Abstract
Dengue virus (DV) is a flavivirus and infects mammalian cells through mosquito vectors. This study investigates the roles of domain III of DV type 2 envelope protein (EIII) in DV binding to the host cell. Recombinant EIII interferes with DV infection to BHK21 and C6/36 cells by blocking dengue virion adsorption to these cells. Inhibition of EIII on BHK21 cells was broad with no serotype specificity; however, inhibition of EIII on C6/36 cells was relatively serotype specific. Soluble heparin completely blocks binding of EIII to BHK21 cells, suggesting that domain III binds mainly to cell surface heparan sulfates. This suggestion is supported by the observation that EIII binds very weakly to gro2C and sog9 mutant mammalian cell lines that lack heparan sulfate. In contrast, heparin does not block binding of EIII to mosquito cells. Furthermore, a synthetic peptide that includes amino acids (aa) 380 to 389 of EIII, IGVEPGQLKL, inhibits binding of EIII to C6/36 but not BHK21 cells. This peptide corresponds to a lateral loop region on domain III of E protein, indicating a possible role of this loop in binding to mosquito cells. In summary, these results suggest that EIII plays an important role in binding of DV type 2 to host cells. In addition, EIII interacts with heparan sulfates when binding to BHK21 cells, and a loop region containing aa 380 to 389 of EIII may participate in DV type 2 binding to C6/36 cells.
Dengue virus (DV) is an arthropod-borne human pathogen that causes a serious public health threat in tropical and subtropical regions of the world (58). The World Health Organization reports that there are approximately 500,000 cases of dengue fever per year and that the infection rate is approximately 50 million per year (see reference 15 and literature cited therein). DV has four serotypes (DEN-1 to DEN-4) that cause diseases ranging from mild dengue fever to severe symptoms such as dengue hemorrhagic fever and dengue shock syndrome (17, 25, 27, 70).
The dengue viral genome is a single-stranded, positive-strand RNA with genome organization similar to those of other flaviviruses (47). DV infects a broad range of mammalian cell lines from several species in vitro but is transmitted to humans in vivo by mosquito vectors such as Aedes aegypti and Aedes albopictus (7). Primary human cells such as peripheral blood leukocytes, blood monocytes/macrophages, dendritic cells, and B lymphocytes could also be infected by DV (9, 12, 13, 28, 32, 33, 46, 53, 55, 75, 76, 82).
Previous studies indicate that cell surface heparan sulfates (HS) are involved in attachment of DV to mammalian cells including Vero, CHO, and human hepatoma cells (11, 23, 31, 35, 44, 54). HS are repeating disaccharides composed of uronic acid or l-iduronic acid and a derivative of glucosamine that is variably O-sulfated (21). Extensive sulfate modification causes cell surface HS to be highly negatively charged. The biological roles of HS are quite diverse, including cell attachment and migration, compressive resilience of cartilage, control of fibrinogenesis, cell signaling, and virus infection (3). Many pathogenic microorganisms, including viruses, gram-positive and gram-negative bacteria, and parasites, attach to HS during entry into host cells (14, 69, 74). Since HS is ubiquitously expressed on many cell types and is commonly used by other pathogens to gain access into cells, an additional coreceptor has been postulated to explain the limited cell tropism of DV. This coreceptor may be related to a trypsin-sensitive protein or protein complex that was shown to play a role in virion binding to mammalian cells (18, 52). Several candidate coreceptor proteins for DV have been suggested for many mammalian cell lines. These proteins are between 20 to 40 kDa and 60 to 90 kDa in size and bind to dengue virions in vitro (5, 31, 56, 59, 66). Recently, DC-SIGN, a dendritic cell surface lectin, was shown to mediate DV infection to primary dendritic cells (61, 81). Nevertheless, the molecular mechanism by which DV enters these cells remains poorly characterized.
Besides mammalian cells, mosquito cell lines that express cell surface proteins capable of binding to DV were also reported. In C6/36 cells, 40- to 45-kDa cell surface proteins were identified to bind DEN-4 (72, 83). Additional candidate coreceptors on C6/36 cells were reported to be 67 and 80 kDa (60). Up to now, no receptor has been identified on mosquito cells.
Flavivirus virions contain a major viral envelope (E) protein that has about 40% homology with different members of the family (7). The crystal structures of E protein of tick-borne encephalitis virus (TBEV) and, more recently, of DV revealed that E protein is composed of three domains (I, II, and III) that exhibit significant structural conservation (57, 68). Studies mainly with TBEV E protein indicate that E protein exists as dimers on the native virion and forms trimers at low pH to allow fusion of the viral envelope with membranes of the target cells (1, 30, 78). Recently, the virion structures of mature and immature DV particles were also obtained by cryoelectron microscopy (39, 84). Comparison of the virion structures of TBEV, DV, and alphavirus showed that these viruses are structurally similar and employ a unique fusion mechanism distinct from those of other enveloped viruses such as human immunodeficiency virus and herpes simplex virus (22, 41, 65, 67).
Mutations in the domain III region of flavivirus E protein (EIII) are associated with attenuated virulence or the ability of virus to escape immune neutralization, suggesting that EIII plays a role in receptor recognition (19, 26, 43, 49, 62, 71, 73). Furthermore, EIII of Langart virus binds to target Vero cells and blocked virus infection (4). Monoclonal antibodies against the EIII region strongly inhibit DV adsorption to Vero cells (16). Finally, the EIII region contains positive charged residues within amino acids (aa) 284 to 310 and 386 to 411 that constitute putative HS-binding sites in DV E protein (11). However, it is not known whether DV EIII directly binds to HS during virion attachment to host cells, and consequently, the role of EIII in DV infection remains elusive.
This study analyzes the role of EIII of DEN-2 in DV infection of mammalian BHK21 and mosquito C6/36 cells. Binding assays were performed with mammalian and mosquito cells, and the structural requirements for EIII for binding to each of the cell lines were investigated. The results suggest that EIII is important for DV entry into BHK21 and C6/36 cells and that different structural elements of EIII are required for DEN-2 entry into these two types of cells.
MATERIALS AND METHODS
Viruses, cells, and reagents.
DV type 1 (DEN-1) prototype strain Hawaii, DV type 2 (DEN-2) strains 16681 and PL046, DV type 3 (DEN-3) prototype strain H87, and DV type 4 (DEN-4) strain H241 were described previously (63, 77, 85). Local Taiwanese isolates of DVs of four serotypes were obtained from patients with dengue fever or dengue hemorrhagic fever and cultured in C6/36 cells with minimal passages (<3): 766733 (DEN-1), 766635 (DEN-2), 990360 (DEN-3), and 466088 (DEN-4) (45). Japanese encephalitis virus (JEV) (RP9) was obtained from Y.-L. Lin (10). Wild-type vaccinia virus (VV) of WR strain was described previously (14).
BHK21 cells were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). C6/36, a mosquito cell line established from Aedes albopictus, was maintained in DMEM-M&M supplemented with 10% FBS (36). L cells and gro2C and sog9 cells have been described previously and were obtained from F. Tufaro (2, 24).
Soluble heparin (HP), chondroitin sulfate (CS), and dermatan sulfate (DS) were purchased from Sigma, Inc. HiTrap chelating HP columns and an ECL protein biotinylation module were purchased from Amersham Pharmacia Biotech, Inc. Peptides were synthesized from United Biochemical Research, Inc. Chemicals for electrophoresis were purchased from Bio-Rad, Inc. Other reagents were obtained from Merck or Sigma, Inc. A monoclonal antibody (MAb) against T7 tag sequences was purchased from Novagen, Inc. and used at a dilution of 1:10,000. A rabbit serum recognizing purified dengue virions was generated in this study and used at a dilution of 1:1,000. A MAb (4G2), which was originally raised to recognize E protein of DEN-2, recognized E protein of other three serotypes and was obtained from H.-C. Wu (National Taiwan University) and used at 1:1,000 dilution.
Protein expression and purification.
An expression construct was prepared to express EIII of DEN-2 virus. A DNA fragment corresponding to EIII was PCR amplified with the following reverse transcription-PCR primers: 5′ primer, 5′-AAGGAATTCTCAAAGGAATGTCATAC-3′ (EcoRI site is underlined); 3′ primer, 5′-AGAAAGCTTTTTCTTAAACCAGTTGAG-3′ (HindIII site is underlined). The primers were hybridized to viral RNA purified from DEN-2 (PL046) virions and amplified for 25 cycles of 94°C for 1 min, 50°C for 1 min, and 72°C for 40 s. The amplified DNA was ligated into pCRII-TOPO plasmid (Invitrogen, Inc.), and the DNA sequences encoding EIII were determined. The predicted EIII amino acid sequences were identical to the sequences in DEN-2 16681 and New Guinea-C strains (37, 77). The DNA was digested with EcoRI and HindIII and cloned into pET21c (Novagen, Inc.). The resulting plasmid expresses EIII with a T7 tag at the N terminus and a hexahistidine tag at the C terminus for affinity purification.
The EIII expression plasmid was transformed into Escherichia coli strain BL21(DE3), and the bacterial culture was induced with 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 30 min at 30°C. The bacterial pellets were harvested, sonicated, and centrifuged. The supernatant was loaded onto a nickel column, and the bound protein was eluted with 0.3 M imidazole buffer. The fractions containing soluble EIII were dialyzed against phosphate-buffered saline (PBS) at 4°C overnight as described previously (14).
Blocking of DV infection to BHK21 and C6/36 cells by soluble EIII.
To test whether soluble EIII blocks DV infection of BHK21 (or C6/36) cells, soluble EIII or control bovine serum albumin (BSA) protein at different concentrations (shown at the bottom of each of the figures) was incubated with 5 × 105 BHK21 (or C6/36) cells in a 60-mm dish at 4°C for 2 h. Cells were subsequently infected with DEN-1,-2, -3, or -4, JEV, or VV at a multiplicity of infection (MOI) of 5 PFU per cell at 4°C for 2 h. The infected cultures were washed three times with PBS and incubated in RPMI medium with 2% FBS at 37°C. At 24 h postinfection (p.i.), supernatant from cells infected with DEN-1,-2, -3, -4 or JEV or cell lysates from cells infected with VV were collected for virus titer determination by plaque assay on BHK21 cells as described below.
Plaque assays of DV, JEV, and VV on BHK21 cells.
BHK21 cells were seeded (3 × 105 per well) in six-well plates. The next day, BHK21 cells were infected at 37°C for 2 h with 0.2 ml of serial dilutions of culture supernatant (collected from cells infected with DEN-1 to -4 and JEV) or cell lysates (collected from cells infected with VV) from infected cell cultures as described in the previous paragraph. After infection, BHK21 cells were washed, overlaid with 1% top agarose in RPMI 1640 medium, supplemented with 2% FBS, and incubated at 37°C for 4 to 5 days until plaques became visible. These infected cells were fixed under agarose overlay in 10% formaldehyde in PBS for 1 h at room temperature. Top agarose was removed, and the infected BHK21 cells were stained with 0.25% crystal violet in 50% ethanol for 60 min to make the plaques visible for counting.
Blocking of DV binding to BHK21 and C6/36 cells by soluble EIII.
To test whether soluble EIII blocks DV binding to BHK21 (or C6/36) cells, BHK21 (or C6/36) cells were preincubated with soluble EIII protein (300 μg/ml) at 4°C for 2 h and subsequently infected with DEN-1, -2, -3, -4, JEV, or VV at an MOI of 5 PFU per cell at 4°C for 2 h. The infected cell cultures were washed with PBS and immediately harvested in sodium dodecyl sulfate (SDS)-containing buffer. The cell lysates were loaded onto an SDS-10% polyacrylamide gel electrophoresis gel, and the amount of cell-associated virions was determined by monitoring the amount of virion protein that reacted with anti-DEN E MAb 4G2 (1:1,000), anti-JEV serum (1: 1,000), or anti-VV (1:1,000) in immunoblot analyses, as described below. The blots were scanned and quantitated with software, Image Gauge, version 3.45, that was installed on a FUJIFILM LAS-1000 plus pictrography 3000 (Fuji, Inc.).
Immunoblot analyses.
Cell lysates harvested after viral infection were fractionated on appropriate SDS-polyacrylamide gels and transferred to nitrocellulose membranes according to the manufacturer's protocol (Bio-Rad). After incubation with 3% nonfat milk in TTBS (0.5% Tween 20, 20 mM Tris-HCl [pH 7.4], 0.5 M NaCl) for 60 min, the membranes were washed once with TTBS and incubated with the appropriate primary antibody, i.e., anti-E MAb 4G2 (1:1,000), rabbit anti-JEV serum (1:1,000), anti-VV serum (1:1,000), or anti-T7 tag MAb (1: 10,000), at room temperature for 12 h. Membranes were then washed three times with TTBS and incubated with alkaline phosphatase-conjugated goat anti-mouse secondary antibody (1:3,000) for 2 h. Blots were washed three times again with TTBS and developed with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (Promega, Inc.) or CDP-Star (Applied Biosystems, Inc.) as described by the manufacturers. When we used anti-JEV and anti-VV antisera in immunoblots, we measured and averaged the three most abundant viral bands in each blot for our quantitative measurement of virion binding.
Biotinylation of soluble EIII.
Soluble EIII was biotinylated with the ECL protein biotinylation module (Amersham Pharmacia Biotech, Inc.). In brief, purified EIII (1 mg/ml) was mixed with 40 μl of biotinylation reagent N-hydroxysuccinamide ester in 40 mM bicarbonate buffer (pH 8.6) at room temperature for 1 h according to the manufacturer's instructions. The mixture was loaded onto a Sephadex G25 column preequilibrated with 20 ml of PBS-1% BSA. Biotinylated EIII was eluted with 10 ml of PBS and collected in 500-μl aliquots. The extent of biotinylation was confirmed by immunoblot analysis with alkaline phosphatase-conjugated streptavidin (1:3,000).
Soluble EIII binding assays to BHK21 and C6/36 cells.
BHK21 or C6/36 cells (5 × 105) were washed with cold PBS and incubated with biotinylated EIII protein at 5, 50, 100, and 250 μg/ml (for BHK21 cells) or at 5, 25, 125, and 625 μg/ml (for C6/36 cells) in staining buffer (PBS-4% FBS-10 mM HEPES [pH 7.2]) for 1 h at 4°C. In competition experiments, soluble glycosaminoglycans (GAGs), such as HP, CS, and DS, at 1, 10, and 100 μg/ml or synthetic peptides, such as DV1-1, DV2-1, DV3-1, DV4-1, DV2-2, or DV2-3, at 100 μg/ml were added as competitors prior to the addition of EIII protein. Cells were then washed three times with cold PBS and incubated with phycoerythrin-conjugated streptavidin (1:500) for 60 min at 4°C, washed with PBS, detached with 5 mM EDTA in PBS, and analyzed with a fluorescence-activated cell sorter (FACS) (excitation, 488 nm; emission, 578 nm) as described previously (42).
Protein structure graphics.
The three-dimensional structures of DV (ID, 10AM) and TBEV (ID, ISVB) were published and obtained from www.rcsb.org/pdb (57, 68). The graphics were displayed with WebLab ViewPro 4.0 software.
RESULTS
Soluble EIII protein binds to BHK21 and C6/36 cells.
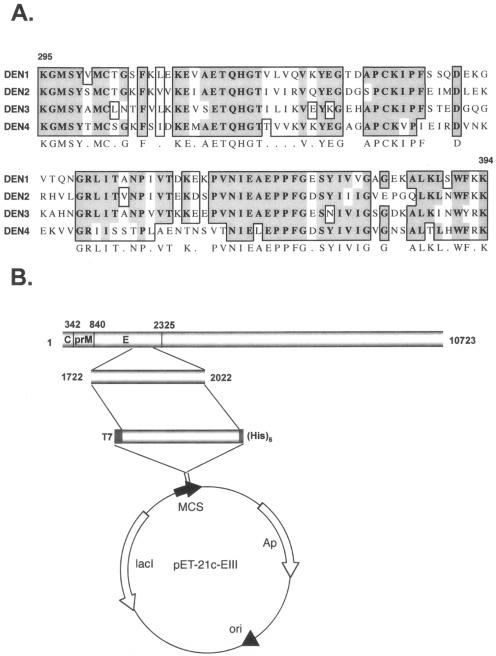
A DNA fragment encoding the domain III region (EIII) of E protein, aa 295 to 394, was amplified from reverse transcription-PCR of purified DEN-2 virion RNA. The deduced amino acid sequences were aligned with the corresponding regions of other serotypes (Fig. 1A). When EIII was expressed in E. coli as a glutathione S-transferase N-terminal fusion protein, the recombinant protein formed inclusion bodies and became insoluble aggregates (data not shown). However, when EIII was expressed with an N-terminal T7 tag and C-terminal hexahistidine tag by using a pET expression vector, as shown in Fig. 1B, EIII was in the soluble fraction. The recombinant EIII was purified by nickel affinity chromatography (Fig. 1C, lane 5) and could be recognized by antibody to T7 tag sequences (Fig. 1C, lane 6) and antibody to DV E protein (Fig. 1C, lane 7).
FIG. 1.
Sequence, construction, and expression of recombinant EIII from DEN-2 (PL046). (A) Multiple-sequence alignment of domain III region (aa 295 to 394) of four DV serotypes. Identical residues are shaded as dark gray areas, and conserved residues are shaded as light gray areas. (B) Expression vector construct showing the fragment of DEN-2 virus genome that was cloned into pET-21c vector for EIII production in bacteria. The nucleotide numbers represent the domain III region in DEN-2 viral genome. The MCS, multiple cloning site, is flanked by a T7 tag at the N terminus and a hexahistidine tag at the C terminus. (C) Recombinant EIII analyzed by SDS-12% polyacrylamide gel electrophoresis and stained with Coomassie blue (lanes 1 to 5) or transferred for immunoblot analyses (lanes 6 and 7). M (lane 1), mock-induced bacterial lysates; IPTG (lane 2), IPTG-induced bacterial lysates; Sup. (lane 3), postsonication supernatant fraction containing soluble EIII protein; Pel. (lane 4), postsonication insoluble pellet fraction; EIII (lane 5), purified EIII protein; α-T7 (lane 6), immunoblot of purified EIII with anti-T7 MAb (Novagen Inc.); α-E (lane 7): immunoblot of purified EIII with rabbit anti-DV Abs.
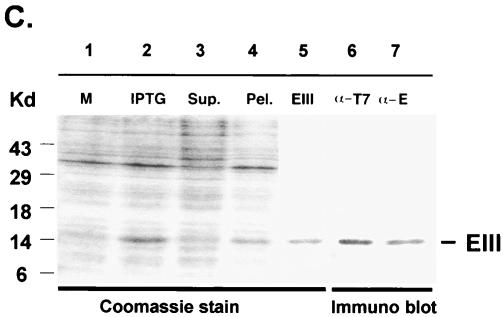
Recombinant EIII was biotinylated and tested for its ability to bind to mammalian BHK21 and mosquito C6/36 cells (Fig. 2). BHK21 cells were incubated with biotinylated EIII for 1 h at 4°C and analyzed by FACS. A significant shift in fluorescence intensity was observed at a low concentration of EIII (5 μg/ml), which increased and saturated when cells were incubated with 100 to 250 μg/ml of EIII, indicating that EIII binds to BHK21 cells (Fig. 2A). EIII also bound to several other types of human cell lines including SW48, LS180, TT, and HT1376, indicating that it could be a general feature to many mammalian cells (data not shown). In addition, EIII bound to mosquito C6/36 cells in a dose-dependent manner, and the binding became saturated at 125 to 625 μg/ml, within a lower fluorescence range (Fig. 2B). It thus appeared that recombinant EIII alone is capable of cell binding.
FIG. 2.
Binding of soluble EIII to BHK21 and C6/36 cells. BHK21 cells (A) or C6/36 cells (B) were mock treated (black line) or treated with various amounts of biotinylated EIII protein as indicated on the figure and analyzed by FACS as described previously (14).
Soluble EIII blocks DV binding to BHK21 and C6/36 cells.
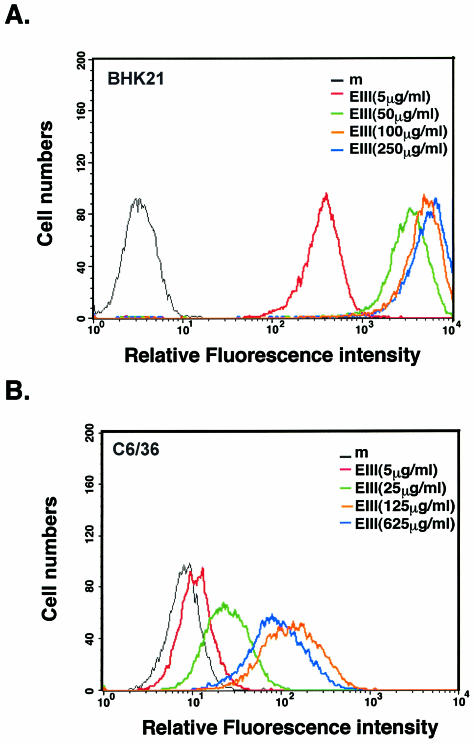
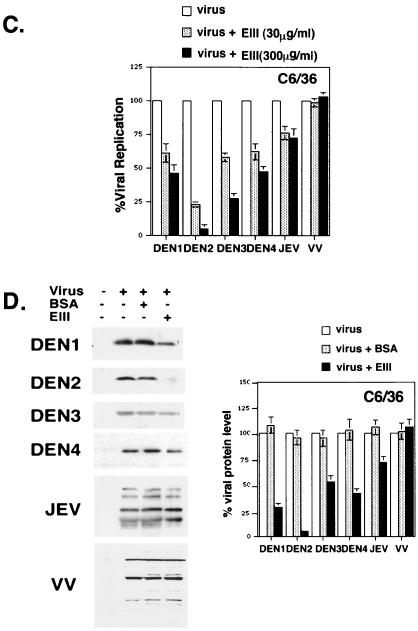
If domain III of E protein mediates DV entry into mammalian and mosquito cells, soluble EIII should interfere with virus infection of these cells. As shown in Fig. 3A (left panel), preincubation of BHK21 cells with EIII protein prior to DEN-2 infection exhibited a dosage-dependent inhibition on BHK21 cells. More than 95% reduction of plaque number formed by DEN-2 was specifically observed when EIII was added to cells at the concentration of 300 μg/ml, since BSA, even at 1,200 μg/ml, did not inhibit DEN-2 plaque formation. Therefore, we used EIII at the concentration of 300 μg/ml in all subsequent blocking experiments. To test whether EIII also block infections of DEN-1, DEN-3, and DEN-4 on BHK21 cells, we incubated BHK21 cells with EIII at 30 and 300 μg/ml prior to virus infection and assayed for plaque reduction (Fig. 3A, right panel). The results indicated that EIII reduced plaque formation by DEN-1, DEN-3, and DEN-4 on BHK21 cells as completely as for DEN-2. In contrast, EIII slightly inhibited JEV plaque formation by 25%, and there was no inhibition to VV plaque formation on BHK21 cells. These results showed that soluble EIII specifically inhibits DV infection of BHK21 cells in a serotype-independent manner.
FIG. 3.
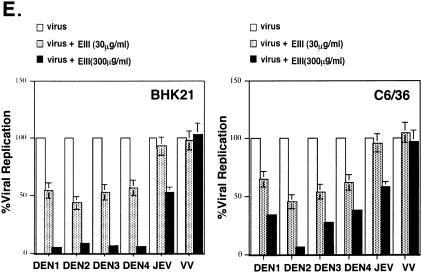
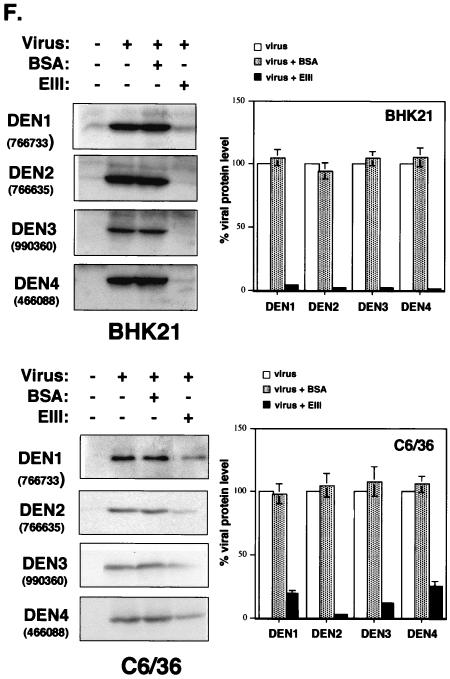
Soluble EIII inhibits DV infection to BHK21 and C6/36 cells by blocking virion binding to cells. (A) EIII blocked DEN-2 infection on BHK21 cells. (Left panel) BHK21 cells were mock treated or treated with various concentrations of BSA or EIII, as shown at the bottom of the figure, and infected with DEN-2 (16681) at an MOI of 5 PFU per cell. At 24 h p.i., the cell cultures were harvested and the virus titers were determined by plaque assays on BHK21 cells. (Right panel) BHK21 cells were treated with EIII (30 or 300 μg/ml) and infected with different viruses, at an MOI of 5 PFU per cell, as shown at the bottom of the figure. At 24 h p.i., cell cultures were harvested and the respective virus titers were determined by plaque assays on BHK21 cells. % Viral replication = (the number of plaques produced with EIII/the number of plaques produced without EIII treatment) × 100. (B) EIII blocked DV binding to BHK21 cells. BHK21 cells were treated with EIII (300 μg/ml) for 2 h and then infected with DEN-1, -2, -3, or -4, JEV, or VV at an MOI of 5 PFU per cell. Cells were immediately harvested after virus infection and used for immunoblot analyses with either MAb 4G2 against E protein (for DEN1 to DEN4) or a rabbit serum against whole virions (for JEV and VV). The blots were scanned and quantitated with Image Gauge, version 3.45, installed on a FUJIFILM LAS-1000 plus pictrography 3000 (Fuji Inc.). For JEV and VV blots, the most abundant three bands were averaged for quantitation. (C) EIII blocked DV infection on C6/36 cells. C6/36 cells were treated with EIII (30 or 300 μg/ml) as described for panel A. At 24 h p.i., cell cultures were harvested and the respective virus titers in supernatant were determined on BHK21 cells. (D) EIII blocked DV binding to C6/36 cells. C6/36 cells were treated with EIII (300 μg/ml) for 2 h and then infected at an MOI of 5 PFU per cell with DEN-1, -2, -3, or -4, JEV, or VV. Cells were immediately harvested after virus infection, and the amount of virus bound to cells was determined by immunoblot analyses as described for panel B. (E) EIII blocked infection of four low-passage DV isolates on BHK21 and C6/36 cells. BHK21 and C6/36 cells were treated with EIII (30 or 300 μg/ml) and infected with low-passage DVs. At 24 h p.i., cell cultures were harvested and the respective virus titers in supernatant were determined on BHK21 cells. (F) Binding of four low-passage DV isolates on BHK21 and C6/36 cells was blocked by EIII. BHK21 and C6/36 cells were treated with BSA or EIII (300 μg/ml) for 2 h and infected with each of the four low-passage DV isolates and harvested for immunoblot analyses as described for panel B. The low-passage DV isolates used for panels E and F are identical.
We next investigated whether the inhibitory effect of EIII was at the virion binding step. BHK21 cells was preincubated with EIII and infected with DEN-2 at an MOI of 5 PFU per cell. Cells were harvested immediately after virus infection for immunoblot analyses to determine the amount of bound virions on cells. As shown in Fig. 3B, preincubation of cells with soluble EIII significantly reduced the amounts of virion E protein bound to cells, indicating that EIII blocked virion adsorption to cells. EIII not only blocked binding of DEN-2 virion but also blocked binding of DEN-1, DEN-3, and DEN-4 virions, indicating that EIII blocked adsorption of DV of all four serotypes to BHK21 cells.
EIII was also tested for its ability to block DEN-2 infection of C6/36 cells (Fig. 3C). Consistent with the results with BHK21 cells, EIII, at 300 μg/ml, effectively blocked DEN-2 virus infection down to 4.5% of control infection. However, EIII exhibited less inhibition to DV of other serotypes than to DEN-2. EIII did not significantly inhibit virus infection by control JEV or VV on C6/36 cells. Thus, EIII blocks DV infection of C6/36 cells in a serotype-specific manner.
To determine whether EIII blocks virus binding to C6/36 cells, blocking experiments similar to those described above were performed and the results are shown in Fig. 3D. The amounts of cell-associated DEN-2 virions were significantly reduced if C6/36 cells were pretreated with EIII prior to virus infection. Interestingly, EIII blocked binding of DEN-1, -3, and -4 to C6/36 cells to different extents, consistent with the plaque reduction results shown in Fig. 3C. We concluded that EIII reduced dengue virion binding to C6/36 cells in a serotype-specific manner.
We obtained several clinical DVs isolated from local hospitals, and these viruses were grown up with minimal passages (<3) to minimize mutations generated from long-term passages. We tested whether DV replication (Fig. 3E) or virion binding of these DV isolates on BHK21 and C6/36 cells (Fig. 3F) is affected by EIII. Consistent with results obtained from the four prototype viruses, infection and adsorption of these low-passage viruses were also reduced by EIII on BHK21 and C6/36 cells. In addition, the inhibitory effect of EIII on BHK21 cells was broad, whereas on C6/36 cells remained serotype specific.
In summary, the results in Fig. 3 indicate that EIII interferes with dengue virion adsorption to both BHK21 and C6/36 cells. In addition, the blocking effect of EIII on BHK21 cells is serotype independent, whereas on C6/36 cells, it appeared serotype specific, indicating the mode of EIII binding to these two cell lines may not be identical.
EIII binds to HSs on BHK21 cells.
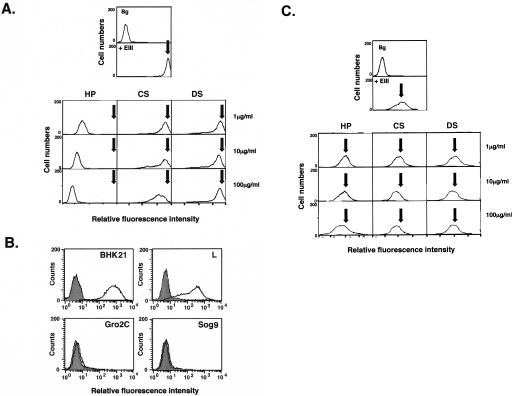
Previous studies indicate that cell surface HS is involved in attachment of DV to Vero and CHO cells. Furthermore, EIII sequences contain part of the putative HS-binding sequencers, i.e., residues 284 to 310 and 386 to 411 (11). The role of putative HS-binding sites on EIII in binding to BHK21 and C6/36 cells was examined with a binding competition assay. Biotinylated EIII was incubated with BHK21 cells in the presence or absence of soluble GAGs, and the level of EIII binding to cells was analyzed by FACS (Fig. 4). Binding of EIII to BHK21 cells caused a significant increase in fluorescence intensity (Fig. 4A, top panel), but HP (1 μg/ml) reduced fluorescence significantly (Fig. 4A, left panel). Higher HP concentrations (10 or 100 μg/ml) decreased and completely blocked EIII binding to BHK21 cells. However, CS and DS did not inhibit binding of EIII to BHK21 cells (Fig. 4A, middle and right panels), indicating that EIII binds specifically to cell surface HS on BHK21 cells.
FIG. 4.
The role of cell surface HS in EIII binding to BHK21 cells and C6/36 cells. (A) EIII binding to BHK21 cells was competed by HP. BHK21 cells were incubated with PBS (Bg), biotinylated EIII alone (+ EIII), or biotinylated EIII with addition of 1, 10, or 100 μg of each of the soluble GAGs (HP, CS, and DS)/ml. Cells were analyzed by FACS as described previously (42). (B) HS is required for EIII binding. BHK21, L, gro2C, and sog9 cells were incubated with PBS (gray area) or biotinylated EIII (black line) and analyzed by FACS as described previously (42). (C) EIII binding to C6/36 cells is HS independent. C6/36 cells were incubated with PBS (Bg), biotinylated EIII alone (+ EIII), or biotinylated EIII with addition of 1, 10, or 100 μg of each of the soluble GAGs (HP, CS, and DS)/ml and analyzed by FACS. The arrows indicate the positions of the peak of cell fluorescence intensity obtained with EIII binding alone (+ EIII).
Although the above-described competition assays with soluble GAGs have been widely used in cell binding analyses, they are, nevertheless, indirect measurements of cell surface interaction. Direct binding of EIII to cell surface HS was confirmed by measuring binding of EIII to cell lines expressing different GAGs. Mouse L cells express HS and CS on their surfaces, whereas a mutant cell line derived from L cells, gro2C, expresses only CS (24). Sog9 cells, which were selected from gro2C, lack expression of HS and CS (2). These mutant cells were used previously to study interaction between cell surface GAGs and envelope proteins of herpes simplex virus type 1 and VV (20, 34, 38, 42, 80). Biotinylated EIII bound to BHK21 and L cells, causing a detectable fluorescence shift, as expected (Fig. 4B, upper panels). In contrary, EIII did not bind to gro2C or sog9 cells (Fig. 4B). These results are consistent with the fact that soluble HP interferes with EIII binding to BHK21 cells and provide direct evidence that EIII interacts with cell surface HS.
EIII binding to mosquito C6/36 cells is GAG independent.
GAG competition assays were also performed with EIII and C6/36 cells to determine whether GAGs interfere with EIII binding to these cells (Fig. 4C). Soluble HP did not block EIII binding to C6/36 cells at the 1-, 10-, or 100-μg/ml level (Fig. 4C, left panel). Furthermore, CS and DS had no effect on binding of EIII to mosquito cells (Fig. 4C, middle and right panels). Thus, EIII binds to C6/36 cells in a GAG-independent manner. This difference of EIII in GAG dependence may reflect the fact that EIII utilizes distinct structural elements or motifs when binding to BHK21 and C6/36 cells.
EIII binds to mosquito cells through a lateral loop region containing aa 380 to 389.
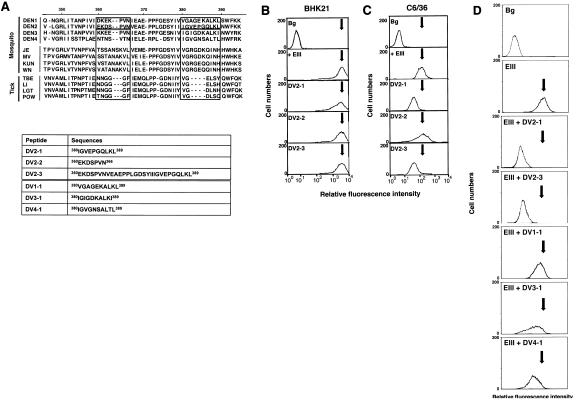
Computer modeling of the TBEV E protein has been used to identify solvent-accessible regions in the lateral loops of EIII (68). One of these regions at aa 360 to 366, corresponding to DV2-2 sequences in DEN-2, is hypervariable and could be subdivided into distinct groups based on vector usage (Fig. 5A). Another loop region corresponds to aa 380 to 389 of DV2-1 sequences, which includes an insertion of four amino acids forming an extended loop structure (57). This extended loop region is conserved in DV and other mosquito-borne viruses but is absent in tick-borne viruses (68). Additional computation analysis of DV E protein for identification of functional loops also predicted DV2-1 to be a potential ligand binding site on EIII (T. Cheng and C. Lim, unpublished data). To test the role of these two regions of EIII in host cell interaction, two peptides, DV2-1 and DV2-2, were synthesized. The third peptide, DV2-3, was synthesized with residues from aa 360 to 389, which includes both DV2-1 and DV2-2 sequences, and used in the competition studies (Fig. 5A, lower panel).
FIG. 5.
Synthetic peptides containing an external loop region blocked EIII binding to C6/36 but not BHK21 cells. (A) Alignment of amino acid sequences of EIII region among flaviviruses. Residues 360 to 366 and 380 to 389 of DV and the corresponding regions of other flaviviruses are boxed. The amino acid sequences of DV2-1 and DV2-2 are double underlined. Synthetic peptides are shown in the lower panel. DV2-1, DV2-2, and DV2-3 are derived from EIII of DEN-2, and DV1-1, DV3-1, and DV4-1 are derived from EIII of DEN-1, DEN-3, and DEN-4. (B and C) BHK21 cells (B) and C6/36 cells (C) were incubated with PBS (Bg) and biotinylated EIII in the absence (+ EIII) or presence (DV2-1, DV2-2, and DV2-3) of synthetic peptides and analyzed by FACS. (D) C6/36 cells were incubated with PBS (Bg), biotinylated EIII alone (EIII), or biotinylated EIII with addition of each synthetic peptide (EIII + DV2-1, EIII + DV2-3, EIII + DV1-1, EIII + DV3-1, and EIII + DV4-1) and analyzed by FACS. The arrows indicate the positions of the peak of cell fluorescence intensity obtained with EIII binding alone (+ EIII).
BHK21 or C6/36 cells were pretreated with DV2-1, DV2-2, or DV2-3 at 100 μg/ml and subsequently incubated with biotinylated EIII protein. The amount of EIII binding to cells was measured by FACS. None of these peptides inhibited EIII binding to BHK21 cells (Fig. 5B). However, DV2-1, but not DV2-2, interfered with binding of EIII to C6/36 cells (Fig. 5C). DV2-3, a longer peptide comprising both DV2-1 and DV2-2 sequences, inhibited EIII binding to approximately the same extent as DV2-1 alone. These data suggest that the lateral loop region of aa 380 to 389 may play a role in EIII binding to C6/36 but not to BHK21 cells.
To investigate whether DV2-1 blocks EIII binding to C6/36 cells in a serotype-specific manner, we synthesized three additional peptides, DV1-1, DV3-1, and DV4-1, which contain residues corresponding to aa 380 to 389 in the DEN-1, DEN-3, and DEN-4 serotypes, respectively (Fig. 5A, lower panel). These peptides were tested for their ability to block DEN-2 EIII binding to C6/36 cells as described above. The results showed that while the control DV2-1 and DV2-3 peptides significantly blocked biotinylated EIII binding to C6/36 cells, DV3-1 and DV4-1 showed moderate inhibitory activity and DV1-1 showed minor competition activity in FACS analysis (Fig. 5D). The results thus revealed that peptides synthesized from the loop region blocked EIII binding to C6/36 cells in a serotype-specific manner.
DISCUSSION
This study analyzes structural elements of EIII of DV type 2 E protein that are required for binding to mammalian and mosquito cells. The results show that EIII plays an important role in DV infection of mammalian BHK21 and mosquito C6/36 cells. Soluble EIII binds to and blocks infection of DV on BHK21 and C6/36 cells. These results are consistent with previous reports that mutations within domain III of flavivirus exhibited reduced virulence (8, 29, 40, 48, 50, 51, 64, 73, 79). This study also demonstrated that EIII interacts differently with mammalian BHK21 and mosquito C6/36 cells.
The crystal structure of DV E protein has recently been solved and showed a high degree of conservation with that of TBEV E protein (57, 68). The EIII regions of TBEV and DV are shown in Fig. 6. There are six conserved lysine residues in DV that lie in the putative HS-binding sites of EIII. Four lysines at K295, K305, K310, and K394 of DV E protein correspond to K300, K311, K315, and K395 in TBEV and are conserved in all four serotypes of DV. Additionally, K307 is conserved in DEN-1, -2, and -4, and K393/R393 is conserved in all four serotypes. These lysine residues could directly contribute to highly positive charges important for EIII binding to HS moiety on cells. Since there is little serotype-specific variation for these lysine residues, it is expected that EIII of all serotypes binds to cell surface HS. Attachment to HS is not just an adaptation process as a consequence of continuous laboratory passages in vitro, because EIII also blocked infections of virus isolates of low passage numbers (<3) to BHK21 cells. Although our data showed that DEN-2 EIII mainly binds to HS on BHK21 cells, it remains possible that EIII may bind to other coreceptors on the cell surface through postbinding conformational change. It is also likely that other regions of viral E protein, i.e., domains I and II, will participate in coreceptor recognition and fusion steps during DV entry into mammalian cells.
FIG. 6.
The putative HS-binding site and the external loop region in EIII of DV. The monomer structures of domain III of TBEV and DV, based on the crystal structures of E protein of TBEV and DV, are shown in detail (57, 68). Conserved lysine residues within the putative HS-binding sites of TBEV and DV are shown in cyan. Two lysine residues within the putative HS-binding sites that are conserved in DV but not in TBEV, K307 and K393, are shown in dark blue. The orange line represents the external loop region, i.e., DV2-1, from aa 380 to 389, including the central core sequences shown in red. The green line shows the DV2-2 region from aa 360 to 366.
HS-binding sites do not appear to play a role in the interaction between DV and mosquito C6/36 cells. Instead, this study suggests that a lateral loop region in DEN-2 EIII could play an important role during DV infection of C6/36 cells. The length of the external loop region is conserved in DVs, but certain residues within this loop region vary in a serotype-specific manner in DV. For example, the central different core residues within the loops are AGEK, VEPG, IGDK, and VGNS in DEN-1, -2, -3, and -4, respectively. These four residues are very different from the corresponding core residues, i.e., RG(D/E)(K/Q), present in other flaviviruses transmitted by mosquitoes, such as JEV.
The fact that DEN-2 EIII inhibits infection of DV on C6/36 cells in a serotype-dependent manner could be explained by several hypotheses. The simplest explanation is that DV of each serotype binds to different molecules on C6/36 cells. Indeed, 40- to 45-kDa cell surface proteins were observed to bind DEN-4 in C6/36 cells, whereas proteins of 67 and 80 kDa in size were reported to bind DEN-2 (60, 72, 83). Alternatively, it is possible that EIII has a different affinity for the same molecule on C6/36 cells, depending on the exact core sequence of the loop region in DEN-1 to -4. Thus, soluble DEN-2 EIII would most effectively inhibit infection by DEN-2, if EIII from other serotypes has a higher affinity for its cell surface target. This idea is consistent with a previous report suggesting that DV binds HL60 cells in an HS-independent manner and varies for different DV serotypes (6). We have not expressed EIII from other serotypes of DV yet. In the future, comparative studies of EIII of all four serotypes for binding to mosquito cells may provide direct measurement to clarify this issue. It has been suggested that E protein-mediated membrane fusion requires postbinding conformational changes that promote a dimer-to-trimer transition (22, 39, 65). Whether domain III, in addition to blocking virion adsorption, plays any additional role in homophilic dimer-to-trimer transition is currently unknown. We cannot exclude any of the above hypotheses at this point, and more work is needed in the future to help clarify this important issue.
The synthetic peptides that blocked EIII binding to cells contain residues located in the extended portion of a lateral loop of EIII (Fig. 6). The length of this loop is longer in flaviviruses transmitted by mosquitoes (i.e., DV, JEV) than in viruses transmitted by other vectors (i.e., TBEV). It is tempting to speculate that this extended loop binds specifically to a putative receptor on mosquito cells, and the search for such molecules is currently under investigation. Finally, DC-SIGN, a cell surface lectin, was recently shown to mediate DV infection of human dendritic cells (61, 81). Currently, it is not known how DC-SIGN interacts with virion E protein during DV infection. Whether EIII could block DC-SIGN-mediated DV entry will be investigated in the near future.
The mechanism by which a virus infects its target host cell is a major determinant of the cellular tropism of the virus and is critical for viral pathogenesis. This study suggests that DV has evolved different structural regions on E protein to infect mammalian and mosquito cells. This could confer a selective advantage for DV during evolution. For example, if an identical structural motif is required to infect mammalian and mosquito host cells, any mutation in E protein that reduces virulence or pathogenicity in the mammalian host would also be likely to affect virus transmission in the mosquito vector. On the other hand, if separate structures of E protein are used for infecting different hosts, mutations of E protein to escape neutralization in human cells could occur without affecting virus infection of mosquito vectors. Additional studies are needed to elucidate the molecular mechanism of DV entry into different host cells.
.
Acknowledgments
We thank H.-C. Wu for providing anti-DEN2-E MAb (4G2) and L.-Y. Lin for providing JEV.
This work is supported by grants from Academia Sinica (AS91IMB7PP) and National Health Research Institute (NRHI-CN-CL8903P) of the Republic of China.
REFERENCES
- 1.Allison, S. L., J. Schalich, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banfield, B. W., Y. Leduc, L. Esford, K. Schubert, and F. Tufaro. 1995. Sequential isolation of proteoglycan synthesis mutants by using herpes simplex virus as a selective agent: evidence for a proteoglycan-independent virus entry pathway. J. Virol. 69:3290-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernfield, M., M. Gotte, P. W. Park, O. Reizes, M. L. Fitzgerald, J. Lincecum, and M. Zako. 1999. Functions of cell surface heparan sulfate proteoglycans. Annu. Rev. Biochem. 68:729-777. [DOI] [PubMed] [Google Scholar]
- 4.Bhardwaj, S., M. Holbrook, R. E. Shope, A. D. Barrett, and S. J. Watowich. 2001. Biophysical characterization and vector-specific antagonist activity of domain III of the tick-borne flavivirus envelope protein. J. Virol. 75:4002-4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bielefeldt-Ohmann, H. 1998. Analysis of antibody-independent binding of dengue viruses and dengue virus envelope protein to human myelomonocytic cells and B lymphocytes. Virus Res. 57:63-79. [DOI] [PubMed] [Google Scholar]
- 6.Bielefeldt-Ohmann, H., M. Meyer, D. R. Fitzpatrick, and J. S. Mackenzie. 2001. Dengue virus binding to human leukocyte cell lines: receptor usage differs between cell types and virus strains. Virus Res. 73:81-89. [DOI] [PubMed] [Google Scholar]
- 7.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1126. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 8.Cecilia, D., and E. A. Gould. 1991. Nucleotide changes responsible for loss of neuroinvasiveness in Japanese encephalitis virus neutralization-resistant mutants. Virology 181:70-77. [DOI] [PubMed] [Google Scholar]
- 9.Chaturvedi, U. C., E. A. Elbishbishi, R. Agarwal, R. Raghupathy, R. Nagar, R. Tandon, A. S. Pacsa, O. I. Younis, and F. Azizieh. 1999. Sequential production of cytokines by dengue virus-infected human peripheral blood leukocyte cultures. J. Med. Virol. 59:335-340. [DOI] [PubMed] [Google Scholar]
- 10.Chen, L. K., Y. L. Lin, C. L. Liao, C. G. Lin, Y. L. Huang, C. T. Yeh, S. C. Lai, J. T. Jan, and C. Chin. 1996. Generation and characterization of organ-tropism mutants of Japanese encephalitis virus in vivo and in vitro. Virology 223:79-88. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Y., T. Maguire, R. E. Hileman, J. R. Fromm, J. D. Esko, R. J. Linhardt, and R. M. Marks. 1997. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat. Med. 3:866-871. [DOI] [PubMed] [Google Scholar]
- 12.Chen, Y. C., and S. Y. Wang. 2002. Activation of terminally differentiated human monocytes/macrophages by dengue virus: productive infection, hierarchical production of innate cytokines and chemokines, and the synergistic effect of lipopolysaccharide. J. Virol. 76:9877-9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, Y. C., S. Y. Wang, and C. C. King. 1999. Bacterial lipopolysaccharide inhibits dengue virus infection of primary human monocytes/macrophages by blockade of virus entry via a CD14-dependent mechanism. J. Virol. 73:2650-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung, C. S., J. C. Hsiao, Y. S. Chang, and W. Chang. 1998. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 72:1577-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clarke, T. 2002. Dengue virus: break-bone fever. Nature 416:672-674. [DOI] [PubMed] [Google Scholar]
- 16.Crill, W. D., and J. T. Roehrig. 2001. Monoclonal antibodies that bind to domain III of dengue virus E glycoprotein are the most efficient blockers of virus adsorption to Vero cells. J. Virol. 75:7769-7773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.da Fonseca, B. A., and S. N. Fonseca. 2002. Dengue virus infections. Curr. Opin. Pediatr. 14:67-71. [DOI] [PubMed] [Google Scholar]
- 18.Daughaday, C. C., W. E. Brandt, J. M. McCown, and P. K. Russell. 1981. Evidence for two mechanisms of dengue virus infection of adherent human monocytes: trypsin-sensitive virus receptors and trypsin-resistant immune complex receptors. Infect. Immun. 32:469-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunster, L. M., H. Wang, K. D. Ryman, B. R. Miller, S. J. Watowich, P. D. Minor, and A. D. Barrett. 1999. Molecular and biological changes associated with HeLa cell attenuation of wild-type yellow fever virus. Virology 261:309-318. [DOI] [PubMed] [Google Scholar]
- 20.Dyer, A. P., B. W. Banfield, D. Martindale, D. M. Spannier, and F. Tufaro. 1997. Dextran sulfate can act as an artificial receptor to mediate a type-specific herpes simplex virus infection via glycoprotein B. J. Virol. 71:191-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esko, J. D., and S. B. Selleck. 2002. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 71:435-471. [DOI] [PubMed] [Google Scholar]
- 22.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7:593-602. [DOI] [PubMed] [Google Scholar]
- 23.Germi, R., J. M. Crance, D. Garin, J. Guimet, H. Lortat-Jacob, R. W. Ruigrok, J. P. Zarski, and E. Drouet. 2002. Heparan sulfate-mediated binding of infectious dengue virus type 2 and yellow fever virus. Virology 292:162-168. [DOI] [PubMed] [Google Scholar]
- 24.Gruenheid, S., L. Gatzke, H. Meadows, and F. Tufaro. 1993. Herpes simplex virus infection and propagation in a mouse L cell mutant lacking heparan sulfate proteoglycans. J. Virol. 67:93-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gubler, D. J. 1998. Dengue and dengue hemorrhagic fever. Clin. Microbiol. Rev. 11:480-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hahn, C. S., J. M. Dalrymple, J. H. Strauss, and C. M. Rice. 1987. Comparison of the virulent Asibi strain of yellow fever virus with the 17D vaccine strain derived from it. Proc. Natl. Acad. Sci. USA 84:2019-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halstead, S. B. 1984. Selective primary health care: strategies for control of disease in the developing world. XI. Dengue. Rev. Infect. Dis. 6:251-264. [DOI] [PubMed] [Google Scholar]
- 28.Hase, T., P. L. Summers, and K. H. Eckels. 1989. Flavivirus entry into cultured mosquito cells and human peripheral blood monocytes. Arch. Virol. 104:129-143. [DOI] [PubMed] [Google Scholar]
- 29.Hasegawa, H., M. Yoshida, T. Shiosaka, S. Fujita, and Y. Kobayashi. 1992. Mutations in the envelope protein of Japanese encephalitis virus affect entry into cultured cells and virulence in mice. Virology 191:158-165. [DOI] [PubMed] [Google Scholar]
- 30.Heinz, F. X., and S. L. Allison. 2000. Structures and mechanisms in flavivirus fusion. Adv. Virus Res. 55:231-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hilgard, P., and R. Stockert. 2000. Heparan sulfate proteoglycans initiate dengue virus infection of hepatocytes. Hepatology 32:1069-1077. [DOI] [PubMed] [Google Scholar]
- 32.Ho, L. J., J. J. Wang, M. F. Shaio, C. L. Kao, D. M. Chang, S. W. Han, and J. H. Lai. 2001. Infection of human dendritic cells by dengue virus causes cell maturation and cytokine production. J. Immunol. 166:1499-1506. [DOI] [PubMed] [Google Scholar]
- 33.Hotta, H., A. S. Wiharta, S. Hotta, and M. Homma. 1984. Dengue type 2 virus infection in human peripheral blood monocyte cultures. Microbiol. Immunol. 28:1099-1109. [DOI] [PubMed] [Google Scholar]
- 34.Hsiao, J.-C., C.-S. Chung, and W. Chang. 1999. Vaccinia envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 73:8750-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hung, S. L., P. L. Lee, H. W. Chen, L. K. Chen, C. L. Kao, and C. C. King. 1999. Analysis of the steps involved in Dengue virus entry into host cells. Virology 257:156-167. [DOI] [PubMed] [Google Scholar]
- 36.Igarashi, A. 1978. Isolation of a Singh's Aedes albopictus cell clone sensitive to Dengue and Chikungunya viruses. J. Gen. Virol. 40:531-544. [DOI] [PubMed] [Google Scholar]
- 37.Irie, K., P. M. Mohan, Y. Sasaguri, R. Putnak, and R. Padmanabhan. 1989. Sequence analysis of cloned dengue virus type 2 genome (New Guinea-C strain). Gene 75:197-211. [DOI] [PubMed] [Google Scholar]
- 38.Karger, A., A. Saalmuller, F. Tufaro, B. W. Banfield, and T. C. Mettenleiter. 1995. Cell surface proteoglycans are not essential for infection by pseudorabies virus. J. Virol. 69:3482-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leitmeyer, K. C., D. W. Vaughn, D. M. Watts, R. Salas, I. Villalobos de Chacon, C. Ramos, and R. Rico-Hesse. 1999. Dengue virus structural differences that correlate with pathogenesis. J. Virol. 73:4738-4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 42.Lin, C. L., C. S. Chung, H. G. Heine, and W. Chang. 2000. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 74:3353-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin, C. W., and S. C. Wu. 2003. A functional epitope determinant on domain III of the Japanese encephalitis virus envelope protein interacted with neutralizing-antibody combining sites. J. Virol. 77:2600-2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin, Y., H. Lei, T. Yeh, S. Chen, and H. Liu. 2002. Heparin inhibits dengue-2 virus infection of five human liver cell lines. Antivir. Res. 56:93. [DOI] [PubMed] [Google Scholar]
- 45.Lin, Y. L., C. L. Liao, L. K. Chen, C. T. Yeh, C. I. Liu, S. H. Ma, Y. Y. Huang, Y. L. Huang, C. L. Kao, and C. C. King. 1998. Study of Dengue virus infection in SCID mice engrafted with human K562 cells. J. Virol. 72:9729-9737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lin, Y. W., K. J. Wang, H. Y. Lei, Y. S. Lin, T. M. Yeh, H. S. Liu, C. C. Liu, and S. H. Chen. 2002. Virus replication and cytokine production in dengue virus-infected human B lymphocytes. J. Virol. 76:12242-12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1042. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 1. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 48.Lobigs, M., R. Usha, A. Nestorowicz, I. D. Marshall, R. C. Weir, and L. Dalgarno. 1990. Host cell selection of Murray Valley encephalitis virus variants altered at an RGD sequence in the envelope protein and in mouse virulence. Virology 176:587-595. [DOI] [PubMed] [Google Scholar]
- 49.Mandl, C. W., S. L. Allison, H. Holzmann, T. Meixner, and F. X. Heinz. 2000. Attenuation of tick-borne encephalitis virus by structure-based site-specific mutagenesis of a putative flavivirus receptor binding site. J. Virol. 74:9601-9609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mandl, C. W., F. X. Heinz, and C. Kunz. 1988. Sequence of the structural proteins of tick-borne encephalitis virus (western subtype) and comparative analysis with other flaviviruses. Virology 166:197-205. [DOI] [PubMed] [Google Scholar]
- 51.Mandl, C. W., H. Kroschewski, S. L. Allison, R. Kofler, H. Holzmann, T. Meixner, and F. X. Heinz. 2001. Adaptation of tick-borne encephalitis virus to BHK-21 cells results in the formation of multiple heparan sulfate binding sites in the envelope protein and attenuation in vivo. J. Virol. 75:5627-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marianneau, P., F. Megret, R. Olivier, D. M. Morens, and V. Deubel. 1996. Dengue 1 virus binding to human hepatoma HepG2 and simian Vero cell surfaces differs. J. Gen. Virol. 77:2547-2554. [DOI] [PubMed] [Google Scholar]
- 53.Marianneau, P., A. M. Steffan, C. Royer, M. T. Drouet, D. Jaeck, A. Kirn, and V. Deubel. 1999. Infection of primary cultures of human Kupffer cells by dengue virus: no viral progeny synthesis, but cytokine production is evident. J. Virol. 73:5201-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marks, R. M., H. Lu, R. Sundaresan, T. Toida, A. Suzuki, T. Imanari, M. J. Hernaiz, and R. J. Linhardt. 2001. Probing the interaction of dengue virus envelope protein with heparin: assessment of glycosaminoglycan-derived inhibitors. J. Med. Chem. 44:2178-2187. [DOI] [PubMed] [Google Scholar]
- 55.Marovich, M., G. Grouard-Vogel, M. Louder, M. Eller, W. Sun, S. J. Wu, R. Putvatana, G. Murphy, B. Tassaneetrithep, T. Burgess, D. Birx, C. Hayes, S. Schlesinger-Frankel, and J. Mascola. 2001. Human dendritic cells as targets of dengue virus infection. J. Investig. Dermatol. Symp. Proc. 6:219-224. [DOI] [PubMed] [Google Scholar]
- 56.Martinez-Barragan, J. J., and R. M. del Angel. 2001. Identification of a putative coreceptor on Vero cells that participates in dengue 4 virus infection. J. Virol. 75:7818-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Monath, T. P. 1994. Dengue: the risk to developed and developing countries. Proc. Natl. Acad. Sci. USA 91:2395-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moreno-Altamirano, M. M., F. J. Sanchez-Garcia, and M. L. Munoz. 2002. Non Fc receptor-mediated infection of human macrophages by dengue virus serotype 2. J. Gen. Virol. 83:1123-1130. [DOI] [PubMed] [Google Scholar]
- 60.Munoz, M. L., A. Cisneros, J. Cruz, P. Das, R. Tovar, and A. Ortega. 1998. Putative dengue virus receptors from mosquito cells. FEMS Microbiol. Lett. 168:251-258. [DOI] [PubMed] [Google Scholar]
- 61.Navarro-Sanchez, E., R. Altmeyer, A. Amara, O. Schwartz, F. Fieschi, J.-L. Virelizier, F. Arenzano-Seisdedos, and P. Despres. 2003. Dendritic-cell-specific ICAM3 grabbing non-integrin is essential for the productive infection of human dendritic cells by mosquito-cell-derived dengue viruses. EMBO Rep. 4:723-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ni, H., K. D. Ryman, H. Wang, M. F. Saeed, R. Hull, D. Wood, P. D. Minor, S. J. Watowich, and A. D. Barrett. 2000. Interaction of yellow fever virus French neurotropic vaccine strain with monkey brain: characterization of monkey brain membrane receptor escape variants. J. Virol. 74:2903-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Osatomi, K., and H. Sumiyoshi. 1990. Complete nucleotide sequence of dengue type 3 virus genome RNA. Virology 176:643-647. [DOI] [PubMed] [Google Scholar]
- 64.Pletnev, A. G., M. Bray, and C. J. Lai. 1993. Chimeric tick-borne encephalitis and dengue type 4 viruses: effects of mutations on neurovirulence in mice. J. Virol. 67:4956-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pletnev, S. V., W. Zhang, S. Mukhopadhyay, B. R. Fisher, R. Hernandez, D. T. Brown, T. S. Baker, M. G. Rossmann, and R. J. Kuhn. 2001. Locations of carbohydrate sites on alphavirus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105:127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ramos-Castaneda, J., J. L. Imbert, B. L. Barron, and C. Ramos. 1997. A 65-kDa trypsin-sensible membrane cell protein as a possible receptor for dengue virus in cultured neuroblastoma cells. J. Neurovirol. 3:435-440. [DOI] [PubMed] [Google Scholar]
- 67.Rey, F. A. 2003. Dengue virus envelope glycoprotein structure: new insight into its interactions during viral entry. Proc. Natl. Acad. Sci. USA 100:6899-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 69.Rostand, K. S., and J. D. Esko. 1997. Microbial adherence to and invasion through proteoglycans. Infect. Immun. 65:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rothman, A. L., and F. A. Ennis. 1999. Immunopathogenesis of dengue hemorrhagic fever. Virology 257:1-6. [DOI] [PubMed] [Google Scholar]
- 71.Ryman, K. D., T. N. Ledger, G. A. Campbell, S. J. Watowich, and A. D. Barrett. 1998. Mutation in a 17D-204 vaccine substrain-specific envelope protein epitope alters the pathogenesis of yellow fever virus in mice. Virology 244:59-65. [DOI] [PubMed] [Google Scholar]
- 72.Salas-Benito, J. S., and R. M. del Angel. 1997. Identification of two surface proteins from C6/36 cells that bind dengue type 4 virus. J. Virol. 71:7246-7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sanchez, I. J., and B. H. Ruiz. 1996. A single nucleotide change in the E protein gene of dengue virus 2 Mexican strain affects neurovirulence in mice. J. Gen. Virol. 77:2541-2545. [DOI] [PubMed] [Google Scholar]
- 74.Schneider-Schaulies, J. 2000. Cellular receptors for viruses: links to tropism and pathogenesis. J. Gen. Virol. 81:1413-1429. [DOI] [PubMed] [Google Scholar]
- 75.Scott, R. M., A. Nisalak, U. Cheamudon, S. Seridhoranakul, and S. Nimmannitya. 1980. Isolation of dengue viruses from peripheral blood leukocytes of patients with hemorrhagic fever. J. Infect. Dis. 141:1-6. [DOI] [PubMed] [Google Scholar]
- 76.Sittisombut, N., N. Maneekarn, A. Kanjanahaluethai, W. Kasinrerk, K. Viputtikul, and J. Supawadee. 1995. Lack of augmenting effect of interferon-gamma on dengue virus multiplication in human peripheral blood monocytes. J. Med. Virol. 45:43-49. [DOI] [PubMed] [Google Scholar]
- 77.Sriburi, R., P. Keelapang, T. Duangchinda, S. Pruksakorn, N. Maneekarn, P. Malasit, and N. Sittisombut. 2001. Construction of infectious dengue 2 virus cDNA clones using high copy number plasmid. J. Virol. Methods 92:71-82. [DOI] [PubMed] [Google Scholar]
- 78.Stiasny, K., S. L. Allison, J. Schalich, and F. X. Heinz. 2002. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 76:3784-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sumiyoshi, H., G. H. Tignor, and R. E. Shope. 1995. Characterization of a highly attenuated Japanese encephalitis virus generated from molecularly cloned cDNA. J. Infect. Dis. 171:1144-1151. [DOI] [PubMed] [Google Scholar]
- 80.Tal-Singer, R., C. Peng, M. Ponce De Leon, W. R. Abrams, B. W. Banfield, F. Tufaro, G. H. Cohen, and R. J. Eisenberg. 1995. Interaction of herpes simplex virus glycoprotein gC with mammalian cell surface molecules. J. Virol. 69:4471-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tassaneetrithep, B., T. H. Burgess, A. Granelli-Piperno, C. Trumpfheller, J. Finke, W. Sun, M. A. Eller, K. Pattanapanyasat, S. Sarasombath, D. L. Birx, R. M. Steinman, S. Schlesinger, and M. A. Marovich. 2003. DC-SIGN (CD209) mediates dengue virus infection of human dendritic cells. J. Exp. Med. 197:823-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu, S. J., G. Grouard-Vogel, W. Sun, J. R. Mascola, E. Brachtel, R. Putvatana, M. K. Louder, L. Filgueira, M. A. Marovich, H. K. Wong, A. Blauvelt, G. S. Murphy, M. L. Robb, B. L. Innes, D. L. Birx, C. G. Hayes, and S. S. Frankel. 2000. Human skin Langerhans cells are targets of dengue virus infection. Nat. Med. 6:816-820. [DOI] [PubMed] [Google Scholar]
- 83.Yazi Mendoza, M., J. S. Salas-Benito, H. Lanz-Mendoza, S. Hernandez-Martinez, and R. M. del Angel. 2002. A putative receptor for dengue virus in mosquito tissues: localization of a 45-kDa glycoprotein. Am. J. Trop. Med. Hyg. 67:76-84. [DOI] [PubMed] [Google Scholar]
- 84.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao, B., E. Mackow, A. Buckler-White, L. Markoff, R. M. Chanock, C. J. Lai, and Y. Makino. 1986. Cloning full-length dengue type 4 viral DNA sequences: analysis of genes coding for structural proteins. Virology 155:77-88. [DOI] [PubMed] [Google Scholar]