Abstract
Epstein-Barr virus (EBV) nuclear antigen 3C (EBNA3C) is critical for EBV immortalization of infected B lymphocytes and can coactivate the EBV LMP1 promoter with EBNA2. EBNA3C amino acids 365 to 545 are necessary and sufficient for coactivation and are required for SUMO-1 and SUMO-3 interaction. We found that EBNA3C but not EBNA3CΔ343-545 colocalized with SUMO-1 in nuclear bodies and was modified by SUMO-2, SUMO-3, and SUMO-1. EBNA3C amino acids 545 to 628 and amino acids 30 to 365 were also required for EBNA3C sumolation and nuclear body localization but were dispensable for coactivation, indicating that EBNA3C sumolation is not required for coactivation. Furthermore, EBNA3C amino acids 476 to 992 potently coactivated with EBNA2 but EBNA3C amino acids 516 to 922 lacked activity, indicating that amino acids 476 to 515 are critical for coactivation. EBNA3C amino acids 476 to 515 include DDDVIEV507-513, which are similar to SUMO-1 EEDVIEV84-90. EBNA3C m1 and m2 point mutations, DDD507-509 mutated to AAA and DVIEVID509-513 mutated to AVIAVIA, respectively, diminished SUMO-1 and SUMO-3 interaction in directed yeast two-hybrid and glutathione S-transferase pulldown assays. Furthermore, EBNA3C m1 and m2 did not coactivate the LMP1 promoter with EBNA2. Overexpression of wild-type SUMO-1, SUMO-3, and the SUMO-conjugating enzyme UBC9 coactivated the LMP1 promoter with EBNA2. Since EBNA2 activation is dependent on p300/CBP, the possible effect of EBNA3C on p300-mediated transcription was assayed. EBNA3C potentiated transcription of p300 fused to a heterologous DNA binding domain, whereas EBNA3C m1 and m2 did not. All of these data are consistent with a model in which EBNA3C upregulates EBNA2-mediated gene activation by binding to a sumolated repressor and inhibiting repressive effects on p300/CBP and other transcription factor(s) at EBNA2-regulated promoters.
On infection of human B lymphocytes, Epstein-Barr virus (EBV) expresses six nuclear (EBNAs) and two integral membrane (LMPs) proteins, resulting in lymphoproliferation (for review, see reference 23). EBNA2 upregulates transcription of cell and viral promoters through association with a cellular DNA-specific transcription factor, RBP-Jκ (CBF-1) (15, 23). EBNA2's effects are upregulated by EBNALP (17, 23). EBNA3A, EBNA3B, and EBNA3C are encoded by three related genes that are in tandem in the EBV genome; their transcription is regulated by EBNA2 and EBNALP. Like EBNA2, EBNA3A, EBNA3B, and EBNA3C associate at a high level with RBP-Jκ, limiting transcription from some promoters, such as the EBNA Cp promoter, and coactivating transcription of other promoters, such as the LMP1 promoter (24, 26, 36-38, 52, 57-59).
EBNA3C is a potent coactivator of the LMP1 promoter with EBNA2 in transient assays and can be critically important in maintaining LMP1 levels, as is evident from the low LMP1 levels in growth-arrested Raji cells (1-3, 24, 59). In Raji cells, a Burkitt's lymphoma cell line infected with an EBV episome lacking the EBNA3C open reading frame, expression of EBNA3C restores LMP1 levels. EBNA3C domains can also affect transcription alone and when fused to the GAL4 DNA binding domain (5, 24, 26, 35, 58, 59). EBNA3C amino acids 365 to 545 are necessary and sufficient for coactivation of the LMP1 promoter with EBNA2 (24), whereas EBNA3C amino acids 280 to 525 repress promoter activity when fused to the GAL4 DNA binding domain (5).
The common roles of EBNA3A, EBNA3B, and EBNA3C in transcriptional regulation do not explain the need for three distinct genes and the unique requirement for EBNA3C and EBNA3A for lymphoblastoid cell (LCL) outgrowth (50, 51). Some clues to EBNA3C function have come from the identification of interacting cell proteins implicated in transcriptional regulation and cell survival. These include DP103 (gemin3), the metastatic tumor suppressor Nm-23H1, CtBP, Rint-1, and prothymosin α (7, 16, 24, 42, 43, 51). Other clues come from differential effects of EBNA3C versus EBNA3A and EBNA3B in EBNA2 coactivation (24).
SUMO-1 and SUMO-3 interact with EBNA3C in yeast two-hybrid screens. EBNA3C amino acids 365 to 545, which are necessary and sufficient for coactivation of the LMP1 promoter with EBNA2, are also required for binding SUMO-1 and SUMO-3 (24). SUMO-1 is a 101-amino-acid ubiquitinlike protein which is conjugated to target proteins in a series of enzymatic steps requiring a heterodimeric E1 activation complex (SAE1/SAE2) and an E2-like protein (UBC9). E3 ligases for SUMO include ring finger proteins that differ in target protein specificity (10, 13, 14, 34, 41, 47, 56). Unlike polyubiquitination, which generally targets proteins for degradation, sumolation can affect intracellular localization, protein stability, and transcriptional effects (reviewed in references 56 and 30). SUMO-1 modification of the promyelocytic leukemia protein PML is required for targeting to nuclear bodies (also known as ND10 bodies) and for recruitment of other NB proteins such as Sp100 and Daxx. SUMO-1 modification of IκBα increases its half-life by preventing ubiquitination and inhibits NF-κB signaling. SUMO-1 modification of HDAC-1 is required for repression, while sumolation of the transcription factor Sp3 represses activation (8, 9, 20, 39, 60). SUMO-1 has 48% identity with SUMO-2 and 46% identity with SUMO-3, whereas SUMO-2 and SUMO-3 are 97% identical to each other. In contrast to SUMO-1, which lacks an internal sumolation consensus motif, SUMO-2 and SUMO-3 can form polymeric chains. Unique functions for SUMO-2 and SUMO-3 have not been clearly delineated (40, 48). Since EBNA3C transcriptional activation is mediated by amino acids 365 to 545, which are required for binding to SUMO, EBNA3C interaction with SUMO and modification by SUMO could be important for EBNA3C transcriptional effects. The experiments reported here investigated this hypothesis.
MATERIALS AND METHODS
Cell culture.
293-T and HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. The EBV-negative Burkitt's lymphoma cell line BJAB was maintained in RPMI with 10% fetal calf serum.
Plasmids.
Plasmid pSG5-flagEBNA3C encoding EBNA3C amino acids 11 to 992 and deletion constructs of EBNA3C have been described previously (24). Expression plasmids pSG5-EBNA3Cm1, which has codons for DDD507-509 mutated to encode AAA, and pSG5-EBNA3Cm2, which has codons for DVIEVID511-515 mutated to encode AVIAVIA, were constructed in pSG5-flagEBNA3C with the Quikchange site-directed mutagenesis system (Stratagene, La Jolla, Calif.) and the following primer sets: 5′ GTTGTGTACGACGATGCTGTCATAGCGGTGATTGCTGTTGAAACCACC 3′; 5′ GGTGGTTTCAACAGCAATCACCGCTATGACAGCATCGTCGTACACAAC 3′; 5′ GCGTGTGTTGTGTACGCCGCTGCTGTCATAGAGGTG 3′; and 5′ CACCTCTATGACAGCAGCGGCGTACACAACACACGC 3′.
Plasmids pcDNA-3, encoding processed forms of hemagglutinin (HA)-tagged SUMO-1, SUMO-2, and SUMO-3 and green fluorescent protein (GFP)-tagged SUMO-1 were kindly provided by Ron Hay (University of St. Andrews, Scotland). pGEX-2TK SUMO-3 was created by subcloning a BamHI-EcoRI fragment from pcDNA3-HA-SUMO-3 into the BamHI and EcoRI sites of the PGEX-2TK vector (Amersham Pharmacia Biotech, Piscataway, N.J.). Plasmids pAS1-CYH2 and pACT were kindly provided by S. Elledge, and pAS1-EBNA3C11-992 has been described (24). pAS1-EBNA3Cm1 and pAS1-EBNA3Cm2 were constructed by subcloning the SpeI-BsiWI fragment from pSG5 EBNA3Cm1 and pSG5 EBNA3Cm2 into pAS1-EBNA3C11-992. Plasmid p(−512/+72) LMP1-luc has been described (24).
In vivo sumolation assays.
Monolayer 293-T cells at 60 to 80% confluence in 60-mm dishes were transfected with 4 μg of wild-type and mutant pSG5-EBNA3C expression plasmids and control vector and with 4 μg of pcDNA3 HA-SUMO-2 and HA-SUMO-3 expression plasmids and control vector with Superfect reagent (Qiagen, Valencia, Calif.). After 48 h, the cells were dislodged with ice-cold PBS, cell pellets were resuspended in 100 μl of PBS and cells were disrupted by addition of 100 μl of 2% sodium dodecyl sulfate in Tris-buffered saline (pH 7.5) (6). Cell lysates were then boiled for 10 min, followed by the addition of 5 volumes of 0.5% NP-40 in Tris-buffered saline with the protease inhibitors phenylmethylsulfonyl fluoride and aprotinin. Diluted lysates were sonicated for 30 s, precleared with protein G-agarose beads, and immune precipitated with antibody and protein G-agarose beads.
Transfections and reporter assays.
BJAB cells (≈107) were transfected in 0.4 ml of RPMI 1640 supplemented with 10% fetal calf serum with a Bio-Rad Gene Pulser at 220 V and 960 μF. For reporter assays, each transfection contained 10 μg of reporter plasmid, the indicated amounts of expression plasmids, and 0.5 μg of pGK-βgal as a normalization control. Total plasmid DNA was made constant by the addition of pSG5 vector DNA. After transfection, the cells were resuspended in 10 ml of complete medium and incubated at 37°C for 24 to 48 h. Cells were collected, washed in phosphate-buffered saline, lysed in reporter buffer (Luciferase Assay System; Promega, Madison, Wis.) by one freeze-thaw cycle, and assayed for luciferase and β-galactosidase activities (Galacto-Light; Tropix) with an Optocomp I luminometer (MGM Instruments).
Yeast two-hybrid analysis.
Saccharomyces cerevisiae strain Y190 (MATa gal4Δ gal80Δ his3-200 trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GAL1-lacZ LYS::GAL-HIS3 cyhR) containing HIS3 and lacZ genes with upstream GAL4 DNA binding sites was transformed with the bait plasmid pAS1-EBNA3C11-992 together with either pACT-RBP-Jκ, pACT-SUMO-1, pACT-SUMO-3, or pACT-UBC9. Transformants were selected on medium without Trp or Leu (Bio101, Inc, Vista, Calif.). Three days after transformation, six individual colonies were selected, streaked onto medium without Leu, Trp, or His and containing 50 mM 3-aminotriazole (Sigma), and grown for 6 days. Interaction was assessed by growth on His-deficient medium and by a β-galactosidase filter lift assay.
Confocal immunofluorescence studies.
HeLa cells were seeded on 9.6-cm2 single-chamber Lab-Tek slides (Nalgene, Naperville, Ill.) and transfected 24 h after seeding at a confluence of approximately 80% with 2 μg of pSG5-EBNA3C and deletion mutants together with 2 μg each of pSG5 and pSG5-GFP-SUMO-1. After 48 h, the cells were washed with phosphate-buffered saline (PBS), fixed with 3% paraformaldehyde for 20 min, and incubated with blocking buffer (3% bovine serum albumin in PBS) for 45 min prior to incubation with primary antibody for 1 to 2 h. After three washes with PBS, cells were incubated with Texas red-conjugated anti-mouse immunoglobulin secondary antibody (EBNA3C) and Alexa Fluor 546-conjugated anti-rabbit immunoglobulin secondary antibody (PML) (Molecular Probes, Madison, Wis.) for 45 min. Cells were washed extensively and coverslips were mounted onto Prolong antifade medium (Molecular Probes, Madison, Wis.), for microscopy with a Zeiss axioscope confocal microscope fitted with a Nikon confocal laser.
GST pulldowns.
For glutathione S-transferase (GST) fusion protein pulldown assays, 293-T cells were transfected with wild-type and mutant pSG5-EBNA3Cs. After 48 h, cells were collected in PBS and lysed in 1 ml of GST binding buffer (0.5% NP-40, 150 mM NaCl, 10% glycerol, 50 mM Tris, pH 7.4) in the presence of protease inhibitors (phenylmethylsulfonyl fluoride and aprotinin). Lysates were clarified by centrifugation, precleared with GST-coated beads for 45 min at 4°C, and incubated for 1 h at 4°C with GST fusion protein beads that had been blocked with 3% bovine serum albumin in PBS for 1 h. Approximately 100 μg of GST or GST-SUMO-3 was used in each adsorption.
RESULTS
EBNA3C colocalizes with GFP-SUMO-1 and disperses PML.
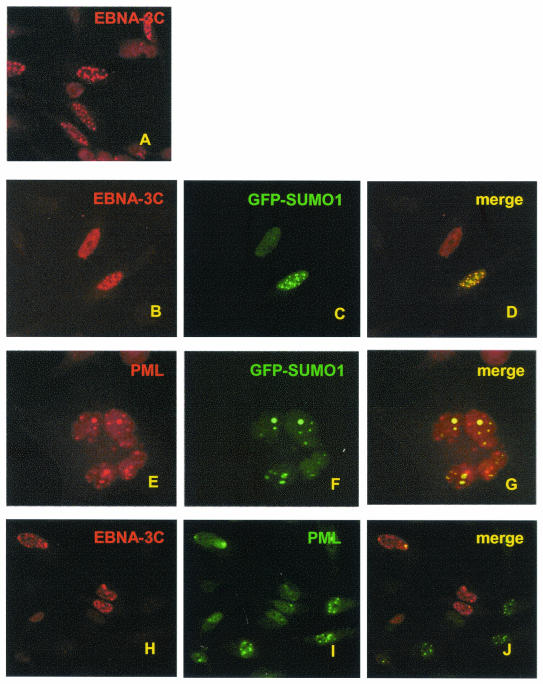
To determine whether EBNA3C can colocalize with SUMO-1, EBNA3C was expressed together with GFP-SUMO-1 in HeLa cells. EBNA3C expressed alone was either localized to nuclear dots or diffusely distributed in the nucleus (Fig. 1A). When cotransfected, EBNA3C and GFP-SUMO-1 completely colocalized to nuclear dots (Fig. 1B to D). To determine whether GFP-SUMO-1 foci corresponded to PML nuclear bodies, GFP-SUMO-1 was transfected alone, and endogenous PML was visualized with an anti-PML rabbit polyclonal antibody (Fig. 1E to G). The transfected GFP-SUMO-1 localized to a subset of PML nuclear dots.
FIG. 1.
EBNA3C colocalizes with GFP-SUMO-1 in nuclear bodies and disperses endogenous PML. HeLa cells were transfected with 2 μg of EBNA3C (A to D, H to J) and/or GFP-SUMO-1 (B to G) expression vectors. Endogenous PML and transfected EBNA3C were visualized by staining with anti-PML rabbit polyclonal antibody and A10 EBNA3C monoclonal antibody, followed by fluorophore-conjugated secondary antibody.
To further determine if EBNA3C colocalizes with PML nuclear bodies, transfected EBNA3C and endogenous PML were visualized by dual-color immunofluorescence (Fig. 1H to J). Immunostaining for EBNA3C again revealed either diffuse nuclear or punctuate nuclear staining (Fig. 1H). Although EBNA3C colocalized with PML bodies at the periphery of the nucleus in one or two dots (Fig. 1J), the number of PML-positive nuclear bodies was greatly reduced in EBNA3C-expressing cells (Fig. 1I). In other experiments, EBNA3C colocalized with overexpressed GFP-PML without reduced GFP-PML foci, compared to untransfected cells (data not shown). Thus, EBNA3C can colocalize with overexpressed GFP-PML in nuclear bodies, overexpressed EBNA3C disperses endogenous PML from most nuclear bodies, and EBNA3C consistently localizes with overexpressed SUMO-1.
EBNA3C and EBNA3B are modified by SUMO-3, but EBNA3A is not.
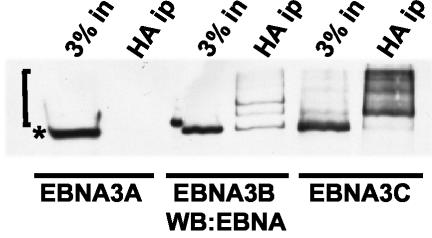
To determine whether EBNA3C, EBNA3B, and EBNA3A are modified by SUMO-3, HA-tagged SUMO-3 was coexpressed with EBNA3A, EBNA3B, and EBNA3C in 293-T cells. HA-tagged proteins were immunoprecipitated with anti-HA-conjugated agarose beads from sodium dodecyl sulfate-treated cell lysates that had been diluted in radioimmunoprecipitation assay buffer before immune precipitation. After electrophoresis of the immune precipitates, EBNAs were blotted and detected with EBV immune human serum (Fig. 2). Comparison of the EBNA Western blot of the cell lysate with that of HA-SUMO-3 immunoprecipitated proteins revealed abundant SUMO-3-modified forms of EBNA3C. SUMO-3-modified forms of EBNA3B were less evident, and modified EBNA3A was not detected. These data indicate that EBNA3C and, to a lesser extent, EBNA3B are modified by SUMO-3, while EBNA3A is not.
FIG. 2.
EBNA3C and EBNA3B are modified by SUMO-3, but EBNA3A is not. 293-T cells were transfected with 4 μg of HA-tagged SUMO-3 expression plasmid and 4 μg of EBNA3A, EBNA3B, or EBNA3C expression plasmid. After 48 h HA-tagged proteins were immunoprecipitated (HA ip) from clarified cell lysates with HA antibody. Immunoprecipitates were separated on 8% polyacrylamide gels, transferred to nitrocellulose, and blotted with EBV-immune human serum, which recognizes all the EBNA proteins. The square bracket indicates the position of SUMO-3-modified EBV nuclear proteins, while the asterisk indicates the position of unmodified protein.
EBNA3C amino acids 343 to 545 are critical for SUMO-2 and SUMO-3 modification.
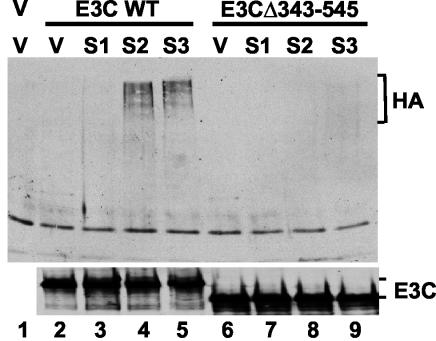
The role of EBNA3C amino acids 343 to 545 in SUMO modification was evaluated by comparing HA-tagged SUMO-1, SUMO-2, and SUMO-3 modification of wild-type EBNA3C with that of EBNA3C Δ343-545 in transfected 293-T cells (Fig. 3). Both EBNA3C and EBNA3CΔ343-545 were efficiently immunoprecipitated by monoclonal EBNA3C-specific antibody (Fig. 3, lower panel). EBNA3C modification by SUMO-2 and SUMO-3 polymers was readily detectable with anti-HA antibody in EBNA3C immunoprecipitates, while EBNA3CΔ343-545 modification by SUMO-2 and SUMO-3 was only detectable on very long exposure and SUMO-1 modification was not detected (Fig. 3, upper panel). Therefore, EBNA3C amino acids 343 to 545 are important for efficient SUMO-2 and SUMO-3 modification.
FIG. 3.
Modification by SUMO-2 and SUMO-3 polymers requires EBNA3C amino acids 343 to 545. Cell extracts were prepared from 293-T cells transfected with 2 μg of empty pSG5 vector (lane 1) or 2 μg of plasmid expressing either wild-type (WT) EBNA3C (lanes 2 to 5) or EBNA3C Δ343-545 (lanes 6 to 9). In addition, 2 μg of expression vectors encoding HA-tagged SUMO-1 (S1, lanes 3 and 7), SUMO-2 (S2, lanes 4 and 8), and SUMO-3 (S3, lanes 5 and 9) and pSG5 vector (V, lanes 1, 2, and 6) were cotransfected where indicated. Proteins were immunoprecipitated with A10 EBNA3C antibody, resolved on a sodium dodecyl sulfate-6% polyacrylamide gel, transferred to nitrocellulose, and immunoblotted with HA and EBNA3C antibodies.
EBNA3C amino acids 30 to 365 and 545 to 628 are also required for SUMO-3 modification.
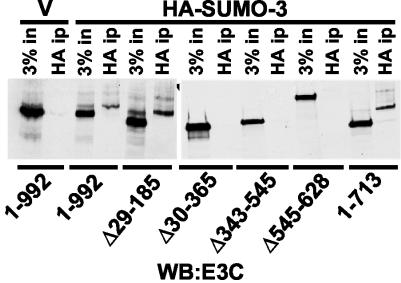
To investigate whether other domains of EBNA3C outside of amino acids 343 to 545 are also required for SUMO-3 modification, we transfected 293-T cells with wild-type EBNA3C and an EBNA3C deletion mutant together with either vector or HA-tagged SUMO-3 (Fig. 4). HA-modified proteins were retrieved from cell lysates by immunoprecipitation with anti-HA conjugated to agarose, and the presence of EBNA3C and SUMO-3-modified EBNA3C was detected by immunoblot with anti-EBNA3C monoclonal antibody (Fig. 4). Modified forms of EBNA3C, EBNA3C Δ29-185, and EBNA3C amino acids 1 to 713 were clearly detected in the HA immune precipitate lane, as were the more rapidly migrating unmodified EBNA3C proteins in the adjacent input lanes. Identically migrating forms of EBNA3C were also evident in cells expressing EBNA3C without added HA-SUMO-3 (Fig. 4 vector, input lane), indicating that EBNA3C could be modified by endogenous SUMO.
FIG. 4.
EBNA3C amino acids 185 to 365 and 545 to 628 are required for SUMO-3 modification. 293-T cells were transfected with wild-type or mutant EBNA3C expression vectors and with HA-SUMO-3 expression vector and empty pSG5 vector (V). HA-SUMO-3 modification of EBNA3C was assessed by HA antibody-specific immunoprecipitation and immunoblot for the indicated EBNA3C deletion mutants with the EBNA3C-specific A10 monoclonal antibody. A 3% input (3% in) was loaded adjacent to the HA immunoprecipitation (ip).
EBNA3C Δ30-365, Δ343-545, and Δ545-628 were expressed at nearly wild-type levels but were not detectably modified. These data indicate that amino acids 186 to 365, which partially overlap amino acids 343 to 545, and amino acids 545 to 628, which are C-terminal to amino acids 343 to 545, are required for SUMO modification; residues N-terminal to amino acid 185 and C-terminal to amino acid 713 are not required. The harsh conditions under which these immunoprecipitations were performed and the dependence of the SUMO-modified forms on the size of the input EBNA3C deletion mutant (Fig. 4, compare EBNA3C Δ1-992 with EBNA3C Δ29-185, for example) indicates that EBNA3C rather than an associated protein is SUMO-3 modified. In similar experiments with the same assay, SUMO-1-modified EBNA3C was detected, but to a much lesser degree than was observed with HA-SUMO-3 (data not shown).
Wild-type EBNA3C and mutants that can be SUMO modified localize to nuclear dots, whereas some EBNA3C mutants that are not SUMO modified can still coactivate transcription with EBNA2.
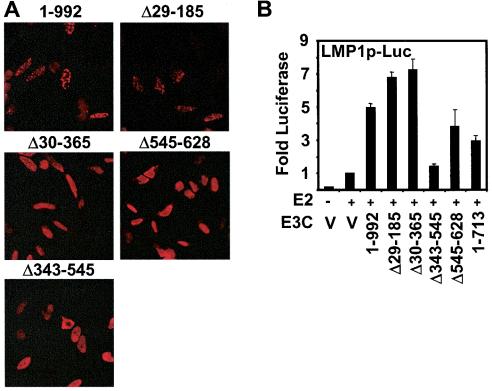
To determine if SUMO-3 modification is required for EBNA3C assembly into nuclear dots, wild-type EBNA3C and EBNA3C Δ29-185, which can be SUMO-3 modified, and EBNA3C Δ30-365, Δ343-545, and Δ545-628, which are not modified were expressed in HeLa cells and localization was determined. Wild-type EBNA3C and EBNA3C Δ29-185 were in nuclear dots or diffuse in the nucleus (Fig. 5A). However, EBNA3C Δ30-365, Δ343-545, and Δ545-628, which are not significantly SUMO-3 modified, were diffusely localized in cell nuclei with peripheral nuclear accentuation. These data indicate that EBNA3C SUMO-3 modification correlates with EBNA3C localization in nuclear dots.
FIG. 5.
EBNA3C sumolation is required for assembly into nuclear dots but not for LMP1 promoter coactivation with EBNA2. (A) HeLa cells were transfected with 4 μg of plasmid DNA encoding wild-type EBNA3C (1 to 992) or EBNA3C deletion mutants. EBNA3C was detected with the EBNA3C A10 monoclonal antibody and Texas Red-conjugated anti-mouse immunoglobulin secondary antibody. (B) BJAB cells were transfected with10 μg of the (−512/+72) LMPp-Luc reporter plasmid, 1 μg of vector (V), or 1 μg of EBNA2 (E2) expression vector and with 2 μg of the indicated EBNA3C deletion mutant plasmid. Transfection efficiency was normalized by cotransfected β-galactosidase expression, and results are reported as activation above that with EBNA2 alone. A representative experiment is shown in which the mean of three replicates was calculated ± standard error.
To determine if SUMO modification is required for LMP1 promoter coactivation with EBNA2, wild-type EBNA3C and EBNA3C deletion mutants were tested for their ability to coactivate the LMP1 promoter with EBNA2 in transient BJAB assays (Fig. 5B). EBNA3C deletion mutants Δ30-365 and Δ545-628, which were not SUMO-3 modified, coactivated the LMP1 promoter with EBNA2, and their activity was similar to that of wild-type EBNA3C. EBNA3C Δ29-185 and EBNA3C amino acids 1 to 713 were also similar to wild-type EBNA3C in coactivating with EBNA2, whereas EBNA3C Δ343-545 was null for both sumolation and coactivation assays. These data indicate that EBNA3C modification by SUMO-3 is not required for coactivation with EBNA2 in LMP1 reporter assays but is required for nuclear dot localization.
EBNA3C amino acids 476 to 515 are critical for LMP1 promoter coactivation with EBNA2.
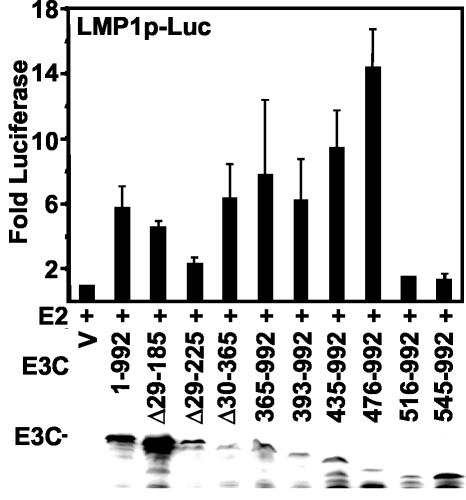
To identify the residues within EBNA3C amino acids 365 to 545 that are critical for LMP1 promoter coactivation with EBNA2, we tested a series of EBNA3C mutants that were N-terminally truncated (Fig. 6). Surprisingly, N-terminal truncation to amino acid 475 enhanced LMP1 promoter coactivation with EBNA2 approximately two- to threefold over wild-type EBNA3C. However, N-terminal deletion to amino acid 516 resulted in complete loss of coactivation with EBNA2 without affecting protein expression level (Fig. 6) or nuclear localization (not shown). These data indicate that EBNA3C amino acids 476 to 516 are critical for EBNA2 coactivation.
FIG. 6.
EBNA3C amino acids 476 to 516 are critical for EBNA3C coactivation with EBNA2. BJAB cells were transfected with 10 μg of the p(−512/+72)LMP-Luc reporter, 1 μg of EBNA2 (E2) expression vector, or 2 μg of wild-type EBNA3C, the indicated EBNA3C deletion mutant, or vector control plasmid (V). Activity above that with EBNA2 alone after β-galactosidase normalization is indicated. The average and standard error of two replicates from two independent experiments are reported. The panel below shows an immunoblot with A10 EBNA3C-specific monoclonal antibody.
EBNA3C amino acids 476 to 516 include amino acids found in SUMO-1.
EBNA3C amino acids 509 to 513 are identical to SUMO-1 amino acids 86 to 90, are preceded by EE, DE, and DD in SUMO-1, SUMO3, and EBNA3C, and are conserved among primate lymphocryptoviruses (21, 57) (Fig. 7). Since SUMO-1 amino acids 86 to 90 bind to UBC9, the SUMO-conjugating enzyme (25), the potential role of these EBNA3C amino acids in interaction with UBC9, SUMO-1, and SUMO-3 was examined in directed yeast two-hybrid and GST pulldown assays. Wild-type EBNA3C, EBNA3C Δ343-545, EBNA3C Δ501-545, and EBNA3C with DDDVIEVID507-515 mutated to AAAVIEVID (m1) and DDAVIAVIA (m2) (Fig. 7) were cloned into the yeast two-hybrid bait vector pAS1 and tested for interaction with UBC9, SUMO-1, and SUMO-3 cDNAs which were cloned into pACT (Table 1). GAL4 DNA-binding domain fusions with EBNA3C, EBNA3C Δ501-545, EBNA3C m1, and EBNA3C m2 were expressed in S. cerevisiae at similar levels by Western blot (not shown) and interacted at a high level with RBP-Jκ, which binds to EBNA3C amino acids 183 to 363. EBNA3C Δ343-545 was expressed at the same level as EBNA3C, but, as expected, did not interact with RBP-Jκ.
FIG. 7.
EBNA3C has colinear homology with SUMO-1. B95-8 (type I) EBNA3C and human SUMO-1 and SUMO-3 amino acid sequences are shown. Residues conserved between human and herpesvirus papio EBNA3C are underlined.
TABLE 1.
Focused yeast two-hybrid assaysa
| Bait plasmid | EBNA3C prey plasmid | Growth on His− medium | Relative β-galactosidase activity |
|---|---|---|---|
| RBP-Jκ | 1-992 | +++ | +++ |
| Δ501-545 | +++ | +++ | |
| m1 | +++ | +++ | |
| m2 | +++ | +++ | |
| 365-545 | − | − | |
| Δ343-545 | − | − | |
| SUMO-1 | 1-992 | +++ | +++ |
| Δ501-545 | ++ | ++ | |
| m1 | +/− | +/− | |
| m2 | +/− | +/− | |
| 365-545 | +++ | +++ | |
| Δ343-545 | − | − | |
| SUMO-3 | 1-992 | +++ | +++ |
| Δ501-545 | − | − | |
| m1 | − | − | |
| m2 | − | − | |
| 365-545 | +++ | +++ | |
| Δ343-545 | − | − |
Yeast strain Y190 was transformed with the prey (pAS1) and bait (pACT) plasmids encoding the indicated proteins. Interaction was scored by growth on His−/Leu−/Trp− synthetic medium in the presence of 50 mM 3-aminotriazole and by a β-galactosidase assay. +++, ++, +/−, X-Gal conversion in 30 min, 1 to 2 h, or greater than 2 h.
Although the DVIEV sequence in SUMO-1 that is important for its binding to UBC9 is also found in EBNA3C, neither wild-type nor mutant EBNA3Cs interacted with UBC9 in directed yeast two-hybrid assays (data not shown). However, EBNA3C and EBNA3C amino acids 365 to 545 did interact with SUMO-1 and SUMO-3. Furthermore, EBNA3C interaction with SUMO-1 and SUMO-3 was dependent on EBNA3C amino acids 507 to 515 because EBNA3C Δ343-545, EBNA3C Δ501-545 and EBNA3C m1 and m2 interacted at a lower level or failed to interact with SUMO-1 and SUMO-3. These data indicate that EBNA3C does not interact with UBC9 in S. cerevisiae and that EBNA3C interactions with SUMO-1 and SUMO-3 are dependent on EBNA3C amino acids 507 to 515.
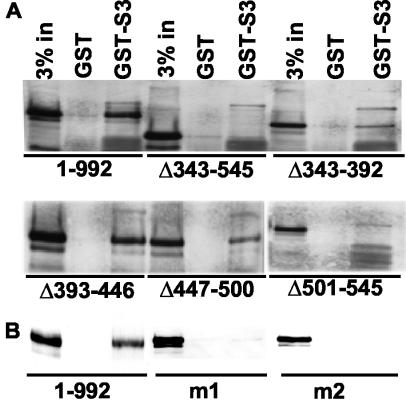
To further evaluate the binding of wild-type and mutant EBNA3C to UBC9, SUMO-1, and SUMO-3, wild-type and mutant EBNA3Cs were expressed in 293-T cells, and lysates were incubated with GST and GST fused to UBC9, SUMO-1, and SUMO-3. Although a small amount of EBNA3C bound to GST-UBC9, the binding did not map to a particular domain and was almost completely inhibited by the addition of albumin to the binding reactions (not shown). In contrast, approximately 1% of wild-type EBNA3C reproducibly bound to GST-SUMO-3. Binding to SUMO-3 was dependent on EBNA3C amino acids 343 to 545 and on the residues homologous to SUMO. EBNA3C Δ343-545, EBNA3C Δ501-545, EBNA3C m1, and EBNA3C m2 did not bind to GST-SUMO-3, whereas EBNA3C Δ343-392, EBNA3C Δ393-446, and EBNA3C Δ447-500 were intermediate in binding (Fig. 8A and B). These data confirm that EBNA3C amino acids 507 to 515 are important for binding to SUMO-3.
FIG. 8.
EBNA3C amino acids 507 to 515 are important for SUMO-3 binding. (A) Wild-type EBNA3C and deletion mutants were expressed in 293-T cells. After 24 h, the cells were lysed in GST binding buffer. Lysates were precleared with GST beads and then incubated with either GST or GST-SUMO-3 beads for 1 h at 4°C. The beads were washed three times with binding buffer. Bound proteins were eluted, resolved on an 8% polyacrylamide gel, transferred to nitrocellulose, and immune blotted with rabbit polyclonal anti-EBNA3C antibody (ExAlpha Corporation). (B) The EBNA3C and EBNA3C m1 and m2 mutants were tested in the same assay with A10 EBNA3C-specific monoclonal antibody.
EBNA3C amino acids 507 to 515 are critical for LMP1 promoter activation with EBNA2.
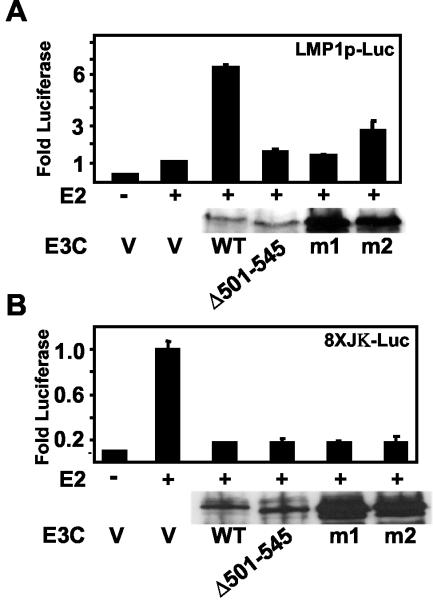
The EBNA3C amino acids that are critical for SUMO binding were tested for their role in EBNA2 coactivation reporter assays (Fig. 9). While wild-type EBNA3C coactivated the LMP1 promoter with EBNA2 approximately sixfold above the activation observed with EBNA2 alone, EBNA3C Δ501-545 and EBNA3C m1 and m2 had substantially reduced activity in LMP1 promoter assays (Fig. 9A). These EBNA3C mutants still blocked EBNA2 transactivation of a luciferase reporter downstream of a promoter with eight RBP-Jκ binding sites (Fig. 9B). The EBNA3C Δ501-545 mutant was expressed at levels comparable to that of the wild-type and m1 and m2 at a higher level; all three mutants localized to the nucleus when expressed in HeLa cells (not shown). Thus, EBNA3C amino acids 507 to 515 are critical for coactivation of the LMP1 promoter with EBNA2; EBNA3C m1 and m2 are specifically impaired in coactivation of the LMP1 promoter with EBNA2.
FIG. 9.
EBNA3C amino acids 507 to 515 are critical for coactivation of the LMP1 promoter with EBNA2. (A) BJAB cells were transfected with 10 μg of (−512/+72) LMPp-Luc reporter construct, 1 μg of EBNA2 (E2) expression vector, or 1 μg of wild-type and mutant EBNA3C expression vector. Activity above that with EBNA2 is indicated after β-galactosidase normalization. The average and standard error of duplicates in a representative experiment are reported. (B) EBNA3C and EBNA3C mutants were tested for repression of EBNA2 activity with a promoter with eight upstream copies of the Cp RBP-Jκ binding site. Quantities of transfected plasmids are the same as in A except that 10 μg of EBNA3C expression plasmids and 2 μg of EBNA2 expression plasmid were used in the repression assay. Results are the average of two replicates from a representative experiment. Expression of EBNA3C and EBNA3C mutants was detected with the A10 EBNA3C monoclonal antibody.
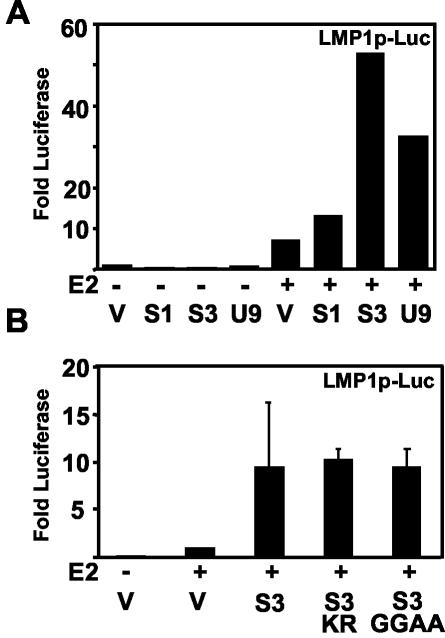
SUMO-1, SUMO-3, and UBC9 overexpression coactivates the LMP1 promoter with EBNA2.
To further investigate the potential role of SUMO in coactivation of the LMP1 promoter with EBNA2, the effect of SUMO-1, SUMO-3, and UBC9 overexpression on coactivation was evaluated. SUMO-1, SUMO-3, and UBC9 had no effect on the level of basal promoter activation in the absence of EBNA2 (Fig. 10A). However, EBNA2 activation of the LMP1 promoter was substantially enhanced by cotransfection of SUMO-1, SUMO-3, and UBC9 (Fig. 10A). SUMO-3 had the greatest effect. The UBC9 C93S mutant, which is defective for conjugating activity (14), coactivated similarly to wild-type UBC9 (not shown). The SUMO-3 mutant K11R, which cannot assemble into multimeric chains, and SUMO-3 GG93-94AA, which cannot covalently modify proteins, also coactivated with EBNA2 and were similar to wild-type SUMO-3, indicating that SUMO-3 and UBC9 can mediate coactivation but covalent protein modification is not required (Fig. 10B). These data are consistent with overexpressed SUMO-3 and UBC9 enhancing transcriptional activation and squelching repression.
FIG. 10.
SUMO-1, SUMO-3, and UBC9 can coactivate with EBNA2 at the LMP1 promoter. Coactivation does not depend on SUMO-3 modification. (A) LMP1 promoter (−512/+72) LMPp-Luc reporter was transfected with β-galactosidase into BJAB cells with 10 μg of empty vector (V) or plasmid expressing SUMO-1 (S1), SUMO-3 (S3), or UBC9 (U9) alone and with 1 μg of EBNA2 (E2) expression vector. Luciferase values were normalized for cotransfected β-galactosidase reporter. Activity above that with the vector alone is shown. Results are representative of three independent experiments. (B) Luciferase reporter assays were done as in A with 10 μg of cotransfected SUMO-3 (S3), SUMO-3 K11R (S3 KR), or SUMO-3 with terminal diglycines mutated to alanines (S3 GGAA). Luciferase above that with EBNA2 alone is shown. Values represent the average and standard error of duplicates from a representative experiment.
EBNA3C binding domain for SUMO, which is required for coactivation with EBNA2, is also required for coactivation with p300.
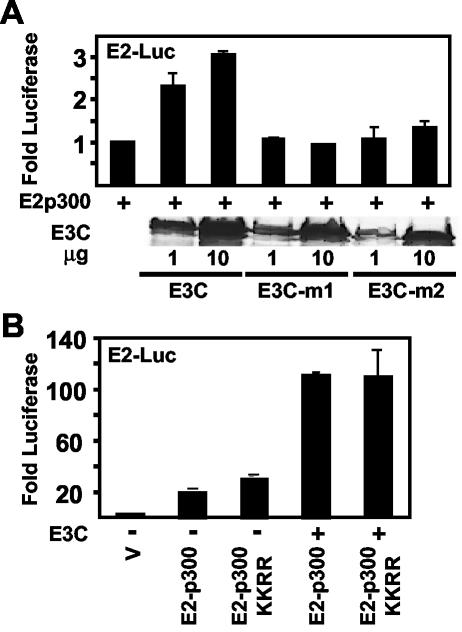
Since p300 has a crucial role in EBNA2-mediated transcriptional activation (53), EBNA3C coactivation with EBNA2 could be mediated by an effect on p300 activity. Indeed, EBNA3C binding to SUMO-conjugated p300 could enhance p300 effects on EBNA2 by blocking HDAC-6 association with p300 (12). We tested this hypothesis by first investigating whether EBNA3C could potentiate p300 transcriptional effects with p300 fused to the DNA binding domain of the papillomavirus E2 protein (E2-p300) in assays with a reporter having upstream multimerized E2 DNA binding sites (4) (Fig. 11). EBNA3C coactivated E2-p300 three- to fourfold, and the effect was dependent on the level of EBNA3C expression. Furthermore, the EBNA3C m1 and m2 mutants, which cannot bind to SUMO, failed to coactivate despite equivalent expression levels (Fig. 11A). These data indicate that EBNA3C can potentiate p300 transcriptional effects and that this potentiation requires EBNA3C residues that are critical for SUMO interaction and EBNA2 coactivation.
FIG. 11.
EBNA3C SUMO homology domain is required for EBNA3C coactivation with p300, but p300 sumolation in the cell cycle regulatory domain is not required. (A) BJAB cells were transfected with a plasmid containing multimerized papillomavirus E2 binding sites upstream of a promoter and the luciferase reporter (5 μg), with pGK-βgal expression vector control, with 5 μg of vector (V) and E2-p300, and with the indicated amounts of EBNA3C expression plasmid. The mean activity above E2-p300 activity after β-galactosidase normalization of duplicates from a representative experiment is reported. Expression levels from one of the two replicates for wild-type EBNA3C and mutant EBNA3C transfection are shown in the immune blot with A10 antibody. (B) Same as A except that 2 μg of E2-p300 and E2-p300 K1020R, K1024R and 5 μg of EBNA3C expression plasmid and vector were cotransfected where indicated. Values are normalized relative to that with the reporter alone (V).
We proceeded to specifically evaluate the hypothesis that EBNA3C potentiates p300 by binding to sumolated p300 (12). EBNA3C activation of wild-type p300 was compared to its activation of the nonsumolated p300 double point mutant K1020R K1024R (12). Surprisingly, p300 K1020R K1024R was only modestly increased in basal activity relative to wild-type p300 (Fig. 11B). Furthermore, EBNA3C equivalently coactivated both the wild-type and double mutant p300 (Fig. 11B). Thus, EBNA3C coactivation of p300-mediated transcription is independent of p300 K1020 and K1024 sumolation. However, since EBNA3C can potentiate p300 and the EBNA3C SUMO binding domain is critical for this effect as well as for EBNA3C effects on EBNA2 coactivation, EBNA3C could be coactivating with EBNA2 and p300 by competing with p300 for association with a sumolated transcriptional repressor.
DISCUSSION
EBNA3C is essential for B-lymphocyte conversion to LCLs (49) and can affect EBNA2-mediated transcription through RBP-Jκ (26, 37, 38, 52) and PU.1 (59). EBNA3C amino acids 365 to 545 are essential and sufficient for EBNA3C coactivation of the LMP1 promoter with EBNA2 in reporter assays and are required for SUMO-1 and SUMO-3 interaction in yeast two-hybrid assays (24). However, little was known about the significance of SUMO binding for EBNA3C transcriptional regulation. The experiments described here identify, within the EBNA3C coactivation domain, a SUMO-homologous sequence, DDDVIEV, which is necessary for EBNA3C association with SUMO and for transcriptional activation. These residues are well conserved between EBV type 1 and type 2 EBNA3C and are also largely conserved in the related rhesus and herpesvirus papio lymphocryptovirus EBNA3Cs, compatible with a critical role in EBNA3C function (21, 57). Mutations of DDDVIEV to AAAVIEV and DDDVIEVID to DDAVIAVIA abolished SUMO binding and EBNA3C coactivation of the LMP1 promoter with EBNA2, tightly linking EBNA3C SUMO binding to EBNA3C transcriptional coactivation. The data presented here suggest a mechanism whereby EBNA3C uses these residues, which are critical for SUMO interaction, to modulate transcription.
EBNA3C transcriptional regulation is largely in concert with EBNA2 transcriptional activation (26, 37, 38, 52, 53, 57), which is dependent on p300/CBP and PCAF recruitment to the EBNA2 acidic domain. In LCLs, p300/CBP and PCAF histone acetylase activities may be limiting for gene transcription (32). EBNA3C may act to relieve that limitation, in a promoter-specific fashion, by potentiating the effects of p300/CBP recruited by EBNA2 at the LMP1 promoter while competing for RBP-Jκ binding at the Cp promoter. At the LMP1 promoter, EBNA3C may stably accumulate and coactivate transcription with EBNA2 through an interaction with a sumolated protein(s). HDAC-1 has been shown to bind to EBNA3C and is activated by sumolation (8, 36). EBNA3C may bind to a SUMO-activated repressor like HDAC-1 and block histone deacetylation, indirectly enhancing p300/CBP-mediated acetylation. In support of this model, EBNA3C coactivation with EBNA2 and with E2-p300 is abolished by m1 and m2 mutations. Our data also indicate that SUMO-1 and SUMO-3 as well as wild-type and mutant UBC9 overexpression can replace EBNA3C in coactivation with EBNA2. We speculate that excess SUMO-1 and SUMO-3 may distract histone deacetylases from binding p300 and thereby coactivate the LMP1 promoter with EBNA2. Since SUMO modification of proteins has usually been associated with enabling transcriptional repression, EBNA3C binding to SUMO on a potentiated repressor could more generally activate transcription.
The EBNA3C SUMO-homologous sequence is highly charged and likely to be available for protein-protein interaction on the surface of EBNA3C. The corresponding SUMO-1 and SUMO-3 residues are important in SUMO binding to UBC9 and ULP-1 by nuclear magnetic resonance spectroscopy (25, 29) and biochemical studies (13). The nuclear magnetic resonance structure of SUMO-1 with UBC9 predicts that these residues contact positively charged side chains at the N terminus of UBC9 (25). These residues on the surface of EBNA3C and SUMO may also be a docking motif for SUMO and sumolated proteins. The presence of a similar motif (DDDVIEK) in Arabidopsis thaliana and wheat E1 proteins is consistent with the potential importance of this domain in mediating noncovalent interactions with SUMO and sumolated proteins (18, 19). DDDVIEV is also found at the C terminus of a likely human ortholog of a steroid receptor-interacting SNF2 domain-containing protein (KIAA0809/gi|124307995), and a similar motif (DEETIEV) is found in another SNF2 domain protein, SRCAP. The SNF2 domain is required for chromatin remodeling and is contained in a variety of proteins involved in transcriptional regulation (22, 28). EBNA2 associates with hSNF5, and chromatin immunoprecipitates of SWI/SNF complexes are enriched for sequences that contain EBNA2 response elements (54, 55). Coactivators such as EBNA3C and SRCAP may also use a (D/E)(D/E)(D/E)(V/T)IEV motif to associate with sumolated proteins.
We also note that EBNA3C can be SUMO modified and associate with PML bodies, partially displacing PML. Modification and PML body localization require EBNA3C amino acids 30 to 365 and 545 to 628 as well as amino acids 343 to 545. Since EBNA3C amino acids 30 to 365 and amino acids 545 to 628 are dispensable for coactivation, SUMO modification and PML body interaction are not essential for transcriptional coactivation of the LMP1 promoter with EBNA2. However, these properties of EBNA3C may have a role in transcription and cell growth regulation in EBV growth-transformed lymphoblastoid cell lines. EBNA3C can partially substitute for EBNALP in maintaining LMP1 levels, and both proteins interact with PML bodies (17, 24, 59). While EBNA3C localizes diffusely in LCL nuclei, EBNALP, the principal EBNA2 coactivator, is a stable component of PML bodies in LCLs (33, 44, 46). EBNALP moves out of PML nuclear bodies and into the nucleolus when lymphoblasts reach high density (45). The EBV genome in Raji cells is deleted for EBNA3C, and LMP1 levels decline as cells reach high density. Expression of EBNA3C restores LMP1 levels (2, 3). EBNA3C may therefore substitute for EBNALP to maintain LMP1 levels as cells reach saturation.
Most EBNA3C-containing foci in transfected HeLa cells were surprisingly depleted of PML, and EBNA3C colocalized with PML in only one or two points at nuclear poles, yet EBNA3C localized very well with SUMO-1. EBNA3C may compete with PML for SUMO-1 and may mediate PML desumolation since PML requires sumolation to localize to nuclear bodies (11, 31). Alternatively, EBNA3C may mimic herpes simplex virus type 1 ICP0 (Vmw110), which induces proteasome-dependent degradation of SUMO-modified PML and counteracts interferon effects on herpes simplex virus type 1-infected fibroblasts (6, 12). By targeting PML, EBNA3C may partially counteract interferon effects on EBV replication in LCLs. Disruption of PML bodies in acute promyelocytic leukemia causes differentiation arrest and proliferation, whereas retinoic acid treatment in acute promyelocytic leukemia stabilizes the PML-retinoic acid receptor α fusion protein, reconstitutes PML bodies, and causes leukemic blasts to differentiate to myelocytes (reviewed in reference 27). EBNA3C may inhibit EBV-infected B-lymphocyte differentiation by altering PML association with nuclear bodies.
Acknowledgments
We thank Roger Everett for many helpful discussions, Ron Hay and his group for advice and for SUMO expression plasmids, Donald Bloch for rabbit anti-PML antibody and advice, Steve Grossman for the E2-p300 reporter system, and Ellen Cahir-McFarland for helpful discussions and assistance with confocal microscopy. We also thank Bo Zhao and Micah Luftig for helpful suggestions.
E.J. is supported by grant 1K-08 AI49943-02 from the National Institutes of Health. A.R. is supported by research fellow grant 5056-03 from the Leukemia and Lymphoma Society of America. This research was supported by grant CA47006 from the National Cancer Institute of the USPHS, DHHS.
REFERENCES
- 1.Allday, M. J., D. H. Crawford, and J. A. Thomas. 1993. Epstein-Barr virus (EBV) nuclear antigen 6 induces expression of the EBV latent membrane protein and an activated phenotype in Raji cells. J. Gen. Virol. 74:361-369. [DOI] [PubMed] [Google Scholar]
- 2.Allday, M. J., D. H. Crawford, and J. A. Thomas. 1993. Epstein-Barr virus (EBV) nuclear antigen 6 induces expression of the EBV latent membrane protein and an activated phenotype in Raji cells. J. Gen. Virol. 74:361-369. [DOI] [PubMed] [Google Scholar]
- 3.Allday, M. J., and P. J. Farrell. 1994. Epstein-Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J. Virol. 68:3491-3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arany, Z., D. Newsome, E. Oldread, D. M. Livingston, and R. Eckner. 1995. A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature 374:81-84. [DOI] [PubMed] [Google Scholar]
- 5.Bain, M., R. J. Watson, P. J. Farrell, and M. J. Allday. 1996. Epstein-Barr virus nuclear antigen 3C is a powerful repressor of transcription when tethered to DNA. J. Virol. 70:2481-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buschmann, T., D. Lerner, C. G. Lee, and Z. Ronai. 2001. The Mdm-2 amino terminus is required for Mdm2 binding and SUMO-1 conjugation by the E2 SUMO-1 conjugating enzyme Ubc9. J. Biol. Chem. 276:40389-40395. [DOI] [PubMed] [Google Scholar]
- 7.Cotter, M. A., 2nd, and E. S. Robertson. 2000. Modulation of histone acetyltransferase activity through interaction of Epstein-Barr nuclear antigen 3C with prothymosin κ. Mol. Cell. Biol 20:5722-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David, G., M. A. Neptune, and R. A. DePinho. 2002. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J. Biol. Chem. 277:23658-23663. [DOI] [PubMed] [Google Scholar]
- 9.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 10.Desterro, J. M., J. Thomson, and R. T. Hay. 1997. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 417:297-300. [DOI] [PubMed] [Google Scholar]
- 11.Duprez, E., A. J. Saurin, J. M. Desterro, V. Lallemand-Breitenbach, K. Howe, M. N. Boddy, E. Solomon, H. de The, R. T. Hay, and P. S. Freemont. 1999. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci. 112:381-393. [DOI] [PubMed] [Google Scholar]
- 12.Girdwood, D., D. Bumpass, O. A. Vaughan, A. Thain, L. A. Anderson, A. W. Snowden, E. Garcia-Wilson, N. D. Perkins, and R. T. Hay. 2003. p300 transcriptional repression is mediated by SUMO modification. Mol. Cell 11:1043-1054. [DOI] [PubMed] [Google Scholar]
- 13.Gong, L., T. Kamitani, K. Fujise, L. S. Caskey, and E. T. Yeh. 1997. Preferential interaction of sentrin with a ubiquitin-conjugating enzyme, Ubc9. J. Biol. Chem. 272:28198-28201. [DOI] [PubMed] [Google Scholar]
- 14.Gong, L., B. Li, S. Millas, and E. T. Yeh. 1999. Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 448:185-189. [DOI] [PubMed] [Google Scholar]
- 15.Grossman, S. R., E. Johannsen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc. Natl. Acad. Sci. USA 91:7568-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grundhoff, A. T., E. Kremmer, O. Tureci, A. Glieden, C. Gindorf, J. Atz, N. Mueller-Lantzsch, W. H. Schubach, and F. A. Grasser. 1999. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 274:19136-19144. [DOI] [PubMed] [Google Scholar]
- 17.Harada, S., and E. Kieff. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA2 acidic domain-mediated transcriptional activation. J. Virol. 71:6611-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatfield, P. M., J. Callis, and R. D. Vierstra. 1990. Cloning of ubiquitin activating enzyme from wheat and expression of a functional protein in Escherichia coli. J. Biol. Chem. 265:15813-15817. [PubMed] [Google Scholar]
- 19.Hatfield, P. M., M. M. Gosink, T. B. Carpenter, and R. D. Vierstra. 1997. The ubiquitin-activating enzyme (E1) gene family in Arabidopsis thaliana. Plant J. 11:213-226. [DOI] [PubMed] [Google Scholar]
- 20.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss, 3rd, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, H., Y. G. Cho, and F. Wang. 2000. Structural, functional, and genetic comparisons of Epstein-Barr virus nuclear antigen 3A, 3B, and 3C homologues encoded by the rhesus lymphocryptovirus. J. Virol. 74:5921-5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnston, H., J. Kneer, I. Chackalaparampil, P. Yaciuk, and J. Chrivia. 1999. Identification of a novel SNF2/SWI2 protein family member, SRCAP, which interacts with CREB-binding protein. J. Biol. Chem. 274:16370-16376. [DOI] [PubMed] [Google Scholar]
- 23.Kieff, E., and A. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2573. In H. D. Knipe, D. Griffin, R. Lamb, M. Martin, B. Roizman, and S. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 24.Lin, J., E. Johannsen, E. Robertson, and E. Kieff. 2002. Epstein-Barr virus nuclear antigen 3C putative repression domain mediates coactivation of the LMP1 promoter with EBNA2. J. Virol. 76:232-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, Q., C. Jin, X. Liao, Z. Shen, D. J. Chen, and Y. Chen. 1999. The binding interface between an E2 (UBC9) and a ubiquitin homologue (UBL1). J. Biol. Chem. 274:16979-16987. [DOI] [PubMed] [Google Scholar]
- 26.Marshall, D., and C. Sample. 1995. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J. Virol. 69:3624-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Melnick, A., and J. D. Licht. 1999. Deconstructing a disease: RARκ, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 93:3167-3215. [PubMed] [Google Scholar]
- 28.Monroy, M. A., D. D. Ruhl, X. Xu, D. K. Granner, P. Yaciuk, and J. C. Chrivia. 2001. Regulation of cAMP-responsive element-binding protein-mediated transcription by the SNF2/SWI-related protein, SRCAP. J. Biol. Chem. 276:40721-40726. [DOI] [PubMed] [Google Scholar]
- 29.Mossessova, E., and C. D. Lima. 2000. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5:865-876. [DOI] [PubMed] [Google Scholar]
- 30.Muller, S., C. Hoege, G. Pyrowolakis, and S. Jentsch. 2001. SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell. Biol. 2:202-210. [DOI] [PubMed] [Google Scholar]
- 31.Muller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishioka, K., and D. Reinberg. 2001. Transcription. Switching partners in a regulatory tango. Science 294:2497-2498. [DOI] [PubMed] [Google Scholar]
- 33.Petti, L., C. Sample, and E. Kieff. 1990. Subnuclear localization and phosphorylation of Epstein-Barr virus latent infection nuclear proteins. Virology 176:563-574. [DOI] [PubMed] [Google Scholar]
- 34.Pichler, A., A. Gast, J. S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109-120. [DOI] [PubMed] [Google Scholar]
- 35.Radkov, S. A., M. Bain, P. J. Farrell, M. West, M. Rowe, and M. J. Allday. 1997. Epstein-Barr virus EBNA3C represses Cp, the major promoter for EBNA expression, but has no effect on the promoter of the cell gene CD21. J. Virol. 71:8552-8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radkov, S. A., R. Touitou, A. Brehm, M. Rowe, M. West, T. Kouzarides, and M. J. Allday. 1999. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J. Virol. 73:5688-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robertson, E. S., S. Grossman, E. Johannsen, C. Miller, J. Lin, B. Tomkinson, and E. Kieff. 1995. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J. Virol. 69:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson, E. S., J. Lin, and E. Kieff. 1996. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJ(κ). J. Virol. 70:3068-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross, S., J. L. Best, L. I. Zon, and G. Gill. 2002. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell 10:831-842. [DOI] [PubMed] [Google Scholar]
- 40.Saitoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252-6258. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt, D., and S. Muller. 2002. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. USA 99:2872-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian, C., S. Hasan, M. Rowe, M. Hottiger, R. Orre, and E. S. Robertson. 2002. Epstein-Barr virus nuclear antigen 3C and prothymosin κ interact with the p300 transcriptional coactivator at the CH1 and CH3/HAT domains and cooperate in regulation of transcription and histone acetylation. J. Virol. 76:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian, C., and E. S. Robertson. 2002. The metastatic suppressor Nm23-H1 interacts with EBNA3C at sequences located between the glutamine- and proline-rich domains and can cooperate in activation of transcription. J. Virol. 76:8702-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szekely, L., W. Q. Jiang, K. Pokrovskaja, K. G. Wiman, G. Klein, and N. Ringertz. 1995. Reversible nucleolar translocation of Epstein-Barr virus-encoded EBNA-5 and hsp70 proteins after exposure to heat shock and cell density congestion. J. Gen. Virol. 76:2423-2432. [DOI] [PubMed] [Google Scholar]
- 45.Szekely, L., W. Q. Jiang, K. Pokrovskaja, K. G. Wiman, G. Klein, and N. Ringertz. 1995. Reversible nucleolar translocation of Epstein-Barr virus-encoded EBNA-5 and hsp70 proteins after exposure to heat shock and cell density congestion. J. Gen. Virol. 76:2423-2432. [DOI] [PubMed] [Google Scholar]
- 46.Szekely, L., K. Pokrovskaja, W. Q. Jiang, H. de The, N. Ringertz, and G. Klein. 1996. The Epstein-Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J. Virol. 70:2562-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi, Y., T. Kahyo, E. A. Toh, H. Yasuda, and Y. Kikuchi. 2001. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 276:48973-48977. [DOI] [PubMed] [Google Scholar]
- 48.Tatham, M. H., E. Jaffray, O. A. Vaughan, J. M. Desterro, C. H. Botting, J. H. Naismith, and R. T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368-35374. [DOI] [PubMed] [Google Scholar]
- 49.Tomkinson, B., and E. Kieff. 1992. Second-site homologous recombination in Epstein-Barr virus: insertion of type 1 EBNA 3 genes in place of type 2 has no effect on in vitro infection. J. Virol. 66:780-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA3A and EBNA3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Touitou, R., M. Hickabottom, G. Parker, T. Crook, and M. J. Allday. 2001. Physical and functional interactions between the corepressor CtBP and the Epstein-Barr virus nuclear antigen EBNA3C. J. Virol. 75:7749-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waltzer, L., M. Perricaudet, A. Sergeant, and E. Manet. 1996. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-J. κ-EBNA2-activated transcription by inhibiting the binding of RBP-J.κ to DNA. J. Virol. 70:5909-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, L., S. R. Grossman, and E. Kieff. 2000. Epstein-Barr virus nuclear protein 2 interacts with p300, CBP, and PCAF histone acetyltransferases in activation of the LMP1 promoter. Proc. Natl. Acad. Sci. USA 97:430-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu, D. Y., G. V. Kalpana, S. P. Goff, and W. H. Schubach. 1996. Epstein-Barr virus nuclear protein 2 (EBNA2) binds to a component of the human SNF-SWI complex, hSNF5/Ini1. J. Virol. 70:6020-6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu, D. Y., A. Krumm, and W. H. Schubach. 2000. Promoter-specific targeting of human SWI-SNF complex by Epstein-Barr virus nuclear protein 2. J. Virol. 74:8893-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yeh, E. T., L. Gong, and T. Kamitani. 2000. Ubiquitin-like proteins: new wines in new bottles. Gene 248:1-14. [DOI] [PubMed] [Google Scholar]
- 57.Zhao, B., R. Dalbies-Tran, H. Jiang, I. K. Ruf, J. T. Sample, F. Wang, and C. E. Sample. 2003. Transcriptional regulatory properties of Epstein-Barr virus nuclear antigen 3C are conserved in simian lymphocryptoviruses. J. Virol. 77:5639-5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao, B., D. R. Marshall, and C. E. Sample. 1996. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J. Virol. 70:4228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao, B., and C. E. Sample. 2000. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an spi-1/Spi-B binding site. J. Virol. 74:5151-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong, S., S. Muller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748-2752. [PubMed] [Google Scholar]