Abstract
Evidence suggests that the herpes simplex virus regulatory protein ICP27 mediates the nuclear export of viral transcripts; however, the extent of this activity during infection is unclear. ICP27 is required for efficient expression of the long, leaky-late UL24 transcripts, but not for that of the short, early UL24 transcripts. We found that infection by an ICP27-null mutant resulted in undetectable UL24 protein expression, which represented at least a 70-fold decrease relative to that of wild-type virus. Because lack of ICP27 had a greater effect on levels of UL24 protein than on transcripts, we examined its effect on subcellular localization of UL24 transcripts. In wild-type-infected cells, both short and long UL24 transcripts fractionated predominantly with the cytoplasm. However, in the absence of ICP27, greater than 50% of long UL24 transcripts were nuclear, while the percentage of short UL24 transcripts that were cytoplasmic was not reduced. These results also imply that the short UL24 transcripts are translated poorly. The effect of ICP27 on cytoplasmic localization of the long UL24 transcripts did not extend to other transcripts with which it shared a common 3′ end or to other transcripts tested, including gC and UL42, whose overall expression is highly dependent on ICP27. Thus, the dual effects of ICP27 on mRNA accumulation and cytoplasmic localization are not always linked. These results identify viral transcripts that are dependent on ICP27 for efficient cytoplasmic localization during infection, but they also indicate the existence of ICP27-independent nuclear export pathways that are accessible to many viral transcripts during infection.
Eukaryotic gene expression is a highly complex process in which the multiple stages of transcription and translation are linked, with each stage potentially subject to regulation (reviewed in reference 29). When herpes simplex virus type 1 (HSV-1) infects a cell, viral genes are expressed in an ordered temporal pattern of immediate-early, early, and late genes, which results from the combined activity of viral and cellular factors, including RNA polymerase II (reviewed in reference 42). Like cellular mRNAs, viral mRNAs must be exported from the nucleus via RNA export factors to be translated in the cytoplasm. mRNA export factors are recruited cotranscriptionally and in a splicing-dependent manner to nascent transcripts and direct them to the nuclear pore complex for export (26, 27; reviewed in reference 29). The nuclear export of many cellular mRNAs is mediated by the REF(Aly)/TAP pathway (41; reviewed in reference 8). However, not all mRNAs use this pathway. For example, the intronless transcripts of human immunodeficiency virus type 1 access a CRM1-dependent export pathway through the virally encoded Rev protein (28, 54).
The HSV-1 protein ICP27 (512 amino acids) is a critical regulatory factor mediating the efficient expression of certain early and most late genes (30, 31, 39, 52). ICP27 stimulates viral gene expression at least in part at the level of transcription (23, 30). ICP27 associates with the RNA polymerase II holoenzyme and the viral transcription factor ICP4 (34, 55). It can also function to repress expression from certain viral promoters (46, 49). Through the stimulation of expression of early genes that encode replication proteins, ICP27 promotes viral DNA replication (30, 52). However, ICP27 can also function at the posttranscriptional level. ICP27 has been reported to stimulate the usage of certain polyadenylation [poly(A)] signals (31, 32) and to inhibit host gene expression, in part, by inhibiting splicing (3, 15, 16). ICP27 has been demonstrated to shuttle between the nucleus and cytoplasm, suggesting a role for it in the nuclear export of viral transcripts (38, 45, 47). Consistent with these observations, overexpression of the REF/Aly nuclear export factor increases the ICP27-dependent nuclear export of certain viral transcripts in the context of infection (4), and in Xenopus laevis oocyte microinjection experiments ICP27 can stimulate the nuclear export of certain viral transcripts through the REF/Aly cellular mRNA nuclear export pathway (24). However, it is not clear how important this particular activity of ICP27 is to the efficient nuclear export of viral transcripts during infection.
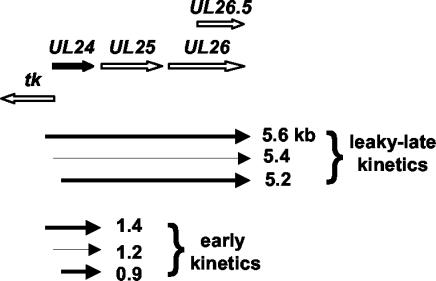
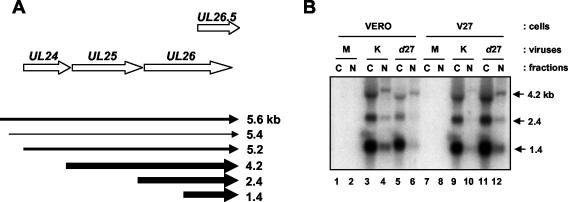
The HSV UL24 gene encodes a nuclear-associated protein (18, 35) that appears to inhibit cell-to-cell fusion during infection (22, 44, 50). The expression of UL24 is quite complex (Fig. 1). Six UL24 transcripts have been identified which originate from three transcription start sites and terminate either at the UL24 poly(A) signal and exhibit early kinetics (1.4, 1.2, and 0.9 kb; short UL24 transcripts) or at the UL26 signal and exhibit leaky-late kinetics (5.6, 5.4, and 5.2 kb; long UL24 transcripts) (6, 14). The mRNAs originating from both the first and second transcription start site can be translated into full-length UL24 protein (35). In the absence of ICP27, levels of the long UL24 transcripts are greatly diminished, while levels of the short UL24 transcripts are affected little, if at all (14).
FIG. 1.
Diagram of the UL24 transcription unit. The relative positions and orientations of the UL24 and flanking ORFs are indicated by the fat arrows at the top of the figure. The six transcripts containing UL24 sequence that have been identified are illustrated below. The three long transcripts that terminate at the end of UL26 are expressed with leaky-late kinetics. The three short transcripts that terminate at the end of UL24 are expressed with early kinetics. Transcripts beginning at the second UL24 transcription start site are less abundant than those beginning at the other two start sites, which is reflected qualitatively by the relative thickness of the arrows.
To investigate further how ICP27 regulates UL24 gene expression, we examined the effect of an ICP27-null mutation on UL24 protein expression. These experiments led us to test whether ICP27 functions to regulate expression of UL24 transcripts at the level of mRNA nuclear export. We have found that ICP27 is required for efficient cytoplasmic localization of the long UL24 transcripts but not of the short UL24 transcripts or many other viral mRNAs. Our results provide evidence that translation of the short UL24 transcripts is inefficient and suggest the existence of multiple nuclear export pathways for viral transcripts during infection, including ICP27-independent pathways.
MATERIALS AND METHODS
Cells and viruses.
HSV-1 wild-type strain KOS1.1 (20) and the ICP27-null mutant d27 (39) were propagated on Vero cells (American Type Culture Collection) or V27 cells (39), respectively, as described previously (5). For analysis of gene expression, cells in 60-mm-diameter dishes were infected at a multiplicity of infection (MOI) of 10 PFU per cell, and the inocula were back-titrated on V27 cells (an ICP27-complementing cell line) to confirm that equal PFU of each virus were used.
Western blotting.
The harvesting of lysates from infected cells and Western blotting analysis using rat UL24 antiserum were carried out as described previously (35). Quantification of UL24 signals was done using a standard curve derived from a dilution series of infected cell lysate that was run on the same gel as the experimental samples. The antibody directed against HSV thymidine kinase (TK) protein was generously provided by W. C. Summers (Yale University). The TK signal was detected by chemiluminescence using the SuperSignal West Femto kit (Pierce) according to the manufacturer's instructions.
RNA isolation.
RNA was isolated using the Qiagen RNeasy kit following the manufacturer's instructions for adherent cells. To isolate RNA from cytoplasmic and corresponding nuclear fractions, cells were scraped and collected in 1 ml of phosphate-buffered saline. The RNA from the cytoplasmic fractions was isolated following the Qiagen RNeasy protocol for isolation of cytoplasmic RNA. RNA was also isolated from the nuclear pellets, which were resuspended in lysis buffer provided by the kit and processed according to the protocol for isolation of total RNA from cells.
Northern blot hybridization.
[32P]dCTP- or [32P]dATP-labeled DNA probes (New England Nuclear) were synthesized by using the DNA Random Primed DNA Labeling kit (Roche) following the manufacturer's instructions. The DNA templates used to make various DNA probes were as follows: for UL24 (7) and gB (7) the templates have been described previously. For VP16 the 993-bp template was synthesized by PCR from viral DNA using the primers 5′ CTATGTACCATGCTCGATAC and 5′ CGTCTAGCGCGTCGGCA. For UL42 the 991-bp template was synthesized by PCR using the primers 5′ CGTTTCGCACGCTGGTTC and 5′ AGGTCGCGAAAGTAACAC. For gC the 1,062-bp template was synthesized by PCR using the primers 5′ CGTGTGGTGCGACCGC and 5′ TCAACCGACAGATGTACTC. For UL26 the 790-bp BamHI fragment containing the UL26 sequence was isolated from the EcoRIF clone (13) and ligated into pSK+ (Stratagene) (plasmid constructed and graciously provided by David Wensel). The BamHI fragment of this plasmid was subsequently excised and used as a template. All oligonucleotides were obtained from Integrated DNA Technologies. RNA was resolved on a 0.9% denaturing agarose gel (SeaKem) containing formaldehyde in a 3[N-morpholino] propanesulfonic acid buffering system (43). Northern blot hybridization was carried out as described previously (14) except for the experiment shown in Fig. 3, where PerfectHyb Plus hybridization solution (Sigma) and Hybond N+ membrane (Amersham Pharmacia) were used. Signals were detected by autoradiography. For quantification, blots were exposed to phosphor storage screens and signals were analyzed using the Personal Molecular Imaging FX system (Bio-Rad) and Quantity One software (Bio-Rad). Background signals were subtracted from each measured experimental value. To visualize particularly faint bands for quantification, the images were transformed to make the bands appear darker (a process that does not alter their measured values), which allowed for their accurate delineation.
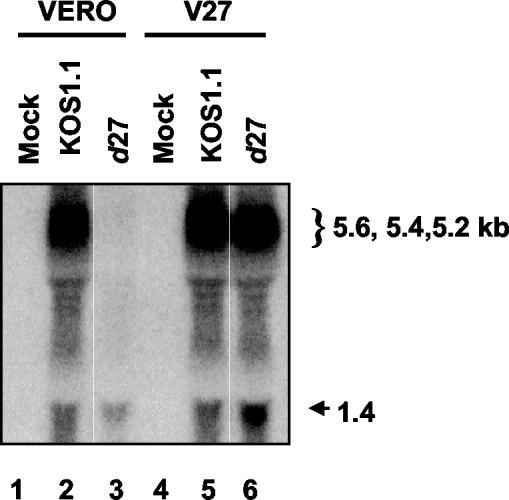
FIG. 3.
Effect of an ICP27-null mutation on the expression of UL24 transcripts in infected cells. Vero or V27 cells were either mock infected (lanes 1 and 4) or infected at an MOI of 10 with either KOS1.1 (lanes 2 and 5) or d27 (lanes 3 and 6) cells, and at 13 h p.i. total RNA was isolated. RNA was analyzed by Northern blot hybridization with a probe corresponding to the UL24 sequence. The position of the 1.4-kb short UL24 transcript is indicated by an arrow to the right of the panel. The position of the broad band corresponding to the 5.2-, 5.4-, and 5.6-kb-long UL24 transcripts is indicated by a bracket to the right of the panel.
RESULTS
ICP27 is required for the expression of UL24 protein.
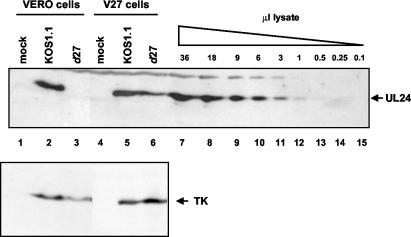
In previous experiments our laboratories found that an ICP27-null mutation resulted in a marked decrease in levels of the long UL24 transcripts that arise from utilization of the UL26 poly(A) signal but had little, if any, effect on the levels of the short UL24 transcripts that arise from the use of the UL24 poly(A) signal (14). We wanted to compare the effect of ICP27 on UL24 protein levels with that on levels of UL24 transcripts. Vero or V27 cells were either mock infected (Fig. 2, top panel, lanes 1 and 4) or infected with either KOS1.1 (lanes 2 and 5) or with d27 (lanes 3 and 6), which contains a null mutation in the ICP27 gene, and at 15 h postinfection (p.i.) cell lysates were harvested, resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and analyzed in Western blot experiments. UL24 was clearly detected in lysates from Vero cells infected with the parental strain, KOS1.1 virus (top panel, lane 2). In contrast, we were unable to detect any UL24 protein in lysates from Vero cells infected with d27 virus (lane 3). A standard curve was constructed based on a dilution series of infected cell lysate run on the same gel (lanes 7 to 15), which allowed us to calculate that in this experiment there was at least 70-fold less UL24 expressed in the absence of ICP27 than in its presence. UL24 protein levels were restored to nearly wild-type levels when the d27 virus was grown on the ICP27-complementing cell line V27 (lanes 5 and 6), indicating that the defect in UL24 protein levels seen in Vero cells was indeed a consequence of the ICP27 mutation. As a loading control, the blot was stripped and incubated with an antibody directed against the HSV TK protein (Fig. 2, bottom panel). We found that levels of this viral protein varied less than threefold between samples. We also quantified UL24 transcript levels in total RNA isolated from cells infected with KOS1.1 or d27 (Fig. 3). In this particular experiment there was a 10-fold decrease in levels of UL24 transcripts (short and long combined) (compare lanes 2 and 3). This effect varied among experiments from as low as 5-fold to as high as 40-fold, although we consistently observed that the effect on levels of the short UL24 transcripts was quite modest, as opposed to the much more pronounced decrease in levels of the long UL24 transcripts. Thus, even though close-to-wild-type levels of the short UL24 transcripts and some, though clearly reduced, levels of the long UL24 transcripts were expressed, we were unable to detect any UL24 protein in the absence of ICP27.
FIG. 2.
Western blot showing the effect of an ICP27-null mutation on the expression of UL24 protein in infected cells. Vero or V27 cells were either mock infected (lanes 1 and 4) or infected at an MOI of 10 with either KOS1.1 (lanes 2 and 5) or d27 (lanes 3 and 6) cells, and total cell lysates were harvested at 15 h p.i. and concentrated 10-fold. A 20-μl aliquot of each concentrated lysate was analyzed by Western blotting with antiserum raised against UL24, the position of which is indicated to the right of the top panel. In lanes 7 to 15, a dilution series of concentrated KOS1.1-infected cell lysate was loaded. The volumes (in microliters) are indicated above the panel. The membrane was subsequently stripped and incubated with an antibody directed against the HSV TK, the position of which is indicated to the right of the bottom panel.
ICP27 affects the nucleocytoplasmic distribution of a subset of UL24 transcripts.
Because levels of the short UL24 transcripts are affected little in cells infected with an ICP27-null virus and, although reduced in amount, some long UL24 transcripts are still detected, we investigated the possibility that the lack of UL24 protein expression was a consequence of a defect in mRNA nuclear export. Vero and V27 cells were either mock infected or infected with KOS1.1 or d27. At 13 h p.i. cells were harvested, and RNA was isolated from the corresponding cytoplasmic and nuclear fractions, resolved on an agarose gel, and stained with ethidium bromide (Fig. 4A). As expected, the precursor 45S and 32S rRNAs were detected only in the nuclear and not in the cytoplasmic fractions. Likewise, the 28S and 18S rRNAs, which are processed in the nucleus and then quickly exported to the cytoplasm (9), were predominantly in the cytoplasmic fractions. Thus, we concluded that the fractionation into, and subsequent isolation of RNA from, the cytoplasmic and nuclear fractions was successful. Nucleic acids from this gel were transferred to nitrocellulose for analysis by Northern blotting and probed for UL24 sequences. The blot presented in Fig. 4B is overexposed so that it is possible to clearly see the long UL24 transcripts in the d27 samples. The migration of the UL24 long transcripts, particularly in the cytoplasmic lanes, is altered because of the comigration of the broad band of 28S rRNA (Fig. 4A). However, despite these complications Northern blot analysis allowed us to differentiate between and quantify (see Materials and Methods) the short and long UL24 transcripts. Where percentages of nuclear and cytoplasmic transcripts are reported, 100% represents the total amount of transcript detected in both fractions together, which varied from transcript to transcript.
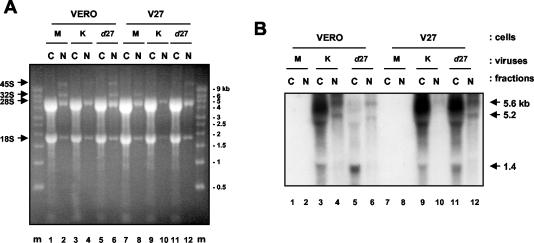
FIG. 4.
ICP27 affects the cytoplasmic versus nuclear distribution of the long, leaky-late UL24 transcripts. Vero cells (lanes 1 to 6) or V27 cells (lanes 7 to 12) were either mock infected (M) (lanes 1, 2, 7, and 8) or infected with KOS1.1 cells (lanes 3, 4, 9, and 10) or the ICP27-null virus d27 (lanes 5, 6, 11, and 12). (A) RNA was isolated from nuclear (N) and cytoplasmic (C) fractions at 13 h p.i., resolved on a denaturing formaldehyde agarose gel, and stained with ethidium bromide to allow for visualization of the major rRNA species. The positions of the various rRNAs are indicated by arrows to the left of the panel. RNA molecular mass markers (Ambion) were resolved in the first and last lanes of the gel (lanes m). The sizes of the markers are located to the right of the panel. (B) Nucleic acids from the agarose gel were analyzed by Northern blot hybridization for UL24 transcripts, the positions of which are indicated by arrows to the right of the panel. The bands corresponding to the long UL24 transcripts in the cytoplasmic fractions are distorted due to the comigration of the 28S rRNA.
In Vero cells infected with the parental strain KOS1.1 (Fig. 4B, lanes 3 and 4), most of both the long and short UL24 transcripts were cytoplasmic. However, in Vero cells infected with d27 (lanes 5 and 6), we observed different effects of the lack of ICP27 on the intracellular distribution of the long and the short UL24 transcripts. In Vero cells infected with d27, the fraction of 1.4-kb UL24 transcripts, which are the most clearly visible of the short UL24 transcripts, that was cytoplasmic was slightly more than that in KOS1.1-infected cells (87 versus 76%). (In this experiment we also observed increased levels of short UL24 transcripts in Vero cells infected with d27 compared to KOS1.1. This was also observed for the early transcripts ICP8 [data not shown] and tk [reference 10 and data not shown]. These modest increases likely reflect the role of ICP27 in DNA replication and in the transition from early to late gene expression [30, 31, 39]. Presumably, in the absence of ICP27, early gene expression is not down-regulated efficiently, leading to somewhat higher levels of expression of the early, short UL24 transcripts at late times in infection.)
In contrast, the percentage of the remaining long UL24 transcripts that was cytoplasmic dropped from 70% in KOS1.1-infected Vero cells to 47% in d27-infected cells. This defect in cytoplasmic localization was highly reproducible. Furthermore, when the d27 virus was grown on the ICP27-complementing cell line V27 (lanes 11 and 12), cytoplasmic localization of the majority of long UL24 transcripts was restored. Although the defect in cytoplasmic localization of the long UL24 transcripts in d27-infected cells was not complete, these results show that during infection ICP27 is important for the efficient cytoplasmic localization of the long UL24 transcripts, but not of the short UL24 transcripts. In addition, the observation that levels of long UL24 transcripts in the nucleus are reduced in d27-infected cells compared to wild-type-infected cells suggests a defect in synthesis of these transcripts.
Interestingly, we also observed a slight increase in the cytoplasmic localization of both the short and long UL24 transcripts (to 96 and 89%, respectively) when levels of ICP27 were unusually high as a result of infecting an ICP27-complementing cell line (V27) with an ICP27-competent virus (KOS1.1) (52) such as seen in Fig. 4B (compare lanes 3 and 4 to lanes 9 and 10), an observation that we have made for several other viral transcripts analyzed (see subsequent figures).
The effect of ICP27 on the nucleo-cytoplasmic distribution of the long UL24 transcripts does not correlate with utilization of the UL26 poly(A) signal.
One possible explanation for the differential effect of an ICP27 defect on the levels and subcellular distribution of various UL24 transcripts is that the long UL24 transcripts utilize the UL26 poly(A) signal while the short UL24 transcripts utilize the poly(A) signal immediately 3′ to the UL24 open reading frame (ORF). The long UL24 transcripts (5.6, 5.4, and 5.2 kb) share the UL26 poly(A) signal with the UL25 (4.2 kb), UL26 (2.4 kb), and UL26.5 (1.4 kb, not to be confused with the 1.4-kb UL24 transcript) transcripts (Fig. 5A). We therefore investigated whether the effects of ICP27 on expression and nucleocytoplasmic distribution of the long UL24 transcripts were also observed for those transcripts with which it shares a poly(A) signal. We stripped the Northern blot shown in Fig. 4 and probed it with sequences contained within UL26.5, which would hybridize to the six transcripts illustrated in Fig. 5A. We found that levels of UL26.5, UL26, and UL25 transcripts (Fig. 5B) were reduced somewhat in d27-infected cells (less than twofold). However, the percentages of cytoplasmic UL26.5 and UL26 transcripts were similar in d27-infected Vero cells compared to KOS1.1-infected Vero cells (76 to 70% and 84 to 89%, respectively). In this experiment it was not possible to clearly differentiate between the long UL24 and the UL25 transcripts. However, the UL25 transcripts have been reported to be more abundant than any of the long UL24 transcripts (17). Therefore, because greater than 50% of the long UL24 transcripts from cells infected with d27 were nuclear (Fig. 4B), the observation that the 4.2-kb signal was stronger in the cytoplasmic fraction than in the nuclear one in Fig. 5B implied that the UL25 transcripts were efficiently localized to the cytoplasm in cells infected with the d27 virus. Thus, it appears that the 3′ end of the long UL24 transcripts was not sufficient to mediate ICP27-dependent cytoplasmic localization.
FIG. 5.
The 3′ UTR of the long UL24 transcripts is not sufficient to confer ICP27-dependent cytoplasmic localization. (A) Diagram illustrating the six 3′ coterminal transcripts that utilize the UL26 polyadenylation signal. The 5.6-, 5.4-, and 5.2-kb transcripts represent the long UL24 transcripts, the 4.2-kb transcripts originate from the UL25 promoter, the 2.4-kb transcripts originate from the UL26 promoter, and the 1.4-kb transcript corresponds to UL26.5. (B) The membrane analyzed in Fig. 4 was stripped and hybridized with a radioactive probe corresponding to the UL26.5 sequence. The positions of the UL25 (4.2-kb), UL26 (2.4-kb), and UL26.5 (1.4-kb) transcripts are indicated by arrows to the right of the panel. M, mock; K, KOS1.1; N, nuclear; C, cytoplasmic.
Effect of ICP27 on the nucleocytoplasmic distribution of other viral transcripts.
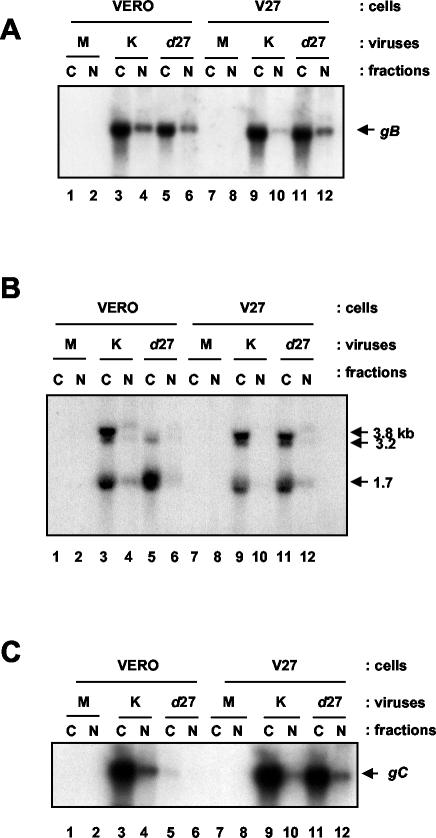
ICP27 is reported to associate preferentially with certain viral mRNAs in the cytoplasmic compartment of cells (45). ICP27 interacts with REF (4, 24), a component of the REF(Aly)/TAP cellular mRNA nuclear export pathway, and stimulates the nuclear export of certain viral transcripts in Xenopus oocyte microinjection experiments (24). Furthermore, overexpression of REF through transient transfection of mammalian cells stimulates the nuclear export of certain viral transcripts during infection in an ICP27-dependent manner (4). We therefore asked what effect the absence of ICP27 had on the cytoplasmic-to-nuclear ratio of some of these viral transcripts during a typical infection in cell culture. The blot from Fig. 4 was stripped and reprobed for gB transcripts, which have been reported to become more cytoplasmic upon overexpression of REF in an ICP27-dependent manner (4). Total levels of gB transcripts were approximately twofold lower in cells infected with d27 than in cells infected with the parental virus KOS1.1 (Fig. 6A). To our surprise, we did not observe a meaningful difference in the ratio of cytoplasmic to nuclear gB transcripts between the KOS1.1- and d27-infected Vero cells (83 to 17% and 82 to 18%, respectively). We did note, however, that similar to what we observed for UL24 transcripts, there was a slight increase in the cytoplasmic localization of gB transcripts when levels of ICP27 were unusually high, as a result of infecting an ICP27-complementing cell line (V27) with an ICP27-competent virus (KOS1.1) (93%) (compare lanes 3 and 4 to lanes 9 and 10).
FIG. 6.
Effect of ICP27 on the cytoplasmic localization of gB, VP16, and gC transcripts. (A) The membrane analyzed in Fig. 4 was stripped and hybridized with a radioactive probe corresponding to the gB sequence. The position of the gB transcripts is indicated to the right of the panel. (B) The membrane in Fig. 4 was stripped and hybridized with a VP16-specific probe (UL48). The positions of the 1.7-kb transcripts originating at the UL48 promoter, the 3.2-kb transcripts originating at the UL49 promoter, and the 3.8-kb transcripts originating at the UL49.5 promoter are indicated to the right of the panel. (C) The membrane in Fig. 4 was stripped and hybridized with a gC-specific probe. The position of the gC transcripts is indicated to the right of the panel. Abbreviations for all three panels: M, mock; K, KOS1.1; N, nuclear; C, cytoplasmic.
We also examined transcripts that contain the VP16 ORF. In Xenopus oocyte experiments, coinjection of recombinant ICP27 stimulated the nuclear export of synthetic VP16 transcripts (24). We used a probe corresponding to sequences within the VP16 ORF to probe the same membrane analyzed in the previous figures. This probe detected three coterminal transcripts corresponding to transcripts originating at the UL48 (VP16) promoter (1.7 kb), the UL49 promoter (3.2 kb), and the UL49.5 promoter (3.8 kb) (36) (Fig. 6B). Levels of the UL48 and UL49 transcripts were similar in d27- and KOS1.1-infected cells, although levels of UL49.5 transcripts were clearly reduced in the absence of ICP27. Once again, we were surprised to find that VP16 transcripts localized efficiently to the cytoplasm in Vero cells infected with either KOS1.1 or d27 (lanes 3 to 6). The band corresponding to the UL49.5 (3.8-kb) transcripts was distorted in the cytoplasmic fraction due to comigration of the 28S rRNA; however, it does appear that there was a modest increase in the relative percentage of 3.8-kb transcripts present in the nuclear fraction of d27-infected Vero cells compared with the KOS1.1-infected cells (30 and 19%, respectively). The significance of this observation is difficult to ascertain, although it could be consistent with a role for ICP27 in the nucleocytoplasmic distribution of transcripts containing VP16 sequences. Similar to what we observed previously, there appeared to be a modest increase in cytoplasmic localization of each of these transcripts when the wild-type virus was grown on the ICP27-complementing cell line (compare lanes 3 and 4 to lanes 9 and 10).
We decided to examine a transcript that is known to be highly dependent on ICP27 for its synthesis in case there was a link between a dependency on ICP27 for expression and a dependency on ICP27 for efficient cytoplasmic localization. Thus, we analyzed gC, whose transcription has been demonstrated to be highly dependent on ICP27 (23, 40). As expected, in the absence of ICP27 levels of gC transcripts were greatly reduced, in this particular experiment greater than 50-fold (Fig. 6C) and, thus, in order to visualize them the blot was overexposed. However, despite this critical dependency on ICP27 for expression, those gC transcripts that were expressed were predominantly cytoplasmic. Therefore, it would appear that a dependency on ICP27 for synthesis is not necessarily indicative of a dependency for cytoplasmic localization.
Effect of ICP27 on the nucleocytoplasmic distribution of transcripts at earlier times in infection.
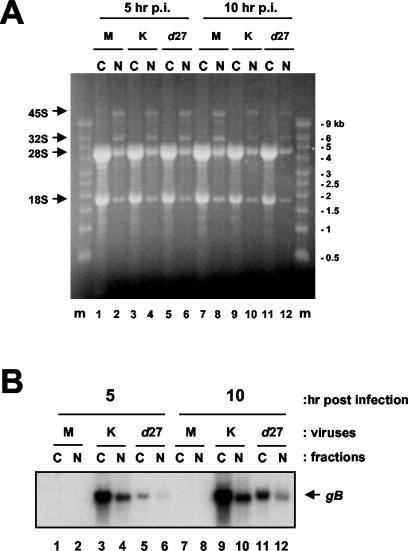
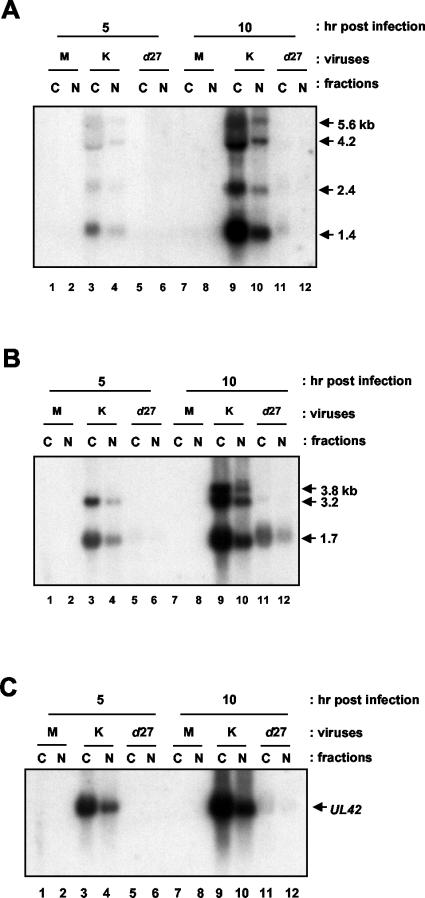
We considered the possibility that the reason we did not detect a dependency on ICP27 for the cytoplasmic location of any transcripts tested other than the long UL24 transcripts was related to the relatively late time point we had chosen. This choice had been made to ensure that we would be able to detect the reduced levels of the long UL24 transcripts in d27-infected cells well enough to draw conclusions about their cytoplasmic versus nuclear localization. However, the experiments showing a stimulation of nuclear export upon overexpression of REF used a time point of 6 h p.i. (4). Therefore, we either mock infected cells or infected Vero cells with KOS1.1 or d27, and at 5 and 10 h p.i. total RNA was isolated from nuclear and cytoplasmic fractions. Once again, the ethidium bromide-stained gel indicated that the fractionation was successful, that the RNA isolated was of a high quality, and that similar amounts of RNA were loaded for each sample (Fig. 7A). We probed the corresponding Northern blot for UL24 transcripts. In the presence of ICP27, most of the UL24 transcripts were predominantly cytoplasmic; however, we could not compare the ratios of cytoplasmic to nuclear RNAs in Vero cells infected with d27 because, as expected, levels of these transcripts were barely detectable at these early times and could not be quantified with confidence (data not shown). We next stripped and probed the blot for gB transcripts (Fig. 7B). At these earlier time points, we did detect a greater effect on overall levels of gB transcripts such that they were reduced 5- to 10-fold in the absence of ICP27. However, we did not observe a dependency on ICP27 for the efficient cytoplasmic localization of gB transcripts at either 5 or 10 h p.i. (Fig. 7B). In each combination of viruses and cells tested, between 75 and 82% of gB transcripts were present in the cytoplasmic fractions. We also stripped and reprobed the blot for UL26 transcripts (Fig. 8A). At these earlier times, levels of transcripts in the d27 fraction were too low to quantify accurately, but upon visual inspection it was clear that for the UL26.5 transcripts the band in the lane corresponding to the cytoplasmic fraction at 10 h p.i. was more intense than in the corresponding nuclear fraction (Fig. 8A, compare lanes 11 and 12). Thus, it appears that there was no major defect in cytoplasmic localization of UL26.5 transcripts in the absence of ICP27 expression during infection.
FIG. 7.
Effect of ICP27 on the cytoplasmic localization of gB transcripts at earlier times in infection. (A) Vero cells were either mock infected (lanes 1, 2, 7, and 8) or infected with KOS1.1 (lanes 3, 4, 9, and 10) or the ICP27-null virus d27 (lanes 5, 6, 11, and 12). (A) At 5 h (lanes 1 to 6) or 10 h (lanes 7 to 12) p.i., total RNA was isolated from nuclear (N) and cytoplasmic (C) fractions, resolved on a denaturing formaldehyde agarose gel, and stained with ethidium bromide. The positions of precursor 45S and 32S rRNAs and fully processed 28S and 18S rRNAs are indicated by arrows to the left of the panel. RNA molecular mass markers are resolved in the first and last lanes of the gel (lanes m). The sizes of the markers are located to the right of the panel. (B) The membrane was hybridized with a gB-specific probe. The position of gB transcripts is indicated by an arrow to the right of the panel. M, mock; K, KOS1.1; N, nuclear; C, cytoplasmic.
FIG. 8.
Effect of ICP27 on the cytoplasmic localization of various viral transcripts at earlier times in infection. (A) The membrane in Fig. 7 was stripped and hybridized with a UL26-specific probe. The positions of the UL26.5 (1.4-kb), UL26 (2.4-kb), UL25 (4.2-kb), and UL24 (5.6-kb) transcripts are indicated to the right of the panel. (B) The membrane in Fig. 7 was stripped and hybridized with a VP16 (UL48)-specific probe. The positions of the UL48 (1.7-kb), UL49 (3.2-kb), and UL49.5 (3.8-kb) transcripts are indicated to the right of the panel. (C) The membrane in Fig. 7 was stripped and hybridized with a UL42-specific probe. The position of the UL42 transcripts is indicated to the right of the panel. M, mock; K, KOS1.1; N, nuclear; C, cytoplasmic.
We next probed the blot for VP16 transcripts (Fig. 8B). Levels of all transcripts were clearly reduced at these earlier times in the absence of ICP27. At 5 h p.i. levels of transcripts were too low to be clearly detected in Vero cells infected with d27 lanes (lanes 5 and 6). At 10 h p.i., the UL48 (1.7-kb) transcripts were detectable in Vero cells infected with either KOS1.1 or d27; however, they were efficiently localized to the cytoplasm in both the absence and presence of ICP27 (lanes 9 to 12).
Finally, we looked at UL42 transcripts as an example of an early gene whose expression is highly dependent on ICP27 (52). The blot was overexposed so that we could detect the low levels of transcripts in the d27 samples, at least at 10 h p.i. (Fig. 8C). Here again, although the dependency on ICP27 for expression was clearly observed, in that there was an approximately 50-fold reduction in levels of UL42 transcripts in Vero cells infected with d27 compared to that with the parental strain, the remaining transcripts were still predominantly cytoplasmic, suggesting that even at earlier times in infection, transcripts whose expression is highly dependent on ICP27 can be efficiently localized to the cytoplasm of infected cells in the absence of this regulatory protein.
In this experiment we observed a slight retardation in the electrophoretic mobility of several different viral transcripts from Vero cells infected with d27 compared to that with wild-type virus (Fig. 5B, 7B, and 8A to C). These effects are similar to the effect that has been seen for gC transcripts in transfected cells with or without ICP27 (37). Further study will be required to determine the significance of these observations; however, one possible explanation might be modifications in length of poly(A) tails, similar to what has been observed for the α-globin gene during HSV-1 infection (10) and consistent with previous reports that ICP27 affects polyadenylation of viral transcripts (32).
Thus, we have found that for those genes whose expression is dependent on ICP27, this stimulatory effect is more pronounced at earlier than at late times in infection. However, transcripts whose expression is dependent on ICP27, even highly dependent, are not necessarily dependent on ICP27 for their efficient cytoplasmic localization. Of those transcripts we have analyzed, only the long UL24 transcripts exhibited a clear defect in cytoplasmic localization in the absence of ICP27.
DISCUSSION
Regulation of gene expression can occur at multiple levels simultaneously, which can complicate the interpretation of results obtained from mutational analyses of multifunctional proteins such as ICP27. Despite sharing common transcription start sites, the long and short UL24 transcripts are affected differently by ICP27 defects and, thus, they are very useful for investigating the function of this regulatory factor. Conversely, the differential effect of ICP27 on the short and long UL24 transcripts can be exploited to further our understanding of the role of the different UL24 transcripts during infection.
The effect of ICP27 on levels of UL24 protein.
ICP27 is crucial for the expression of UL24 protein, because we were unable to detect any UL24 protein (>70-fold decrease) in the absence of ICP27 expression during infection. Both the long and short UL24 transcripts are expressed during productive infection; however, we have now identified several instances where the expression of UL24 protein does not correlate with levels of the short UL24 transcripts but rather with levels of the long UL24 transcripts that encode multiple genes. For example, inhibition of viral DNA replication has little effect on levels of the short UL24 transcripts; however, levels of UL24 protein are reduced by fivefold (35), which is similar to the sixfold reduction in levels of the long UL24 transcripts observed under such conditions (6, 14; A. Pearson and D. Coen, unpublished data). Similarly, here we report that the absence of ICP27 had little effect on the levels and cytoplasmic localization of the short UL24 transcripts, but levels of UL24 protein were reduced greater than 70-fold. These results suggest that the short UL24 transcripts are poorly translated. This observation raises the question of whether translation of the short UL24 transcripts is aborted somehow, or whether these transcripts ever associate with ribosomes. It is interesting that in a large-scale analysis of polyadenylated, ribosome-associated transcripts corresponding to this region of the viral genome, the long UL24 transcripts, but not the short UL24 transcripts, were detected (17). We have previously found evidence suggesting that there is antisense regulation of the short but not of the long UL24 transcripts by overlapping tk mRNA (7), which may contribute to the down-regulation of translation of the short UL24 transcripts, perhaps, for example, by double-stranded RNA-mediated RNA editing (reviewed in references 2 and 25). We cannot rule out the possibility that translation of the short UL24 transcripts is cell-type dependent. However, it is also possible that the short UL24 transcripts may play a non-protein-coding regulatory role or that inhibiting their translation is important. That said, it is not clear what function, if any, the short UL24 transcripts may play during infection.
Possible importance of late expression of UL24.
As reviewed above, translation of the short UL24 transcripts, which are expressed with early kinetics, appears to be inhibited. ICP27 helps promote the transition from early to late gene expression (31). Thus, a requirement for ICP27 in the expression of UL24 protein and other late proteins may serve to delay their expression until late times in infections. Kinetics of expression can have an impact on pathogenesis, as has been demonstrated for VP5 (ICP5) (51). UL24 shares several features with viral genes that have been demonstrated to play a role in egress of newly synthesized viral particles (18, 35), such as gK (UL53) (12, 21) and UL20 (1, 53). If indeed UL24 functions at the stage of membrane fusion during viral egress, or at some other late stage in the viral life cycle, then perhaps it is advantageous for the virus to delay expression of UL24 until the assembly of viral capsids is well under way.
Differential regulation of the short and long UL24 transcripts by ICP27.
ICP27 regulates gene expression at multiple levels. Our results indicate that in addition to regulating the expression level of the long UL24 transcripts (14), ICP27 regulates the expression of UL24 at the level of mRNA cytoplasmic localization. Notwithstanding the formal possibility that in the absence of ICP27 the long UL24 transcripts that are cytoplasmic have a shorter half-life than those present in the nucleus, given the extensive literature on ICP27 and nuclear export, for the remaining discussion we will assume that our results were due to effects on nuclear export.
It is not obvious what features of the long UL24 transcripts differentiate them from the short UL24 transcripts with regard to ICP27-regulated nuclear export. Given that the long and short UL24 transcripts share common transcription start sites, it would appear that promoter elements alone are not mediating this difference. Similarly, because the long UL24 transcripts and the UL25, UL26, and UL26.5 transcripts share a common 3′ end, it would also appear that elements shared with these latter transcripts are not sufficient to mediate ICP27-dependent nuclear export of the long UL24 transcripts. Perhaps it is a combination of sequence elements that affect the nuclear export of these transcripts. Links between different aspects of RNA metabolism are well documented (reviewed in reference 29). Thus, 5′ elements of UL24 may specifically direct transcripts that utilize the UL26 poly(A) signal, but not the UL24 signal, to a nuclear export pathway distinct from that of the other transcripts we have analyzed.
One possible difference between the long and the short UL24 transcripts may lie in a portion of the 3′ untranslated region (3′ UTR) of UL24. Depending on where the exact start site of transcription for UL25 is located (it has not, to our knowledge, been mapped yet), there may be sequences between it and the cleavage and polyadenylation site of the UL24 short transcripts further upstream that would be unique to the long UL24 transcripts. Regardless, our results with UL24 transcripts do not support a model whereby ICP27 functions to compensate for a missing RNA element in HSV transcripts (24), because the long UL24 transcripts that do apparently depend on ICP27 for their efficient nuclear export would appear to contain all of the sequences found within the short UL24 transcripts that are efficiently exported from the nucleus independent of ICP27. It is possible, however, that depending on the cells being used for the experiments, the pathways and mechanisms being studied could be different.
ICP27-dependent HSV mRNA nuclear export.
We have found that ICP27 is required for efficient nuclear export of long UL24 transcripts. There is evidence that ICP27 associates with the RNA nuclear export factor REF and that under certain experimental conditions ICP27 can function in the nuclear export of several late viral transcripts (4, 24). Our observation that artificially high levels of ICP27 resulting from the infection of ICP27-complementing cells with an ICP27-competent virus seemed to correlate with small increases in the cytoplasmic localization of several viral transcripts appears to mirror the results obtained from experiments looking at the stimulatory effect of overexpressing REF on nuclear export of viral transcripts (4). The long UL24 transcripts, which exhibit a defect in cytoplasmic localization in the absence of ICP27, may access the ICP27/REF pathway (4, 24), or they may access a different nuclear export pathway that is regulated indirectly by ICP27. For example, the expression of ICP27 may bolster the activity of a nuclear export pathway that is accessed by the long UL24 transcripts in some manner, and in the absence of ICP27 this pathway is less active.
ICP27-independent HSV mRNA nuclear export.
Regardless of the mechanism of ICP27-dependent nuclear export, we must invoke the existence of another export pathway to explain the residual cytoplasmic localization of the long UL24 transcripts in the absence of ICP27 and that of all the other HSV transcripts that were efficiently localized to the cytoplasm under those conditions. Even transcripts such as gB, whose nuclear export is stimulated by the overexpression of REF in an ICP27-dependent manner (4), and VP16, whose nuclear export in Xenopus oocytes is stimulated by the coinjection of recombinant ICP27 protein (24), were efficiently localized to the cytoplasm during infection with either an ICP27-competent or an ICP27-null virus. One possible explanation for the different results regarding the importance of ICP27 in the cytoplasmic localization of viral transcripts may be the use of different cells in the experiments. Nevertheless, our results demonstrate that during infection of Vero cells ICP27-independent nuclear export pathways do exist for viral transcripts, including late and intronless transcripts. One obvious candidate is the CRM1 pathway that mediates nuclear export of human immunodeficiency virus intronless transcripts (11). HSV replication is inhibited by leptomycin B, a pharmacological inhibitor of the CRM1 nuclear export pathway, and the cytoplasmic localization of several viral transcripts is reduced upon treatment of cells with this drug (33, 48). Although there is evidence against ICP27-mediated nuclear export functioning through the CRM1 pathway (4, 24), the evidence does not rule out the possibility that viral transcripts access a CRM1-nuclear export pathway independent of ICP27. Interestingly, ICP27 mutations can confer resistance to leptomycin B (33). This effect might conceivably be due to a gain of ICP27 function that can compensate for the loss of CRM1 function. Another possibility is that one of the newly identified TAP-binding proteins, 9G8 and SRp20, may be mediating the nuclear export of viral transcripts though the TAP nuclear export pathway (19).
Like many biological systems, it appears that there is redundancy built into the system of HSV mRNA nuclear export. The relative importance of the possible different pathways may be dependent on the cellular environment. The UL24 transcription unit has a ready-made set of transcripts with various combinations of promoters and 3′ ends, making it a particularly good system with which to study gene regulation during HSV infection. Thus, one can distinguish between promoter-dependent and -independent events. Similarly, the relative importance of 3′ UTRs can also be investigated in the context of similar promoter elements. This system will be particularly useful in the study of multifunctional regulatory factors such as ICP27. ICP27 clearly has, at a minimum, dual roles in viral gene expression including (i) the stimulation of transcription of at least some true late genes and (ii) increased cytoplasmic localization of some transcripts. This work raises the need to look at more early and late genes to better understand the gene-specific role of ICP27 in promoting transcription of viral genes and their cytoplasmic accumulation. In addition, the possible effect of cell type on the role of ICP27 during infection must be investigated.
Acknowledgments
We thank D. Wensel for the plasmid containing UL26 sequences. We thank W. C. Summers for the generous gift of TK antibody. We thank S. A. Rice for helpful discussions.
This work was supported by grant R01 AI26126 from the National Institutes of Health to D.M.C. and by grant RO1 AI20530 to D.M.K.
REFERENCES
- 1.Avitabile, E., P. L. Ward, C. Di Lazzaro, M. R. Torrisi, B. Roizman, and G. Campadelli-Fiume. 1994. The herpes simplex virus UL20 protein compensates for the differential disruption of exocytosis of virions and viral membrane glycoproteins associated with fragmentation of the Golgi apparatus. J. Virol. 68:7397-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass, B. L. 1997. RNA editing and hypermutation by adenosine deamination. Trends Biochem. Sci. 22:157-162. [DOI] [PubMed] [Google Scholar]
- 3.Bryant, H. E., S. E. Wadd, A. I. Lamond, S. J. Silverstein, and J. B. Clements. 2001. Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J. Virol. 75:4376-4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, I. H., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coen, D. M., H. E. Fleming, Jr., L. K. Leslie, and M. J. Retondo. 1985. Sensitivity of arabinosyladenine-resistant mutants of herpes simplex virus to other antiviral drugs and mapping of drug hypersensitivity mutations to the DNA polymerase locus. J. Virol. 53:477-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook, W. J., and D. M. Coen. 1996. Temporal regulation of herpes simplex virus type 1 UL24 mRNA expression via differential polyadenylation. Virology 218:204-213. [DOI] [PubMed] [Google Scholar]
- 7.Cook, W. J., K. K. Wobbe, J. Böni, and D. M. Coen. 1996. Regulation of neighboring gene expression by the herpes simplex virus type 1 thymidine kinase gene. Virology 218:193-203. [DOI] [PubMed] [Google Scholar]
- 8.Cullen, B. R. 2000. Nuclear RNA export pathways. Mol. Cell. Biol. 20:4181-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darnell, J. E., Jr. 1968. Ribonucleic acids from animal cells. Bacteriol. Rev. 32:262-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellison, K. S., S. A. Rice, R. Verity, and J. R. Smiley. 2000. Processing of alpha-globin and ICP0 mRNA in cells infected with herpes simplex virus type 1 ICP27 mutants. J. Virol. 74:7307-7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 12.Foster, T. P., and K. G. Kousoulas. 1999. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J. Virol. 73:8457-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldin, A. L., R. M. Sandri-Goldin, M. Levine, and J. C. Glorioso. 1981. Cloning of herpes simplex virus type 1 sequences representing the whole genome. J. Virol. 38:50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hann, L. E., W. J. Cook, S. L. Uprichard, D. M. Knipe, and D. M. Coen. 1998. The role of herpes simplex virus ICP27 in the regulation of UL24 gene expression by differential polyadenylation. J. Virol. 72:7709-7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardwicke, M. A., and R. M. Sandri-Goldin. 1994. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J. Virol. 68:4797-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardy, W. R., and R. M. Sandri-Goldin. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 68:7790-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holland, L. E., R. M. Sandri-Goldin, A. L. Goldin, J. C. Glorioso, and M. Levine. 1984. Transcriptional and genetic analyses of the herpes simplex virus type 1 genome: coordinates 0.29 to 0.45. J. Virol. 49:947-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong-Yan, Z., T. Murata, F. Goshima, H. Takakuwa, T. Koshizuka, Y. Yamauchi, and Y. Nishiyama. 2001. Identification and characterization of the UL24 gene product of herpes simplex virus type 2. Virus Genes 22:321-327. [DOI] [PubMed] [Google Scholar]
- 19.Huang, Y., R. Gattoni, J. Stevenin, and J. A. Steitz. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11:837-843. [DOI] [PubMed] [Google Scholar]
- 20.Hughes, R. G., Jr., and W. H. Munyon. 1975. Temperature-sensitive mutants of herpes simplex virus type 1 defective in lysis but not in transformation. J. Virol. 16:275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutchinson, L., and D. C. Johnson. 1995. Herpes simplex virus glycoprotein K promotes egress of virus particles. J. Virol. 69:5401-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobson, J. G., S. L. Martin, and D. M. Coen. 1989. A conserved open reading frame that overlaps the herpes simplex virus thymidine kinase gene is important for viral growth in cell culture. J. Virol. 63:1839-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jean, S., K. M. LeVan, B. Song, M. Levine, and D. M. Knipe. 2001. Herpes simplex virus 1 ICP27 is required for transcription of two viral late (gamma 2) genes in infected cells. Virology 283:273-284. [DOI] [PubMed] [Google Scholar]
- 24.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20:5769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar, M., and G. G. Carmichael. 1998. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol. Mol. Biol. Rev. 62:1415-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lei, E. P., H. Krebber, and P. A. Silver. 2001. Messenger RNAs are recruited for nuclear export during transcription. Genes Dev. 15:1771-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo, M. J., and R. Reed. 1999. Splicing is required for rapid and efficient mRNA export in metazoans. Proc. Natl. Acad. Sci. USA 96:14937-14942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malim, M. H., J. Hauber, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254-257. [DOI] [PubMed] [Google Scholar]
- 29.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 30.McCarthy, A. M., L. McMahan, and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J. Virol. 63:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGregor, F., A. Phelan, J. Dunlop, and J. B. Clements. 1996. Regulation of herpes simplex virus poly(A) site usage and the action of immediate-early protein IE63 in the early-late switch. J. Virol. 70:1931-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLauchlan, J., A. Phelan, C. Loney, R. M. Sandri-Goldin, and J. B. Clements. 1992. Herpes simplex virus IE63 acts at the posttranscriptional level to stimulate viral mRNA 3′ processing. J. Virol. 66:6939-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murata, T., F. Goshima, T. Koshizuka, H. Takakuwa, and Y. Nishiyama. 2001. A single amino acid substitution in the ICP27 protein of herpes simplex virus type 1 is responsible for its resistance to leptomycin B. J. Virol. 75:1039-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Panagiotidis, C. A., E. K. Lium, and S. J. Silverstein. 1997. Physical and functional interactions between herpes simplex virus immediate-early proteins ICP4 and ICP27. J. Virol. 71:1547-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pearson, A., and D. M. Coen. 2002. Identification, localization, and regulation of expression of the UL24 protein of herpes simplex virus type 1. J. Virol. 76:10821-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pellett, P. E., J. L. McKnight, F. J. Jenkins, and B. Roizman. 1985. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing alpha genes. Proc. Natl. Acad. Sci. USA 82:5870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perkins, K. D., J. Gregonis, S. Borge, and S. A. Rice. 2003. Transactivation of a viral target gene by herpes simplex virus ICP27 is posttranscriptional and does not require the endogenous promoter or polyadenylation site. J. Virol. 77:9872-9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phelan, A., and J. B. Clements. 1997. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J. Gen. Virol. 78:3327-3331. [DOI] [PubMed] [Google Scholar]
- 39.Rice, S. A., and D. M. Knipe. 1990. Genetic evidence for two distinct transactivation functions of the herpes simplex virus alpha protein ICP27. J. Virol. 64:1704-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rice, S. A., V. Lam, and D. M. Knipe. 1993. The acidic amino-terminal region of herpes simplex virus type 1 alpha protein ICP27 is required for an essential lytic function. J. Virol. 67:1778-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodrigues, J. P., M. Rode, D. Gatfield, B. J. Blencowe, M. Carmo-Fonseca, and E. Izaurralde. 2001. REF proteins mediate the export of spliced and unspliced mRNAs from the nucleus. Proc. Natl. Acad. Sci. USA 98:1030-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 43.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 44.Sanders, P. G., N. M. Wilkie, and A. J. Davison. 1982. Thymidine kinase deletion mutants of herpes simplex virus type 1. J. Gen. Virol. 63:277-295. [DOI] [PubMed] [Google Scholar]
- 45.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sekulovich, R. E., K. Leary, and R. M. Sandri-Goldin. 1988. The herpes simplex virus type 1 alpha protein ICP27 can act as a trans-repressor or a trans-activator in combination with ICP4 and ICP0. J. Virol. 62:4510-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soliman, T. M., R. M. Sandri-Goldin, and S. J. Silverstein. 1997. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 71:9188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soliman, T. M., and S. J. Silverstein. 2000. Herpesvirus mRNAs are sorted for export via Crm1-dependent and -independent pathways. J. Virol. 74:2814-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Su, L., and D. M. Knipe. 1989. Herpes simplex virus alpha protein ICP27 can inhibit or augment viral gene transactivation. Virology 170:496-504. [DOI] [PubMed] [Google Scholar]
- 50.Tognon, M., R. Guandalini, M. G. Romanelli, R. Manservigi, and B. Trevisani. 1991. Phenotypic and genotypic characterization of locus Syn 5 in herpes simplex virus 1. Virus Res. 18:135-150. [DOI] [PubMed] [Google Scholar]
- 51.Tran, R. K., P. T. Lieu, S. Aguilar, E. K. Wagner, and D. C. Bloom. 2002. Altering the expression kinetics of VP5 results in altered virulence and pathogenesis of herpes simplex virus type 1 in mice. J. Virol. 76:2199-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Uprichard, S. L., and D. M. Knipe. 1996. Herpes simplex ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J. Virol. 70:1969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ward, P. L., G. Campadelli-Fiume, E. Avitabile, and B. Roizman. 1994. Localization and putative function of the UL20 membrane protein in cells infected with herpes simplex virus 1. J. Virol. 68:7406-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 55.Zhou, C., and D. M. Knipe. 2002. Association of herpes simplex virus type 1 ICP8 and ICP27 proteins with cellular RNA polymerase II holoenzyme. J. Virol. 76:5893-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]