Abstract
In this work, we sought to determine whether the components of the murine polyomavirus capsid establish specific interactions with the minichromosome encapsidated into the mature viral particles by using the cis-diamminedichloroplatinum(II) cross-linking reagent. Our data indicated that VP1, but not minor capsid proteins, interacts with the viral genome in vivo. In addition, semiquantitative PCR assays performed on cross-linked DNA complexes revealed that VP1 binds to all regions of the viral genome but significantly more to the regulatory region. The implications of such an interaction for viral infectivity are discussed.
The murine polyomavirus (Py) genome is organized into a minichromosome that is formed from a supercoiled circular double-stranded 5.3-kb DNA molecule with which cellular histones are associated (15). This chromatin is encapsidated into an icosahedral capsid composed of 72 pentamers of the major capsid protein VP1, with each pentamer being associated with minor capsid protein VP2 or VP3 (15). Several functions in the lytic life cycle, other than the structural one, have been assigned to the capsid proteins. The VP1 protein mediates the initial attachment and entry into host cells through interaction with sialic acids and integrins (2, 8, 20, 42, 43), and VP2 has been proposed to participate in viral cell entry (11, 28, 39). In addition, previous characterizations of Py mutants of the VP1 DE loop have also suggested a role for VP1 in viral growth control (22, 29, 38). Indeed, at early times postinfection, VP1 appears to colocalize with the infecting viral minichromosome in the cell nucleus and, in particular, at the nuclear matrix (NM), where viral transcription and replication take place (5, 10, 14). The finding that VP1 interacts with the NM regulatory protein YY1 early after infection suggests that VP1 may be required to anchor viral genomes to the NM for the formation of protein complexes that are necessary for early transcription (35). In accordance with this hypothesis, previous reports aimed at studying the potential interactions between capsid proteins and DNA have demonstrated that VP1, but not the minor coat proteins, is able to bind DNA through a DNA-binding domain (DBD) mapped within its first seven N-terminal residues (9, 31). These experiments were performed with an in vitro method and thus did not allow the attribution of any DNA sequence specificity to the DBD of VP1, as VP1 binds all Py and non-Py DNA sequences with equivalent high affinities (9, 31). The aim of the present work was to determine whether the components of the Py capsid establish interactions with the minichromosome encapsidated into the mature viral particles and, if so, with what specificity. For this purpose, we decided to treat mature Py virions with the cis-diamminedichloroplatinum(II) (cis-DDP) reagent, a molecule that produces inter- and intra-DNA strand cross-links independently of the sequence (6, 25, 37) and preferentially induces cross-links between DNA and nonhistone proteins located within a distance of 4 Å (16, 27). In contrast to formaldehyde, cis-DDP does not form protein-protein complexes (40). Furthermore, with respect to other cross-linking agents such as UV, the cross-linking reaction promoted by cis-DDP is fully reversible and allows for the recovery and characterization of the cross-linked proteins and DNA sequences (7, 17, 30, 32).
cis-DDP cross-links viral DNA in virions.
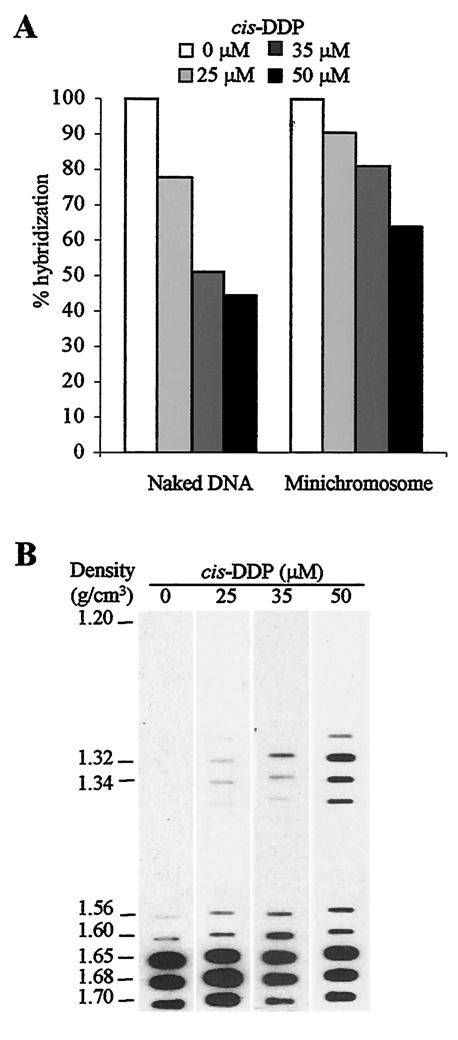
To set up the experimental conditions, mature viral particles or a control naked DNA (Py genome cloned into plasmid pAT153 [22]) was incubated with different concentrations (25, 35, and 50 μM) of cis-DDP (Sigma) for 90 min at 37°C. The efficiency of cross-linking was first tested by analyzing the ability of the DNA that was extracted from the cross-linked samples to be hybridized in Southern blot experiments with the Py genome used as the probe (22). Quantification of the hybridization efficiency was performed with an Instant-Imager. As shown in Fig. 1A, the cis-DDP reagent reduced the hybridization efficiency of both the control naked DNA and the encapsidated minichromosome in a dose-dependent manner (with a maximum of 35%), indicating that the drug had penetrated the viral particles and cross-linked the viral DNA. The same kind of results were obtained by PCR analysis when DNA extracted from cross-linked samples was used as the template; this DNA appeared less amplified than that extracted from non-cross-linked samples (data not shown). Next, to isolate cross-linked DNA complexes and to test which drug concentration was the most appropriate to recover them with the highest efficiency, viral particles (approximately 5 μg of VP1) treated with different cis-DDP concentrations were dissociated with 10 mM EDTA and 3 mM dithiothreitol and separated on a denaturing gradient composed of 0.5% Sarkosyl and 1.42 g of CsCl/cm3 by ultracentrifugation (35,000 rpm for 72 h [Optima LE-80K ultracentrifuge; Beckman Coulter]) (34). This type of denaturing gradient abolishes the interactions between DNA and non-cross-linked proteins and fully disassembles nucleosomes (34). Free DNA would be expected to sediment at a density of ≥1.65 g/cm3, whereas DNA-protein complexes should sediment in fractions of lower density. We analyzed the distribution of viral DNA in the gradient with a slot blot experiment by hybridizing an equal volume (1/30) of each fraction with the whole Py genome as a probe. As anticipated, in both cis-DDP-treated and untreated samples, free DNA sedimented at a density of ≥1.65 g/cm3 (Fig. 1B). Additionally, two types of signals were detected at lower densities in cis-DDP-treated virions at all of the concentrations used (Table 1). In fractions with densities of 1.32 to 1.34 g/cm3, signals increased with drug concentrations and represented up to 18.2% of total DNA at 50 μM (67% of the cross-linked DNA). The signal corresponded to stabilized, undissociated viral particles as confirmed by electron microscopy (data not shown). In addition, another signal that was detected at densities of 1.56 to 1.60 g/cm3 (hypothesized to contain DNA-protein complexes) appeared consistently more intense in virions that were treated with 35 μM cis-DDP than in those that were treated with 25 μM cis-DDP or left untreated (16.2% versus 3.38 and 0.38% of total DNA, respectively). But higher drug concentrations (50 μM cis-DDP) resulted in reduced viral dissociation (Table 1) due to a further stabilization of undissociated virions and consequently decreased the recovery of DNA-protein complexes (8.62% of total DNA versus 16.2% at 35 μM). Therefore, the next experiments were performed with the intermediate concentration of cis-DDP (35 μM).
FIG. 1.
cis-DDP cross-links encapsidated minichromosomes. (A) cis-DDP penetrates viral particles. Naked viral DNA and viral particles were incubated with or without cis-DDP at different concentrations, and treated DNA was analyzed by Southern blotting with the whole Py genome used as a probe. The level of hybridization was quantified by phosphorimager analysis. One hundred percent hybridization corresponds to the level of hybridization detected in cis-DDP-untreated samples. (B) Effect of cis-DDP concentration on the recovery of DNA-protein complexes. Viral particles were incubated with different concentrations of cis-DDP. After virus dissociation, samples were ultracentrifuged in denaturing CsCl gradients. The distribution of viral DNA in the fractions of the gradient was analyzed by slot blotting with the whole Py genome used as a probe.
TABLE 1.
Efficiency of viral DNA cross-linking
| cis-DDP (μM) | Cross-linked DNA (%)a | Stabilized virions (%)b | DNA-protein complexes (%)c |
|---|---|---|---|
| 0 | 0.38 | 0 | 0.38 |
| 25 | 4.14 | 0.76 | 3.38 |
| 35 | 23.9 | 7.7 | 16.2 |
| 50 | 26.82 | 18.2 | 8.62 |
Percentage of the total DNA of the gradient based on densitometry of the signals in slot blots from Fig. 1B.
Stabilized virions correspond to fractions with a density of 1.28 to 1.34 g/cm3.
DNA-protein complexes correspond to fractions with a density of 1.56 to 1.60 g/cm3.
VP1, but not VP2 or VP3, binds viral DNA.
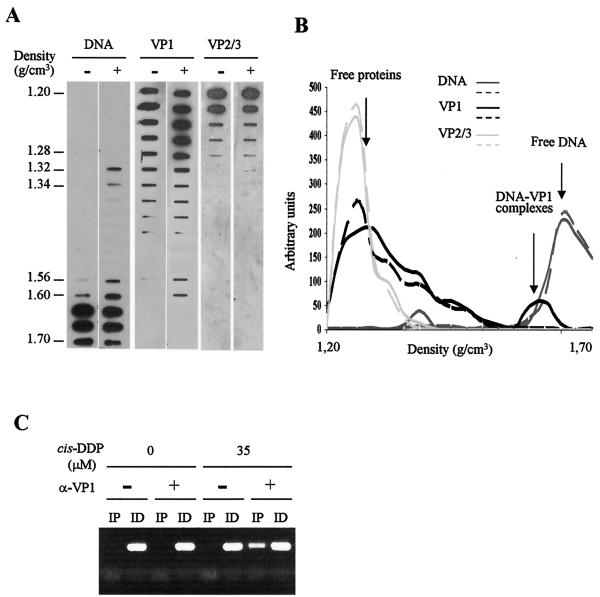
In order to determine if Py capsid proteins bind to the genome in the mature particles, the presence of these proteins in cross-linked DNA complexes was examined as follows. The same viral preparation was divided into two samples: one was treated with cis-DDP (35 μM), and the other was left untreated. Then, viral particles were dissociated and loaded onto a denaturing CsCl gradient as described above. After centrifugation, fractions were analyzed for both Py DNA (see above) and capsid proteins by slot blotting by using anti-VP1 or anti-VP2/3 antisera (18). The distribution of VP1 and VP2/3 in the gradient was revealed by using horseradish peroxidase-conjugated secondary anti-mouse immunoglobulin G and an enhanced chemiluminescence reaction (Pierce) and was then quantified by densitometry (ImageMaster 2D Elite; Pharmacia Biotech). As shown in Fig. 2A and B, different patterns of distribution of the VP1 protein were found in cis-DDP-treated and -untreated samples. Indeed, whereas under both experimental conditions the VP1 protein was detected mainly in the lower-density fractions, in cis-DDP-treated samples, a strong signal (arbitrary units, an approximate 25-fold increase with respect to cis-DDP-untreated samples) was evidenced in the fractions of higher density (1.56 to 1.60 g/cm3) that were presumed to contain cross-linked DNA complexes. In contrast, the distributions of the VP2/3 proteins were very similar in cis-DDP-treated and -untreated samples, i.e., the minor capsid proteins were detected in the lowest-density fractions (1.20 to 1.28 g/cm3), but no signal at all could be detected in the higher-density fractions (1.56 to 1.60 g/cm3) (Fig. 2A). In fractions corresponding to the undissociated virions (density, 1.32 to 1.34 g/cm3), VP2/3 proteins were found exclusively in the cis-DDP-treated samples. These data suggest that VP1, but not the minor proteins, is associated with the viral genome in mature virions. To confirm this hypothesis, fractions from cis-DDP-treated or -untreated samples with buoyant densities of 1.56 to 1.60 g/cm3 were pooled, dialyzed against a Tris-EDTA buffer to remove CsCl, and then subjected to immunoprecipitation with anti-VP1 antibodies or without antibodies (as a control) in a radioimmunoprecipitation assay buffer (36). Immunoprecipitated and immunodepleted samples were treated with 1.5 M thiourea (Sigma) to reverse cross-links, digested with proteinase K, and extracted with phenol-chloroform. The presence of viral DNA in the extracted samples was assessed by PCR with primer pairs specific for the Py genome (see Fig. 4A) as follows. The PCR buffer was comprised of 1× Taq polymerase buffer (Promega), 3 mM MgCl2, 0.3 mM each deoxynucleoside triphosphate (Roche), 0.5 U of Taq polymerase (Promega), and 0.5 pmol of each primer/μl, and the PCR was carried out for 30 cycles of 92°C for 30 s, 50°C for 1 min, and 74°C for 2 min. The initial cycle had a denaturation step of 3 min at 94°C. As shown in Fig. 2C, in a representative PCR assay performed with primer pair 9, only anti-VP1-immunoprecipitated samples from cis-DDP-treated virions gave an amplification signal, confirming the physical association between VP1 and the viral minichromosome.
FIG. 2.
VP1 binds viral genome in mature particles. Qualitative (A) and quantitative (B) densitometry analysis of the distribution of viral DNA and capsid proteins VP1 and VP2/3 in the fractions of the gradient. Virions treated with (+) or without (−) the cis-DDP reagent are indicated at the top of each lane in panel A. Aliquots of each fraction were analyzed by slot blotting. Viral DNA was detected by hybridization as described in the legend to Fig. 1. Viral proteins were detected by using monoclonal antibodies directed against VP1 or VP2/3 (13) followed by incubation with horseradish peroxidase-conjugated secondary antibodies and an enhanced chemiluminescence reaction. Solid and broken lines represent cis-DDP-treated and cis-DDP-untreated virions, respectively, in panel B. (C) VP1 is associated with the viral genome. Complexes containing fractions of cis-DDP-treated (35 μM) or cis-DDP-untreated (0 μM) virions were immunoprecipitated with (+) or without (−) anti-VP1 antibodies. After reversal of cross-links, the DNA purified from the immunoprecipitated (IP) and immunodepleted (ID) samples was used as a template in PCRs (with primer pair 9) to amplify the Py origin of replication region.
FIG. 4.
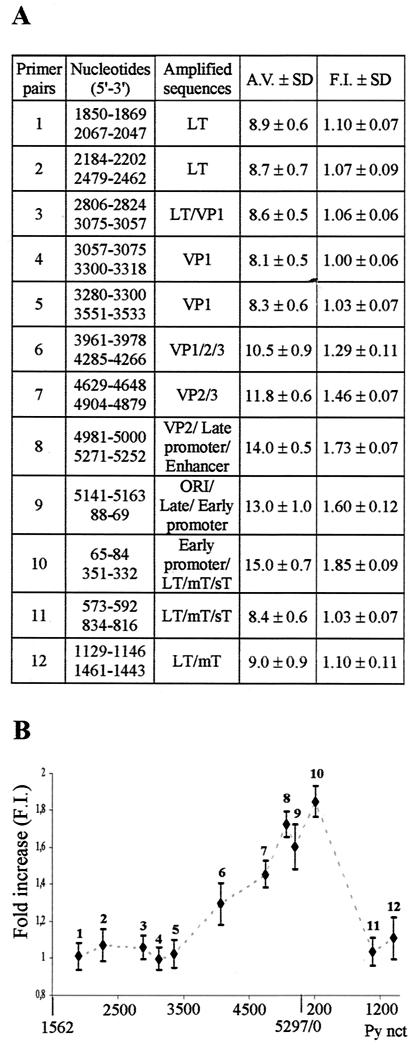
Semiquantitative PCR analysis of VP1 cross-linked DNA fragments. (A) Description of the primer pairs used to amplify target Py sequences. For a statistical analysis, each PCR was done in triplicateand repeated at least six times. The relative amplification value (A.V.) was calculated for each primer pair as the ratio of (Py/ref)IP to [(Py/ref)IP + (Py/ref)ID]. The fold increase (F.I.) value was calculated as the ratio of the amplified value of each primer pair to the amplified value of primer pair 4 (lowest amplified value). ORI, origin of replication; LT, large T antigen; mT, middle T antigen; sT, small T antigen. (B) Graphical representation of the semiquantitative PCR analysis. The numbering of the Py genome starts at the EcoRI site (nt 1562). nct, nucleotide.
Analysis of the viral DNA sequences cross-linked to Py VP1.
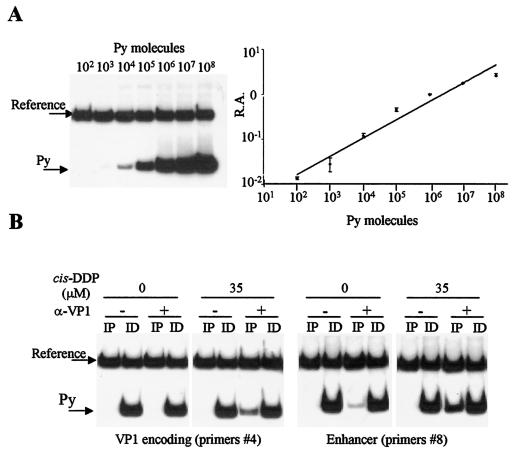
We next investigated whether viral DNA that was cross-linked to VP1 had any sequence specificity. Since cis-DDP modifies some bases, rendering impossible the use of DNA endonucleases (4) to fragment DNA, viral genomes were broken mechanically by sonication. We first determined the time that was necessary to induce homogeneous DNA fragmentation. Viral particles treated with cis-DDP were divided into equal volumes and sonicated for 10, 20, 30, 60, or 90 s (amplitude, 80). Cross-links were reversed, and DNA was extracted for Southern blot analysis. The results of hybridization which were quantified with an Instant-Imager indicated that 10, 20, or 30 s was not long enough to obtain a complete fragmentation. Instead, 60 s was sufficient to give rise to DNA fragments ranging from 250 to 660 bp (mean, approximately 430 bp), while a longer sonication time (90 s) did not significantly reduce the size of fragments and only increased variability (data not shown). Therefore, a sonication time of 60 s was chosen for our analysis. Then, to characterize the cross-linked viral DNA sequences, the DNA from fractions at buoyant densities of 1.56 to 1.60 g/cm3 was sonicated, immunoprecipitated with anti-VP1 antibodies, and used as a template in semiquantitative differential PCRs. This method relies on the coamplification of both the Py target sequence (variable amount) and a reference sequence (fixed amount) in the same reaction vessel (21). Equal volumes of the immunoprecipitated samples, which were designated the target sequences, were coamplified with the pLITMUS plasmid (New England Biolabs), which was used as the reference sequence. pLITMUS sequences were amplified with the primers corresponding to nucleotides (nt) 2095 to 2112 (forward) and nt 2660 to 2645 (reverse). The PCRs were performed as described above in the presence of 106 cpm of [α-32P]dATP (NEN Life Science Products, Boston, Mass.). Twelve primer pairs were designated to cover all of the viral genome and to amplify segments of approximately 300 bp (see Fig. 4A) with a similar annealing temperature. PCR products were electrophoresed on 7% nondenaturing polyacrylamide gels; the gels were fixed and dried, and the bands were quantified. The choice of 107 molecules per tube as a fixed amount of reference sequence was determined in preliminary PCRs in which increasing amounts of the target sequence were used (Fig. 3A, left panel). The ratio of amplification of the Py and pLITMUS sequences appeared as a function of the initial amount of the template Py sequence, thus validating our calculation method (Fig. 3A, right panel). The results of representative experiments obtained with primer pairs 4 and 8 are shown in Fig. 3B (left and right panels, respectively), and results obtained with all of the primer pairs are reported in Fig. 4 . To take into consideration any potential differential efficiency of amplification among primer pairs, the amplification value was calculated for each primer pair as the ratio (Py/ref)IP/[(Py/ref)IP + (Py/ref)ID], where Py is the target sequence, ref is the reference sequence, IP is immunoprecipitation, and ID is immunodepletion. The finding that all the primer pairs that were tested on VP1-immunoprecipitated complexes gave positive amplification signals indicated that VP1 is associated with all segments of the Py genome. Some viral regions appeared to be more abundantly represented among the complexes. However, the region encompassing the enhancer, the origin of replication, and the early and late promoters and their transcription start sites (nt 4900 to 190) was found to be 1.8-fold more amplified than the nonregulatory region. These results suggest that Py VP1 is associated with the entire Py genome but significantly more with the control region.
FIG. 3.
Experimental design of semiquantitative PCR analysis of VP1 cross-linked DNA fragments. (A) The relative amplification value (R.A.), measured as the ratio of amplification of Py to amplification of reference sequences, is a function of the initial amount of Py molecules present in the samples. (B) Representative PCR experiments performed in the presence of [α-P32]ATP with primer pairs 4 and 8 amplifying the VP1-encoding and enhancer regions, respectively.
Conclusions.
Previous reports have analyzed the structural features of the Py and simian virus 40 (SV40) minichromosomes as model systems for studying eukaryotic gene structure-function relationships. These studies were done with viral genomes that were extracted from infected cells before encapsidation (70S) and capsid assembly intermediates (180S) but not with the mature virions (240S) (12). In this work, we used the cis-DDP reagent to cross-link intact murine Py particles in order to analyze the potential interactions between the minichromosome and the proteins (histones excluded) present in viral particles. We found that VP1 interacts with the viral genome but that the minor coat proteins VP2 and VP3 do not. In addition, at a level of sensitivity of 100 pg, no other nonhistone proteins that were associated with viral DNA could be detected (data not shown). Our data are in accord with previous in vitro experiments that have mapped the DBD of VP1 to the N terminus, a disordered region predicted by crystallography to be oriented internally towards the minichromosome core (9, 31, 44). Although the affinity of the VP1-DNA interaction could not be measured by the present method, this association appeared to be very strong. In fact, we could detect the VP1-DNA complexes in un-cross-linked samples at very low levels in the radiolabeled semiquantitative PCR assays (Fig. 3B, right panel). The present work has demonstrated that, in mature viral particles, VP1 interacts with all regions of the genome. Such a broad association may suggest that VP1 plays a structural role in the protection of the encapsidated DNA from the extracellular medium. This hypothesis seems to be sustained by the observation that the fixation of the DNA-VP1 interactions with cis-DDP generates totally stabilized and undissociated viral particles. In the related virus SV40, such a role has also been proposed, and mutations in the DBD of the VP1 are indeed associated with altered virion formation and viability (26). In addition, VP1 may contribute to modulating or maintaining the chromatin compaction of the encapsidated viral DNA, promoted initially by the linker histone H1 once it is displaced from the minichromosome during the final maturation of viral particles (45). The other finding of this report is that the viral sequences extending over the regulatory region (i.e., the origin of replication, the enhancer, and early and late promoters) are 60 to 80% more highly represented in the sonication fragments that were immunoprecipitated with anti-VP1 antibodies than in the sonicated fragments covering the coding Py sequences. We found that the VP1-rich region was quite large (approximately 1,200 bp), but this may be ascribed in part to the size of the sonication fragments (mean, 430 bp) used for the PCR assays. This region is the one described earlier as nucleosome free in viral genomes extracted from infected cells, which correspond to the 70S chromatin form (13, 23). For SV40, a comparative analysis of the nucleosome spacing of minichromosomes showed an increase in length distribution of nucleosomes in assembly intermediates of 180S with respect to the preencapsidated chromatin of 70S, suggesting the nucleosome redistribution of the nucleosome-free region during encapsidation (12). In addition, studies of SV40 temperature-sensitive mutants have led to the proposal that VP1 has a role in this process (1, 3). Our present data show that VP1 is preferentially associated with this particular region of the Py genome. Although we cannot conclude that nucleosomes are totally absent in the VP1-rich region, our data certainly indicate that this region has greater accessibility by VP1 for interaction than does the rest of the genome. We propose that the binding of VP1 to this region represents more of a structural preference than a sequence-specific interaction with the regulatory region, which is consistent with studies performed in vitro with murine Py and SV40 that failed to detect any sequence preference for the VP1 interaction with viral DNA (9, 31, 41). In contrast to SV40 (33), no specific encapsidation initiation site has been reported for the Py genome. We suggest that the binding of VP1 to the noncoding region is used as the signal for the encapsidation of murine viral DNA. Furthermore, since it is well known that the accessibility of regulatory factors by chromatin conditions gene expression, it is likely that the preferential association of VP1 with the regulatory region may have a role in the initial phases of viral infection. Indeed, previous evidence has shown that VP1 may be required to control viral expression and to anchor viral genomes to the NM through interactions with NM factors such as YY1 (35). The higher (and/or stronger) association of VP1 with the enhancer region may favor the binding of host factors to this region by influencing the nucleosome organization and may consequently influence viral gene transcription. In addition, a direct role in the formation of the complexes necessary for early transcription is also possible for VP1 that is bound to the enhancer region. This hypothesis is supported by the previous observation that cis-dominant mutations of VP1 change the Py host range and early transcription ability (22, 29, 38). Further cross-linking studies performed with infected cells should help to identify qualitative and/or quantitative variations in the association of VP1 with the viral genome before early transcription occurs and therefore help to verify the hypothesized multifunctional role of VP1 in the Py life cycle. It has been demonstrated that Py capsids and pseudocapsids can transfer heterologous DNA to mammalian cells but that this transfer is quite inefficient (19, 24). This may be due in part to an incorrect chromatin structure of the cloned DNA associated with VP1. Therefore, a better knowledge of the DNA-binding properties of VP1 during the infectious process seems to be of primary interest for the further improvement of pseudocapsid vehicles for gene transfer.
Acknowledgments
We thank Massimo Gentile for electron microscopy analysis and Carlo Turano, Rossella Maione, and Maddalena Caruso for useful suggestions and critical discussions.
This work was supported by MIUR funds. Istituto Pasteur-Fondazione Cenci Bolognetti finances both departments.
REFERENCES
- 1.Ambrose, C., V. Blasquez, and M. Bina. 1986. A block in initiation of simian virus 40 assembly results in the accumulation of minichromosomes containing an exposed regulatory region. Proc. Natl. Acad. Sci. USA 83:3287-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer, P. H., C. Cui, T. Stehle, S. C. Harrison, J. A. De Caprio, and T. L. Benjamin. 1999. Discrimination between sialic acid-containing receptors and pseudoreceptors regulates polyomavirus spread in the mouse. J. Virol. 73:5826-5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasquez, V., A. Stein, C. Ambrose, and M. Bina. 1986. Simian virus 40 protein VP1 is involved in spacing nucleosomes in minichromosomes. J. Mol. Biol. 191:97-106. [DOI] [PubMed] [Google Scholar]
- 4.Brabec, V., and Z. Balcarova. 1993. Restriction-enzyme cleavage of DNA modified by platinum(II) complexes. Eur. J. Biochem. 216:183-187. [DOI] [PubMed] [Google Scholar]
- 5.Buckler-White, A. J., G. W. Humphrey, and V. Pigiet. 1980. Association of polyoma T antigen and DNA with the nuclear matrix from lytically infected 3T6 cells. Cell 22:37-46. [DOI] [PubMed] [Google Scholar]
- 6.Burstyn, J. N., W. J. Heiger-Bernays, S. M. Cohen, and S. J. Lippard. 2000. Formation of cis-diamminedichloroplatinum(II) 1, 2-intrastrand cross-links on DNA is flanking-sequence independent. Nucleic Acids Res. 28:4237-4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carr, A., and M. D. Biggin. 1999. An in vivo UV crosslinking assay that detects DNA binding by sequence-specific transcription factors. Methods Mol. Biol. 119:497-508. [DOI] [PubMed] [Google Scholar]
- 8.Caruso, M., L. Belloni, O. Sthandier, P. Amati, and M.-I. Garcia. 2003. α4β1 integrin acts as a cell receptor for murine polyomavirus at the postattachment level. J. Virol. 77:3913-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, D., X. Cai, and R. A. Consigli. 1993. Characterization of the DNA binding properties of polyomavirus capsid proteins. J. Virol. 67:6327-6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, L.-F., K. Ito, Y. Murakami, and Y. Ito. 1998. The capacity of polyomavirus enhancer binding protein 2αB (AML1/Cbfa2) to stimulate polyomavirus DNA replication is related to its affinity for the nuclear matrix. Mol. Cell. Biol. 18:4165-4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, X. S., T. Stehle, and S. C. Harrison. 1998. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J. 17:3233-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coca-Prados, M., H. Y.-H. Yu, and M.-T. Hsu. 1982. Intracellular forms of simian virus 40 nucleoprotein complexes. IV. Micrococcal nuclease digestion. J. Virol. 44:603-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cremisi, C., P. F. Pignatti, O. Croissant, and M. Yaniv. 1976. Chromatin-like structures in polyoma virus and simian virus 40 lytic cycle. J. Virol. 17:204-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deppert, W., and R. Schirmbeck. 1995. The nuclear matrix and virus function. Int. Rev. Cytol. 162:485-537. [DOI] [PubMed] [Google Scholar]
- 15.Eckhart, W. 1990. Polyomavirinae and their replication, p. 1593-1607. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology. Raven, New York, N.Y.
- 16.Filipski, J., K. W. Kohn, and W. M. Bonner. 1983. Differential crosslinking of histones and non-histones in nuclei by cis-Pt(II). FEBS Lett. 152:105-108. [DOI] [PubMed] [Google Scholar]
- 17.Filipski, J., K. W. Kohn, R. Prather, and W. M. Bonner. 1979. Thiourea reverses cross-links and restores biological activity in DNA treated with dichlorodiaminoplatinum(II). Science 204:181-183. [DOI] [PubMed] [Google Scholar]
- 18.Forstová, J., N. Krauzewicz, S. Wallace, A. J. Street, S. M. Dilworth, S. Beard, and B. E. Griffin. 1993. Cooperation of structural proteins during late events in the life cycle of polyomavirus. J. Virol. 67:1405-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forstovà, J., N. Krauzewicz, V. Sandig, J. Elliott, Z. Palkovà, M. Strauss, and B. E. Griffin. 1995. Polyoma virus pseudocapsids as efficient carriers of heterologous DNA into mammalian cells. Hum. Gene Ther. 6:297-306. [DOI] [PubMed] [Google Scholar]
- 20.Fried, H., L. D. Cahan, and J. C. Paulson. 1981. Polyoma virus recognizes specific sialyligosaccharide receptors on host cells. Virology 109:188-192. [DOI] [PubMed] [Google Scholar]
- 21.Frye, R. A., C. C. Benz, and E. Liu. 1989. Detection of amplified oncogenes by differential polymerase chain reaction. Oncogene 4:1153-1157. [PubMed] [Google Scholar]
- 22.Garcia, M. I., M. Perez, M. Caruso, O. Sthandier, R. Ferreira, M. Cermola, C. Macchia, and P. Amati. 2000. A mutation in the DE loop of the VP1 protein that prevents polyomavirus transcription and replication. Virology 272:293-301. [DOI] [PubMed] [Google Scholar]
- 23.Jakobovits, E. B., S. Bratosin, and Y. Aloni. 1980. A nucleosome-free region in SV40 minichromosomes. Nature 285:263-265. [DOI] [PubMed] [Google Scholar]
- 24.Krauzewicz, N., and B. E. Griffin. 2000. Polyoma and papilloma virus vectors for cancer gene therapy. Adv. Exp. Med. Biol. 465:73-82. [DOI] [PubMed] [Google Scholar]
- 25.Lemaire, M. A., A. Schwartz, A. R. Rahmouni, and M. Leng. 1991. Interstrand cross-links are preferentially formed at the d(GC) sites in the reaction between cis-diamminedichloroplatinum(II) and DNA. Proc. Natl. Acad. Sci. USA 88:1982-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, P. P., A. Nakanishi, D. Shum, P. C.-K. Sun, A. M. Salazar, C. F. Fernandez, S.-W. Chan, and H. Kasamatsu. 2001. Simian virus 40 Vp1 DNA-binding domain is functionally separable from the overlapping nuclear localization signal and is required for effective virion formation and full viability. J. Virol. 75:7321-7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lippard, S. J., and J. D. Hoeschele. 1979. Binding of cis- and trans-dichlorodiammineplatinum(II) to the nucleosome core. Proc. Natl. Acad. Sci. USA 76:6091-6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mannova, P., D. Liebl, N. Krauzewicz, A. Fejtova, J. Stokrova, Z. Palkova, B. E. Griffin, and J. Forstovà. 2002. Analysis of mouse polyomavirus mutants with lesions in the minor capsid proteins. J. Gen. Virol. 83:2309-2319. [DOI] [PubMed] [Google Scholar]
- 29.Mezes, B., and P. Amati. 1994. Mutations of polyomavirus VP1 allow in vitro growth in undifferentiated cells and modify in vivo tissue replication specificity. J. Virol. 68:1196-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Micetich, K., L. A. Zwelling, and K. W. Kohn. 1983. Quenching of DNA:platinum(II) monoadducts as a possible mechanism of resistance to cis-diamminedichloroplatinum(II) in L1210 cells. Cancer Res. 43:3609-3613. [PubMed] [Google Scholar]
- 31.Moreland, R. B., L. Montross, and R. L. Garcea. 1991. Characterization of the DNA-binding properties of the polyomavirus capsid protein VP1. J. Virol. 65:1168-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olinski, R., A. Wedrychowski, W. N. Schmidt, R. C. Briggs, and L. S. Hnilica. 1987. In vivo DNA-protein cross-linking by cis- and trans-diamminedichloroplatinum(II). Cancer Res. 47:201-205. [PubMed] [Google Scholar]
- 33.Oppenheim, A., Z. Sandalon, A. Peleg, O. Shaul, S. Nicolis, and S. Ottolenghi. 1992. A cis-acting DNA signal for encapsidation of simian virus 40. J. Virol. 66:5320-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orlando, V., and R. Paro. 1993. Mapping Polycomb-repressed domains in the bithorax complex using in vivo formaldehyde cross-linked chromatin. Cell 75:1187-1198. [DOI] [PubMed] [Google Scholar]
- 35.Palková, Z., D. Hollanderová, V. Gottifredi, H. Španielová, J. Forstová, and P. Amati. 2000. The polyomavirus major capsid protein VP1 interacts with the host nuclear matrix regulatory protein YY1. FEBS Lett. 467:359-364. [DOI] [PubMed] [Google Scholar]
- 36.Pelicci, G., L. Lanfrancone, A. E. Salcini, A. Romano, S. Mele, M. Grazia Borrello, O. Segatto, P. P. Di Fiore, and P. G. Pelicci. 1995. Constitutive phosphorylation of Shc proteins in human tumors. Oncogene 11:899-907. [PubMed] [Google Scholar]
- 37.Pinto, A. L., and S. J. Lippard. 1985. Binding of the antitumor drug cis-diamminedichloroplatinum(II) (cisplatin) to DNA. Biochim. Biophys. Acta 780:167-180. [DOI] [PubMed] [Google Scholar]
- 38.Ricci, L., R. Maione, C. Passananti, A. Felsani, and P. Amati. 1992. Mutations in the VP1 coding region of polyomavirus determine differentiating stage specificity. J. Virol. 66:7153-7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sahli, R., R. Freund, T. Dubensky, R. L. Garcea, R. Bronson, and T. L. Benjamin. 1993. Defect in entry and altered pathogenicity of a polyoma virus mutant blocked in VP2 myristylation. Virology 192:142-153. [DOI] [PubMed] [Google Scholar]
- 40.Solomon, M. J., and A. Varshavsky. 1985. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc. Natl. Acad. Sci. USA 82:6470-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Soussi, T. 1986. DNA-binding properties of the major structural protein of simian virus 40. J. Virol. 59:740-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stehle, T., Y. Yan, T. L. Benjamin, and S. C. Harrison. 1994. Structure of polyomavirus complexed with an oligosaccharide receptor fragment. Nature 369:160-163. [DOI] [PubMed] [Google Scholar]
- 43.Stehle, T., and S. C. Harrison. 1997. High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding. EMBO J. 16:5139-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan, Y., T. Stehle, R. C. Liddington, H. Zhao, and S. C. Harrison. 1996. Structure determination of simian virus 40 and murine polyomavirus by a combination of 30-fold and 5-fold electron-density averaging. Structure 4:157-164. [DOI] [PubMed] [Google Scholar]
- 45.Yuen, L. K., and R. A. Consigli. 1985. Identification and protein analysis of polyomavirus assembly intermediates from infected primary mouse embryo cells. Virology 144:127-138. [DOI] [PubMed] [Google Scholar]