Abstract
Severe acute respiratory syndrome (SARS) is a deadly form of pneumonia caused by a novel coronavirus, a viral family responsible for mild respiratory tract infections in a wide variety of animals including humans, pigs, cows, mice, cats, and birds. Analyses to date have been unable to identify the precise origin of the SARS coronavirus. We used Bayesian, neighbor-joining, and split decomposition phylogenetic techniques on the SARS virus replicase, surface spike, matrix, and nucleocapsid proteins to reveal the evolutionary origin of this recently emerging infectious agent. The analyses support a mammalian-like origin for the replicase protein, an avian-like origin for the matrix and nucleocapsid proteins, and a mammalian-avian mosaic origin for the host-determining spike protein. A bootscan recombination analysis of the spike gene revealed high nucleotide identity between the SARS virus and a feline infectious peritonitis virus throughout the gene, except for a 200- base-pair region of high identity to an avian sequence. These data support the phylogenetic analyses and suggest a possible past recombination event between mammalian-like and avian-like parent viruses. This event occurred near a region that has been implicated to be the human receptor binding site and may have been directly responsible for the switch of host of the SARS coronavirus from animals to humans.
Severe acute respiratory syndrome (SARS) is a potentially fatal atypical pneumonia that arose in Guangdong Province of the People's Republic of China in November 2002. Within 6 months SARS spread to over 30 countries and killed over 700 people (27, 37). This outbreak has had a profound impact on public health and economies worldwide and reinforced the danger of emerging infectious diseases in densely populated societies.
SARS is believed to be a viral zoonotic disease (9). Viral zoonoses are responsible for most of the emerging infectious diseases (8, 21) and often originate due to genetic exchange between viruses with different host specificities. Shifts in the host range of the influenza virus have been shown to be due to the genetic exchange of the host-determining neuraminidase and hemagglutinin genes (17, 36). One such genetic exchange event is believed to have been responsible for the 1918 Spanish influenza pandemic which killed over 20 million people. There is also some evidence that such genetic reassortment may have occurred in the virus that causes SARS.
SARS is caused by a novel coronavirus (6, 19, 24, 40). Coronaviruses are enveloped, positive-strand RNA viruses that cause localized but mild respiratory tract infections in a wide variety of animals including pigs, cows, mice, horses, and birds (35). In humans, coronaviruses normally cause symptoms associated with the common cold (12). The SARS coronavirus is similar in composition and genetic organization to previously described coronaviruses. Its genome includes genes encoding a replicative polyprotein (PP1ab), surface spike glycoprotein (S), envelope, matrix (M), and nucleocapsid (N) proteins (5, 22). The SARS coronavirus also encodes an additional nine predicted open reading frames whose precise function is unknown (22). The S protein is of particular importance as it is a key virulence and host range determinant (3, 4, 14, 25, 26, 31, 34). The region between amino acids (aa) 417 and 547 of the human coronavirus 229E S protein has been shown to be the binding region for the human aminopeptidase N receptor (3, 38). Targeted recombination between feline and mouse S proteins has been shown to enable feline-specific coronaviruses to infect mice (10). Comparative analyses of the SARS coronavirus genomes revealed two amino acid substitutions within the S protein, signaling possible positive selection due to immunological pressures (29).
The origin of the SARS coronavirus has been the subject of intense speculation. Enserink (7) recently reported that the SARS virus could be isolated from masked palm civets available in the food markets of southern China. The civet family (Viverridae) is in the suborder Fissipedia and is a close relative of the cat family (Felidae) (32). Despite this hopeful lead, phylogenetic analyses have been unable to definitively identify the evolutionary origin of this virus. Previous studies have reported that the SARS coronavirus is phylogenetically distinct and not a composite of other coronaviruses (13, 22, 28). Rota et al. (28) were unable to identify any partitions in the pattern of polymorphism along the SARS genome consistent with recombination (13). There are also claims that the SARS virus is not a host range mutant of any previously described coronaviruses due to its low sequence identity to known coronaviruses (13, 29).
To further investigate the origins of the SARS coronavirus, we performed a detailed phylogenetic analysis of the PP1ab, S, M, and N proteins with Bayesian, neighbor-joining, and split decomposition methods. Additionally, we examined the gene that encodes the host-determining S protein for evidence of past recombination events that may have resulted in a new ability to infect human hosts. Our results indicate that the SARS virus is in fact a mosaic of mammalian and avian-like viruses and that recombination between the parent viruses may have occurred in the host-determining S gene.
MATERIALS AND METHODS
Sequence retrieval.
Homologous sequences for the PP1ab, S, M, and N proteins of the SARS coronavirus strain Urbani (AY278741) were acquired by BLASTP against the GenBank nr database. Redundant sequences or sequences sharing ≥95% protein similarity (as determined by a pairwise BLAST search) were discarded. An attempt was made to include sequences from all three taxonomically described groups of coronaviruses, and whenever possible, the same representative taxa (accessions) were used for all gene genealogies. Noncoronavirus sequences that were similar to the SARS virus were identified for the SARS PP1ab polyprotein by performing BLASTP with the following Entrez limit: “polyprotein OR PP1ab OR replicase NOT coronavirus.” This search resulted in the identification of two domains of the SARS PP1ab polyprotein (aa 4751 to 5024 and aa 5276 to 5851) that show significant similarity (98.6 bits; E = 6e − 18; 37% similarity) to the gill-associated okavirus from the Roniviridae, a sister family to the Coronaviridae. Imposing a similar Entrez limit on BLASTP and BLASTN searches of the S, M, and N proteins did not yield any significant hits to viruses in other families.
Phylogenetic analyses.
Amino acid alignments used in the phylogenetic analyses were made with ClustalX version 1.83 (33) with default parameters. An exception to this protocol was the M protein alignment, where a multiple alignment gap opening penalty of 5 was used. Alignments were imported into Genedoc version 2.5 (http://www.psc.edu/biomed/genedoc) where gaps and ambiguously aligned regions were deleted. The two domains of the gill-associated okavirus, totaling 880 aa, were concatenated and edited in a similar manner. The multiple sequence alignments and phylogenetic trees are available at http://www.botany.utoronto.ca/ResearchLabs/guttmanLab.
Phylogenetic analyses were made on 840, 162, and 345 aa of the S, M, and N proteins, respectively. The rooted PP1ab phylogeny was based on an 883-aa region to include the gill-associated okavirus, whereas the unrooted PP1ab phylogeny was generated using a 322-aa region for which there was a feline coronavirus sequence available. Phylogenetic trees were constructed only with the amino acid data due to the extremely high level of nucleotide diversity.
Neighbor-joining trees were made with PHYLIP version 3.6a (J. Felsenstein, Phylogeny inference package, Department of Genetics, University of Washington, Seattle) using SEQBOOT, PROTDIST, NEIGHBOR, and CONSENSE programs. Protein distance calculations were based on the Jones-Taylor-Thornton protein weight matrix with 1,000 bootstrap (BS) replicates. All other variables were set as default. Bayesian trees were constructed with MRBAYES version 3.04b (15) using Jones-Taylor-Thornton protein weight matrix with 500,000 generations and a burn-in of 100. All other values were set as default. Consensus trees were generated by majority rule and viewed with TREEVIEW version 1.6.6 (23). The topologies of the Bayesian and neighbor-joining trees for each protein were identical; therefore, only the Bayesian trees are shown.
Split decomposition analyses were performed with SplitsTrees version 3.2 (16) using Hamming correction and are presented with equal edge length for clarity. Recombination effectively causes lineages to coalesce forward in time, resulting in trees that have reticulations or a network structure rather than the simple branching structure seen with most phylogenies. Split decomposition does not force evolutionary relationships to be strictly bifurcating or multibranching but permits network relationships. A split decomposition graph will look less tree-like and more net-like as the influence of recombination becomes more important in the history of a set of taxa.
Bootscanning.
A nucleotide alignment of the gene encoding the S protein was generated with ClustalX version 1.83 with a pairwise and multiple sequence gap opening penalty of 10 and a gap extension penalty of 2. The alignment was edited to remove all gaps and regions that could not be aligned. One representative sequence was used from both the avian infectious bronchitis coronavirus group and the group 1 coronaviruses (feline). Two representatives were chosen from the group 2 coronaviruses, representing the two major divisions in that group (murine and human). Bootscanning (30) was performed by using Simplot version 3.2b (20) with the F84 model (window size, 200 bp; step, 20 bp) and with the SARS S nucleotide sequence as the query. Bootscanning uses a sliding window approach to detect recombination. At each window position a phylogenetic analysis is performed and the data are bootstrapped. The stronger the BS support for the clade that includes the query sequence is, the higher the bootscan value will be. Recombination may change the branching order of phylogenies, thereby potentially moving the query sequence to another clade. Significant changes in phylogenetic relationships from window to window will result in changes in bootscan values and are indicative of probable recombination events.
Bootscanning was performed on the nucleotide sequences since there is no application available to perform this analysis on amino acid sequences. Given the extensive nucleotide diversity of the sample, we could perform a bootscan analysis only on a relatively small region of the SARS genome and had to restrict our analysis to a much smaller group of taxa.
Relative rate test.
Tajima's relative rate test was performed with the MEGA2 (18) program on the amino acid alignments. Homogeneity in the rate of evolution among lineages supports the use of midpoint rooting for the phylogenetic trees.
RESULTS AND DISCUSSION
We examined the phylogenetic relationships of the SARS coronavirus and related viruses by using Bayesian, neighbor-joining, and split decomposition phylogenetic methods. Separate phylogenies were constructed from each protein sequence. With the exception of the PP1ab replicase polyprotein (discussed below), all trees are presented as unrooted dendrograms since there was no objective means to root the separate trees.
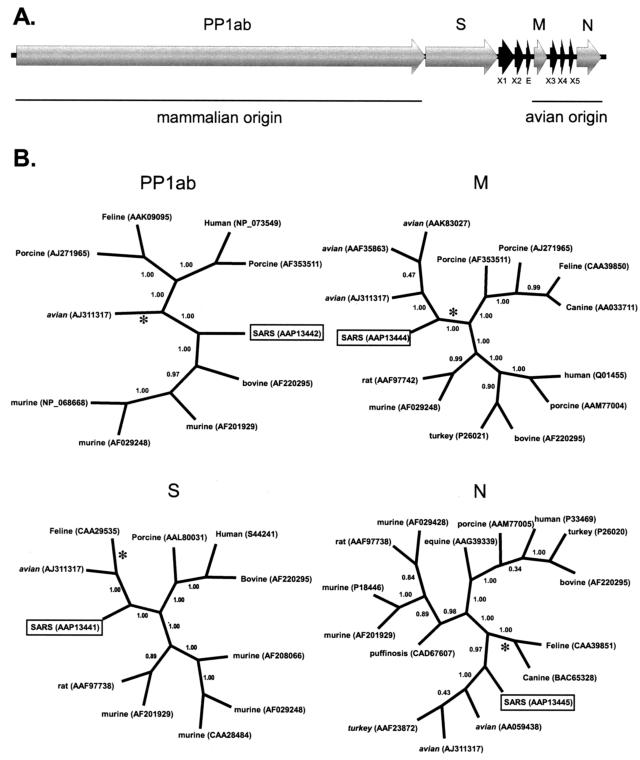
The M and N genes, which encode two of the primary structural proteins of the SARS virus, are on the 3′ end of the genome. Phylogenetic analyses show that both of these proteins diverge from the lineage leading to the avian infectious bronchitis coronaviruses. Support for an avian-like viral ancestor for these proteins rests upon the branch separating SARS and avian viruses from all of the other coronaviruses (Fig. 1B). This branch has extremely strong support, with Bayesian posterior probability (PP) and neighbor-joining BS values for the avian-SARS M protein clade at 100 and 89%, respectively, and support for the avian-SARS N protein clade at 100 and 97%, respectively. Bayesian PP values are generally considered significant when they are greater than 95% (15), while BS scores are considered significant at 70% (11). No noncoronavirus sequences with any similarity to either the M or N proteins were found in GenBank; therefore, there is no objective way to root these trees. Nevertheless, the most exclusive monophyletic group (with respect to accepted coronavirus taxonomy) that includes the two SARS proteins also includes the avian viruses and excludes any mammalian (groups 1 and 2) coronaviruses. These relationships are also supported by the midpoint rooting of the two trees. The M protein tree midpoint roots on the branch linking SARS to the group 1 (porcine, feline, and canine) and group 2 (bovine and murine) coronaviruses. The N protein tree midpoint roots on the branch leading to the group 1 coronaviruses. The appropriateness of midpoint rooting is supported by a Tajima's relative rate test, which finds no evolutionary rate heterogeneity among the major coronavirus groups (data not shown). Trees presented by Marra et al. (22) also support the grouping of the SARS virus M and N proteins with the avian clade. These results are also consistent with the characteristic avian-like elements of the SARS genome, including a stem-loop II-like motif at the 3′ end that is found only in avian coronaviruses (22). Marra et al. (22) suggest that this motif has been acquired through horizontal transfer, a theory supported by our study.
FIG. 1.
(A) Organization of the SARS coronavirus genome. (B) Bayesian-inferred unrooted phylogenies of the SARS proteins, generated with MRBAYES version 3.04b using Jones-Taylor-Thornton protein weight matrix. Tree sampling was performed for 500,000 generations, and a majority-rule consensus tree was built with a burn-in of 100. The PP1ab phylogeny was based on a conserved 322-aa region, corresponding to aa 3230 to 3552 of the polyprotein. The S, M, and N genealogies are based on edited, whole-protein alignments of 840, 162, and 345 aa, respectively. Branch confidence values are given as PPs. Congruent trees were obtained by neighbor joining, with critical nodes being supported by bootstrap values that are >89%. Asterisks denote the location of midpoint rooting. Group 1 coronaviruses have initial capital letters. Group 2 coronaviruses are in lowercase letters. Group 3 coronaviruses are in italics.
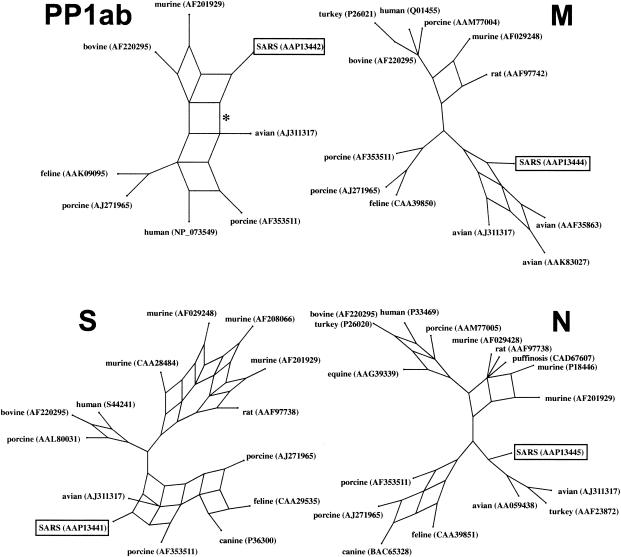
Split decomposition analyses of the SARS M and N proteins also support descent from an avian-like coronavirus ancestor (Fig. 2). The N protein split decomposition graph largely mirrors the Bayesian and neighbor-joining trees. The SARS N protein diverges from the lineage leading to the avian infectious bronchitis coronaviruses. Since there is no reticulation in this part of the graph, there is little evidence of recombination between these sequences. BS support for the SARS-avian clade is marginal at 71.2%. The M protein split decomposition graph is more informative. SARS again forms a significant clade with the avian coronaviruses (BS, 74.1%), but unlike the N protein, the M protein shows reticulations in the SARS-avian clade that are clear evidence for recombination between these viruses.
FIG. 2.
Split decomposition graph of the SARS coronavirus genome. The PP1ab phylogeny was based on a conserved 322-aa region, corresponding to aa 3230 to 3552 of the polyprotein. The S, M, and N genealogies are based on edited, whole-protein alignments of 840, 162, and 345 aa, respectively. The asterisk on the PP1ab protein denotes the branch that is lost when the whole protein is analyzed.
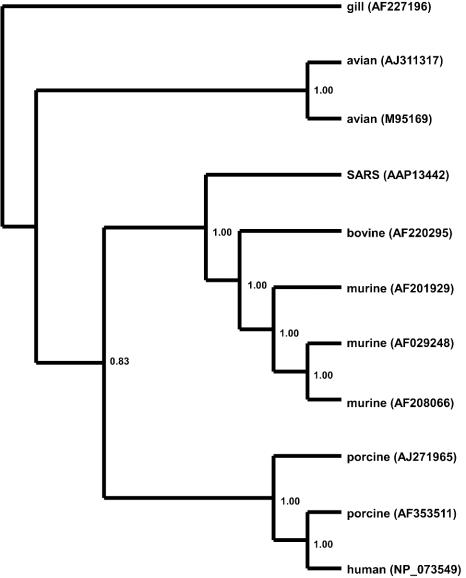
The phylogeny of the PP1ab polyprotein is dramatically different from that of the M and N proteins. The tree shown in Fig. 1B is from a 322-aa region of the PP1ab protein, corresponding to aa 3230 to 3552, for which a feline coronavirus sequence was available. The branching structure of the tree generated from this subsection of the PP1ab protein is identical to the structure of the tree generated from the entire 7,073-aa PP1ab protein. Unlike the previous two proteins, the SARS PP1ab protein does not clearly fall into any clade. SARS diverges near the center of the unrooted dendrogram shown in Fig. 1B. Its position makes phylogenetic clustering problematic, since it could cluster significantly (BS, 100%; PP, 100%) with either the group 2 (murine and bovine) coronaviruses or, just as significantly, with the group 1 (feline, porcine, and human) and avian coronaviruses, depending on how the tree is rooted. To clarify this issue, we searched GenBank for more distantly related sequences that could be used to root this tree. By restricting the BLASTP similarity search to noncoronavirus sequences, we were able to identify a gill-associated okavirus with significant similarity (98.6 bits; E = 6e − 18; 37% similarity) to the SARS PP1ab protein. There were two conserved domains of the okavirus, totaling 880 aa, which showed similarity to the SARS PP1ab protein. These regions were concatenated, edited, and used to root the PP1ab polyprotein phylogeny. Unfortunately, this region does not overlap with any region for which the feline infectious peritonitis virus showed similarity to the PP1ab sequence data; therefore, the feline coronavirus sequence was excluded from the rooted phylogeny. The rooted PP1ab phylogeny (Fig. 3) shows strong support for monophyly of the SARS virus with the group 2 murine-bovine coronaviruses (BS, 83%; PP, 100%), indicating that the SARS PP1ab protein originated from a mammalian-like ancestral virus. This topology is also supported by midpoint rooting (Fig. 1B, asterisk). A phylogenetic analysis conducted by Peiris et al. (24) on approximately 215 aa of the PP1ab polyprotein of the SARS Hong Kong 03/2003 isolate also supports the monophyly of SARS with the murine-bovine group. This conclusion is consistent with the view of Anand et al. (2), who consider the SARS virus a variant of the murine-bovine coronaviruses even though the SARS coronavirus lacks the hemagglutinin esterase gene that characterizes this group (22). In addition, the core transcription-regulating sequence of SARS, which serves to regulate the discontinuous transcription of subgenomic RNAs (39), has been suggested to be more like those of the murine-bovine coronavirus group (22).
FIG. 3.
Bayesian-inferred rooted phylogeny of the PP1ab polyprotein. Two conserved regions of the polyprotein, homologous to sequences found in the gill-associated okavirus, were concatenated and edited to eliminate gaps and ambiguously aligned regions. A phylogeny was generated on the resulting 880-aa alignment as described in the legend to Fig. 1. The tree was rooted on the gill-associated okavirus, a member of the Roniviridae, which is a sister family to the Coronaviridae. Branch confidence values are given as PPs.
The split decomposition analysis of the SARS PP1ab replicase polyprotein also supports a mammalian origin for this protein. The split decomposition graph shown in Fig. 2 was derived from the same region of SARS-feline similarity shown in Fig. 1B. This graph is similar to the Bayesian and neighbor-joining trees in showing that SARS is positioned between the avian infectious bronchitis coronaviruses and the group 2 (murine and bovine) coronaviruses. When the entire 7,073-aa protein is analyzed, the branch connecting the SARS and avian viruses (Fig. 2, asterisk) is lost, and the group 2-SARS clade is supported by a BS score of 100%.
The genes encoding the mammalian-like PP1ab protein and avian-like M and N proteins are separated by the gene encoding the host-determining S protein. A phylogenetic analysis of the S protein places the SARS coronavirus within a clade that includes homologous proteins from the avian infectious bronchitis virus and the group 1 coronaviruses, which includes the feline infectious peritonitis virus (Fig. 1B). Support for this grouping is highly significant (BS, 100%; PP, 100%). A relative rate test again showed no rate heterogeneity between major coronavirus groups, supporting the use of midpoint rooting. The midpoint root of this tree is on the branch leading to the group 1 coronaviruses.
The split decomposition analysis of the S protein again strongly supports the Bayesian and neighbor-joining analyses (Fig. 2). SARS forms a highly significant clade (BS, 100%) with the avian infectious bronchitis coronaviruses and the group 1 (feline, canine, and porcine) coronaviruses. The highly reticulate nature of this clade supports the occurrence of extensive recombination in the evolutionary history of these samples. These findings are consistent with the view that the SARS S protein is a mosaic of coronaviruses from two distinct groups—the avian and group 1 coronaviruses.
The phylogenetic relationships defined above are not simply dependent on the region or domain chosen for phylogenetic analysis. Phylogenetic analyses on whole-protein alignments, both edited and unedited, yield similar tree topologies for all proteins. Focusing on smaller, more conserved regions that have adequate phylogenetic signals permitted these phylogenies to be supported with greater confidence.
As there is clearly phylogenetic incongruence across the SARS genome, we sought to identify the precise point of recombination between the mammalian-like and avian-like parent viruses that gave rise to the present SARS virus. Previous studies that used a bootscanning method have found no evidence of recombination within the SARS genome (28). This lack of evidence is surprising since frequent recombination and mosaic evolution are more the rule than the exception in RNA viral evolution (for a review, see reference 1). Given that the two phylogenetically distinct partitions meet at the SARS S protein and that this protein is a critical host determinant, we focused on the S gene as a candidate locus for recombination. We performed a bootscan analysis on a gapless nucleotide alignment of the S proteins from four taxa with the SARS S protein as the query.
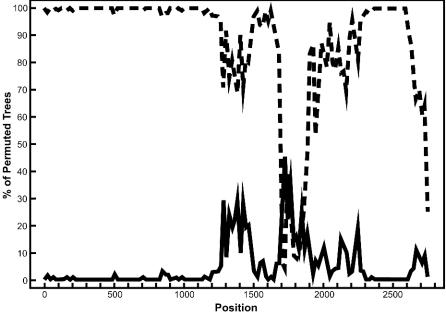
Bootscanning revealed one prominent region of the S gene where recombination was likely to have taken place. High nucleotide identity between the SARS and feline infectious peritonitis virus S genes is evident in the first 1,800 nucleotides of the alignment (Fig. 4). This finding is particularly intriguing given recent reports that the SARS virus has been isolated from masked palm civets (7), which are phylogenetically and immunologically closely related to cats (32). The sequence identity between the SARS and feline coronaviruses is significantly reduced between positions 1800 and 2000, where the sequence clearly becomes more similar to the avian coronavirus sequence. This region roughly corresponds to the region between nucleotides 2472 and 2694 of the SARS S protein but does not appear to be within the domain previously shown to bind the human aminopeptidase N receptor (3, 38). The region corresponding to the binding domain of the S protein lies between nucleotides 1253 and 1641 of the S gene. However, as this binding region is hypervariable among all coronaviruses (data not shown), large portions of it were eliminated from the alignment when gap-containing columns were removed. Despite this, it has been noted that different regions of the S protein may serve as a binding region for different coronaviruses (22). It is therefore plausible that a recombination event in the middle of this gene may have resulted in a new viral variant with a modified host range. It should be noted that other taxa were included in the bootscan analysis but had percentages of permuted trees that were less than 10%, suggesting low similarity to the SARS coronavirus S gene. As a result, these taxa did not register on the bootscan output. Alternative alignments of the S gene analyzed by bootscanning yielded similar results, with the SARS S gene being a composite of predominantly avian and feline coronavirus sequences (data not shown). These bootscan results support our phylogenetic analysis of the S protein that places SARS in a monophyletic group with the avian and feline sequences.
FIG. 4.
Bootscan analysis of the S gene to detect recombination. Bootscanning was conducted with Simplot on a gapless nucleotide alignment, generated with ClustalX, with the SARS S gene as the query sequence. The dashed line denotes the feline infectious peritonitis virus (X06170), while the solid line denotes the avian infectious bronchitis virus (M95169). The greater the percentage of permuted trees, the greater the sequence identity to the SARS S sequence. Other taxa that were included in the alignment but had less than 10% permuted trees were murine hepatitis virus (AF208066) and human coronavirus OC43 (L14643).
Conclusion.
Despite the fact that the SARS virus does cluster with existing coronavirus groups, its genome sequence is substantially divergent, and a new taxonomic group has already been proposed (22, 24, 28). Irrespective of its taxonomic grouping, it is clear that the SARS coronavirus is a mosaic, with at least two distinct evolutionary histories. Our analyses indicate that the SARS coronavirus is mammalian-like through the replicase protein, and avian-like through the M and N proteins and that there is a mammalian-avian mosaic in the S protein. The gene encoding the S protein lies between the replicase gene and the M and N genes. We propose that a recombination event likely occurred within the S gene, as demonstrated by our bootscan analysis. Since the S protein is responsible for host specificity, this event may have been the critical step in the switch to a human host and the subsequent emergence of this new pathogen. The testing of this hypothesis will require extensive sampling of coronaviruses from a broad range of hosts. Identifying the source of the SARS coronavirus will not only lead to better public health practices but will also contribute to our understanding of how these important pathogens evolve.
Acknowledgments
We thank Jean-Marc Moncalvo, Pauline Wang, and Sara Sarkar for their valuable critique, suggestions, and comments.
D.S.G. is supported by grants from the Natural Sciences and Engineering Research Council of Canada and the Canadian Foundation for Innovation.
REFERENCES
- 1.Alejska, M., A. Kurzynska-Kokorniak, M. Broda, R. Kierzek, and M. Figlerowicz. 2001. How RNA viruses exchange their genetic material. Acta Biochim. Pol. 48:391-407. [PubMed] [Google Scholar]
- 2.Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. [DOI] [PubMed] [Google Scholar]
- 3.Bonavia, A., B. D. Zelus, D. E. Wentworth, P. J. Talbot, and K. V. Holmes. 2003. Identification of a receptor-binding domain of the spike glycoprotein of human coronavirus HCoV-229E. J. Virol. 77:2530-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang, S. H., J. L. Bae, T. J. Kang, J. Kim, G. H. Chung, C. W. Lim, H. Laude, M. S. Yang, and Y. S. Jang. 2002. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cell 14:295-299. [PubMed] [Google Scholar]
- 5.de Haan, C. A. M., H. Volders, C. A. Koetzner, P. S. Masters, and P. J. M. Rottier. 2002. Coronaviruses maintain viability despite dramatic rearrangements of the strictly conserved genome organization. J. Virol. 76:12491-12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. M. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 7.Enserink, M. 2003. Clues to the animal origins of SARS. Science 300:1351. [DOI] [PubMed] [Google Scholar]
- 8.Feldmann, H., M. Czub, S. Jones, D. Dick, M. Garbutt, A. Grolla, and H. Artsob. 2002. Emerging and re-emerging infectious diseases. Med. Microbiol. Immunol. 191:63-74. [DOI] [PubMed] [Google Scholar]
- 9.Guan, Y., B. J. Zheng, Y. Q. He, X. L. Liu, Z. X. Zhuang, C. L. Cheung, S. W. Luo, P. H. Li, L. J. Zhang, Y. J. Guan, K. M. Butt, K. L. Wong, K. W. Chan, W. Lim, K. F. Shortridge, K. Y. Yuen, J. S. M. Peiris, and L. L. M. Poon. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302:276-278. (First published 4 September 2003; 10.1126/science.1087139.) [DOI] [PubMed] [Google Scholar]
- 10.Haijema, B. J., H. Volders, and P. J. M. Rottier. 2003. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 77:4528-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hillis, D. M., and J. J. Bull. 1993. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42:182-192. [Google Scholar]
- 12.Holmes, K. V. 1999. Coronaviridae, 2nd ed. Academic Press, New York, N.Y.
- 13.Holmes, K. V. 2003. SARS-associated coronavirus. N. Engl. J. Med. 348:1948-1951. [DOI] [PubMed] [Google Scholar]
- 14.Holmes, K. V., B. D. Zelus, J. H. Schickli, and S. R. Weiss. 2001. Receptor specificity and receptor-induced conformational changes in mouse hepatitis virus spike glycoprotein, p. 173-181. In E. Lavi, S. R. Weiss, and S. T. Hingley (ed.), Nidoviruses (coronaviruses and arteriviruses). Advances in experimental medicine and biology, vol. 494. Kluwer Academic Publishers, Hingham, Mass. [DOI] [PubMed]
- 15.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 16.Huson, D. H. 1998. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14:68-73. [DOI] [PubMed] [Google Scholar]
- 17.Kawaoka, Y., S. Krauss, and R. G. Webster. 1989. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 63:4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence, D. 2003. Coronavirus confirmed as cause of SARS. Lancet 361:1712.12767746 [Google Scholar]
- 20.Lole, K. S., R. C. Bollinger, R. S. Paranjape, D. Gadkari, S. S. Kulkarni, N. G. Novak, R. Ingersoll, H. W. Sheppard, and S. C. Ray. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J. Virol. 73:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ludwig, B., F. B. Kraus, R. Allwinn, H. W. Doerr, and W. Preiser. 2003. Viral zoonoses—a threat under control? Intervirology 46:71-78. [DOI] [PubMed] [Google Scholar]
- 22.Marra, M. A., S. J. M. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. N. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1414. (First published 1 May 2003; 10.1126/science.1085953.)12730501 [Google Scholar]
- 23.Page, R. D. M. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 24.Peiris, J. S. M., S. T. Lai, L. L. M. Poon, Y. Guan, L. Y. C. Yam, W. Lim, J. Nicholls, W. K. S. Yee, W. W. Yan, M. T. Cheung, V. C. C. Cheng, K. H. Chan, D. N. C. Tsang, R. W. H. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phillips, J. J., M. M. Chua, G. F. Rall, and S. R. Weiss. 2002. Murine coronavirus spike glycoprotein mediates degree of viral spread, inflammation, and virus-induced immunopathology in the central nervous system. Virology 301:109-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popova, R., and X. M. Zhang. 2002. The spike but not the hemagglutinin/esterase protein of bovine coronavirus is necessary and sufficient for viral infection. Virology 294:222-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riley, S., C. Fraser, C. A. Donnelly, A. C. Ghani, L. J. Abu-Raddad, A. J. Hedley, G. M. Leung, L.-M. Ho, T.-H. Lam, T. Q. Thach, P. Chau, K.-P. Chan, S.-V. Lo, P.-Y. Leung, T. Tsang, W. Ho, K.-H. Lee, E. M. C. Lau, N. M. Ferguson, and R. M. Anderson. 2003. Transmission dynamics of the etiological agent of SARS in Hong Kong: impact of public health interventions. Science 300:1961-1966. [DOI] [PubMed] [Google Scholar]
- 28.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M.-H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. T. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rassmussen, R. Fouchier, S. Gunther, A. D. M. E. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 29.Ruan, Y. J., C. L. Wei, L. A. Ee, V. B. Vega, H. Thoreau, S. T. S. Yun, J. M. Chia, P. Ng, K. P. Chiu, L. Lim, Z. Tao, C. K. Peng, L. O. L. Ean, N. M. Lee, L. Y. Sin, L. F. P. Ng, R. E. Chee, L. W. Stanton, P. M. Long, and E. T. Liu. 2003. Comparative full-length genome sequence analysis of 14 SARS coronavirus isolates and common mutations associated with putative origins of infection. Lancet 361:1779-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salminen, M. O., J. K. Carr, D. S. Burke, and F. E. McCutchan. 1995. Identification of breakpoints in intergenotypic recombinants of HIV type-1 by bootscanning. AIDS Res. Hum. Retrovir. 11:1423-1425. [DOI] [PubMed] [Google Scholar]
- 31.Sanchez, C. M., A. Izeta, J. M. Sanchez-Morgado, S. Alonso, I. Sola, M. Balasch, J. Plana-Duran, and L. Enjuanes. 1999. Targeted recombination demonstrates that the spike gene of transmissible gastroenteritis coronavirus is a determinant of its enteric tropism and virulence. J. Virol. 73:7607-7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schreiber, A., K. Eulenberger, and K. Bauer. 1998. Immunogenetic evidence for the phylogenetic sister group relationship of dogs and bears (Mammalia, Carnivora: Canidae and Ursidae): a comparative determinant analysis of carnivoran albumin, C3 complement and immunoglobulin micro-chain. Exp. Clin. Immunogenet. 15:154-170. [DOI] [PubMed] [Google Scholar]
- 33.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai, J. C., B. D. Zelus, K. V. Holmes, and S. R. Weiss. 2003. The N-terminal domain of the murine coronavirus spike glycoprotein determines the CEACAM1 receptor specificity of the virus strain. J. Virol. 77:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner, E. K., and M. Hewlett. 1999. Basic virology. Blackwell Science, Malden, Mass.
- 36.Webster, R. G. 2002. The importance of animal influenza for human disease. Vaccine 20:S16-S20. [DOI] [PubMed] [Google Scholar]
- 37.World Health Organization. 2003. SARS. Wkly. Epidemiol. Rec. 78:197-200.12854302 [Google Scholar]
- 38.Yeager, C. L., R. A. Ashmun, R. K. Williams, C. B. Cardellichio, L. H. Shapiro, A. T. Look, and K. V. Holmes. 1992. Human aminopeptidase-N is a receptor for human coronavirus-229e. Nature 357:420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, X. M., C. L. Liao, and M. M. C. Lai. 1994. Coronavirus leader RNA regulates and initiates subgenomic mRNA transcription both in trans and in cis. J. Virol. 68:4738-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zurer, P. 2003. SARS cause found: experiments in monkeys confirm coronavirus is behind deadly disease. Chem. Eng. News 81:9. [Google Scholar]