Abstract
The Epstein-Barr virus (EBV) SM protein is a posttranscriptional regulator of cellular and viral gene expression that binds and stabilizes target mRNAs and shuttles from nucleus to cytoplasm. SM enhances expression of several EBV genes required for lytic replication and is essential for virion production. SM increases accumulation of specific mRNAs but also inhibits expression of several intron-containing transcripts. The mechanism by which SM inhibits gene expression is poorly understood. The experiments described here had several aims: to determine whether specific domains of SM were responsible for activation or inhibition function; whether these functions could be separated; and whether one or more of these functions were essential for virion production. A mutational analysis of SM was performed, focusing on amino acids in SM that are evolutionarily conserved among SM homologs in other herpesviruses. Mutation of the carboxy-terminal region of SM revealed a region that is likely to be structurally important for SM protein conformation. In addition, several amino acids were identified that are critical for activation and inhibition function. A specific mutation of a highly conserved cysteine residue revealed that it was essential for gene inhibition but not for transactivation, indicating that these two functions operate through independent mechanisms. Furthermore, the ability of wild-type SM and the inability of the mutant to inhibit gene expression were shown to correlate with the ability to inhibit splicing of a human target gene and thereby prevent accumulation of its processed mRNA. Surprisingly, some mutations which preserved both activation and inhibition functions in vitro nevertheless abolished virion production, suggesting that other SM functions or protein-protein interactions are also required for lytic replication.
The Epstein-Barr virus (EBV) SM protein is a posttranscriptional regulator of gene expression expressed early in the viral lytic cycle. SM enhances expression of several EBV genes and heterologous genes and is essential for virion production (13, 20, 21, 23, 31, 36). SM protein binds mRNA in vitro and in vivo, shuttles from nucleus to cytoplasm, interacts with components of nuclear export pathways, and enhances the cytoplasmic accumulation of target gene RNA transcripts (3, 4, 10, 29, 36). The presence of introns in the target gene generally leads to inhibition by SM, but SM also activates expression of a few spliced cellular genes (30). In addition, SM displays gene specificity, preferentially activating expression of some but not all intronless genes (20, 29).
Homologs of SM are found in herpes simplex virus, human cytomegalovirus (CMV), varicella-zoster virus, and Kaposi's sarcoma-associated virus and act as transcriptional and posttranscriptional regulators (1, 5, 6, 14, 19, 22, 24). Of these, herpes simplex virus ICP27 has been the most extensively studied and shown to possess both gene inhibition and activation functions. ICP27 inhibits host splicing and host gene expression but also selectively activates specific cellular genes (9, 15, 17). ICP27 activates intronless herpes simplex virus genes and has been shown to bind mRNA (32). The simian rhadinovirus herpesvirus saimiri also encodes a member of the SM/ICP27 family of genes, known as ORF57 (38). Both herpesvirus saimiri ORF57 and the Kaposi's sarcoma-associated virus homolog, also referred to as ORF57 (KS-SM), are most closely related to EBV SM (Fig. 1A).
FIG. 1.
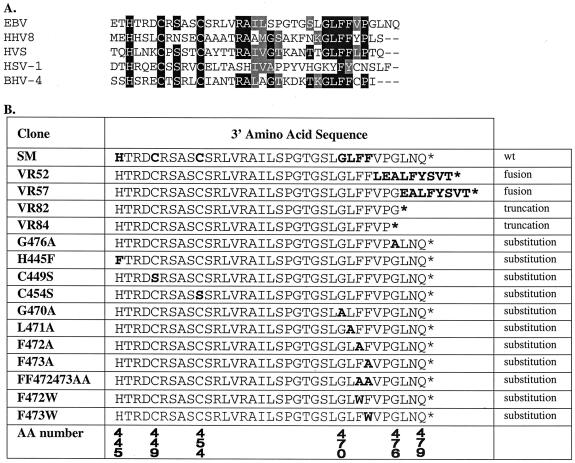
Structures of SM, SM homologs, and SM mutants. (A) Amino acid conservation between SM and homologs in other herpesviruses. The carboxy-terminal portions of SM homologues from human and nonhuman herpesviruses are aligned to display conserved amino acids in the carboxy-terminal regions. HHV8, human herpesvirus 8; HVS, herpesvirus saimiri; HSV-1, herpes simplex virus type 1; BHV-4, bovine herpesvirus 4. (B) Summary of SM mutants. The sequence of the carboxy terminus of wild-type (wt) SM and each mutant is shown. Single and double amino acid substitutions in each mutant are shown in bold. VR52 and VR57 were generated by exonuclease III digestion and include various amounts of exogenous vector sequence appended to the deleted region of SM. The predicted additional amino acids generated by the fusion in VR52 and VR57 are also shown in bold. Amino acid numbers are shown in the last row.
Although these genes are clearly functionally and structurally related, the overall degree of homology is limited. SM can rescue herpes simplex virus mutants defective in ICP27 expression, but the rescued mutants are less replication competent than those rescued with wild-type ICP27 (2). Similarly, CMV UL69 can also rescue replication of SM-deleted EBV in 293 cells, but the degree of functional reconstitution is much less than with SM itself (13). Previous studies of herpesvirus saimiri ORF57 examined the role of specific residues in activation and inhibition functions (12). Mutation of the herpesvirus saimiri ORF57 gene indicated that highly conserved amino acids in the carboxy terminus were essential for both activation and repression functions (12). The carboxy terminus of ICP27 is also essential for function (16). No mutations that specifically affect only the activation or repression functions have been identified (16, 18, 25, 26, 37).
We wished to identify structurally and functionally important amino acids and specifically identify residues of SM that might be required for individual functions. We reasoned that the activation and inhibition functions might operate through separate regions of the molecule and could be distinguished by targeted mutagenesis. By studying such mutants, we also wished to determine the role of these functions in virus replication. We therefore generated SM mutants specifically altered in conserved amino acids and characterized their functional properties.
MATERIALS AND METHODS
Plasmids and cell lines.
CMV-SM consists of SM cDNA cloned in the expression vector pCDNA3 (Invitrogen) and has been described previously (31). CMV-CAT is the chloramphenicol acetyltransferase (CAT) gene cloned in pCDNA3 (31). CMV-i-CAT contains a chimeric intron in the CAT transcript upstream of the CAT gene. BJAB is a cell line derived from an EBV-negative human B lymphoma (27). SV40RL-i contains the Renilla luciferase cDNA originally cloned from Renilla reniformis transcribed from the simian virus 40 immediate-early promoter. RLSV40-i was constructed from pRLSV40 (Promega) by digestion with PstI and NheI, followed by blunt-end repair and ligation to remove the intron present in pRLSV40. CMVRL-i was constructed in an analogous fashion by removing the intron from pRL-CMV (Promega). XGH5 contains the genomic human growth hormone (hGH) gene with four introns, transcribed from a human metallothionein promoter (35). The radiolabeled hGH cDNA probe was generated from cloned hGH cDNA (8).
Mutant SM plasmid construction.
Carboxy-terminal deletions of SM were introduced by exonuclease III digestion as follows. CMV-SM was linearized with XhoI immediately 3′ of the SM termination codon. The digested ends were repaired with Klenow enzyme and a phosphorothioate deoxynucleoside triphosphate. The plasmid was then digested at the XbaI site, which is immediately upstream of the XhoI site. The product was then digested with exonuclease III for various lengths of time, followed by S1 nuclease treatment, Klenow-mediated repair, and ligation. The ligated products were transformed into Escherichia coli, and individual isolates were sequenced to determine the boundaries of the deletion.
Deletions of specific lengths were introduced in the SM carboxy-terminal region by PCR amplification. Briefly, a 5′ primer containing the EcoRI site at nucleotide 1237 in the SM cDNA and flanking sequence was used with a 3′ primer consisting of 16 nucleotides immediately preceding the required deletion site, followed by a stop codon and an XbaI site. The amplification products were purified, cloned into CMV-SM to replace the full-length EcoRI-XbaI fragment, and sequenced.
Individual amino acid substitutions were generated by PCR amplification of two overlapping DNA fragments which contained the mutated nucleotides in the overlapping region. The resulting fragments were column purified, mixed, and used as the template for another round of amplification with only the forward primer from the first reaction and the reverse primer from the second, thereby generating a single amplification product containing the mutation in the central portion. This fragment was then used to replace the corresponding fragment in CMV-SM, and the resulting plasmid was sequenced.
Transfections and enzyme assays.
Cells grown in suspension were transfected by electroporation as previously described (31). HeLa cells grown on glass coverslips were transfected with Lipofectamine Plus (Invitrogen) as per the manufacturer's protocol. CAT assays were performed as described previously. Briefly, 36 h after transfection, cells were harvested, washed in phosphate-buffered saline, and lysed in detergent buffer. Lysates were incubated with acetyl-coenzyme A and [14C]chloramphenicol, extracted with ethyl acetate, and chromatographed, and the percent conversion was directly quantitated with a Packard InstantImager (Packard Instruments). All transfections were performed in triplicate, and the total amount of DNA per transfection was normalized with vector plasmid DNA. Assays were repeated as necessary to obtain results in the linear range of the assay and normalized to protein concentration as determined by the Bradford assay. Renilla luciferase assays were performed with a Renilla luciferase assay system (Promega) as per the manufacturer's instructions.
RNA analysis.
RNA was isolated from cells by a modification of guanidinium-phenol extraction (RNAzol; TelTest, Friendswood, Tex.). For Northern blotting, equal amounts of RNA were electrophoresed, transferred to membranes, and hybridized to the hGH cDNA probe labeled with 32P, followed by autoradiography. Reverse transcription was performed with avian myeloblastosis virus reverse transcriptase followed by PCR amplification with gene-specific primers to detect splicing precursors and spliced hGH mRNA.
Immunofluorescence microscopy and immunoblotting.
Transfected HeLa cells were washed 36 h after transfection and fixed with ice-cold methanol. Coverslips were incubated with polyclonal rabbit anti-SM antibodies, followed by tetramethylrhodamine isothiocyanate-labeled secondary antibodies as previously described (3). Nuclear localization of all mutant proteins and SM was confirmed by 4′,6′-diamidino-2-phenylindole (DAPI) staining. Cells were lysed in cold isotonic Tris-buffered saline containing 0.5% NP-40 and a protease inhibitor cocktail (Sigma). Detergent-soluble and insoluble fractions were separated by centrifugation at 13,000 × g and prepared for electrophoresis. Immunoblotting was performed with polyclonal anti-SM antibodies, anti-gp110 monoclonal antibodies (Chemicon), or anti EA-D monoclonal antibodies (Capricorn), as previously described (3).
Virus production from BMLF1KO-293 cells.
BMLF1KO-293 cells contain recombinant EBV genomes encoding green fluorescent protein and hygromycin resistance in which the SM gene is interrupted (13). Cells were transfected with CMV-HA-Z, which encodes an influenza virus hemagglutinin-tagged cDNA encoding the EBV Z transactivator (kind gift of Paul Lieberman, Wistar Institute) and either wild-type SM or mutant SM plasmids, as previously described (7). At 48 h after transfection, cell supernatant was filtered through a 0.8-μm filter and incubated with an equal volume of Raji cells at a final concentration of 3 × 105 cells/ml. Live cells were examined and counted by fluorescence microscopy or flow cytometry 3 days after infection. Virion yield comparisons were based on a minimum of two independent experiments (3).
RESULTS
Nonconserved amino acids in the carboxy terminus are necessary for activation and repression functions of SM.
In order to map the limits of essential domains in the carboxy terminus of SM, we initially used progressive exonuclease III digestion of CMV-SM to generate a series of random mutations (Fig. 1B). Since these mutations were generated by exonuclease digestion followed by T4 DNA polymerase-mediated blunt-ended repair, they consisted of deletions of SM fused to the distal plasmid vector sequence, resulting in deletion of various lengths of SM sequence and substitution with vector-derived amino acids. The ability of each of these mutants to activate CAT expression from a reporter plasmid, CMV-CAT, when cotransfected in B lymphocytes was measured.
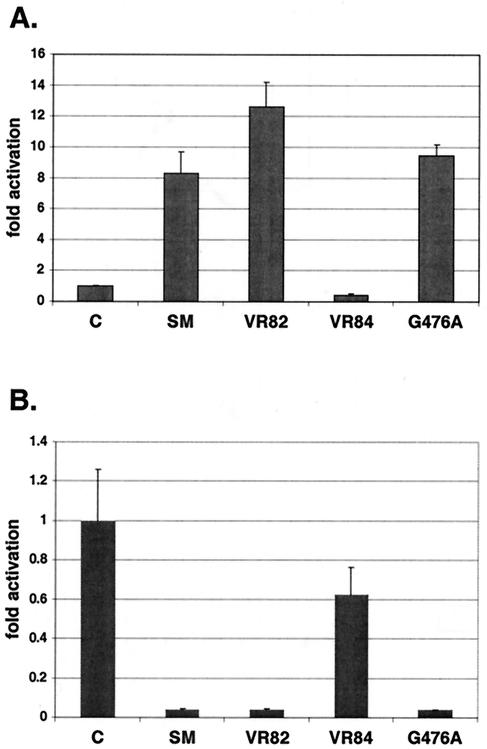
Analysis of several such mutants revealed that a mutant (VR57) which retained SM sequence up to and including amino acid 476 (G) was functional, whereas a mutant (VR52) which had lost amino acids beyond amino acid 472 (F) was completely nonfunctional (data not shown). Although both of these mutants had an additional nine and eight amino acids, respectively, derived from vector sequences appended to the SM sequence, the results suggested that amino acids between 472 and 476 were important for SM transactivation function. We therefore generated specific SM truncation mutants in which successive amino acids were removed, beginning from amino acid 476. A stop codon was also introduced to terminate translation at the desired amino acid. The resulting mutants were similarly analyzed for activation function.
These experiments demonstrated that removal of amino acids 477 to 479 (VR82) did not affect activation, whereas removal of amino acids 476 to 479 (VR84) led to complete loss of activity (Fig. 2A). Although this finding was somewhat surprising, as G476 is not conserved among the gammaherpesvirus homologs of SM, we hypothesized that G476 was essential and that substitution of G476 in the context of the entire SM protein might also lead to loss of activation function. We therefore generated an alanine substitution mutant of G476 and tested its ability to activate gene expression of the CAT reporter construct. Somewhat surprisingly, the G476A mutant was unimpaired in activation function (Fig. 2A). These data therefore suggested that although removal of amino acid 476 could not be tolerated, replacement with alanine was sufficient to permit the conformational structure necessary for full activation function.
FIG. 2.
Functionally essential amino acids in the SM carboxy terminus. Each SM mutant was cotransfected with either (A) an intronless CAT reporter construct to measure activation or (B) an intron-containing CAT reporter to measure repression. Transfections were performed in BJAB cells, and CAT activity in cell lysates was measured 48 h after transfection.
Since we had identified at least one amino acid that was necessary for function, we examined the same mutants for the ability to repress expression of an intron-containing reporter construct, CMV-i-CAT. As shown in Fig. 2B, wild-type SM repressed expression from CMV-i-CAT. The repression function of SM was lost when amino acids 476 to 479 were removed, but not if G476 was replaced with alanine. Removal of amino acids 477 to 479 had no effect on repression function. Thus, both the repression and activation functions were severely affected when SM was truncated beyond amino acid 477, but function was maintained by full-length SM despite substitution of G476 with alanine.
Specific amino acids in the hydrophobic GLFF motif are also essential for function.
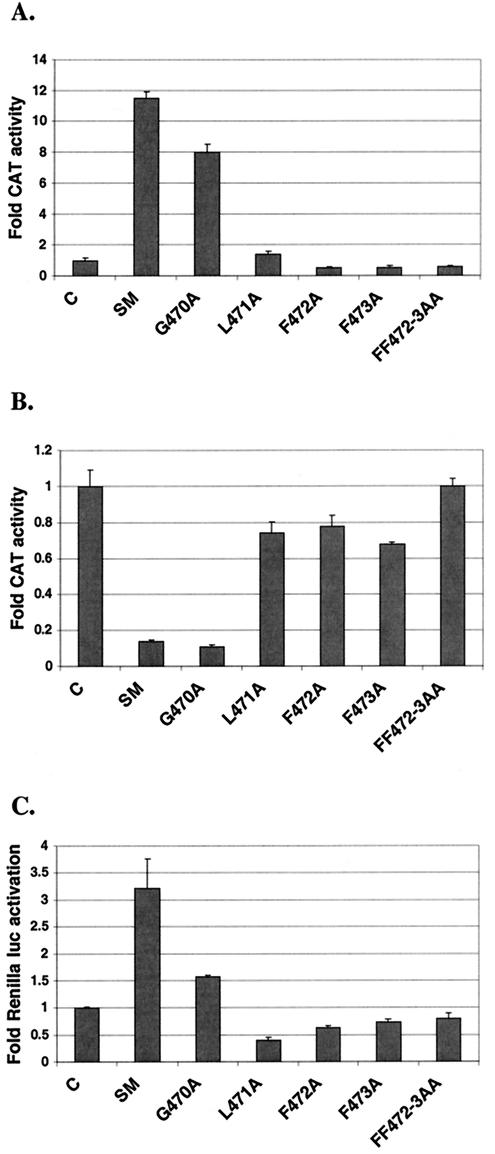
The amino acids GLFF are extremely highly conserved and are found in SM homologs in Kaposi's sarcoma-associated virus and herpesvirus saimiri as well as in bovine herpesvirus 4 (Fig. 1A). These amino acids have been determined to be essential for the activation but not the repression function of the herpesvirus saimiri ORF57 protein (12). Alanine substitution of any of these four amino acids in herpesvirus saimiri ORF57 was sufficient to reduce activation by 70 to 80% but did not affect repression. We therefore wished to determine whether the GLFF amino acids played a similar role in SM function.
Alanine substitutions of each of these four amino acids or of both phenylalanines simultaneously were generated by PCR, and the resulting mutant SM genes were sequenced. Each mutant was compared to wild-type SM for its ability to activate intronless reporter gene expression and inhibit intron-containing reporter CAT gene expression. Mutation of L471, F472, F473, or both F472 and F473 all led to complete loss of activation and repression functions (Fig. 3). Mutant G470A, however, maintained both activation and repression functions, although activation by G470A was reduced to approximately 75% of wild-type SM levels. These data are therefore distinctly different from those reported for herpesvirus saimiri ORF57. In herpesvirus saimiri ORF57, mutation of any of the amino acids in the GLFF motif led to loss of the activation but not the inhibition function. In EBV SM, however, mutation of any of the critical amino acids L471, F472, or F473 led to loss of both activation and inhibition rather than selective loss of activation. Moreover, the glycine residue in the GLFF motif appeared to be nonessential for activation by EBV SM.
FIG. 3.
Effect of mutating the GLFF motif on activation and repression. Each SM mutant was tested for its ability to activate (A) or inhibit (B) reporter CAT gene expression in BJAB cells. Transfections were also performed with an intronless Renilla luciferase reporter plasmid and each SM mutant (C).
In addition to exerting opposite effects on intronless genes versus those containing introns, SM also displays target gene-dependent differences in activation function (29). Therefore, in order to confirm the results obtained with the mutants described above, we repeated cotransfection experiments with another reporter gene which is responsive to SM, Renilla luciferase. As shown in Fig. 3C, the results were consistent with those obtained with a CAT reporter gene. Mutants altered in each of the amino acids found to be essential for CAT activation, L471, F472, and F473, were also unable to activate Renilla luciferase expression, whereas G470A remained active, although at a reduced level compared to wild-type SM.
Intracellular localization of SM mutants.
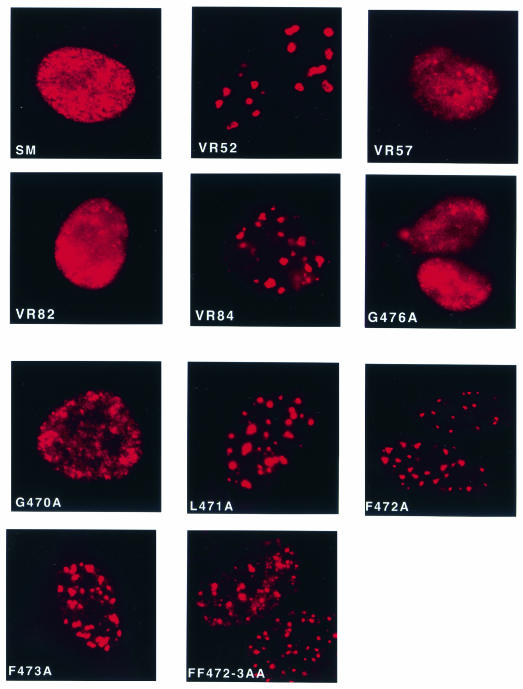
Since mutation of several of the carboxy-terminal amino acids led to loss of both activation and inhibition functions, it appeared likely that the structure of the protein had been altered, globally affecting function. It was therefore of interest to determine whether the intracellular location of these mutants was affected. Visualization of the mutant proteins would also allow verification that each mutant protein was expressed stably in amounts similar to wild-type SM. Each mutant was transfected into HeLa cells, in which wild-type SM localizes in diffuse nuclear speckles, sparing the nucleoli (Fig. 4). Each nonfunctional mutant (VR52, VR84, L471A, F472A, F473A, and FF472/473AA) had an abnormal intranuclear distribution, whereas each functional mutant (VR82, VR57, G476A, and G470A) had a distribution similar to that of wild-type SM (Fig. 4). All nonfunctional mutants formed distinct foci in the nucleus and exhibited little of the normal diffuse nucleoplasmic and speckled distribution. These data therefore suggested that the carboxy-terminal mutations resulted in global perturbations of structure that altered protein conformation and therefore affected SM activity.
FIG. 4.
Nuclear localization of wild-type SM and carboxy-terminal mutant SM proteins. HeLa cells were transfected with each plasmid, fixed 48 h after transfection, stained with anti-SM antibodies and tetramethylrhodamine isothiocyanate-conjugated goat anti-mouse immunoglobulin antibodies, and visualized by immunofluorescence microscopy. The transfected plasmid is shown in each panel.
Functional role of the hydrophobic amino acids in the GLFF motif.
Examination of the predicted structure and amino acid composition of the carboxy terminus of SM revealed a highly hydrophobic region centered around the GLFF motif and flanked by coiled regions. The predicted secondary structure suggests that the GLFFV residues are likely to assume an extended strand or sheet conformation flanked by loops on either side (Fig. 5A) (28). As might be expected, this hydrophobic region is predicted to be relatively solvent inaccessible as opposed to the flanking regions. Each mutation which resulted in a nonfunctional protein and abnormal nuclear localization is also predicted to alter this arrangement of hydrophobic residues flanked by two loop regions. In particular, substitution of alanine for F472 or F473 has the potential of introducing a helical turn into the internal extended strand region.
FIG. 5.
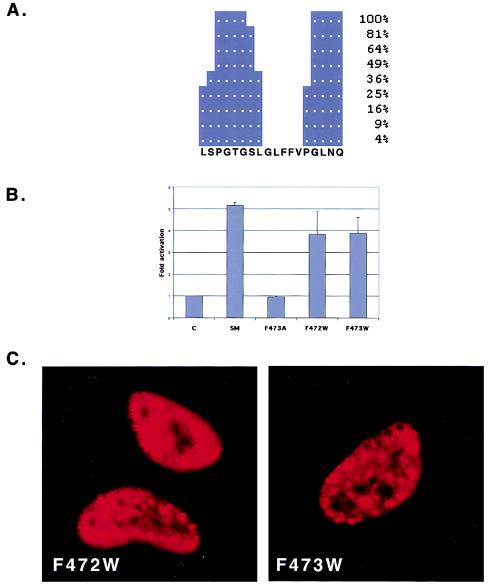
Effect of tryptophan substitution for phenylalanine in the GLFF motif. (A) Secondary structure of the carboxy-terminal region containing the conserved GLFF motif. The predicted relative solvent accessibility of each amino acid is shown as a percentage on the right. (B) The activation function of each mutant was compared to that of wild-type SM in CAT cotransfection assays performed in BJAB cells. (C) Intranuclear localization of W472F and W473F in transfected HeLa cells. Cells were fixed and immunostained for SM as in Fig. 4.
Since it appeared likely that the deleterious effect of these mutations on SM function was structural, we reasoned that substitution of another amino acid for F472 and F473 with less likelihood of disrupting the secondary structure in this region would result in preservation of function and normal intracellular localization. We predicted that substitution of phenylalanine with tryptophan would result in a structure more closely resembling that of wild-type SM. We therefore generated SM mutants in which tryptophan was substituted for each of the two relevant phenylalanine residues. These mutants (F472W and F473W) were then used in activation assays. Both mutants were functional, albeit slightly less so than wild-type SM (Fig. 5B). Examination of the intracellular localization of F472W and F473W by immunofluorescence demonstrated that both mutants, unlike F472A and F473A, were found diffusely throughout the nucleus (Fig. 5C). However, the distribution of both mutants was not completely normal, as large intracellular foci were also observed, suggesting that the mutations cause subtle alterations in structure and function.
Association of SM with the detergent-insoluble nuclear matrix fraction.
We have previously reported that a large proportion of SM is tightly associated with nuclear matrix components and is resistant to solubilization with nonionic detergents (3). Since mutation of amino acids in the carboxy terminus appeared to affect SM function primarily by altering structure, we wished to determine whether loss of function also correlated with a change in detergent solubility. We therefore solubilized cells transfected with either wild-type SM or one of the mutants with lysis buffer containing 1% NP-40, which we had previously determined to be effective in solubilizing SM without mechanical disruption of the nuclei (3). Such treatment in isotonic buffer results in solubilization of approximately 50% of the total SM present in the cell.
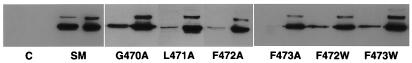
The detergent-soluble and -insoluble fractions, the latter containing nuclear matrix and envelope fractions, were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). As shown in Fig. 6, all the nonfunctional mutants were almost completely insoluble and remained in the nuclear fraction. However, their stability was not impaired, as the total amounts of the various mutant SM proteins were not significantly different from that of wild-type SM. The functional mutants G470A, W472F, and W473F were somewhat more soluble than the nonfunctional mutants. However, all three of these mutants, which retained function and relatively normal intranuclear localization, were nevertheless significantly less soluble than wild-type SM. These data confirmed that mutation of the carboxy terminus of SM results in alterations in structure that markedly affect their physical properties. However, although tight association with the nuclear fraction and detergent insolubility correlated with functional impairment, some mutants retained function despite being relatively insoluble by detergent.
FIG. 6.
Detergent solubility of wild-type SM and carboxy-terminal mutants. Each mutant was transfected into HeLa cells, and transfected cells were harvested 48 h later. Cells were lysed, and detergent-soluble and -insoluble fractions were separated by centrifugation and analyzed by SDS-PAGE and immunoblotting with anti-SM antibodies.
Conserved histidine/cysteine motif is important for both activation and repression functions.
All the SM homologs among human herpesviruses as well as some primate, bovine, equine, and alcelaphine herpesviruses exhibit conservation of an HX3CX4C motif. Mutation of any of these individual amino acids in herpesvirus saimiri ORF57 led to loss of both activation and repression functions (12). UL69, the human CMV homolog of SM, is thought to act by a transcriptional rather than posttranscriptional mechanism and does not repress intron-containing genes (40). The first cysteine in UL69, the counterpart of C449 in EBV SM, has been shown to be required for activation and for interaction with hSPT6, a chromatin-regulatory protein (39). We therefore wished to determine whether this conserved motif was also essential for SM function.
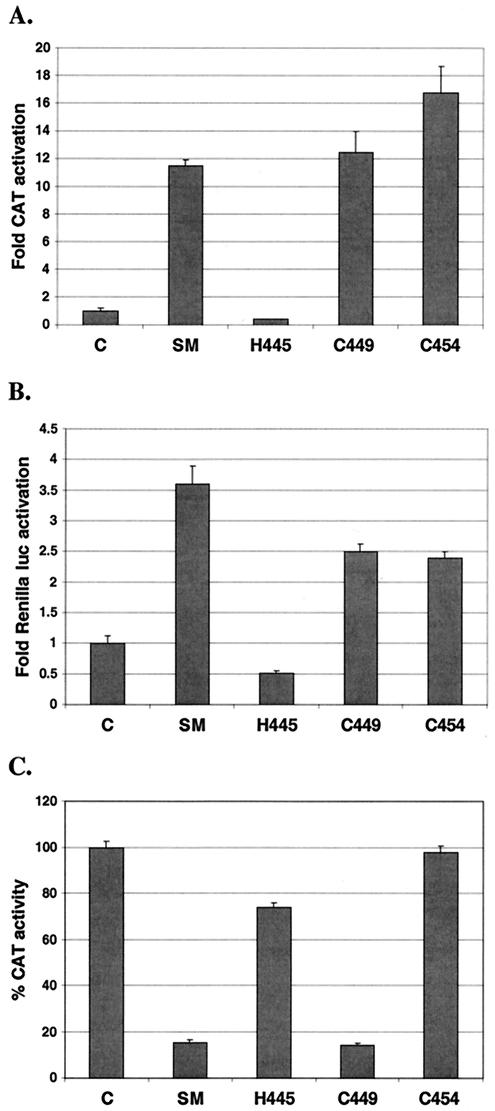
SM mutants were generated in which each of the three amino acids was replaced by PCR-based mutagenesis. The histidine at position 445 was replaced with phenylalanine and the cysteines at positions 449 and 454 were replaced with serine in an effort to minimize overall structural changes which could affect protein conformation. These substitutions are the same as those that were made in the previous study on herpesvirus saimiri ORF57 referred to above (12). The ability of each mutant to activate gene expression was then tested with CAT and Renilla luciferase as reporter genes (Fig. 7A, B). The repression function was also tested with an intron-containing CAT reporter gene construct (Fig. 7C). As shown in Fig. 7, mutation of H445 led to loss of both activation and repression. Conversely, C449S retained both activation and repression functions. In contrast, C454S retained activating function but lost repression function. The properties of the C454 mutant therefore demonstrate that the activation and inhibition functions of SM are distinct and separable.
FIG. 7.
Role of the conserved histidine/cysteine motif in activation and repression by SM. (A) Activation of CAT expression by each mutant was compared to that by wild-type SM in reporter cotransfection assays in BJAB cells. (B) Activation of Renilla luciferase by each mutant in BJAB cells. (C) Repression of intron-containing CAT gene by each mutant in BJAB cells.
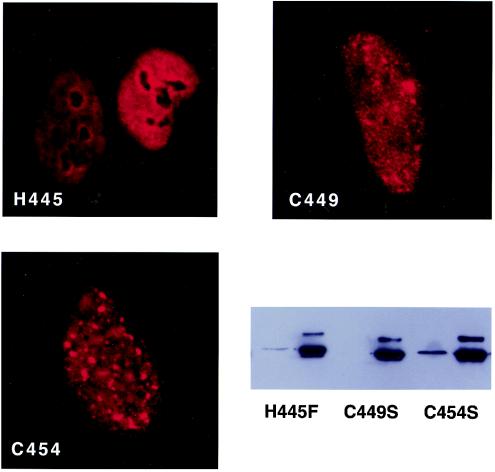
These results are also different from those reported for herpesvirus saimiri ORF57 and human CMV UL69 in that the cysteines do not play an essential role in activation by EBV SM in vitro. Furthermore, it appeared that the second cysteine, C454, may play an essential role in inhibiting but not activating target gene expression. In order to determine if these effects were perhaps also due to conformational changes, we examined the intracellular localization of each mutant in transfected cells. The nuclear staining pattern of H445F was diffuse and finely speckled. However, there was an unusual and distinctive increased accumulation around the nucleoli of the H445F mutant, which was clearly visible and led to an accentuation of the nucleoli (Fig. 8). Both C449S and C454S exhibited relatively normal intranuclear distribution, albeit with some slightly larger aggregates than are observed with wild-type SM. These results also suggested that the loss of function of H445F and selective loss of repression of C454S were not due to gross alterations in structure but perhaps due to disruption of essential protein-protein interactions.
FIG. 8.
Intranuclear localization and detergent solubility of histidine and cysteine mutants. The intranuclear localization of each mutant in HeLa cells was visualized by immunofluorescence microscopy. Each mutant was also transfected into HeLa cells and harvested 48 h after transfection, and detergent-soluble and -insoluble fractions were analyzed by immunoblotting (bottom right panel).
In order to determine whether the alterations in intranuclear distribution and function of the histidine and cysteine mutants correlated with their solubility, we examined the fraction of each mutant protein that was extractable by detergent treatment (Fig. 8). Unlike what was observed with the mutations in the GLFF motif, there was no clear correlation of function with solubility. Although the nonfunctional H445F mutant was relatively insoluble, both of the activation-competent mutants C449S and C454 were also poorly soluble compared to wild-type SM. These data therefore suggest that detergent solubility of SM from the nuclear matrix may reflect not only overall solubility but also functional interactions with other intranuclear proteins and structures.
SM inhibits splicing and the C454S mutant is defective in splicing inhibition.
It has been demonstrated previously that SM inhibits expression of target genes that contain synthetic introns as well as genomic intron-containing genes (31). Such repression by SM has been demonstrated to occur at the posttranscriptional level, since SM does not affect the rates of transcript initiation but does prevent the accumulation of spliced mRNA. ICP27 also inhibits intron-containing gene expression and has been shown to globally inhibit host cell splicing (17, 34). Whether SM directly affects host cell splicing has not been established. Although regions of ICP27 required for repression and activation have been mapped, these appear to overlap, and point mutations that discretely affect transrepression have not been identified (16, 33, 37) (18, 24). It was therefore of interest to determine whether the C454S mutation affected the normally observed SM-mediated decrease in steady-state levels of mRNA transcribed from genes with introns.
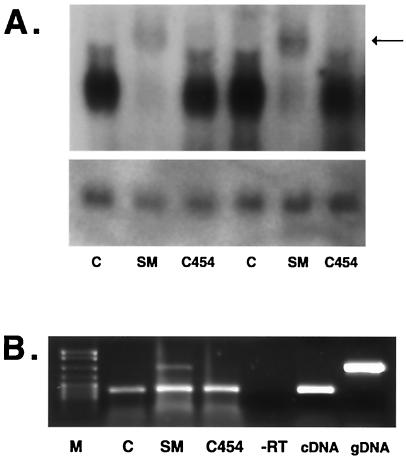
We have previously demonstrated that expression of the hGH gene, which contains four introns, is dramatically inhibited by cotransfection of SM in B lymphocytes (31). Cotransfection of SM with plasmid pXGH5, which encodes the genomic hGH gene, leads to reduction of secreted hGH to undetectable levels. hGH mRNA also fails to accumulate when SM is coexpressed. We have previously demonstrated by nuclear run-on transcription assays that SM does not affect the rate of hGH transcript initiation (31), indicating that inhibition of hGH expression by SM is posttranscriptional. In order to evaluate the effect of C454S on hGH mRNA levels, BJAB cells were transfected with pXGH5 and either a control plasmid, SM, or C454S. Northern blotting of RNA from these cells indicated that whereas hGH mRNA failed to accumulate in the presence of SM, it did so in the presence of the C454S mutant (Fig. 9A). Close examination of the Northern blot results suggested that small amounts of unspliced or partially spliced forms of hGH pre-mRNA accumulated in the presence of SM, suggesting that wild-type SM inhibited splicing and that the C454S mutant was deficient in this function, thereby allowing greater expression of the spliced hGH gene.
FIG. 9.
Effect of SM on splicing of a gene with multiple introns and role of C454 in splicing inhibition. (A) The effect of wild-type SM, C454S, and control plasmid on a cotransfected hGH gene was analyzed by Northern blotting. BJAB cells were transfected with each plasmid, and RNA was harvested 40 h after transfection. RNA was electrophoresed, blotted, and probed with 32P-labeled hGH cDNA. The arrow marks the position of higher-molecular-weight hGH RNA seen in the presence of SM. Blots were stripped and reprobed with a glyceraldehyde-3-phosphate dehydrogenase probe as a control for loading (bottom panel). (B) The effect of each plasmid on splicing was assessed by reverse transcription-PCR. Equal amounts of total RNA from each transfection were reverse transcribed and PCR amplified with hGH primers. Lanes contained size markers (M), vector plasmid (C), wild-type SM (SM), C454S mutant plasmid (C454), RNA from transfected cells PCR amplified without reverse transcription (−RT), PCR amplification of hGH cDNA (cDNA), and genomic DNA plasmid (gDNA).
To determine if C454 was defective in splicing inhibition, we analyzed splicing of the hGH message in these RNA samples by reverse transcription-PCR. Primers which flanked intron 2 that would yield a 208-bp product from the spliced mRNA and a 393-bp product from the unspliced precursor mRNA were chosen. As shown in Fig. 9B, the shorter PCR product representing spliced hGH mRNA was detectable by reverse transcription-PCR in RNA from all three samples. However, an amplification product corresponding to unspliced hGH mRNA was only detectable with RNA from SM-transfected cells but not with RNA from control plasmid- or C454S-transfected cells. Since spliced hGH mRNA from wild-type SM-transfected cells was not detected by Northern blotting, the reverse transcription-PCR is more sensitive for the spliced product. The latter assay (for the spliced product) is not in the linear range and is therefore not quantitative for the spliced product. However, unspliced product was detectable in wild-type SM- but not C454S-transfected cells. These data therefore indicate that repression of intron-containing genes by SM may be due to splicing inhibition and that defective repression by C454S is due to a failure to inhibit splicing.
Role of specific amino acids in SM function required for EBV virion production.
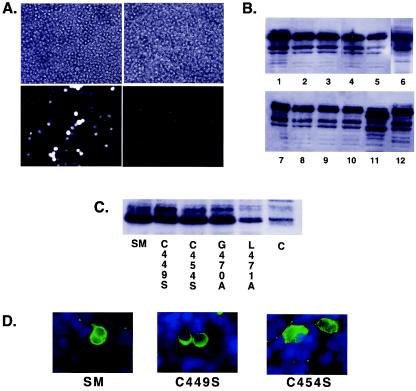
It has been demonstrated that in human 293 cells carrying mutant SM-defective EBV genomes generated by transfection of EBV DNA, transfection of wild-type SM is required to produce and release infectious EBV particles (13). By transfection of a mutant SM plasmid, this system was also used to demonstrate that a region of SM containing RXP amino acid repeats is dispensable for infectious EBV production (13). We therefore used 293 cells carrying SM-defective EBV (BMLF1-KO EBV) to ascertain the effect on EBV production of the mutations that we had introduced into SM. Each mutant SM plasmid was transfected into 293 cells carrying BMLF1-KO EBV (293-BMLF1-KO) along with a plasmid expressing the immediate-early EBV Z transactivator (BZLF1, Zta). The supernatant from transfected cells was harvested, filtered, and used to infect Raji cells as previously described (11). Infected Raji cells, which express recombinant EBV-encoded green fluorescent protein, were detected 72 h after infection by fluorescence microscopy and quantitated by flow cytometry.
293-BMLF1-KO cells transfected with a Z expression vector alone did not yield any green Raji cells, as expected. However, the combination of wild-type SM and Z yielded easily detectable green fluorescent Raji cells by 48 to 72 h (Fig. 10A), as has been reported previously (13). The virus yield measured by the number of fluorescent Raji cells was between 6 × 104 and 1 × 105 per ml of virus supernatant. The G470A mutant, which was functional in vitro, supported EBV production from 293-BMLF1-KO cells indistinguishably from wild-type SM. As might be expected, none of the most structurally and functionally abnormal mutants (L471A, F472A, F473A, and FF472/473AA) were capable of rescuing BMLF1-KO virus production. Also as expected, H445F, which was nonfunctional in vitro, was defective in its ability to rescue BMLF1-KO virus. However, C449S, C454S, F472W, and F473W, which were all functional in activation in vitro, were unable to complement BMLF1-KO EBV.
FIG. 10.
Complementation of SM-deleted EBV by SM transfection in 293 cells. (A) Fluorescence (bottom) and phase-contrast micrographs (top) of Raji cells infected with supernatant from 293-SMKO cells that had been transfected with either Z expression vector alone (right panels) or SM and Z (left panels). (B) Expression of SM and mutant protein in 293-BMLF1-KO cells was confirmed by immunoblotting of lysates of the transfected cells. Lanes: 1, wild-type SM; 2, C449S; 3, C454S; 4, F472W; 5, F472A; 6, H445F; 7, G470A; 8, L471A; 9, F473A; 10, F473W; 11, ΔRXP; 12, wild-type SM. (C) Activation of EA-D expression by SM and SM mutants in 293-BMLF1-KO cells was measured by immunoblotting lysates from cells transfected with each SM plasmid with monoclonal antibodies specific for EA-D. The transfected SM mutant is shown below each lane. Lysate from cells transfected with only Z is shown in the last lane as a control for EA-D expression in the absence of SM (C). (D) Expression of gp110 in 293-BMLF1-KO cells was assessed by immunofluorescence microscopy. 293-BMLF1-KO cells were transfected with the Z expression vector and either SM, C449S, or C454S plasmid, fixed 48 h after transfection, and stained with anti-gp110 antibody.
Although C449S and C454S were similar to wild-type SM in their ability to activate reporter genes in cotransfection assays, it was possible that the mutants were relatively deficient in their ability to activate EBV gene expression in BMLF1KO 293 cells. Expression of the early lytic BMRF1 gene product EA-D, which is essential for lytic EBV replication, is strongly activated by SM (31, 36). We therefore measured EA-D expression in BMLF1KO 293 cells transfected with each of these mutants by immunoblotting with a monoclonal antibody specific for the EA-D gene product. As shown in Fig. 10C, the level of expression of EA-D was similar in the presence of either wild-type SM, C449S, or C454S, indicating that the inability of these mutants to support virus production could not be explained by a quantitative reduction in their activation efficiency. As positive and negative controls, we also examined the ability of the G470A and L471A mutants, respectively, to transactivate EA-D expression. G470A, which is not impaired in transactivation assays in vitro, transactivated EA-D expression, whereas L471A, which is inactive in vitro, was also unable to transactivate EA-D expression.
It remained possible that the cysteine mutants were specifically deficient in the ability to activate late lytic EBV gene expression, which would preclude infectious EBV production. In order to investigate this possibility, we performed a similar transfection of BMLF1KO 293 cells with the Z expression vector and either wild-type SM, C449S, or C454S and assessed gp110 expression by immunofluorescence microscopy of the transfected cells. As shown in Fig. 10D, gp110 expression was detectable in the mutant-transfected cells. The percentage of cells expressing gp110 ranged between 4 and 5% of cells at 48 h after transfection in both wild-type SM- and mutant-transfected cells in three independent experiments. These findings, particularly the inability of C449S (which had relatively normal intranuclear distribution and transactivating and transrepressing function) to support virus production, were unexpected and suggested that these conserved amino acids may play a role in essential SM functions besides their ability to transactivate lytic gene expression or inhibit splicing (see Discussion).
DISCUSSION
In this study we identified structurally and functionally critical amino acids in the EBV gene-regulatory protein SM. Initial experiments were directed towards mapping the limits of the essential domains in the carboxy terminus of SM because this portion of the gene is the most highly conserved among the herpesvirus homologs of SM. Progressive deletion of the carboxy terminus revealed that while the last three amino acids (477 to 479) were dispensable for function, further truncation of SM beyond these amino acids led to complete loss of activity. Substitution of the G476 residue with alanine did not cause loss of function, suggesting that the exact amino acid composition is not critical but that this terminal portion of the SM molecule may play a role in maintaining a functional structure for the whole protein. This conclusion is supported by the intracellular localization of the nonfunctional mutants, which accumulated in several large nuclear aggregates when visualized by immunofluorescence microscopy, whereas the G476A mutant localized normally.
The GLFF amino acid motif found immediately upstream of G476 is highly conserved among gammaherpesviruses. Each of these four residues has been shown to be essential for activation but not inhibition by herpesvirus saimiri ORF57 in vitro, although nuclear localization appeared to be unaffected (12). In the EBV SM protein, however, mutation of G470 was tolerated, and both repression and activation functions were maintained, as was the normal diffuse, speckled SM nuclear distribution (see Table 1 for a summary of the properties of all mutants). Conversely, the other three residues were required for both functions, and mutation led to abnormal nuclear distribution. The detergent solubility of these mutants correlated with the immunofluorescence data, with the inactive and abnormally localized mutants being the least soluble.
TABLE 1.
In vitro activity, solubility, nuclear localization, and virion production of SM mutants
| SM | Activationa | Repressionb | Solubilityc | Nuclear localizationd | Virion productione |
|---|---|---|---|---|---|
| Wild type | ++++ | ++++ | ++++ | N | + |
| H445 | − | − | +/− | A | − |
| C449 | +++ | ++++ | +/− | A | − |
| C454 | +++ | − | + | A | − |
| G470A | +++ | ++++ | +++ | N | + |
| L471A | − | − | + | A | − |
| F472A | − | − | + | A | − |
| F473A | − | − | +/− | A | − |
| FF472473AA | − | − | + | A | ND |
| F472W | +++ | ND | ++ | A | − |
| F473W | +++ | ND | ++ | A | − |
Activation of cotransfected reporter genes in vitro.
Repression of cotransfected reporter genes in vitro. ND, not done.
Relative detergent solubility of each mutant protein compared by SDS-PAGE.
Assessed by immunofluorescence microscopy. A, abnormal; N, normal.
Measured by the abillity of each transfected mutant to rescue recombinant SM-KO virus from 293 cells.
In order to test the hypothesis that mutations which would be less disruptive of the secondary structure of this region would also be functional, we replaced the individual phenylalanine residues with tryptophan and assessed the function, nuclear localization, and detergent solubility of these mutant proteins. The tryptophan-substituted SM mutants more closely resembled wild-type SM in all three respects. Therefore, it is likely that the LFF amino acids, which are predicted to form a hydrophobic pocket in this region, also play a structurally important role. These data indicate that although the GLFF motif is highly conserved among herpesvirus SM homologs, the same amino acids may play different structural and functional roles in different viruses.
The second set of mutants that were studied were altered in the conserved H and two C residues, which have been described as zinc-finger like. These residues have been shown to be required for both activation and inhibition functions in herpesvirus saimiri ORF57 (12). The corresponding first cysteine in CMV UL69 is essential for the activation function (39). Our experiments showed that H445 was essential for both activation and repression by SM. The nuclear distribution of the H445 mutant protein was not grossly abnormal, as was the case with the nonfunctional GLFF mutants. The H445F protein, however, displayed an enhanced perinucleolar distribution compared to wild-type SM, suggesting that mutation of H445 may alter intranuclear trafficking or interactions with other nuclear proteins.
Surprisingly, both cysteines were dispensable for activation in vitro. The nuclear distribution of both of the proteins with mutant cysteines was relatively normal, but their detergent solubility was markedly less than that of wild-type SM, suggesting that both mutations exert subtle effects on structure or protein-protein interactions.
We demonstrate here for the first time that SM inhibits expression of an intron-containing gene by inhibiting splicing. It is likely that C454 is involved in this function since it was found to be specifically important for repression of gene expression by SM and the C454S mutant did not inhibit splicing of an intron-containing reporter construct. This is the first demonstration that the inhibitory and activating properties of SM are separable. These data suggest that SM may interact with and inhibit components of the splicing machinery. Identification of C454 as potentially important in this process should facilitate the identification of cellular proteins involved in inhibition by SM.
We expected in general that, in vitro, the activation function of the SM mutants would correlate with their ability to support EBV virion production. We were also particularly interested in determining whether the gene repression function of SM was required for virus production, as it could potentially play a role in shutting off host gene expression. Surprisingly, however, several mutants which appeared to have relatively preserved function in vitro were essentially incapable of supporting virus production although they were expressed in amounts comparably to wild-type SM in 293 cells, in which the assay for virus production was performed. In addition, activation of the BMRF1 gene product EA-D in BMLF1-KO 293 cells by these mutants was similar in magnitude.
There are several possible explanations for these results. First, although these mutants appeared to activate EBV lytic gene expression in 293 cells, there may be subtle differences in the kinetics of gene expression between the mutants and wild-type SM that are important for virus production. In addition, the complementation experiments were performed in 293 cells, which are currently the most tractable system to perform studies such as these for virion production. Nevertheless, the differences in cellular milieu between transformed epithelial cells such as 293 cells and naturally EBV-infected B cells or epithelial cells may accentuate subtle deficiencies in the function of SM mutants. There may also be a relative deficiency in the ability of the mutants to activate specific EBV genes, leading to an inability to support virus production because of a limiting amount of one or more critical EBV proteins. We consider this less likely because the activity of the various mutants seems to be generally consistent among the few genes we have tested as targets, but this possibility cannot be ruled out without microarrays representing all EBV genes essential for virion production. Finally, the mutations may affect the ability of SM to interact with various cellular proteins, thus affecting intranuclear trafficking and other functions that are not measured by activation assays alone. This possibility is suggested by the differences in intranuclear localization of the H445 mutant and the deficiency of splicing inhibition caused by mutation of C454.
These results open the door to the identification of cellular protein partners of SM that may be important in lytic virus production and may serve as targets for therapeutic intervention in EBV and other herpesvirus infections.
Acknowledgments
This work was supported by NIH grant CA 81133 (S. S.), CR-UK (H.-J.D.), and the Sonderforschungsbereich SFB455, Deutsche Forschungsgemeinschaft, and Bayerische Forschungsstiftung (W.H.).
REFERENCES
- 1.Bello, L. J., A. J. Davison, M. A. Glenn, A. Whitehouse, N. Rethmeier, T. F. Schulz, and J. B. Clements. 1999. The human herpesvirus-8 ORF 57 gene and its properties. J. Gen. Virol. 80:3207-3215. [DOI] [PubMed] [Google Scholar]
- 2.Boyer, J. L., S. Swaminathan, and S. J. Silverstein. 2002. The Epstein-Barr virus SM protein is functionally similar to ICP27 from herpes simplex virus in viral infections. J. Virol. 76:9420-9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle, S. M., V. Ruvolo, A. K. Gupta, and S. Swaminathan. 1999. Association with the cellular export receptor CRM 1 mediates function and intracellular localization of Epstein-Barr virus SM protein, a regulator of gene expression. J. Virol. 73:6872-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buisson, M., F. Hans, I. Kusters, N. Duran, and A. Sergeant. 1999. The C-terminal region but not the Arg-X-Pro repeat of Epstein-Barr virus protein EB2 is required for its effect on RNA splicing and transport. J. Virol. 73:4090-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chee, M., and B. Barrell. 1990. Herpesviruses: a study of parts. Trends Genet. 6:86-91. [DOI] [PubMed] [Google Scholar]
- 6.Davison, A. J., and P. Taylor. 1987. Genetic relations between varicella-zoster virus and Epstein-Barr virus. J. Gen. Virol. 68:1067-1079. [DOI] [PubMed] [Google Scholar]
- 7.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeNoto, F. M., D. D. Moore, and H. M. Goodman. 1981. Hum. growth hormone DNA sequence and mRNA structure: possible alternative splicing. Nucleic Acids Res. 9:3719-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellison, K. S., S. A. Rice, R. Verity, and J. R. Smiley. 2000. Processing of alpha-globin and ICP0 mRNA in cells infected with herpes simplex virus type 1 ICP27 mutants. J. Virol. 74:7307-7319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farjot, G., M. Buisson, M. Duc Dodon, L. Gazzolo, A. Sergeant, and I. Mikaelian. 2000. Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1-independent pathway. J. Virol. 74:6068-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the cooperative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodwin, D. J., K. T. Hall, M. S. Giles, M. A. Calderwood, A. F. Markham, and A. Whitehouse. 2000. The carboxy terminus of the herpesvirus saimiri ORF 57 gene contains domains that are required for transactivation and transrepression. J. Gen. Virol. 81:2253-2265. [DOI] [PubMed] [Google Scholar]
- 13.Gruffat, H., J. Batisse, D. Pich, B. Neuhierl, E. Manet, W. Hammerschmidt, and A. Sergeant. 2002. Epstein-Barr virus mRNA export factor EB2 is essential for production of infectious virus. J. Virol. 76:9635-9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta, A. K., V. Ruvolo, C. Patterson, and S. Swaminathan. 2000. The human herpesvirus 8 homolog of Epstein-Barr virus SM protein (KS-SM) is a posttranscriptional activator of gene expression. J. Virol. 74:1038-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hardwicke, M. A., and R. M. Sandri-Goldin. 1994. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J. Virol. 68:4797-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardwicke, M. A., P. J. Vaughan, R. E. Sekulovich, R. O'Conner, and R. M. Sandri-Goldin. 1989. The regions important for the activator and repressor functions of herpes simplex virus type 1 alpha protein ICP27 map to the C-terminal half of the molecule. J. Virol. 63:4590-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardy, W. R., and R. M. Sandri-Goldin. 1994. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J. Virol. 68:7790-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hibbard, M. K., and R. M. Sandri-Goldin. 1995. Arginine-rich regions succeeding the nuclear localization region of the Herpes Simplex Virus Type 1 regulatory protein ICP27 are required for efficient nuclear localization and late gene expression. J. Virol. 69:4656-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inchauspe, G., S. Nagpal, and J. M. Ostrove. 1989. Mapping of two varicella-zoster virus-encoded genes that activate the expression of viral early and late genes. Virology 173:700-709. [DOI] [PubMed] [Google Scholar]
- 20.Kenney, S., J. Kamine, E. Holley-Guthrie, E. C. Mar, J. C. Lin, D. Markovitz, and J. Pagano. 1989. The Epstein-Barr virus immediate-early gene product BMLF1 acts in trans by a posttranscriptional mechanism which is reporter gene dependent. J. Virol. 63:3870-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kenney, S., J. Kamine, D. Markovitz, R. Fenrick, and J. Pagano. 1988. An Epstein-Barr virus immediate-early gene product trans-activates gene expression from the human immunodeficiency virus long terminal repeat. Proc. Natl. Acad. Sci. 85:1652-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirshner, J. R., D. M. Lukac, J. Chang, and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markovitz, D. M., S. Kenney, J. Kamine, M. S. Smith, M. Davis, and E.-S. Huang. 1989. Disparate effects of two herpesviruses immediate-early gene trans-activators on the HIV LTR. Virology 173:750-754. [DOI] [PubMed] [Google Scholar]
- 24.McCarthy, A. M., L. McMahan, and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J. Virol. 63:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mears, E., and S. A. Rice. 1996. The RGG Box Motif of the Herpes Simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 70:7445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mears, W. E., V. Lam, and S. A. Rice. 1995. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J. Virol. 69:935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menezes, J., W. Liebold, G. Klein, and G. Clements. 1975. Establishment and characterization of an Epstein-Barr virus (EBV)-negative lymphoblastoid B cell line (BJA-B) from an exceptional EBV-genome-negative African Burkitt's lymphoma. Biomedicine 22:276-284. [PubMed] [Google Scholar]
- 28.Rost, B., P. Fariselli, and R. Casadio. 1996. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 5:1704-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruvolo, V., A. K. Gupta, and S. Swaminathan. 2001. Epstein-Barr virus SM protein interacts with mRNA in vivo and mediates a gene-specific increase in cytoplasmic mRNA. J. Virol. 75:6033-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruvolo, V., L. Navarro, C. E. Sample, M. David, S. Sung, and S. Swaminathan. 2003. The Epstein-Barr virus SM protein induces STAT1 and interferon-stimulated gene expression. J. Virol. 77:3690-3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruvolo, V., E. Wang, S. Boyle, and S. Swaminathan. 1998. The Epstein-Barr virus nuclear protein SM is both a posttranscriptional inhibitor and activator of gene expression. Proc. Natl. Acad. Sci. 95:8852-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandri-Goldin, R. M. 1998. ICP27 mediates herpes simplex virus RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandri-Goldin, R. M., M. K. Hibbard, and M. A. Hardwicke. 1995. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J. Virol. 69:6063-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekulovich, R. E., K. Leary, and R. M. Sandri-Goldin. 1988. The herpes simplex virus type 1 alpha protein ICP27 can act as a trans-repressor or a trans-activator in combination with ICP4 and ICP0. J. Virol. 62:4510-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selden, R. F., K. B. Howie, M. E. Rowe, H. G. Goodman, and D. D. Moore. 1986. Human growth hormone as a reporter gene in regulation studies employing transient gene expression. Mol. Cell. Biol. 6:3173-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Semmes, O. J., L. Chen, R. T. Sarisky, Z. Gao, L. Zhong, and S. D. Hayward. 1998. Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein-Barr virus replication gene mRNA. J. Virol. 72:9526-9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, I. L., R. E. Sekulovich, M. A. Hardwicke, and R. M. Sandri-Goldin. 1991. Mutations in the activation region of herpes simplex virus regulatory protein ICP27 can be trans dominant. J. Virol. 65:3656-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitehouse, A., M. Cooper, and D. M. Meredith. 1998. The immediate-early gene product encoded by open reading frame 57 of herpesvirus saimiri modulates gene expression at a posttranscriptional level. J. Virol. 72:857-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winkler, M., T. aus Dem Siepen, and T. Stamminger. 2000. Functional interaction between pleiotropic transactivator pUL69 of human cytomegalovirus and the human homolog of yeast chromatin regulatory protein SPT6. J. Virol. 74:8053-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winkler, M., S. A. Rice, and T. Stamminger. 1994. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J. Virol. 68:3943-3954. [DOI] [PMC free article] [PubMed] [Google Scholar]