Abstract
Many applications require cells to switch between discrete phenotypic states. Here, we harness the FimBE inversion switch to flip a promoter, allowing expression to be toggled between two genes oriented in opposite directions. The response characteristics of the switch are characterized using two-color cytometry. This switch is used to toggle between orthogonal chemosensory pathways by controlling the expression of CheW and CheW*, which interact with the Tar (aspartate) and Tsr* (serine) chemoreceptors, respectively. CheW* and Tsr* each contain a mutation at their protein-protein interface such that they interact with each other. The complete genetic program containing an arabinose-inducible FimE controlling CheW/CheW* (and constitutively-expressed tar/tsr*) is transformed into an E. coli strain lacking all native chemoreceptors. This program enables bacteria to swim towards serine or aspartate in the absence or presence of arabinose, respectively. Thus, the program functions as a multiplexer with arabinose as the selector. This demonstrates the ability of synthetic genetic circuits to connect to a natural signaling network to switch between phenotypes.
Keywords: genetic memory, recombinase, stochastic switching, synthetic biology, systems biology
Introduction
Bacteria have mechanisms by which they can vary their DNA so that individuals within a population are performing different tasks. This heterogeneity is often achieved by enzymes that excise, invert, or rearrange the DNA. In a classical system, the cyanobacterium Anabaena forms filaments and, under nitrogen starvation, roughly every tenth cell differentiates to form nitrogen-fixing heterocysts. This is done via the excision of an 11 bp piece of DNA that causes cells to turn on nitrogenase/hydrogenase, turn off photosystem II and undergo membrane and metabolic changes 1. Similarly, the physical inversion of promoter directionality underlies the phase and antigenic variation that enables pathogens to evade the immune system 2. Some organisms, such as the gut bacterium Bacteriodes, have up to 30 such DNA invertases that form complex “shufflons” 3. In engineering cells, it would be useful to be able to program this heterogeneity such that cells within a population are performing different functions (e.g., different chemical transformations in a bioreactor).
Invertases, which change the orientation of DNA, have been incorporated into a number of synthetic genetic circuits. The cre/lox system has been used since the 1980s as a mechanism for site-directed gene insertion and deletion 4. When the loxP sites are oriented in the same direction, Cre can also function as an invertase. This system forms the core of a synthetic genetic counter, where a cascade of invertases are arranged to record a series of inducer pulses 5. The Hin recombinase was used to construct a genetic program that solves the Hamiltonian path problem 6. These recombinases are reversible and this behavior can lead to instability in the switch. There are also invertases that are irreversible, where different proteins control the inversion in each direction. One of the most studied examples is the FimB/FimE pair that controls the phase variation of type 1 fimbriae in E. coli 7. The FimE protein causes the switch to occur in one direction with nearly 100% fidelity, and FimB switches between both states equally well 8; 9. The FimE protein has been harnessed as part of a synthetic inducible system, where the inversion of promoters produces a switch with no basal activity 10. The memory function can be enhanced by interdigitating the binding sites of multiple orthogonal invertases 11. There are other regulatory interactions, including positive feedback loops and cross repression, which could switch between multiple states, but these mechanisms are less sharp, with higher basal activity in the off state 12; 13.
It would be a useful tool for biotechnology to harness invertases to switch between multiple cellular phenotypes. By changing the orientation of a promoter, the invertase could switch between two phenotypes defined by two genes encoded in opposite orientations. However, it remains challenging to connect synthetic genetic circuits to the control of a cellular phenotype. One difficulty is the identification of simple control points that will produce a distinct phenotype and knowing (and being able to control) the dynamic range required to switch between states. Successful examples include the connection of a toggle switch to the control of biofilm formation 14, a logic gate to the invasion of mammalian cells 15; 16, and quorum sensing to regulated cell death 17. Sensors for cell-cell communication signals and small molecules (theophylline and an herbicide) have been connected to the control of flagellar rotation and swimming 18; 19; 20.
In natural regulatory networks, there are a variety of mechanisms by which signals can be switched between pathways associated with different responses. This switching can occur via a scaffolding protein that aligns signaling proteins into a particular spatial arrangement 21; 22. This scaffolding allows signaling proteins to be shared between pathways while maintaining orthogonality. In eukaryotes, these scaffolds link membrane-bound receptors and ion channels to signal cascades. The pathways themselves often involve shared components. For example, two scaffolding proteins (JIP-1 and MEKK1) link different upstream signals to the activation of stress responses associated with the c-Jun N-terminal-kinases (JNK) 23; 24; 25. This circuit has the potential to perform a function that is analogous to an electronic multiplexer, where multiple inputs can be channeled to a single output based on the state of a third input, referred to as a selector. For example, the selector could be a signal that determines which scaffold protein is expressed. Synthetic scaffolds have been built by recombining protein-protein interaction domains (e.g., SH3 and PDZ domains) to create scaffolds that “rewire” the signaling proteins to build a new pathway 26.
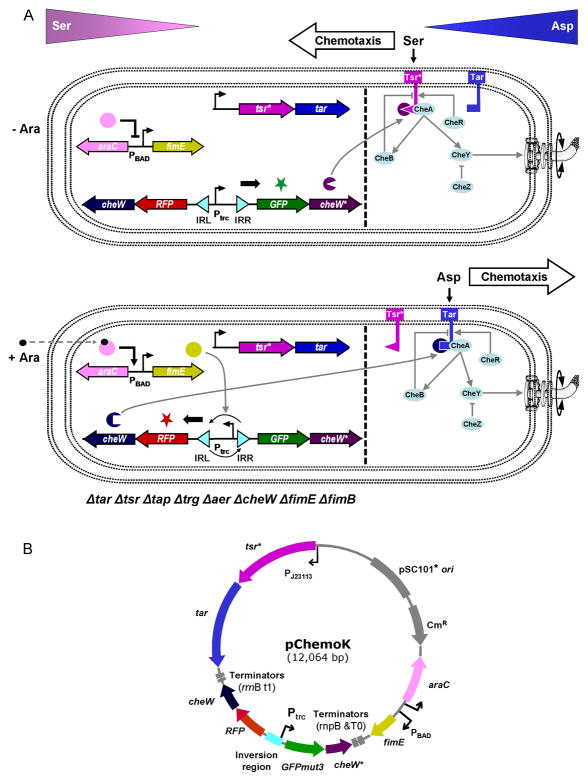
The regulatory network controlling E. coli chemotaxis is one of the well-studied models of signal transduction (Figure 1A). Extracellular signals are sensed by a set of five receptors: Tar (aspartate), Tsr (serine), Trg (galactose/ribose), Tap (dipeptides), and Aer (oxygen) 27. These receptors assemble into large clusters that are predominantly located at the poles of the bacterium 28; 29. The CheW protein binds to all five receptors and acts as a scaffold protein to recruit CheA, whose phosphorylation is stimulated by the receptor. The phosphate is transferred to the cytoplasmic protein CheY, which controls flagellar rotation. In this way, CheW integrates the signal from multiple receptors into a single response 30. Previously, a pair of mutations in CheW* (V108M) and Tsr* (E402A) were shown to produce an orthogonal pair, where the mutated proteins interact with each other but not wild-type Tsr or CheW 31. Note that in this reference, the interactions are functional (i.e., recover activity), but the protein-protein interactions were not measured. In this manuscript, we use this interaction to toggle between activating the serine-responsive Tsr* and aspartate-responsive Tar. An arabinose-inducible system serves as the selector by controlling an invertase that switches between the expression of CheW or CheW*.
Figure 1.
The synthetic switch and sensors can be interfaced with the natural signaling pathways to control bacterial chemotaxis. (A) Without arabinose (top), the engineered strain produces CheW* (along with GFPmut3), making the strain respond to and attracted toward serine (serine chemotaxis). With arabinose addition (bottom), FimE inverts the invertible region, triggering expression of CheW (along with RFP). CheW preferably interacts with Tar, and the strain switches to aspartate chemotaxis. While PBAD is an inducible promoter, Ptrc and PJ23113 used in this paper are constitutive promoters. The dotted line is drawn to show the synthetic parts and devices (left) and the natural signaling pathways (right). CheB (methylesterase), CheR (methyltransferase), and CheZ (phosphatase) are peripheral components of the natural chemotaxis system and affect the concentration of phospho-CheY 27. (B) A map of the pChemoK plasmid is shown.
In this paper, we present a genetic program that controls the activation of two chemosensory pathways by using an invertase-based switch to control the expression of orthogonal CheW adaptors (Figure 1). First, we characterize the kinetics of the arabinose-inducible FimE switch on a low copy plasmid such that it can be reliably connected to control cellular phenotypes. Then, the complete program is constructed and chemotaxis experiments are performed in an E. coli strain where all of the native chemoreceptors (methyl-accepting chemotaxis proteins) are knocked out of the genome. Codon-optimized Tar and Tsr* are expressed from a constitutive promoter, allowing both to be independently optimized and continuously present on the cell membrane. Their activation depends on the expression of either the CheW or CheW* adaptor protein, which changes the attractant to which the bacteria swim.
Results
Characterization of the fim switch using two-color flow cytometry
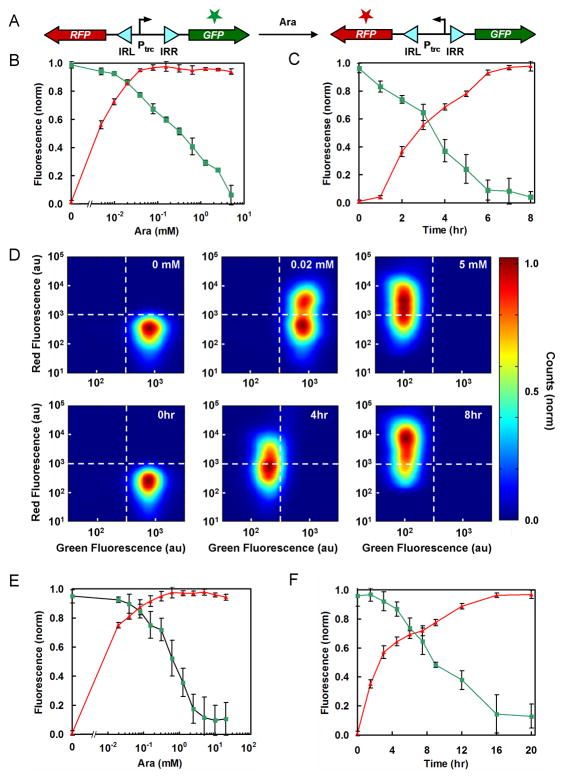
Previously, Arkin and co-workers constructed a synthetic switch based on the FimE invertase 10. The circuit was carried on a high copy plasmid (pFIP, pBR322 ori), and FimE was expressed by the arabinose-inducible PBAD promoter. A constitutively-active version of the Ptrc promoter was placed between the FimE binding sites (IRL and IRR). Its orientation is flipped upon the addition of arabinose and this leads to the expression of green fluorescent protein (GFP). Using a fluorimeter, it was shown that the switch starts to turn on after three hours and is fully induced by six hours. The circuit was also shown to operate as memory, where a pulse of inducer irreversibly turns on the switch, as measured up to eight hours. Surprisingly, the circuit was shown to function in a strain of E. coli where fimBE was still present in the genome. LB medium was used for these experiments.
In theory, the invertase switch should be able to toggle between two genes oriented in opposite directions. To test this, we constructed a plasmid based on pFIP where GFPmut3 is expressed in the initial orientation and RFP after the promoter is inverted (Figure 2A). The plasmid backbone was changed to a lower copy pSC101* origin as toxicity and cloning instability was observed at higher copy. In addition, the fimBE genes were knocked out of E. coli (Methods). As shown in previous work 10, we were able to turn the switch on stably in cells containing the fimBE genes during exponential phase in LB medium. However, we observed reverse switching when the cells were grown in poor medium as well as in exponential phase. Notably, when the cells were plated post-switching, nearly equal numbers of colonies appeared red and green and some colonies had spots of both colors (not shown). All of these issues were resolved once the native fimBE genes were knocked out of the chromosome.
Figure 2.
Characterization of the fim switch using two-color flow cytometry. (A) GFPmut3 (green) and RFP (red) were placed on either side of the fim invertible region. (B–F) FimE-promoted switching occurred more quickly in LB medium than minAaa medium. ▲ = the average RFP fluorescence (normalized); ■ = the average GFPmut3 fluorescence (normalized). Normalization was done by the following formula: [log(F) − log(FB)]/[log(FM) − log(FB)] where F, FB, and FM are sample, background, and maximum fluorescence levels, respectively. The background fluorescence values were obtained from control cells lacking the fluorescent protein genes (CAV8). Data are the averages and standard deviations of three replicates. (B) Cultures in LB medium with 6 hr induction. (C) Cultures in LB medium containing 5 mM arabinose (see Figure S2 for the OD600 data). (D) Two-color flow cytometry data are shown for cultures in LB medium. (Top) Cultures were induced for 6 hr with 0 mM (left), 0.02 mM (middle), and 5 mM (right) of arabinose. (Bottom) Cultures were induced with 5 mM arabinose for 0 hr (left), 4 hr (middle), and 8 hr (right). The dotted lines are drawn as a guide to the eye, separating the green (lower right), red (upper left), and green/red (upper right) regions. (E) Cultures in minAaa medium with 16 hr induction. (F) Cultures in minAaa medium containing 5 mM arabinose (see Figure S2 for the OD600 data).
Experiments were performed where the gain of RFP and loss of GFP were measured after the addition of arabinose to LB medium. The steady-state transfer function of the switch is shown as a function of arabinose concentration (after six hours) (Figure 2B). Full switching occurred at relatively low concentrations of inducer (0.04 mM). However, the decline in GFP signal occurred slowly as a function of inducer concentration. Interestingly, at intermediate concentrations, the two color cytometry distribution showed that there are two distinct populations of cells (Figure 2D center top panel). Both show maximum green fluorescence, but some cells are completely on with respect to red fluorescence whereas others are completely off. This indicates that in some cells there is a mixed population of switched and unswitched plasmids. In contrast, at maximum arabinose induction (5 mM) all of the cells contain fully switched plasmids (Figure 2D right top panel).
The temporal behavior of the switch was also measured (Figure 2C). At maximum induction, the switch begins to turn on after 2 hours and then is fully induced by 6 hours, thus agreeing with the previous measurements 10. The 2-color cytometry distributions also show a graded transition from the green to red states, where the intermediate time point (4 hours) represent intermediate levels of both fluorescent proteins (Figure 2D). This shows that all of the plasmids contain inverted promoters and this time point represents the delay in expression of RFP and degradation of GFP.
The chemotaxis assay requires minimal media because a complex medium, such as LB, could interfere with the serine or aspartate gradient. Many applications would require that the switch operate in microenvironments with similar, less optimal growth conditions. To measure the effect of different media, the switch dynamics were measured in minAaa medium, which is used for the chemotaxis experiments (Methods). The doubling time of the E. coli strain is 130 min in minAaa (5 mM arabinose), as opposed to 50 min in LB (5 mM arabinose). An 8-fold higher inducer concentration (0.3 mM) is required to obtain complete switching at steady-state (Figure 2E). In addition, the switch is much slower, requiring 16 hours for complete conversion (Figure 2F). Complete switch inversion was also confirmed by DNA sequencing of plasmids obtained from randomly sampled cells following a 16 hour induction in minAaa medium. There are several mechanisms that could cause the switch to be slower, including lower expression of FimE or changes in the plasmid copy number.
Genetic program design and optimization
In E. coli, the five chemotaxis receptors (Tar, Tsr, Tap, Trg, Aer) form two-dimensional clusters in the membrane that are preferentially targeted to the poles (or “nose”) of the bacterium 32; 33. The formation of clusters amplifies the signal, produces increased sensitivity and cooperativity in the response, and is involved in signal adaptation 34; 35. As such, the signaling dynamics are sensitive to the ratios of the receptor concentrations (e.g., Tsr signaling depends on the abundance of Tar and Tap) 36; 37. The clusters are also very stable and the exchange of Tar receptors is much slower than cell division, whereas CheW and CheA are exchanged on the order of 5–10 minutes 38. Based on this, our design approach for synthetic chemosensors is to constitutively express the receptors, such that they are intended to form a uniform cluster integrated in the membrane. This allows the expression levels of the receptors and the signaling to be optimized once for the cluster. Which receptors are active is determined by which orthogonal CheW adaptor is expressed. Our system only has two receptors (Tar and Tsr*), but this approach could also scale for the construction of more complex multi-receptor synthetic clusters that incorporate homologous or chimeric receptors 39; 40; 41; 42, or those that have been engineered or evolved to respond to new molecules 43.
The adaptor protein CheW interacts with all five receptors to recruit CheA. Previously, a CheW variant (CheW*) with a single mutation (V108M) was identified that disrupts its interaction with wild-type Tsr 31. The interaction was recovered by making a single compensating mutation in Tsr (E402A) to create Tsr*. The CheW*-Tsr* pair recover 70% of wild-type activity on a swarm plate. The non-cognate CheW-Tsr* interaction only recovers 25%, implying that Tsr* preferentially interacts with CheW*. Homologous recombination is a potential concern when combining CheW and CheW* onto a single plasmid. To avoid this problem, whole gene DNA synthesis was used to build a codon-optimized cheW* that only shares 78% nucleotide identity with the native cheW sequence (79% with the codon-optimized cheW sequence) and the mutations are distributed throughout the sequence (Supplementary Information) 44.
One of the challenges in optimizing this genetic program was obtaining functional expression levels for Tar:CheW and Tsr*:CheW*. High concentrations of CheW or chemoreceptors have been shown to inhibit chemotactic ability 31; 37; 45. The first constructs produced expression levels that were too high, which led to toxicity and instability when cloned into the pFIP plasmid backbone. Moving to a low-copy pSC101* plasmid reduced these problems. The original design also oriented the tsr*-tar operon in the same direction as cheW and it was transcribed by a strong constitutive promoter (PlacIQ). To reduce tar and tsr* expression, a very weak constitutive promoter (PJ23113, BBa_J23113, http://partsregistry.org) was placed upstream of a strong RBS (rbs0 from the Weiss set) 46. Further, toxicity was observed after arabinose induced the FimE-driven promoter flip, which was determined to be due to read-through from the terminators after cheW (rrnB:t1). This was eliminated by orienting the tsr*-tar operon in the opposite direction to cheW. The concentration of CheW also impacts the regulatory network, both through the ratio with CheA and its role in organizing the receptor clusters 47; 48. For CheW and CheW*, a weak RBS (rbs2 from the Weiss set) was paired with the constitutive promoter Ptrc, which gets inverted by the fim switch. The final construct is shown in Figure 1B and sequence details are in the Methods and Supplementary Information.
Wild-type E. coli possesses genes encoding all five native chemoreceptors as well as a gene encoding CheW. To eliminate interference from native chemotaxis pathways, all six genes (tar, tsr, tap, trg, aer, and cheW) were knocked out of the E. coli BW28357 chromosome (Methods). In addition, the wild-type genes encoding the fim recombinase proteins (fimE and fimB) were knocked out to eliminate possible aberrant switching (previous section). Rich medium cannot be used for amino acid chemotaxis assays, because it complicates the creation of an amino acid gradient. However, the strain containing all the knockouts and the pChemoK plasmid did not grow well in minimal A medium. Methionine, histidine, leucine, and threonine do not interfere with chemotaxis, and supplementation with these amino acids restored growth (minAaa medium).
Chemotaxis experiments
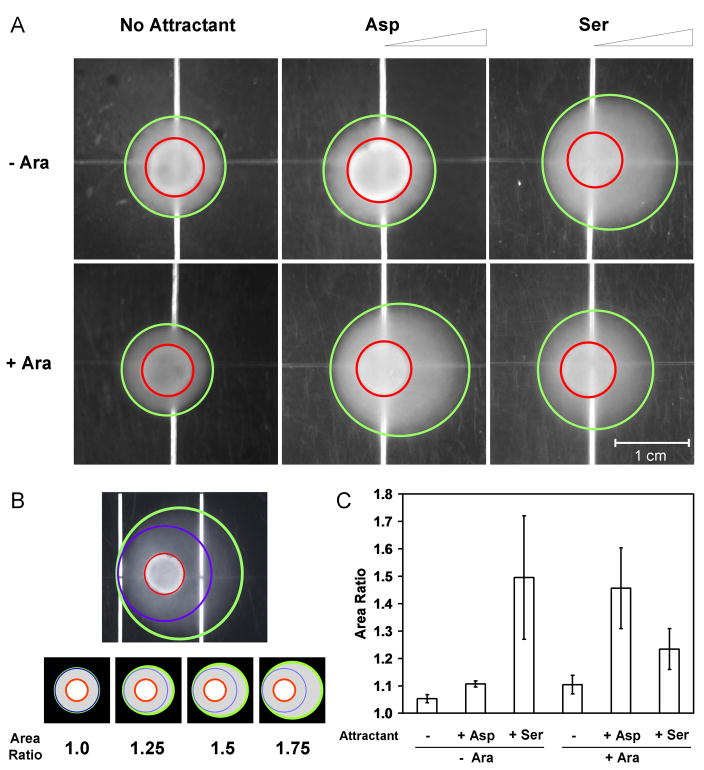
The multiplexer toggles between the activation of Tsr* and Tar with the addition of arabinose, causing the bacteria to change from moving towards serine to aspartate. Chemotaxis is measured using a plate-based assay developed by Goulian and co-workers (Figure 3 and a detailed assay is presented in the Supplementary Information) 43; 49. The only significant change is that more agar is added to the medium, which in our hands produces less dramatic results but reduces day-today variability. Briefly, a gradient of aspartate or serine is created by spotting 10 μl of 10 mM attractant on semisolid agar at 7 mm intervals down the center of a square (81 cm2) plate. The gradient is stabilized by drying the plates at 4°C for 16 hours.
Figure 3.
Assays for bacterial chemotaxis. (A) A gradient of aspartate or serine was created by spotting 10 μl of 10 mM attractant on semisolid agar (on the right sides of these images). Cultures were induced with 5 mM arabinose (bottom) or supplemented with 0.5% (w/v) glucose in order to repress potential leaky expression of PBAD (top). Those shown are representative images of experiments that were performed at least three times. Red circles and green ovals were computationally generated as described in Figure 3B. (B) Image analysis was carried out using a Matlab® algorithm. (Top) Red circles indicate the positions where 10 μl of the culture was placed, and green ovals indicate the entire regions formed by bacterial chemotaxis. The regions surrounded by blue circles are assumed to be formed by random walks only. (Bottom) The area ratio is defined as the green oval area divided by the blue circle area. Bacteria executing random walks only would result in an area ratio of 1, whereas chemotactic bacteria attracted toward aspartate or serine would lead to a higher area ratio. Schematic images are shown for four area ratio values to serve as a guide. (C) Quantitative analysis of bacterial chemotaxis. Data are the averages and standard deviations of three images taken from three experiments performed on three different days. No attractant spotted (−); 10 μl of 10 mM aspartate spotted (+ Asp); 10 μl of 10 mM serine spotted (+ Ser). Uninduced (− Ara) and induced (+ Ara).
The state of the fim switch is set prior to the chemotaxis assay. Cells are grown in minAaa medium in culture tubes for 16 hours at 37°C, either in the presence of 5 mM arabinose (+ Ara) or 0.5% w/v glucose to repress PBAD (− Ara). Then, cells are diluted into minAaa medium in the absence of glucose to promote the expression of flagella 50; 51. After 1 hour of growth, 10 μl of the culture is spotted onto the plate. After 48 hours at 30°C, the plates are photographed and the halos formed by the bacteria are quantified using custom image processing software (Methods). Like the classic swarm plate, a halo is formed by chemotactic bacteria. The halo is circular if the bacteria are swimming in random directions and appears as an oval oriented towards the gradient if the bacteria are swimming biased towards the attractant. This is quantified by the software as the ratio of the surface area of the observed oval to that of a perfect circle (Figure 3B). Bacteria lacking chemotactic abilities would lead to an area ratio of 1, whereas bacteria showing directional bias would result in a higher area ratio. This procedure enables us to quantify the standard deviation between plates.
When the fim switch is not activated (− Ara), the bacteria swim towards serine (Figure 3A and 3C). This is due to the expression of CheW*, which preferentially binds to Tsr*. No response is seen in the absence of attractant or in the presence of aspartate. When the fim switch is activated (+ Ara), then CheW is expressed, which preferentially binds to Tar, and the bacteria move towards aspartate. There is some cross reactivity between CheW and Tsr*, which leads to small, but reproducible chemotaxis to serine. This is consistent with previous results with this orthogonal pair, where the swarm rate of a CheW/Tsr* strain retained 25% of the wild-type rate31. Residual CheW* could also contribute to such chemotaxis to serine.
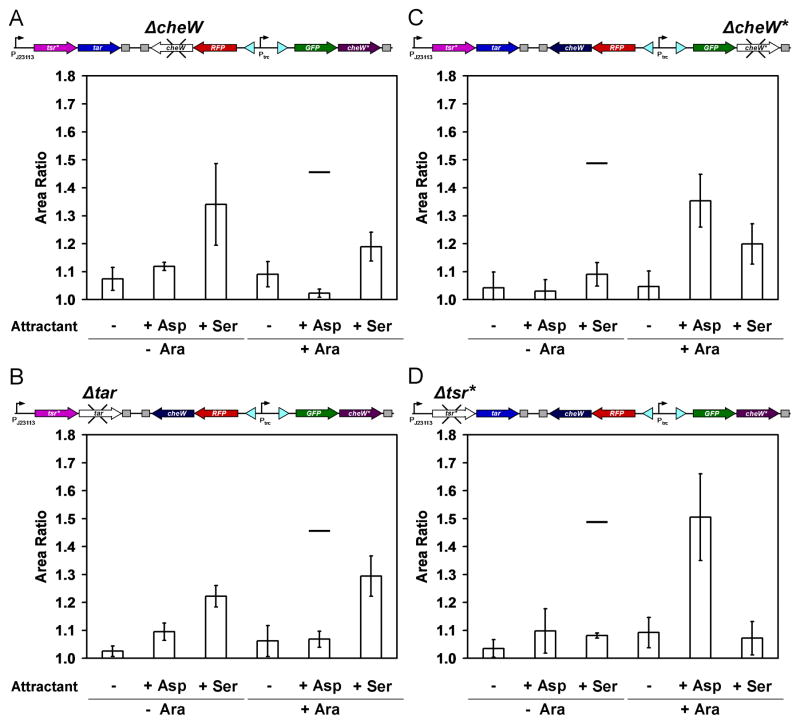
The behavior of the genetic program was further investigated by knocking out individual components and assaying the impact on chemotaxis. The quantitative data are shown in Figure 4 and examples of the plates are shown in the Supplementary Information. When cheW is deleted, Tar is inactive and cells are not able to swim towards aspartate after the addition of arabinose (Figure 4A). In the absence of arabinose, there is no effect on the response, as expected. The elimination of tar has a similar effect, where cells no longer move towards aspartate after the addition of arabinose (Figure 4B).
Figure 4.
Analysis of genetic program with various genes knocked out. The four deletion mutants lack one of the four genetic parts required for the orthogonal chemotaxis signaling, and chemotaxis assays using these mutants were carried out as described in Figure 3. Data are the averages and standard deviations of three images taken from three experiments performed on three different days (for representative images, see Supplementary Information). The black lines, indicating the average values from Figure 3C, are drawn to serve as a comparison guide. No attractant spotted (−); 10 μl of 10 mM aspartate spotted (+ Asp); 10 μl of 10 mM serine spotted (+ Ser). Uninduced (− Ara) and induced (+ Ara). (A) Chemotaxis assay using the cheW deletion mutant. The remaining CheW* after fim switching may result in the higher area ratio than expected (+ Ser + Ara). (B) Chemotaxis assay using the tar deletion mutant. Crosstalk between Tsr* and CheW seems to increase significantly when the cognate partner Tar does not exist (+ Ser + Ara). (C) Chemotaxis assay using the cheW* deletion mutant. Crosstalk between Tsr* and CheW was confirmed in the absence of cheW* (+ Ser + Ara). (D) Chemotaxis assay using the tsr* deletion mutant. No crosstalk was observed between Tar and CheW* (+ Asp − Ara).
When cheW* is deleted, Tsr* is inactive and the cells do not show chemotaxis to serine in the absence of arabinose (Figure 4C). After the addition of arabinose, the response is identical to the complete program. When tsr* is deleted, chemotaxis towards serine is eliminated, both with and without the addition of arabinose (Figure 4D). There is no impact on the remainder of the responses. No crosstalk is observed between Tar and CheW* even when Tsr*, the cognate chemoreceptor for CheW*, is removed.
The chemotaxis towards serine that is observed in the +Ara state in the complete program is Tsr*-dependent. However, chemotaxis towards serine is reduced, but not eliminated, when CheW is deleted (Figure 4A). This implies that residual CheW* remains active following fim inversion. Indeed, even after 16 hour induction, the fim switch is sufficiently slow where a significant fraction of the cells would have recently switched (Figure 2F). Further, growth in this medium is slow and this reduces the dilution rate. To examine this effect, we serially subcultured the cells without induction for additional 39 hours prior to the plate assay. This eliminated the serine response after the 16 hour induction with arabinose (area ratio = 1.10 ± 0.06).
Discussion
Here, we have built a multiplexer using an invertase-based switch that can toggle between chemosensory signaling pathways in response to a chemical inducer. Multiplexers are used in electronic systems to share devices and to reduce resource requirements. This advantage is evident in the genetic program constructed here, where most of the chemotaxis signal proteins are shared between pathways, as opposed to having all of the proteins duplicated to create two orthogonal pathways. We demonstrated that synthetic CheW variants with orthogonal protein-protein interactions with Tar and mutated Tsr can be used to toggle between continuously-expressed surface receptors. When these CheW variants are under differential regulatory control, this forms a 2-to-1 multiplexer, where a selector (in our program, arabinose) toggles between two inputs. Multiplexers are often coupled with demultiplexers (single input switched between multiple outputs) in communications as a cost saving measure to reduce the number of required channels.
E. coli has one of the simpler chemosensory systems, with 5 chemoreceptors and 1 CheW. A survey of the sequenced genomes of microorganisms showed that 65% of 206 organisms with related systems have multiple CheW homologues 52. Geobacter sulfurreducens has one of the more complex systems with 34 Tar, 10 CheW, 4 CheA, and 25 CheY homologues53. The multiple CheW (and CheA) homologues in a single organism could behave as a multiplexer if they are under differential regulatory control. This appears to occur in Rhodobacter, where different CheW homologues mediate chemotaxis during aerobic photoheterotrophic growth 54. In this way, shifts in environmental conditions could lead to switching between active receptors more quickly without having to completely reassemble the stable and highly organized receptor clusters. Following the circuit analogy, the control of the expression of CheW homologues acts as the multiplexer and control over CheY homologues could function as a demultiplexer.
The application of protein engineering to create orthogonal pairs is a generalizable approach to pathway engineering 55. Classically, this is done by randomly mutagenizing one protein, screening for a loss of function, and then randomly mutagenizing the second protein and screening for a suppressor mutation. This approach led to the discovery of the Tsr*/CheW* mutations 31. This pair of single mutations produces orthogonal pathways, but there remains a small, but significant, amount of crosstalk between CheW and Tsr* (which was not explicitly selected against in the original mutagenesis experiments). New approaches in computational design and bioinformatics have yielded orthogonal pairs with reduced cross-reactions. Structure-guided computational methods have been able to predict mutations that alter interaction specificity by designing and repacking side-chains at the protein-protein interface 56; 57. Laub and co-workers have applied mutual information analysis to compare multiple sequence alignments to identify those amino acids at the protein-protein interface that determine specificity 58. Using this approach, they were able to identify a triple mutant of EnvZ that switched its interaction partner from OmpR to RstB. In addition to the thermodynamics of the protein-protein binding interactions, natural pathways have kinetic mechanisms to reduce crosstalk. A histidine kinase can act as a phosphatase, eliminating any inadvertent cross-phosphorylation 59; 60; 61.
Invertases have a number of properties that are desirable for the construction of genetic circuits. They produce discrete switch events with zero basal state, can be irreversible, and represent a mechanism for cellular memory that can be maintained even after cell death. As such, they have been incorporated in a counter 5 and here, a multiplexer. The interdigitation of the binding sites of multiple invertases could be a powerful mechanism to store information. However, there are some drawbacks to these systems. Bidirectional invertases (e.g., cre and flp) could produce instabilities, where continued expression causes the DNA to flip back and forth 62. The unidirectional invertases (e.g., fimE) require a second protein to flip in the opposite direction, but these proteins (e.g., fimB) are not unidirectional 8; 9. This property impedes circuits that need to cycle switching between the two states. The circuits are also very slow to switch, requiring 6 hours in LB and 16 hours in minimal medium. This slow switching behavior limits their ability to be layered into more complex circuits. For example, the counter required two days to count to three 5. Finally, we observe stochastic switching at intermediate induction, where individual cells contain plasmids in different states. Directed evolution has already been used to create orthogonal invertases and mutants with improved thermostability, and to improve the efficiency and unidirectionality of recombinases 63; 64; 65; 66; 67.
Natural populations of bacteria use similar mechanisms to diversify their function in a microenvironment or as a bet hedging strategy. There are a number of potential applications in biotechnology that could harness programmable prokaryotic differentiation. For example, in the division of labor during fermentation, different subpopulations of cells could be dedicated to different tasks such as the breakdown of biomass or the formation or sequestration of product 68. Bacteria could be engineered to seek and destroy various toxic chemicals in response to local concentration gradient 18, and stochastic switching might lead to spontaneous allocation of different pollution areas to different subpopulations of cells. Our work represents a step toward the construction of complex networks, which will be essential for greater control over a broader range of cellular processes and differentiation.
Materials and methods
Strains and growth media
E. coli strain DH10B was used for all molecular biology manipulations. Cells were grown in either LB (Miller, BD Biosciences, San Jose, CA) or minimal A medium 49 supplemented with 0.2% (v/v) glycerol, 1 mM MgSO4, 0.5 mM CaCl2, 0.2 mM ZnSO4, and 2 mg/ml each of methionine, histidine, leucine, and threonine (minAaa medium). Semisolid agar was made from the minAaa medium with 0.3% (w/v) agar (BD Biosciences, San Jose, CA). Kanamycin (20 μg/mL), ampicillin (100 μg/mL), and chloramphenicol (34 μg/mL) were added as appropriate.
E. coli strain BW28357 (F- Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) γ-Δ(rhaD-rhaB)568 hsdR514) was obtained from the Coli Genetic Stock Center at Yale, and the knockout strain (CAV8) with eight chromosomal genes deleted (tar, tsr, tap, trg, aer, cheW, fimE, and fimB) was constructed from BW28357 by γ red-mediated recombination and subsequent P1 transduction 69. The position number of each gene in E. coli MG1655 is as follows: trg (1,490,494 – 1,492,134), tap (1,967,407 – 1,969,008), tar (1,969,054 – 1,970,715), cheW (1,970,860 – 1,971,363), aer (3,215,578 – 3,217,098), fimB (4,538,980 – 4,539,582), fimE (4,540,060 – 4,540,656), and tsr (4,589,680 – 4,591,335). The kanamycin resistance marker was removed using pCP20 69, and each knockout was confirmed by DNA sequencing.
Plasmid Construction
The plasmid pChemoK (Cmr and pSC101* origin), containing the fim switch and the two orthogonal signaling systems for bacterial chemotaxis (Figure 1), was constructed from synthetic DNA (DNA 2.0, Menlo Park, CA) and the one-step isothermal DNA assembly method 70. Construction of the fim switch was based on the pFIP plasmid 10. The two orthogonal signaling systems were constructed using optimized synthetic genes encoding Tar, Tsr*, CheW, and CheW* (for their DNA sequences, see Supplementary Information). The amino acid sequence for RFP was based on monomeric RFP 71 and its codon-optimized gene was synthesized. The gene for GFPmut3 and pSC101* origin were from the psicA_gfp reporter 72 and BBa_I50042 (http://partsregistry.org), respectively. Ribosome binding sites (RBS) and promoters were tuned in order to balance expression levels of chemosensory pathway genes. For cheW and cheW*, a weak synthetic RBS (TCACACAGGAAAGGCCTCG, rbs2 from the Weiss set) 46 was paired with the constitutive promoter Ptrc 10. For tar and tsr* expression, a very weak constitutive promoter PJ23113 (BBa_J23113, CTGATGGCTAGCTCAGTCCTAGGGATTATGCTAGC, http://partsregistry.org) was placed upstream of a strong synthetic RBS (ATTAAAGAGGAGAAATTAAGC, rbs0 from the Weiss set) 46. The full pChemoK plasmid was assembled using the one-step isothermal DNA assembly method 70 and used for both the 2-color cytometry (Figure 2) and chemotaxis experiments (Figure 3). The four deletion mutants that lack one of the four genetic parts required for the orthogonal chemotaxis signaling (tar, tsr*, cheW, and cheW*) were constructed from pChemoK using the one-step isothermal DNA assembly method 70.
Flow cytometry
Cultures induced with arabinose (0 to 20 mM; 0 to 20 hr) were diluted using phosphate buffered saline (pH 7) supplemented with 2 mg/ml kanamycin, and flow cytometry data were obtained using a BD Biosciences LSRII flow cytometer (BD Biosciences, San Jose, CA). The data were gated by forward and side scatter, and each data set consisted of at least 10,000 cells. FlowJo (Tree Star, Inc, Ashland, OR) was used for data analysis and compensation. For compensation, singly-stained samples were collected from green cells (i.e. cells uninduced, top left of Figure 2D) or red cells (i.e. cells fully induced, bottom right of Figure 2D). To measure autofluorescence background, the strain CAV8 was used. Normalization used for Figure 2 was performed by the following formula: [log(F) − log(FB)]/[log(FM) − log(FB)] where F, FB, and FM are sample, background, and maximum fluorescence levels, respectively. Figure 2D was plotted using “smoothhist2D” function of Matlab® (The MathWorks, Inc, Natick, MA) with the following parameters: 30 for lambda smoothing factor, 500×500 for bin size, and 0.001 for outlier cutoff.
Chemotaxis assays and image analysis
Plate-based chemotaxis assays were performed as described by Goulian and co-workers 43; 49. A detailed protocol is included in the Supplementary Information. Briefly, a gradient of aspartate or serine was created by spotting 10 μl of 10 mM attractant (either aspartate or serine) on semisolid agar with ~7 mm spacings down the middle of the square plate (81 cm2) and the plates were then dried at 4°C for 16 hr. Cultures were either induced with 5 mM arabinose or supplemented with 0.5% (w/v) glucose, which served to repress potential sporadic expression of PBAD. After 16-hr culture at 37°C and 250 rpm (in the presence or absence of arabinose), cells were washed and diluted to an OD600 of 0.2 using minAaa medium supplemented with 34 μg/mL chloramphenicol. These diluted cultures were grown (in the absence of glucose and arabinose) for additional 1 hr, and then spotted 13.5, 20, and 27 mm from the center line of attractant. Images were taken using AlphaImager® equipped with Computar TV Zoom Lens (Cell Biosciences, Santa Clara, CA) after 48 hr incubation at 30°C. Distance between lens and plate was 50 cm and exposure time was 0.06 to 0.12 sec. Plate position and zoom lens were adjusted to cover 27×27 mm2 with the spot position to the center of each image.
Image analysis was performed using a Matlab® algorithm that calculates the area of the entire swarm region resulting from bacterial chemotaxis (green ovals in Figure 3B) and compares it to that of a circle corresponding to an equivalent unbiased random walks (blue circles in Figure 3B). The blue circle is centred on the red circle corresponding to the location of the initial spotting (Figure 3B), and the blue circle’s radius reflects the distance from its center to nearest point on the corresponding green oval. The area ratio was defined as the area of the green oval divided by that of the blue circle. Bacteria lacking chemotactic abilities would lead to area ratio of 1, whereas bacteria showing directional bias would result in higher area ratio. To identify the inner red circle, the image was converted into a black and white image (“im2bw”) using a threshold value of 0.7. Holes in the processed image were filled using “imfill” function. The perimeters of the inner colony trace were smoothed out using “imerode” followed by “imdilate” functions. The coordinates of the perimeter boundary were extracted from this processed image using “bwtraceboundary” function. This coordinates were then fitted using least square method to obtain the fitted circle/ellipse equation. The same procedure was repeated (changing the threshold value down to 0.25 in the first step) to identify the “halo” region. All the image analysis functions are from Image Processing toolbox of Matlab®.
Supplementary Material
Acknowledgments
We thank Ala Trusina and Patrick Visperas for some of the initial work as part of the 2006 UCSF iGEM team. We thank Adam Arkin (UC-Berkeley), John Parkinson (Utah), and Mark Goulian (U Penn) for materials and help with assays. DNA 2.0 graciously provided synthesis for the UCSF iGEM project. We are grateful to Brian Caliando and Felix Moser for critical readings of the manuscript. C.A.V. is supported by the Pew and Packard Foundations, Office of Naval Research, National Institutes of Health (NIH) EY016546, NIH AI067699, NSF BES-0547637, and a Sandler Family Opportunity Award. C.A.V. and T.K. are part of the NSF Synthetic Biology Engineering Research Center (SynBERC). T.K. was supported by the Sloan Foundation, an NSF CAREER award, and a Sandler Family Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Golden JW, Robinson SJ, Haselkorn R. Rearrangement of nitrogen fixation genes during heterocyst differentiation in the cyanobacterium Anabaena. Nature. 1985;314:419–23. doi: 10.1038/314419a0. [DOI] [PubMed] [Google Scholar]
- 2.van der Woude MW, Baumler AJ. Phase and antigenic variation in bacteria. Clin Microbiol Rev. 2004;17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, Barron A, Clark L, Corton C, Doggett J, Holden MT, Larke N, Line A, Lord A, Norbertczak H, Ormond D, Price C, Rabbinowitsch E, Woodward J, Barrell B, Parkhill J. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science. 2005;307:1463–5. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 4.Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981;150:467–86. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 5.Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumgardner J, Acker K, Adefuye O, Crowley ST, Deloache W, Dickson JO, Heard L, Martens AT, Morton N, Ritter M, Shoecraft A, Treece J, Unzicker M, Valencia A, Waters M, Campbell AM, Heyer LJ, Poet JL, Eckdahl TT. Solving a Hamiltonian Path Problem with a bacterial computer. J Biol Eng. 2009;3:11. doi: 10.1186/1754-1611-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klemm P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 1986;5:1389–93. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gally DL, Bogan JA, Eisenstein BI, Blomfield IC. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: effects of temperature and media. J Bacteriol. 1993;175:6186–93. doi: 10.1128/jb.175.19.6186-6193.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McClain MS, Blomfield IC, Eisenstein BI. Roles of fimB and fimE in site-specific DNA inversion associated with phase variation of type 1 fimbriae in Escherichia coli. J Bacteriol. 1991;173:5308–14. doi: 10.1128/jb.173.17.5308-5314.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ham TS, Lee SK, Keasling JD, Arkin AP. A tightly regulated inducible expression system utilizing the fim inversion recombination switch. Biotechnol Bioeng. 2006;94:1–4. doi: 10.1002/bit.20916. [DOI] [PubMed] [Google Scholar]
- 11.Ham TS, Lee SK, Keasling JD, Arkin AP. Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS One. 2008;3:e2815. doi: 10.1371/journal.pone.0002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Becskei A, Seraphin B, Serrano L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 2001;20:2528–2535. doi: 10.1093/emboj/20.10.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–42. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi H, Kaern M, Araki M, Chung K, Gardner TS, Cantor CR, Collins JJ. Programmable cells: interfacing natural and engineered gene networks. Proc Natl Acad Sci U S A. 2004;101:8414–9. doi: 10.1073/pnas.0402940101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006;355:619–27. doi: 10.1016/j.jmb.2005.10.076. [DOI] [PubMed] [Google Scholar]
- 16.Anderson JC, Voigt CA, Arkin AP. Environmental signal integration by a modular AND gate. Mol Syst Biol. 2007;3:133. doi: 10.1038/msb4100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.You L, Cox RS, 3rd, Weiss R, Arnold FH. Programmed population control by cell-cell communication and regulated killing. Nature. 2004;428:868–71. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- 18.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat Chem Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Topp S, Gallivan JP. Guiding bacteria with small molecules and RNA. J Am Chem Soc. 2007;129:6807–11. doi: 10.1021/ja0692480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weiss LE, Badalamenti JP, Weaver LJ, Tascone AR, Weiss PS, Richard TL, Cirino PC. Engineering motility as a phenotypic response to LuxI/R-dependent quorum sensing in Escherichia coli. Biotechnol Bioeng. 2008;100:1251–5. doi: 10.1002/bit.21862. [DOI] [PubMed] [Google Scholar]
- 21.Pawson T, Scott JD. Signaling through scaffold, anchoring, and adaptor proteins. Science. 1997;278:2075–80. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 22.Zeke A, Lukács M, Lim WA, Reményi A. Scaffolds: interaction platforms for cellular signalling circuits. Trends in Cell Biology. 2009;19:364–374. doi: 10.1016/j.tcb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickens M, Rogers JS, Cavanagh J, Raitano A, Xia Z, Halpern JR, Greenberg ME, Sawyers CL, Davis RJ. A cytoplasmic inhibitor of the JNK signal transduction pathway. Science. 1997;277:693–6. doi: 10.1126/science.277.5326.693. [DOI] [PubMed] [Google Scholar]
- 24.Garrington TP, Johnson GL. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr Opin Cell Biol. 1999;11:211–8. doi: 10.1016/s0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- 25.Xu S, Cobb MH. MEKK1 binds directly to the c-Jun N-terminal kinases/stress-activated protein kinases. J Biol Chem. 1997;272:32056–60. doi: 10.1074/jbc.272.51.32056. [DOI] [PubMed] [Google Scholar]
- 26.Park SH, Zarrinpar A, Lim WA. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science. 2003;299:1061–1064. doi: 10.1126/science.1076979. [DOI] [PubMed] [Google Scholar]
- 27.Baker MD, Wolanin PM, Stock JB. Signal transduction in bacterial chemotaxis. Bioessays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- 28.Gestwicki JE, Kiessling LL. Inter-receptor communication through arrays of bacterial chemoreceptors. Nature. 2002;415:81–4. doi: 10.1038/415081a. [DOI] [PubMed] [Google Scholar]
- 29.Gosink KK, Buron-Barral MC, Parkinson JS. Signaling interactions between the aerotaxis transducer Aer and heterologous chemoreceptors in Escherichia coli. J Bacteriol. 2006;188:3487–93. doi: 10.1128/JB.188.10.3487-3493.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourret RB, Stock AM. Molecular information processing: lessons from bacterial chemotaxis. J Biol Chem. 2002;277:9625–8. doi: 10.1074/jbc.R100066200. [DOI] [PubMed] [Google Scholar]
- 31.Liu JD, Parkinson JS. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J Bacteriol. 1991;173:4941–51. doi: 10.1128/jb.173.16.4941-4951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maddock JR, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–23. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 33.Greenfield D, McEvoy AL, Shroff H, Crooks GE, Wingreen NS, Betzig E, Liphardt J. Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keymer JE, Endres RG, Skoge M, Meir Y, Wingreen NS. Chemosensing in Escherichia coli: two regimes of two-state receptors. Proc Natl Acad Sci U S A. 2006;103:1786–91. doi: 10.1073/pnas.0507438103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Endres RG, Wingreen NS. Precise adaptation in bacterial chemotaxis through “assistance neighborhoods”. Proc Natl Acad Sci U S A. 2006;103:13040–4. doi: 10.1073/pnas.0603101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 37.Liu JD, Parkinson JS. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc Natl Acad Sci U S A. 1989;86:8703–7. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schulmeister S, Ruttorf M, Thiem S, Kentner D, Lebiedz D, Sourjik V. Protein exchange dynamics at chemoreceptor clusters in Escherichia coli. Proc Natl Acad Sci U S A. 2008;105:6403–8. doi: 10.1073/pnas.0710611105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kwon O, Georgellis D, Lin EC. Rotational on-off switching of a hybrid membrane sensor kinase Tar-ArcB in Escherichia coli. J Biol Chem. 2003;278:13192–5. doi: 10.1074/jbc.M210647200. [DOI] [PubMed] [Google Scholar]
- 40.Michalodimitrakis KM, Sourjik V, Serrano L. Plasticity in amino acid sensing of the chimeric receptor Taz. Mol Microbiol. 2005;58:257–66. doi: 10.1111/j.1365-2958.2005.04821.x. [DOI] [PubMed] [Google Scholar]
- 41.Utsumi R, Brissette RE, Rampersaud A, Forst SA, Oosawa K, Inouye M. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science. 1989;245:1246–9. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- 42.Ward SM, Delgado A, Gunsalus RP, Manson MD. A NarX-Tar chimera mediates repellent chemotaxis to nitrate and nitrite. Mol Microbiol. 2002;44:709–19. doi: 10.1046/j.1365-2958.2002.02902.x. [DOI] [PubMed] [Google Scholar]
- 43.Derr P, Boder E, Goulian M. Changing the specificity of a bacterial chemoreceptor. J Mol Biol. 2006;355:923–32. doi: 10.1016/j.jmb.2005.11.025. [DOI] [PubMed] [Google Scholar]
- 44.Villalobos A, Ness JE, Gustafsson C, Minshull J, Govindarajan S. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics. 2006;7:285. doi: 10.1186/1471-2105-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cardozo MJ, Massazza DA, Parkinson JS, Studdert CA. Disruption of Chemoreceptor Signaling Arrays by High Level of CheW, the Receptor-Kinase Coupling Protein. Mol Microbiol. 2010;75:1171–1181. doi: 10.1111/j.1365-2958.2009.07032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss R. Cellular computation and communications using engineered genetic regulatory networks. Massachusetts Institute of Technology; 2001. [Google Scholar]
- 47.Boukhvalova MS, Dahlquist FW, Stewart RC. CheW binding interactions with CheA and Tar. Importance for chemotaxis signaling in Escherichia coli. J Biol Chem. 2002;277:22251–9. doi: 10.1074/jbc.M110908200. [DOI] [PubMed] [Google Scholar]
- 48.Levit MN, Grebe TW, Stock JB. Organization of the receptor-kinase signaling array that regulates Escherichia coli chemotaxis. J Biol Chem. 2002;277:36748–54. doi: 10.1074/jbc.M204317200. [DOI] [PubMed] [Google Scholar]
- 49.Goldberg SD, Derr P, DeGrado WF, Goulian M. Engineered single- and multi-cell chemotaxis pathways in E. coli. Mol Syst Biol. 2009;5:283. doi: 10.1038/msb.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adler J, Templeton B. The Effect of Environmental Conditions on the Motility of Escherichia coli. J Gen Microbiol. 1967;46:175–184. doi: 10.1099/00221287-46-2-175. [DOI] [PubMed] [Google Scholar]
- 51.Lai HC, Shu JC, Ang S, Lai MJ, Fruta B, Lin S, Lu KT, Ho SW. Effect of Glucose Concentration on Swimming Motility in Enterobacteria. Biochemical and Biophysical Research Communications. 1997;231:692–695. doi: 10.1006/bbrc.1997.6169. [DOI] [PubMed] [Google Scholar]
- 52.Hamer R, Chen PY, Armitage JP, Reinert G, Deane CM. Deciphering chemotaxis pathways using cross species comparisons. BMC Syst Biol. 2010;4:3. doi: 10.1186/1752-0509-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tran HT, Krushkal J, Antommattei FM, Lovley DR, Weis RM. Comparative genomics of Geobacter chemotaxis genes reveals diverse signaling function. BMC Genomics. 2008;9:471. doi: 10.1186/1471-2164-9-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin AC, Wadhams GH, Armitage JP. The roles of the multiple CheW and CheA homologues in chemotaxis and in chemoreceptor localization in Rhodobacter sphaeroides. Mol Microbiol. 2001;40:1261–72. doi: 10.1046/j.1365-2958.2001.02468.x. [DOI] [PubMed] [Google Scholar]
- 55.Mandell DJ, Kortemme T. Computer-aided design of functional protein interactions. Nat Chem Biol. 2009;5:797–807. doi: 10.1038/nchembio.251. [DOI] [PubMed] [Google Scholar]
- 56.Kortemme T, Joachimiak LA, Bullock AN, Schuler AD, Stoddard BL, Baker D. Computational redesign of protein-protein interaction specificity. Nat Struct Mol Biol. 2004;11:371–9. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- 57.Bolon DN, Grant RA, Baker TA, Sauer RT. Specificity versus stability in computational protein design. Proc Natl Acad Sci U S A. 2005;102:12724–9. doi: 10.1073/pnas.0506124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skerker JM, Perchuk BS, Siryaporn A, Lubin EA, Ashenberg O, Goulian M, Laub MT. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–54. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alves R, Savageau MA. Comparative analysis of prototype two-component systems with either bifunctional or monofunctional sensors: differences in molecular structure and physiological function. Mol Microbiol. 2003;48:25–51. doi: 10.1046/j.1365-2958.2003.03344.x. [DOI] [PubMed] [Google Scholar]
- 60.Groban ES, Clarke EJ, Salis HM, Miller SM, Voigt CA. Kinetic buffering of cross talk between bacterial two-component sensors. J Mol Biol. 2009;390:380–93. doi: 10.1016/j.jmb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siryaporn A, Goulian M. Cross-talk suppression between the CpxA-CpxR and EnvZ-OmpR two-component systems in E. coli. Mol Microbiol. 2008;70:494–506. doi: 10.1111/j.1365-2958.2008.06426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voigt K, Izsvak Z, Ivics Z. Targeted gene insertion for molecular medicine. J Mol Med. 2008;86:1205–19. doi: 10.1007/s00109-008-0381-8. [DOI] [PubMed] [Google Scholar]
- 63.Buchholz F, Angrand PO, Stewart AF. Improved properties of FLP recombinase evolved by cycling mutagenesis. Nat Biotechnol. 1998;16:657–62. doi: 10.1038/nbt0798-657. [DOI] [PubMed] [Google Scholar]
- 64.Buchholz F, Stewart AF. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat Biotech. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 65.Santoro SW, Schultz PG. Directed evolution of the site specificity of Cre recombinase. Proc Natl Acad Sci U S A. 2002;99:4185–90. doi: 10.1073/pnas.022039799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sarkar I, Hauber I, Hauber J, Buchholz F. HIV-1 Proviral DNA Excision Using an Evolved Recombinase. Science. 2007;316:1912–1915. doi: 10.1126/science.1141453. [DOI] [PubMed] [Google Scholar]
- 67.Oberdoerffer P, Otipoby KL, Maruyama M, Rajewsky K. Unidirectional Cre-mediated genetic inversion in mice using the mutant loxP pair lox66/lox71. Nucleic Acids Res. 2003;31:e140. doi: 10.1093/nar/gng140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, Del Cardayre SB, Keasling JD. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463:559–62. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 69.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–5. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, 3rd, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 71.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:7877–82. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Temme K, Salis H, Tullman-Ercek D, Levskaya A, Hong SH, Voigt CA. Induction and relaxation dynamics of the regulatory network controlling the type III secretion system encoded within Salmonella pathogenicity island 1. J Mol Biol. 2008;377:47–61. doi: 10.1016/j.jmb.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.