K protein emerging from the ribosome do not allow enough time for the nascent chain to interact with the SRP. Additionally, we noticed that an ATP-dependent factor is necessary for E3-6.7K's translocation but not for its association with membranes.
The E3-6.7K protein adopts various membrane topologies.
The charge distribution is such that all charged residues are basic and are localized at the C terminus of the E3-6.7K protein (Fig. 1). Based on charged residue distribution and the presence of Asn-linked carbohydrates at the N terminus of the protein, it has been proposed that E3-6.7K is a type III protein that has an N-luminal/C-cytoplasmic orientation (54). The SA domain does not, however, act as a stop-transfer sequence when E3-6.7K is fused to E3/19K, directing the fusion protein to the membrane of the ER (53).
We investigated the topology of E3-6.7K by performing translation in vitro in the presence of microsomes followed by digestion of the exposed domain of the in vitro-synthesized and membrane-translocated E3-6.7K with ProtK. We then immunoprecipitated the remaining fragment with the R47-61 antiserum, which recognizes a peptide corresponding to the sequence of residues 47 to 61 of the mature protein (54). Analysis of the topology of E3-6.7K indicates that the 6.7- and 14-kDa forms of the protein adopted different conformations. The molecular form of 6.7 kDa was not glycosylated and appears to be the primary translation product, as seen in Fig. 3A , lane 1, in the absence of microsomes. Digestion of the membrane-bound form of 6.7 kDa with ProtK resulted in the increased mobility of the protected fragment (Fig. 3A, lane 5 versus 2, and B, lane 3 versus 1). The membrane-protected fragment of the 6.7-kDa form contains the C terminus domain, based on the fact that it was recognized by the R47-61 polyclonal antisera. Therefore, we propose that the 6.7-kDa form adopts a type II topology (C-translocated, or CtmE3-6.7K) with its N terminus in the cytoplasm, which precludes the glycosylation of the Asn acceptor sites.
Treatment of the 14-kDa form of the protein with PNGaseF generated the 6.7-kDa polypeptide, demonstrating that the higher molecular weight is Asn linked and glycosylated (Fig. 3A, lane 3). In contrast to the 6.7-kDa form, digestion of the microsome-embedded 14-kDa form did not result in a similar shift in electrophoretic mobility (Fig. 3A, lane 5 versus 2, and B, lane 3 versus 1). Then, deglycosylation of the protected fragment of 14 kDa generated a protein product indistinguishable in size from the primary translation product (6.7-kDa fragment) (Fig. 3A, lane 4 versus 1). This suggests that a subset of the 14-kDa protein is completely protected from exogenously added protease, having both its N and C termini inside the ER lumen (termed NCE3-6.7K). Besides the unaltered change in the electrophoretic mobility of the 14-kDa fragment in Fig. 3A and B, we provided additional arguments for the existence of an NCE3-6.7K topology. By virtue of its glycosylation, we can place the N terminus of the protected fragment of 14 kDa inside the lumen of the ER. We also know that the C terminus is intact because the protected fragment can be immunoprecipitated with the R47-61 C-terminally directed antisera. Based on these facts, we propose that at least some of the 14-kDa glycosylated chains of E3-6.7K are protected from exogenously added ProtK while embedded in the membrane and that both the N and C termini of these proteins are found within the ER lumen. Both the 6.7- and 14-kDa forms of the protein are not susceptible to a high-salt extraction of the microsomes (Fig. 3B, lane 2).
We noticed that there was a noticeable decrease in the intensity of the 14-kDa form following protease digestion (Fig. 3A, lane 5 versus 2, and B, lane 3 versus 1). This suggested that only a subset of 14-kDa proteins assumes the NCE3-6.7K topology and that the loss of signal may be due to a yet unaccounted form of the protein. More specifically, the use of R47-61 C-terminally directed antisera to immunoprecipitate the protein precludes the purification of fragments whose C termini were digested. Consequently, in the results depicted in Fig. 3C, we examined all the membrane-protected proteolytic fragments without further purification. Following translation, the microsomes were treated with ProtK or TPCK-trypsin in the presence or absence of detergent and then examined directly by SDS-PAGE. Following ProtK digestion, we observed a novel 12-kDa proteolytic fragment from the 14-kDa form of E3-6.7K, which we had not detected previously by immunoprecipitation (Fig. 3C, lane 7 versus 6). The novel subset of proteins of the 14-kDa protein accessible to exogenously added ProtK were also digested by exogenous TPCK-trypsin (Fig. 3C, lane 8 versus 6). Based on the sequence of E3-6.7K, the only targets for trypsin (arginine and lysine residues) are found at the C terminus of the protein (Fig. 1). Therefore, we conclude that in addition to the NCE3-6.7K topology, some of the 14-kDa glycosylated proteins have a type III topology, with an N-lumenal/C-cytoplasmic orientation (N-translocated, or NtmE3-6.7K). Predictably, the unglycosylated CtmE3-6.7K form with a type II topology (C terminus inside the lumen) was not susceptible to TPCK-trypsin while embedded in the membrane (6.7-kDa polypeptides) (Fig. 3C, lanes 6 and 8 versus the faster-migrating fragment in lane 10). We tested our protease protection assay by using a protein of known topology, the type I membrane protein Ad2 E3/19K. Both ProtK and TPCK-trypsin cleaved the cytoplasmic domain of E3/19K in Fig. 3C, lanes 2 and 3 versus lane 1.
Even in the presence of detergent, we observed that the core of E3-6.7K exhibits some intrinsic resistance to ProtK (Fig. 3C, lane 9). Protease resistance in the presence of detergent has been observed in other proteins, such as prion protein (Prp), and did not preclude studies of its membrane topology in vitro by protease digestion of exposed hydrophilic domains (18). There is good evidence that both the N and C termini of E3-6.7K are susceptible to ProtK. For example, we observed that the ProtK-resistant fragment detected in Fig. 3C, lane 4, was not recognized by the R47-61 antiserum (Fig. 3B, lane 4), meaning it does not include the C-terminal domain. We also observed that the cytoplasmic domain of the unglycosylated 6.7-kDa form was sensitive to ProtK while embedded in the membrane (Fig. 3A, lanes 4 and 5, and B, lane 3). Since we were able to immunoprecipitate the remaining proteolytic fragment with the R47-61 antiserum, we conclude that the protected fragment retained the C terminus and that the digested cytoplasmic domain contained the N terminus of the protein and was digested by ProtK. This means that both the N and C hydrophilic domains were susceptible to proteolytic digestion and that the fragment exhibiting intrinsic resistance was within the hydrophobic core of the protein and, as such, its presence has no consequence on the conclusions derived from the use of ProtK to digest hydrophilic cytoplasmic domains.
Both N and C termini of E3-6.7K are detectable at the cell surface.
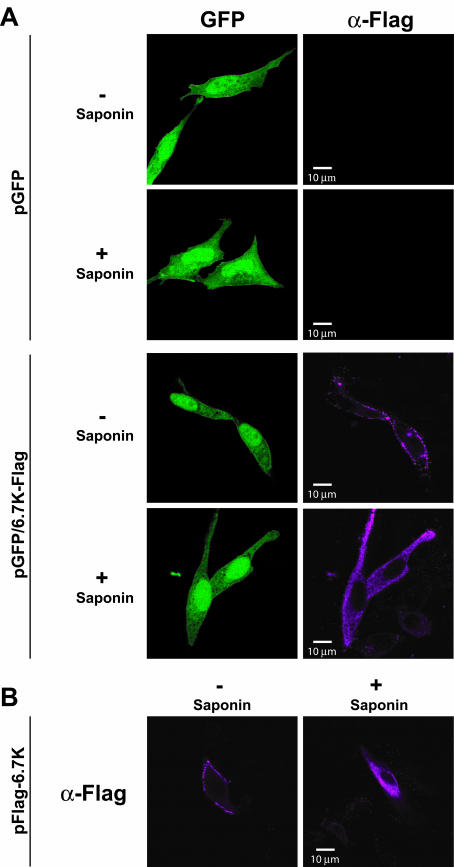
We sought to confirm our observations based on in vitro analysis with studies of the topology of E3-6.7K expressed in cells. The majority of E3-6.7K is intracellular and exhibits sensitivity to endoglycosidase H digestion, indicating that the attached glycans are primarily the ER-specific high-mannose type (54). However, in addition to the ER, there is a subset of the E3-6.7K proteins which localize to the plasma membrane of cells and associate with the adenovirus RID complex, specifically with RIDβ (1). In the previous study, the authors made use of an N-terminally vesicular stomatitis virus-tagged and an N-terminally FLAG-tagged E3-6.7K to demonstrate that the N terminus of the protein is accessible to exogenously added tag-specific antibodies at the cell surface (1). Similarly, in the present study, we use an N-terminally FLAG-tagged and C-terminally FLAG-tagged E3-6.7K to directly study the topology of E3-6.7K in transfected cells. The tagged E3-6.7K cDNA was cloned into a pIRES-GFP vector (Stratagene), which allows us to identify transfected cells by virtue of the presence of the cocistronically expressed GFP. To exclude nonspecific staining, we show that the anti-FLAG monoclonal antibody does not stain cells transfected with the pGFP vector alone (Fig. 4). We analyzed the staining pattern of transfected cells in the presence or absence of saponin (as a control of the plasma membrane integrity). In the presence of saponin, the antibody labeled intracellular membrane compartments (Fig. 4). In contrast, in the absence of saponin, the staining was very specific for the ectoplasmic domain of E3-6.7K (Fig. 4). We demonstrate that for both the N and C termini of E3-6.7K, the FLAG tags are accessible to exogenously added antibodies (Fig. 4). This demonstrates that both N- and C-terminal domains of E3-6.7K can translocate across the membrane and give rise to proteins that are capable of transport to the cell surface. This provides support for both NtmE3-6.7K and CtmE3-6.7 but does not allow us to distinguish between the NCE3-6.7K and the other two forms.
FIG.4.
Detection of FLAG-tagged E3-6.7K on the surface of transfected cells. (A) C-terminal FLAG-tagged E3-6.7K. HeLa cells were transfected with either pGFP vector or pGFP/6.7K-FLAG. GFP exhibits green fluorescence. Cells were costained with M2 anti-FLAG monoclonalantibody and Alexa 568-conjugated anti-mouse secondary antibody (magenta). (B) N-terminal FLAG-tagged E3-6.7K. HeLa cells were transfected with pFLAG-6.7K and were stained with M2 anti-FLAG (α-FLAG) antibody and Alexa 568-conjugated anti-mouse secondary antibody (magenta). Cells not treated with saponin (nonpermeabilized) show staining of the extracellular FLAG epitope while cells treated with saponin (permeabilized) show staining of both extra- and intracellular FLAG epitope. +, compound present; −, compound absent.
DISCUSSION
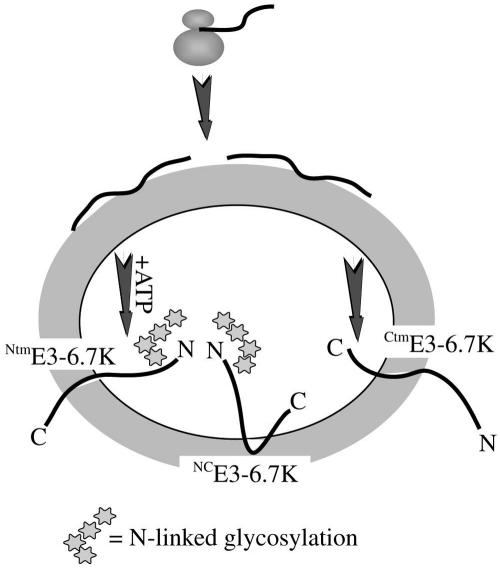
The NtmE3-6.7K and NCE3-6.7K form (demonstrated to occur in vitro in this study) are the first examples of non-TA proteins which translocate across the membrane of the ER in a posttranslational, ribosome-independent manner. A possible scenario for the sequence of events could proceed as outlined in Fig. 5. The nascent polypeptide chain of E3-6.7K is released from the ribosome complex and the nascent polypeptide-associated complex (52), and it interacts with such chaperones as the tailless complex polypeptide 1 ring complex (33) and heat shock proteins 70 and 40 to prevent its folding and aggregation (11, 25). Then an ATP-dependent process, possibly mediated by lumenal BiP, assists the translocation of either the N terminus or the C terminus towards the lumen of the ER, generating the NtmE3-6.7K and CtmE3-6.7K forms, respectively. This mechanism operates through a chaperone-mediated trapping of protein domains (the Brownian ratchet model) as they emerge on the luminal side of the ER (32). Though we do not have evidence that the generation of the CtmE3-6.7K form is also ATP dependent, as the CtmE3-6.7K form is indistinguishable in size from the membrane-associated precursor, the translocation of the similar TA proteins was shown to be ATP dependent (25, 26). Based on our observations of in vitro-synthesized E3-6.7K, a subset of either the NtmE3-6.7K or the CtmE3-6.7K membrane forms or both can undergo a second translocation event of the cytosolic domain to generate a fully translocated NCE3-6.7K protein. Once in the oxidizing environment of the ER lumen, the protein would fold, assisted by chaperones, and allow the attachment of carbohydrate moieties where these domains are accessible to the luminal compartment. The minimum distance between a functional N-terminal glycosylation acceptor site and the luminal end of the transmembrane domain is 14 to 15 residues (37), which suggests that only the most N-proximal Asn residue can be glycosylated out of the three potential sites. The sorting and transport of TA proteins along the secretory pathway following insertion in the ER is not well understood. Results from transfected cells indicate that more than one topological form of E3-6.7K can be transported to the plasma membrane; further studies are necessary to verify the existence and describe the targeting of each of the three different topological forms of E3-6.7K in infected cells.
FIG. 5.
Model of the mode of translocation of E3-6.7K. N and C represent the N and C termini, respectively. The Asn-linked high-mannose carbohydrate modification is shown at the N terminus of the 14-kDa form. The region recognized by the R47-61 rabbit polyclonal antisera is at the C terminus of the protein.
We conclude that the topologies proposed here for the SA-containing protein E3-6.7K challenge the current models of membrane protein conformation in several important ways. To begin with, the fully translocated NCE3-6.7K form and the type II CtmE3-6.7K form of E3-6.7K are exceptions to the positive inside rule, since both forms of the protein have their positively charged C termini in the ER lumen. In addition, there are very few examples of proteins that can adopt more than one membrane topology. These have been observed for the polytopic proteins such as the channel ductin (8), P-glycoprotein (60), aquaporin-1 (31), and some single-spanning proteins such as prion protein (PrP) (57), microsomal epoxide hydrolase (61), and recently, the Newcastle disease virus fusion protein F (34). In addition to two membrane-embedded conformations, represented by a type I and a type II conformation (NtmPrP and CtmPrP, respectively), there is also a fully translocated form of PrP (secPrP), the precursor to the glycolphosphatidyl inositol-anchored protein found at the surface of the cell (PrPC) (18). Though PrP has a cleavable signal sequence, it appears to alternate between the two modes of transport. PrP appears to utilize its signal sequence to cotranslationally generate the NtmPrP and secPrP forms and its transmembrane domain to posttranslationally generate the CtmPrP form (21).
The generation of the various PrP protein forms was proposed to be controlled by trans-acting factors (19). In vitro-synthesized PrP can assume different membrane conformations depending on the composition of the added microsomes (19). Interestingly, when we translated E3-6.7K mRNA in the presence of commercially available canine pancreas microsomes, we observed that the glycosylated forms of E3-6.7K were not successfully generated (results not shown). Canine pancreas microsomal membranes have also been shown not to support the different topologies of aquaporin-1 (31). We are currently pursuing the nature of the trans-acting factors that influence transmembrane protein topology.
As a protein coded by a human pathogen, E3-6.7K belongs to a very small class of mammalian proteins that are posttranslationally transported into the ER (the subject of a recent review, see reference 2). The mechanisms of translocation for synaptobrevin (26, 51), Cx26 connexin (59), and a fragment of the glucose transporter (36) require ATP, whereas for preprocecropin, insertion is both ATP and signal sequence dependent (42). Insertion of the bcl-2 and cytochrome b (5) proteins was shown to require specific residues located at the C terminus (22). However, more recently, a second study reexamined the translocation of cytochrome b (5), demonstrating that translocation as measured by glycosylation was inhibited by apyrase treatment (using a similar assay as described in our study). The same study also demonstrated that sec61-deficient yeast strains support the translocation of cytochrome b (5, 56). The elucidation of the mechanism of translocation of TA proteins will benefit greatly from the use of genetically defined microsomes and from examining a variety of TA proteins in parallel assays. Nevertheless, none of the posttranslationally targeted TA proteins described to date can assume multiple membrane topologies as seen for E3-6.7K.
The variety of functions ascribed to TA proteins show that in many cases the targeting of the protein is functionally relevant. For example, t-SNARE protein Ufe1p (39), syntaxin 3 and 4 (4), and the recently described Slt1 (5) are involved in vesicular traffic. The antiapoptotic protein bcl-2 can be posttranslationally targeted to both the ER and the mitochondria (22), which allows it to inhibit the apoptotic response in both subcellular locations (62). Another TA protein, the herpesvirus Us9 protein (3), localizes to the trans-Golgi network and is necessary for axonal localization and transport of viral membrane proteins (46). E3-6.7K has been implicated in the downregulation of TRAIL-R1 and -R2 by binding to RIDβ (1) (though possibly not required for TRAIL-R1 [45] downregulation). The RID complex is thought to downregulate Fas (9), epidermal growth factor receptor (20), and TRAIL receptors by inducing their internalization and degradation in the endosomal/lysosomal compartment (29). In a separate study, we showed that E3-6.7K alone is sufficient to protect transfected cells against death receptor and thapsigargin-induced apoptosis. In addition, we showed that E3-6.7K maintains calcium ion homeostasis in response to thapsigargin. These effects are reminiscent of the protective role conferred by bcl-2 against thapsigargin-induced calcium flux and apoptosis. In this study, we show that E3-6.7K also shares the topology (the CtmE3-6.7K form) and mode of targeting of bcl-2. Further studies are necessary to establish whether, like bcl-2 (10), E3-6.7K directly or indirectly alters the permeability of ER, hence decreasing the ER calcium stores. It will also be important to establish whether the different topological forms of E3-6.7K are responsible for its distinct functions such as the maintenance of calcium homeostasis in the ER versus downregulation of TRAIL receptors in cooperation with RID at the plasma membrane. We believe that studies of the mechanisms by which E3-6.7K affects the different death receptor and calcium-release pathways will benefit from understanding the topology and targeting of the various forms of E3-6.7K.
Acknowledgments
We are grateful to W. S. M. Wold for the gifts of polyclonal antisera to E3-6.7K and the plasmid bearing the Ad2 E3 region. We thank J. W. Hodgson and L. Matsuuchi for helpful discussions and S. Lok for her help.
This work was supported by operating grants from the NCIC and CIHR to W.A.J. J.R.G. is supported by an NSERC postgraduate scholarship and a BC Science Council GREAT scholarship.
REFERENCES
- 1.Benedict, C. A., P. S. Norris, T. I. Prigozy, J. L. Bodmer, J. A. Mahr, C. T. Garnett, F. Martinon, J. Tschopp, L. R. Gooding, and C. F. Ware. 2001. Three adenovirus e3 proteins cooperate to evade apoptosis by tumor necrosis factor-related apoptosis-inducing ligand receptor-1 and -2. J. Biol. Chem. 276:3270-3278. [DOI] [PubMed] [Google Scholar]
- 2.Borgese, N., S. Colombo, and E. Pedrazzini. 2003. The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J. Cell Biol. 161:1013-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brideau, A. D., B. W. Banfield, and L. W. Enquist. 1998. The Us9 gene product of pseudorabies virus, an alphaherpesvirus, is a phosphorylated, tail-anchored type II membrane protein. J. Virol. 72:4560-4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulbarelli, A., T. Sprocati, M. Barberi, E. Pedrazzini, and N. Borgese. 2002. Trafficking of tail-anchored proteins: transport from the endoplasmic reticulum to the plasma membrane and sorting between surface domains in polarised epithelial cells. J. Cell Sci. 115:1689-1702. [DOI] [PubMed] [Google Scholar]
- 5.Burri, L., O. Varlamov, C. A. Doege, K. Hofmann, T. Beilharz, J. E. Rothman, T. H. Sollner, and T. Lithgow. 2003. A SNARE required for retrograde transport to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 100:9873-9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.da Fonseca, F. G., E. J. Wolffe, A. Weisberg, and B. Moss. 2000. Characterization of the vaccinia virus H3L envelope protein: topology and posttranslational membrane insertion via the C-terminal hydrophobic tail. J. Virol. 74:7508-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denzer, A. J., C. E. Nabholz, and M. Spiess. 1995. Transmembrane orientation of signal-anchor proteins is affected by the folding state but not the size of the N-terminal domain. EMBO J. 14:6311-6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunlop, J., P. C. Jones, and M. E. Finbow. 1995. Membrane insertion and assembly of ductin: a polytopic channel with dual orientations. EMBO J. 14:3609-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elsing, A., and H. G. Burgert. 1998. The adenovirus E3/10.4K-14.5K proteins down-modulate the apoptosis receptor Fas/Apo-1 by inducing its internalization. Proc. Natl. Acad. Sci. USA 95:10072-10077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foyouzi-Youssefi, R., S. Arnaudeau, C. Borner, W. L. Kelley, J. Tschopp, D. P. Lew, N. Demaurex, and K. H. Krause. 2000. Bcl-2 decreases the free Ca2+ concentration within the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 97:5723-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frydman, J., and F. U. Hartl. 1996. Principles of chaperone-assisted protein folding: differences between in vitro and in vivo mechanisms. Science 272:1497-1502. [DOI] [PubMed] [Google Scholar]
- 12.Gilmore, R., G. Blobel, and P. Walter. 1982. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J. Cell Biol. 95:463-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilmore, R., P. Walter, and G. Blobel. 1982. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J. Cell Biol. 95:470-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goder, V., C. Bieri, and M. Spiess. 1999. Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J. Cell Biol. 147:257-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goder, V., and M. Spiess. 2001. Topogenesis of membrane proteins: determinants and dynamics. FEBS Lett. 504:87-93. [DOI] [PubMed] [Google Scholar]
- 16.Gooding, L. R., L. W. Elmore, A. E. Tollefson, H. A. Brady, and W. S. Wold. 1988. A 14,700 MW protein from the E3 region of adenovirus inhibits cytolysis by tumor necrosis factor. Cell 53:341-346. [DOI] [PubMed] [Google Scholar]
- 17.Hartmann, E., T. A. Rapoport, and H. F. Lodish. 1989. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc. Natl. Acad. Sci. USA 86:5786-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hegde, R. S., J. A. Mastrianni, M. R. Scott, K. A. DeFea, P. Tremblay, M. Torchia, S. J. DeArmond, S. B. Prusiner, and V. R. Lingappa. 1998. A transmembrane form of the prion protein in neurodegenerative disease. Science 279:827-834. [DOI] [PubMed] [Google Scholar]
- 19.Hegde, R. S., S. Voigt, and V. R. Lingappa. 1998. Regulation of protein topology by trans-acting factors at the endoplasmic reticulum. Mol. Cell 2:85-91. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman, P., and C. Carlin. 1994. Adenovirus E3 protein causes constitutively internalized epidermal growth factor receptors to accumulate in a prelysosomal compartment, resulting in enhanced degradation. Mol. Cell. Biol. 14:3695-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holscher, C., U. C. Bach, and B. Dobberstein. 2001. Prion protein contains a second endoplasmic reticulum targeting signal sequence located at its C terminus. J. Biol. Chem. 276:13388-13394. [DOI] [PubMed] [Google Scholar]
- 22.Janiak, F., B. Leber, and D. W. Andrews. 1994. Assembly of Bcl-2 into microsomal and outer mitochondrial membranes. J. Biol. Chem. 269:9842-9849. [PubMed] [Google Scholar]
- 23.Jefferies, W. A., and H. G. Burgert. 1990. E3/19K from adenovirus 2 is an immunosubversive protein that binds to a structural motif regulating the intracellular transport of major histocompatibility complex class I proteins. J. Exp. Med. 172:1653-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kida, Y., M. Sakaguchi, M. Fukuda, K. Mikoshiba, and K. Mihara. 2000. Membrane topogenesis of a type I signal-anchor protein, mouse synaptotagmin II, on the endoplasmic reticulum. J. Cell Biol. 150:719-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klappa, P., P. Mayinger, R. Pipkorn, M. Zimmermann, and R. Zimmermann. 1991. A microsomal protein is involved in ATP-dependent transport of presecretory proteins into mammalian microsomes. EMBO J. 10:2795-2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kutay, U., G. Ahnert-Hilger, E. Hartmann, B. Wiedenmann, and T. A. Rapoport. 1995. Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J. 14:217-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 28.Levy, F., R. Larsson, and S. Kvist. 1991. Translocation of peptides through microsomal membranes is a rapid process and promotes assembly of HLA-B27 heavy chain and beta 2-microglobulin translated in vitro. J. Cell Biol. 115:959-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lichtenstein, D. L., P. Krajcsi, D. J. Esteban, A. E. Tollefson, and W. S. Wold. 2002. Adenovirus RIDbeta subunit contains a tyrosine residue that is critical for RID-mediated receptor internalization and inhibition of Fas- and TRAIL-induced apoptosis. J. Virol. 76:11329-11342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lipp, J., and B. Dobberstein. 1986. Signal recognition particle-dependent membrane insertion of mouse invariant chain: a membrane-spanning protein with a cytoplasmically exposed amino terminus. J. Cell Biol. 102:2169-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, Y., I. R. Turnbull, A. Bragin, K. Carveth, A. S. Verkman, and W. R. Skach. 2000. Reorientation of aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol. Biol. Cell. 11:2973-2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matlack, K. E., B. Misselwitz, K. Plath, and T. A. Rapoport. 1999. BiP acts as a molecular ratchet during posttranslational transport of prepro-alpha factor across the ER membrane. Cell 97:553-564. [DOI] [PubMed] [Google Scholar]
- 33.McCallum, C. D., H. Do, A. E. Johnson, and J. Frydman. 2000. The interaction of the chaperonin tailless complex polypeptide 1 (TCP1) ring complex (TRiC) with ribosome-bound nascent chains examined using photo-cross-linking. J. Cell Biol. 149:591-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGinnes, L. W., J. N. Reitter, K. Gravel, and T. G. Morrison. 2003. Evidence for mixed membrane topology of the Newcastle disease virus fusion protein. J. Virol. 77:1951-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moise, A. R., J. R. Grant, T. Z. Vitalis, and W. A. Jefferies. 2002. Adenovirus E3-6.7K maintains calcium homeostasis and prevents apoptosis and arachidonic acid release. J. Virol. 76:1578-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueckler, M., and H. F. Lodish. 1986. Post-translational insertion of a fragment of the glucose transporter into microsomes requires phosphoanhydride bond cleavage. Nature 322:549-552. [DOI] [PubMed] [Google Scholar]
- 37.Nilsson, I. M., and G. von Heijne. 1993. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem. 268:5798-5801. [PubMed] [Google Scholar]
- 38.Persson, H., S. Kvist, L. Ostberg, P. A. Peterson, and L. Philipson. 1980. Adenoviral early glycoprotein E3-19K and its association with transplantation antigens. Cold Spring Harbor Symp. Quant. Biol. 44:509-517. [DOI] [PubMed] [Google Scholar]
- 39.Rayner, J. C., and H. R. Pelham. 1997. Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 16:1832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosch, K., D. Naeher, V. Laird, V. Goder, and M. Spiess. 2000. The topogenic contribution of uncharged amino acids on signal sequence orientation in the endoplasmic reticulum. J. Biol. Chem. 275:14916-14922. [DOI] [PubMed] [Google Scholar]
- 41.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 42.Schlenstedt, G., G. H. Gudmundsson, H. G. Boman, and R. Zimmermann. 1992. Structural requirements for transport of preprocecropinA and related presecretory proteins into mammalian microsomes. J. Biol. Chem. 267:24328-24332. [PubMed] [Google Scholar]
- 43.Shisler, J., C. Yang, B. Walter, C. F. Ware, and L. R. Gooding. 1997. The adenovirus E3-10.4K/14.5K complex mediates loss of cell surface Fas (CD95) and resistance to Fas-induced apoptosis. J. Virol. 71:8299-8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tollefson, A. E., T. W. Hermiston, D. L. Lichtenstein, C. F. Colle, R. A. Tripp, T. Dimitrov, K. Toth, C. E. Wells, P. C. Doherty, and W. S. Wold. 1998. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature 392:726-730. [DOI] [PubMed] [Google Scholar]
- 45.Tollefson, A. E., K. Toth, K. Doronin, M. Kuppuswamy, O. A. Doronina, D. L. Lichtenstein, T. W. Hermiston, C. A. Smith, and W. S. Wold. 2001. Inhibition of TRAIL-induced apoptosis and forced internalization of TRAIL receptor 1 by adenovirus proteins. J. Virol. 75:8875-8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomishima, M. J., and L. W. Enquist. 2001. A conserved alpha-herpesvirus protein necessary for axonal localization of viral membrane proteins. J. Cell Biol. 154:741-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uebel, S., T. H. Meyer, W. Kraas, S. Kienle, G. Jung, K. H. Wiesmuller, and R. Tampe. 1995. Requirements for peptide binding to the human transporter associated with antigen processing revealed by peptide scans and complex peptide libraries. J. Biol. Chem. 270:18512-18516. [DOI] [PubMed] [Google Scholar]
- 48.van Klompenburg, W., I. Nilsson, G. von Heijne, and B. de Kruijff. 1997. Anionic phospholipids are determinants of membrane protein topology. EMBO J. 16:4261-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Heijne, G., and Y. Gavel. 1988. Topogenic signals in integral membrane proteins. Eur. J. Biochem. 174:671-678. [DOI] [PubMed] [Google Scholar]
- 50.Wahlberg, J. M., and M. Spiess. 1997. Multiple determinants direct the orientation of signal-anchor proteins: the topogenic role of the hydrophobic signal domain. J. Cell Biol. 137:555-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitley, P., E. Grahn, U. Kutay, T. A. Rapoport, and G. von Heijne. 1996. A 12-residue-long polyleucine tail is sufficient to anchor synaptobrevin to the endoplasmic reticulum membrane. J. Biol. Chem. 271:7583-7586. [DOI] [PubMed] [Google Scholar]
- 52.Wiedmann, B., H. Sakai, T. A. Davis, and M. Wiedmann. 1994. A protein complex required for signal-sequence-specific sorting and translocation. Nature 370:434-440. [DOI] [PubMed] [Google Scholar]
- 53.Wilson-Rawls, J., S. L. Deutscher, and W. S. Wold. 1994. The signal-anchor domain of adenovirus E3-6.7K, a type III integral membrane protein, can direct adenovirus E3-gp19K, a type I integral membrane protein, into the membrane of the endoplasmic reticulum. Virology 201:66-76. [DOI] [PubMed] [Google Scholar]
- 54.Wilson-Rawls, J., and W. S. Wold. 1993. The E3-6.7K protein of adenovirus is an Asn-linked integral membrane glycoprotein localized in the endoplasmic reticulum. Virology 195:6-15. [DOI] [PubMed] [Google Scholar]
- 55.Wold, W. S., and L. R. Gooding. 1991. Region E3 of adenovirus: a cassette of genes involved in host immunosurveillance and virus-cell interactions. Virology 184:1-8. [DOI] [PubMed] [Google Scholar]
- 56.Yabal, M., S. Brambillasca, P. Soffientini, E. Pedrazzini, N. Borgese, and M. Makarow. 2003. Translocation of the C terminus of a tail-anchored protein across the endoplasmic reticulum membrane in yeast mutants defective in signal peptide-driven translocation. J. Biol. Chem. 278:3489-3496. [DOI] [PubMed] [Google Scholar]
- 57.Yost, C. S., C. D. Lopez, S. B. Prusiner, R. M. Myers, and V. R. Lingappa. 1990. Non-hydrophobic extracytoplasmic determinant of stop transfer in the prion protein. Nature 343:669-672. [DOI] [PubMed] [Google Scholar]
- 58.Zerial, M., P. Melancon, C. Schneider, and H. Garoff. 1986. The transmembrane segment of the human transferrin receptor functions as a signal peptide. EMBO J. 5:1543-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, J. T., M. Chen, C. I. Foote, and B. J. Nicholson. 1996. Membrane integration of in vitro-translated gap junctional proteins: co- and posttranslational mechanisms. Mol. Biol. Cell 7:471-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, J. T., M. Duthie, and V. Ling. 1993. Membrane topology of the N-terminal half of the hamster P-glycoprotein molecule. J. Biol. Chem. 268:15101-15110. [PubMed] [Google Scholar]
- 61.Zhu, Q., P. von Dippe, W. Xing, and D. Levy. 1999. Membrane topology and cell surface targeting of microsomal epoxide hydrolase. Evidence for multiple topological orientations. J. Biol. Chem. 274:27898-27904. [DOI] [PubMed] [Google Scholar]
- 62.Zhu, W., A. Cowie, G. W. Wasfy, L. Z. Penn, B. Leber, and D. W. Andrews. 1996. Bcl-2 mutants with restricted subcellular location reveal spatially distinct pathways for apoptosis in different cell types. EMBO J. 15:4130-4141. [PMC free article] [PubMed] [Google Scholar]