Abstract
Background
Tissue-specific promoters controlling the expression of transgenes in Anopheles mosquitoes represent a valuable tool both for studying the interaction between these malaria vectors and the Plasmodium parasites they transmit and for novel malaria control strategies based on developing Plasmodium-refractory mosquitoes by expressing anti-parasitic genes. With this aim we have studied the promoter regions of two genes from the most important malaria vector, Anopheles gambiae, whose expression is strongly induced upon blood feeding.
Results
We analysed the A. gambiae Antryp1 and G12 genes, which we have shown to be midgut-specific and maximally expressed at 24 hours post-bloodmeal (PBM). Antryp1, required for bloodmeal digestion, encodes one member of a family of 7 trypsin genes. The G12 gene, of unknown function, was previously identified in our laboratory in a screen for genes induced in response to a bloodmeal. We fused 1.1 kb of the upstream regions containing the putative promoter of these genes to reporter genes and transformed these into the Indian malaria vector A. stephensi to see if we could recapitulate the expression pattern of the endogenous genes. Both the Antryp1 and G12 upstream regions were able to drive female-predominant, midgut-specific expression in transgenic mosquitoes. Expression of the Antryp1-driven reporter in transgenic A. stephensi lines was low, undetectable by northern blot analysis, and failed to fully match the induction kinetics of the endogenous Antryp1 gene in A. gambiae. This incomplete conservation of expression suggests either subtle differences in the transcriptional machinery between A. stephensi and A. gambiae or that the upstream region chosen lacked all the control elements. In contrast, the G12 upstream region was able to faithfully reproduce the expression profile of the endogenous A. gambiae gene, showing female midgut specificity in the adult mosquito and massive induction PBM, peaking at 24 hours.
Conclusions
Our studies on two putative blood-meal induced, midgut-specific promoters validate the use of G12 upstream regulatory regions to drive targeted transgene expression coinciding spatially and temporally with pre-sporogonic stages of Plasmodium parasites in the mosquito, offering the possibility of manipulating vector competence or performing functional studies on vector-parasite interactions.
Introduction
Anopheline mosquitoes are the most efficient vectors of the Plasmodium species that cause human malaria, a disease responsible for more than one million deaths per year[1]. Germline transformation technology for both A. gambiae and A. stephensi (the major malaria vectors in Africa and India respectively) now exists, opening the possibility to understand mechanisms of vector competence and the prospect of engineering Plasmodium-refractory anopheline mosquitoes. Irrespective of the molecular mechanism designed to attack the Plasmodium parasite, the development of malaria-refractory mosquitoes ultimately relies on the time- and tissue-regulated expression of anti-parasitic genes in concomitance with the appearance of target parasite developmental stages in order to maximise the effectiveness of transgene expression while minimizing the potential for reduced vigor associated with indiscriminate expression. This goal has driven the successful search for several tissue-specific promoters capable of driving expression in mosquito tissues relevant to parasite transmission, including the midgut, hemolymph, fat body and salivary glands[2], [3], [4], [5], [6]. In particular, blood meal-induced gut-transcribed promoters are ideal candidates to drive the expression of exogenous genes to attack the malaria parasite while it completes its life cycle within the mosquito host. This notion originates from the observation that the pre-sporogonic forms of the malaria parasite that develop within the midgut, from the gametocyte to the ookinetes, represent a major bottleneck in the parasite life cycle[7]. These forms, whose development is complete within 24 hours after a blood meal, expose on their surface a series of antigens that could be targeted by antibodies or other effector molecules before traversal of midgut epithelium is achieved[8]. Indeed, in the two laboratory-selected refractory mosquito strains, malaria parasite development is in fact blocked in the gut, either at the ookinete or the early oocyst developmental stages[9], [10]. Moreover, successful attempts in the past to engineer Plasmodium-refractory mosquitoes have all employed midgut-specific promoters to drive anti-parasitic gene expression[2], [11], [12], [13], [14].
Several strategies have been proposed to create refractory mosquitoes, including silencing the expression of essential molecules through RNAi in host cells invaded by the parasite, expression of toxins or single chain antibodies in close proximity to the invading parasite, and production of peptides to out-compete parasite binding to host epithelial receptors[2], [12], [13], [14], [15], [16]. Given the diversity of Plasmodium stages present within the midgut and the range of possible effector molecules, it is desirable to have a wide-range of promoters to target transgene expression most effectively to each particular stage.
In A. gambiae, a tightly clustered family of 7 gut-specific trypsin genes involved in digestion processes has been extensively studied[17], [18]. These genes are differentially regulated and the two late trypsins, Antryp1 (AGAP008296) and Antryp2 (AGAP008295), are induced upon a blood meal. In particular, Antryp1 is highly expressed in female guts and its induction peaks at 24 hours after blood feeding[17]. The region immediately upstream of Antryp1 has previously been shown as sufficient to direct midgut-specific expression in transgenic Drosophila melanogaster [19], a species evolutionally divergent from Anopheles by 250 million years[20].However, in that model the female-predominant expression was not maintained, and the Antryp1 promoter region did not respond to a “blood meal-related” induction process.
The G12 gene (AGAP006718), of unknown function, is similar to a number of cockroach allergens, and was initially identified in our laboratory in a screen for cDNAs that were enriched in A. gambiae after a bloodmeal. Our detailed analysis of the expression of G12 shows it to be expressed exclusively in the female midgut in response to a bloodmeal.
Here we describe and compare the temporally and spatially directed expression in A. stephensi mosquitoes of reporter genes driven by the putative promoter regions of the A. gambiae Antryp1 (pAntryp1) and G12 (pG12) genes. Both promoters could be of potential use as candidates to drive the time- and tissue-specific expression of transgenes in Anopheles mosquitoes.
Results and Discussion
Validation of pAntryp1 in transgenic mosquitoes
In order to assess the activity of the Antryp1 promoter in A. stephensi mosquitoes, we generated a reporter transcription unit containing the luciferase gene (luc) placed under the control of the 1.1 kb region immediately upstream of Antryp1 (pAntryp1). The selected promoter region spans nucleotides −1 to −1100 upstream of the start codon and encompasses most of the genomic region between the 5′ end of Antryp1 and the 3′ end of Antryp2 lying adjacent in the trypsin cluster. The pAntryp1-luc fragment was cloned, together with an Actin5C-EGFP marker cassette, within the arms of the minos transposable element, to generate transformation vector pMinLuc [21](Figure 1a).
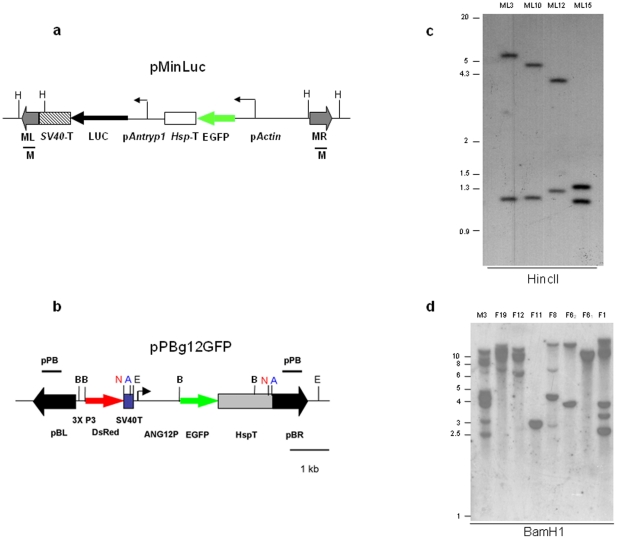
Figure 1. The piggyBac and minos transposable elements integrate into the genome of A. stephensi.
Maps of transformation vectors pMinLuc (a) and pPBg12GFP (b). pActin, D. melanogaster Actin5C promoter; HspT, D. melanogaster Hsp70 terminator sequence; pAntryp1, 1.1 kb of the upstream sequence of the Antryp1 gene; LUC, luciferase gene; SV40T, SV40 polyadenylation signal; 3xP3, artificial neuronal-specific promoter; dsRed, red fluorescent protein; ANG12P, 1.1 kb of putative G12 promoter region pBL, piggyBac left arm; pBR, piggyBac right arm; ML, minos left arm; MR, minos right arm; H, HincII; E, EcoRI; B, BamHI, N, NotI, A, AscI. Black bars represent the probes (pPB and M) used in the Southern Blot analyses. c: Southern Blot analysis of A. stephensi lines AsML3, AsML10, AsML12 and AsML15 digested with HincII and hybridised with probe M (Figure 1a). Each individual transposon integration is expected to yield two hybridising bands. Minos-mediated lines were characterised by single transposon insertions d: Southern blot of A.stephensi lines transformed with pPBg12GFP. Genomic DNA from 8 different transgenic lines, resulting from 7 different founder families, was digested with BamHI and hybridised with probe pPB, expected to give two hybridising bands per transposon insertion.
Transformation experiments yielded four separate founder females that produced transgenic lines, representing a transformation efficiency of 8.5% (Table S1). Segregation of the marker allele in each line indicated the occurrence of an insertion on a single chromosome (data not shown). Southern blot analysis of the 4 lines confirmed that a single integration of the transposable element had occurred in each line (Figure 1b). Two of the lines (AsML10 and AsML15) showed an insertion on the X chromosome, as assessed by both segregation data and in situ hybridisation analysis (data not shown). The minos integration site of transgenic line AsML10 was sequenced and the precise occurrence of minos-mediated integration confirmed (not shown, EMBL accession numbers AJ496323 and AJ496324).
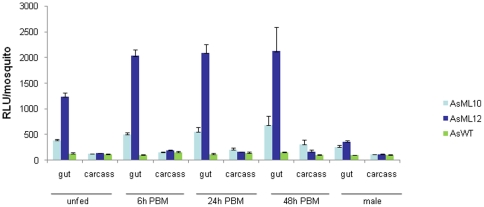
To assess the tissue-specificity of the pAntryp1 transcription unit, luciferase activity was analysed in detail in two A. stephensi transgenic lines, AsML10 and AsML12. Guts and carcasses of transgenic mosquitoes were dissected from males, unfed females and fed females at different time points after a blood meal, and assayed separately. Investigation of pAntryp1 expression revealed luciferase activity in the gut of unfed females of both lines, with levels 3- and 9-fold higher than wild type controls, respectively (Figure 2). In both lines, levels of luciferase activity in the carcass were similar to control levels at all time points analysed, indicating that the Antryp1 promoter is not active in those tissues. Luciferase activity was then assessed in female guts 6 hours after a blood meal. A 1.3- and 1.6-fold induction was observed in AsML10 and AsML12 respectively, when compared to unfed guts. At 24 h after a blood meal, when peak induction of endogenous Antryp1 transcription is observed in A. gambiae, the levels of luciferase in the two A. stephensi lines were not significantly increased over those observed at 6 hours, and they remained constant at the 48 h time point. Preliminary analysis of pAntryp1 activity in the two remaining A. stephensi lines, AsML3 and AsML15, showed luciferase levels similar to those detected in AsML10 (data not shown).
Figure 2. Blood meal inducibility and female predominance of transgenic luciferase activity driven by the Antryp1 promoter.
Comparison of luciferase activity in the transgenic lines AsML10 and AsML12 against wild type A.stephensi host (AsWT). Guts and carcasses from unfed females, females 6 h, 24 h, and 48 h post blood feeding and males were analysed individually. Bars show the mean of 20 samples for each condition. Error bars indicate the standard error of the mean.
In all transgenic lines analysed, there was a significant level of luciferase activity in the guts from transgenic male individuals, although considerably lower than that detected in the unfed females (Figure 2). This observation is in agreement with previous reports documenting low but significant endogenous levels of Antryp1 mRNA in the male guts of A. gambiae mosquitoes[17]. Despite the sex-bias, tissue-bias and bloodmeal induction of expression observed for the pAntryp1-luc cassette, other aspects of the endogenous Antryp1 gene were poorly maintained. Endogenous Antryp1 in A. gambiae is readily detectable by northern blot analysis upon blood feeding, yet we were unable to detect pAntryp1-driven luciferase transcripts in this fashion and the fold induction of luciferase expression was significantly less than that reported for the endogenous Antryp1 [17]. Given that the low level of reporter gene expression was seen in all transgenic lines tested it is unlikely that it is due to positional effects related to the chromosomal location of the transgene. Together these observations suggest either that the pAntryp1 region used in our construct, which contains most of the intervening region between Antryp1 and Antryp2, lacks some control elements for efficient transcription (a frequent problem with promoters isolated from clusters where gene expression may be co-regulated) or that similarly to experiments with Drosophila, the Antryp1 promoter from A. gambiae is not efficiently recognised in A. stephensi.
Interestingly, the endogenous late trypsin gene isolated from A. stephensi (Astryp1)[22] does not exhibit an identical expression profile to the homologous gene from A. gambiae. In particular, the blood-induction of Astryp1 from A. stephensi peaks at 6 hours after a blood meal (Crisanti, unpublished data). Accordingly, the altered induction kinetics of pAntryp1 observed after a blood meal in A. stephensi transgenic lines could reflect the normal activity of the transcriptional machinery in the two vector species. This is in agreement with what has been observed in transgenic Aedes aegypti mosquitoes, where the transcriptional profile of an A. gambiae carboxypeptidase (AgCP) promoter was shifted towards that of the endogenous Ae. aegypti gene [4]. Interestingly, in investigating the ability of the pAntryp1 promoter to drive the luc reporter in its endogenous host, we managed to generate a single transgenic A. gambiae line that showed strong and bloodmeal-induced luciferase expression that was midgut-specific and peaked at 24 h post bloodmeal reminiscent of the endogenous AgAntryp1 expression profile (Figure S1c). This observation lends weight to the hypothesis that some crucial cis and trans regulatory factors necessary for the correct functionality of pAntryp1 differ in A. stephensi. However we cannot rule out the possibility that the superior performance of this promoter construct in A. gambiae is due to position effects associated with the sequence context at the locus of transgene integration in this line.
Validation of pG12 promoter activity in transgenic mosquitoes
To generate a pG12-reporter construct, 1.1 kb of the region immediately 5′ of the putative coding sequence of the G12 gene was cloned upstream of the fluorescent reporter gene EGFP. This reporter construct was included in the piggyBac transformation vector pPB-G12EGFP, containing also the 3xP3-DsRed marker gene (Figure 1c)[23]. Transformation experiments with pPBg12EGFP yielded 7 different founder families, representing a minimum transformation efficiency of ∼13% (Table S2). As has been noted in other reports of piggyBac transformations[24], [25], [26], [27], transformation events were characterised by multiple insertions (Figure 1d, which represents the transgenic lines at generation 5).
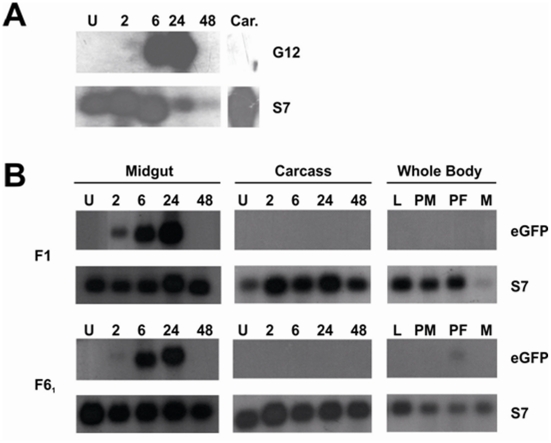
In order to fully characterise the expression pattern of the endogenous G12 gene in A. gambiae, northern blot analysis was carried out at different developmental stages and at various time points after a bloodmeal. We detected a strong midgut-specific expression of G12 in female adults, reaching peak levels of expression at 24 h PBM (Figure 3a). No expression was found in larvae, female carcasses or adult males, although a low level of expression was observed in the pupal stage (data not shown).
Figure 3. Reporter gene expression driven by the G12 promoter closely resembles the transcriptional of the endogenous female-specific, bloodmeal-inducible G12 gene.
(a) Northern blot analysis was performed to profile the expression of the endogenous G12 gene in A. gambiae. We detected maximal expression in midguts 24 hours PBM. No expression was detected in the carcass at this timepoint (Car.), or at any other time point (not shown). As an internal loading control, blots were stripped and re-probed for the ubiquitous ribosomal S7 gene. (b). Northern blot analysis of EGFP transcription under the control of the A. gambiae G12 promoter in transgenic A. stephensi. Two independent transgenic lines (F1 and F61) were tested. Stages and tissues tested were whole males (M), L3/L4 larvae (L), pupae (P), dissected midguts and carcasses unfed (U) and 2,6,24 and 48 hours PBM (2,6,24,48).
We thus examined reporter gene expression to see if the expression pattern of the endogenous A. gambiae G12 gene was recapitulated in the transgenic A. stephensi lines transformed with pPBg12GFP containing the pG12-EGFP reporter cassette. We performed a detailed northern blot analysis of EGFP transcripts in two transgenic lines, one with a single transgene insertion (F61) and one with multiple insertions (F1). In both cases expression was very similar to that of the endogenous AgG12, in terms of tissue- and sex-specificity and temporal profile in response to bloodmeal, with a clear peak at 24 h PBM (Figure 3b). These findings were confirmed in both lines by real time quantitative PCR experiments, which showed that reporter expression rose from being undetectable or less than 2% of the level of the abundant ribosomal S7 control transcript to a peak at 100-fold greater than the same gene at 24 h PBM (Table S3).
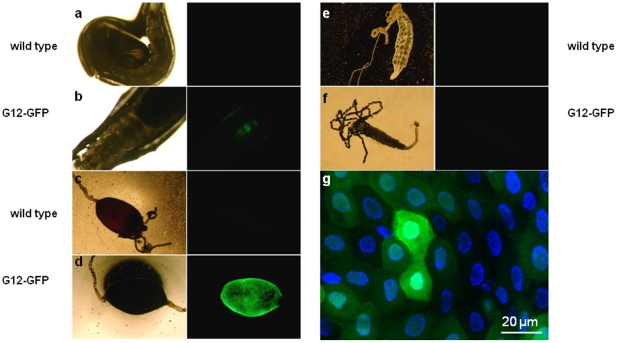
We then examined the tissue-specific accumulation of the EGFP protein expressed by the transgenic construct. Although the temporal pattern of EGFP accumulation will in part depend on the maturation kinetics and stability of the protein, the visualisation of this protein allows a full evaluation at the microscopic level of the tissue-specific localisation afforded by the G12 promoter. The utility of this promoter for directing tissue-specific transgene expression is supported by the fact that each of the 8 separate transgenic lines containing the construct showed a similar pattern of accumulation of the EGFP protein in the midgut in response to bloodmeal. This was clearly visible in midguts dissected at 24 h PBM while no signal was invisible in the unfed state or in any other tissue (see Figure 4a–f). Moreover, EGFP expression was also visible at the pupal stage, mirroring the endogenous G12 transcriptional profile. No expression was detected in the male (not shown). Although the pattern of EGFP accumulation was remarkably consistent, the intensity of the signal showed some variation between lines, presumably due to position effects associated with transgene insertion and/or to differences in number of piggyBac insertions.
Figure 4. EGFP driven by the G12 promoter accumulates in the midgut epithelium.
Pupae (a,b), 24 h PBM midguts (c,d) and unfed female midguts (e,f) from wild type mosquitoes and a pG12::EGFP transgenic line (F61) were visualised under transmission light (left panel) or epifluorescence (right panel). Reconstruction of a series of optical z-sections obtained by the Zeiss Apotome-assisted fluorescence microscopy (g) of 24 h PBM midguts dissected from transgenic mosquitoes reveals patchy distribution of EGFP-positive cells (green) across the epithelium. Cell nuclei are stained with DAPI (blue).
To examine pG12-driven accumulation of EGFP at the cellular level in the midgut epithelia we used fluorescence microscopy at high magnification. The mosquito midgut epithelium is composed of a single layer of cells. Interestingly, the distribution of GFP-expressing cells was non-uniform, with some cells strongly fluorescent surrounded by others showing intermediate or no EGFP expression (Figure 4g). This patchy expression profile suggests that while the high levels of inducible expression afforded by the G12 promoter bode well for the expression of secreted effector proteins active against stages of the Plasmodium parasite present within the midgut lumen, this promoter may not be suitable for the expression of molecules designed to be active intracelullarly against ookinetes as they traverse the midgut epithelium. This observation highlights the importance of microscopic analysis at the level of individual cells in determining the suitability of tissue-specific reporters for their purpose. Given this finding, it will be interesting to determine at the cellular level the expression of previously reported midgut-specific promoters used to drive expression of transgenes[2], [12], [14].
Since many anti-Plasmodium effector molecules are likely to have some deleterious effects on the mosquito as well as the parasite, it is desirable as a strategy to minimise expression of the effector to those tissues that encounter the parasite. In this light the bloodmeal inducibility and strict female midgut-specificity imparted by the pG12 construct give it a potential advantage over constructs containing promoter regions from other bloodmeal-inducible A. gambiae genes such as AgCP and Aper1, which show some constitutive activity in the male gut or the female gut or both[2], or the Antryp1 studied here.
Interestingly, there is evidence that expression of G12 in Anopheles gambiae and Aedes aegypti is massively induced in the midgut in response to a bloodmeal,yet the fold of this induction can be conditioned to some extent both by the presence of Plasmodium in a bloodmeal and by the refractory status of the mosquito[20], [28]. Further studies will assess whether this issue needs to be considered when using pG12 to drive expression of anti-Plasmodium effector molecules. pG12-reporter constructs may therefore be a valuable tool in studying any modulatory effect of the insect immune response on G12 expression.
Conclusions
We have described two promoter regions from A. gambiae that are able to direct, with different characteristics, bloodmeal-induced, midgut-enriched transgene expression in A. stephensi. In particular, pG12 is anticipated to be of great use in Anopheles species for tailoring transgene expression to coincide with midgut stages of the Plasmodium parasite, for the purposes of functional studies on mosquito-parasite interaction and for novel malaria control approaches through the targeted expression of anti-Plasmodium effector molecules.
Methods
Ethics statement
All animal work was conducted according to UK Home Office Regulations and approved under Home Office License PPL 70/6453.
Plasmid construction
Plasmid pMinLuc (Figure 1a) was derived from plasmid pMinEGFP[29] by inserting a Not1 cassette containing 1.1 kb of the region immediately upstream of the start codon of Antryp1 (AGAP008296)[22], amplified from genomic DNA from A.gambiae Suakoko strain, followed by the 1.7 kb Photinus pyralis luciferase cDNA[30] and SV40 polyadenylation signal.
The piggyBac-based plasmid pPBLuc (Figure S1a), containing identical reporter gene and marker elements to MinLuc used to transform A. gambiae was created by sequentially cloning the pAntryp1-luciferase and the Actin5C-EGFP cassettes from pMinLuc as Not1 fragments into the piggyBac-based plasmid pPBKOαNotI24
Plasmid pPBg12GFP was created by amplifying a 1.1 kb of the region immediately upstream of the G12 gene (AGAP006187) from the A. gambiae KWA strain (London School of Tropical Hygiene and Medicine) using the primers pG12forward (5′-cccgaattccacaataccggccctgaa-3′) and pG12reverse (5′-gggggatccgatgctgatgattggattgg-3′), designed to include EcoR1 and BamH1 restriction sites, respectively (italics). The amplified product was ligated as a EcoR1/BamH1 fragment to the EGFP gene in pMinEGFP[29] and a fragment containing the pG12, EGFP and the SV40 terminator sequence was inserted into the final transformation vector pBac[3xP3-DsRedaf][23], which includes the transformation marker DsRed driven by the neuronal-specific synthetic promoter 3xP3.
The helper plasmids pHSS6hsILMi20 and phsp-pBac, respectively providing the Minos and the piggyBac transposase genes, have previously been described[31], [32].
Embryo microinjection
To complete their gonotrophic cycle mosquitoes were fed on anaesthetized mice, in accordance with UK Home Office guidelines. A. stephensi embryos (strain sd500) were injected with a mixture of helper plasmid pHSS6hsILMi20 (100 µg/ml) and transformation vector pMinLuc (400 µg/ml) or a mixture of helper plasmid phsp-pBac (100 µg/ml) and pPBg12GFP (400 µg/ml), as previously described[29]. A. gambiae embryos (KWA strain, London School of Hygiene and Tropical Medicine) with a mixture of helper plasmid phsp-pBac (100 µg/ml) and pPBLuc (400 µg/ml). Hatched larvae were analysed on an inverted microscope under epifluorescence using either a GFP filter or a Texas Red filter to detect dsRED expression, depending on the transformation experiment.
Southern blot analyses
Approximately 4 µg of genomic DNA from transgenic A. stephensi adults was digested with the restriction endonuclease HincII (MinLuc lines), BamHI (PBg12GFP lines) or EcoR1(AgPBLuc line)and blotted according to standard protocols. Digested genomic DNA was separated on a 0.8% agarose gel and transferred onto a nylon membrane. The membranes were hybridised overnight at 65°C with different 32P-labelled probes. In the case of the minos-mediated A. stephensi transformations, genomic DNA was hybridised with the previously described probe M (Figure 1a)[29], encompassing both the left and the right arms of minos. In the case of the piggyBac-mediated PBg12GFP transformations, and the A.gambiae PBLuc line, the previously described probe PB[26], encompassing sequences of the left and right arms of the piggyBac transposon, was utilised (Figure 1c and Figure S1b).
Northern blot analysis
10 µg of total RNA was separated by electrophoresis on a 0.8% denaturing formaldehyde agarose gel and blotted to HYBOND™ nylon membrane (Amersham) through capillary transfer according to manufacturers protocol. Blots were hybridised overnight at 42°C in formamide-based hybridisation buffer with a 32P labelled probe corresponding to the entire G12 or EGFP coding sequence to probe for the endogenous G12 transcripts or the pG12-driven transgenic transcripts, respectively. As a loading control a similar probe was prepared corresponding to the coding sequence of the ribosomal gene S7.
Microscopy
Mosquito tissues were dissected in PBS and EGFP expression was visualised under epifluorescence using a EGFP filter.
Fluorescence microscopy of the midgut epithelium was performed on midguts dissected 24 h PBM in PBS and rinsed to remove blood bolus. Midguts were fixed in methanol-free 4% formaldehyde (Pierce) in PBS for 30 min at room temperature and washed 3 times for 15 min in 0.1% Tween-20 PBS. Midguts were then mounted on slides containing Vectashield mounting medium with DAPI (Vectorlabs. Inc.) with cover slips. Multiplane z-stacks were collected by a Zeiss fluorescence microscope Axiovert 200 M equipped with a Zeiss Apotome module. (www.Zeiss.com).
Luciferase Assay
Four- to five-day old mosquitoes were dissected to separate the midgut from the carcass. Each sample was homogenised in 150 µl of Cell Culture Lysis Reagent (Promega), supplemented with 5 mM phenylmethylsulfonylfluoride and 7% reconstituted milk powder to block degradation of luciferase by endogenous mosquito protease activity. Immediate assay of the samples in the presence of reconstituted milk powder was essential for luciferase detection in female guts. A volume of 30 µl of homogenate was added to 100 µl of Luciferase Assay Substrate (Promega), and light emission measured for 15 seconds with a Berthold luminometer. Twenty guts and carcasses were assayed for each time point.
Supporting Information
Generation and analysis of transgenic A.gambiae line AgPB1. A: map of transformation vector pPBLuc. pActin, D. melanogaster Actin5C promoter; HspT, D. melanogaster Hsp70 terminator sequence; pAntryp1, 1.1 kb of the upstream sequence of the Antryp1 gene; LUC, luciferase gene; SV40T, SV40 polyadenylation signal; pBL, piggyBac left arm; pBR, piggyBac right arm; E, EcoRI. Black bars represent the probe (PB) used in the Southern Blot analyses. B: Southern Blot analysis of genomic DNA from the A. gambiae line AgPB1, digested with EcoRI and hybridised with probe P (Figure 1A). Each single insertion is expected to give two hybridising bands, corresponding to each arm of the piggyBac element. C: Comparison of luciferase activity in A. gambiae line AgPB1 with the transgenic A. stephensi lines AsML10 and AsML12. Guts and carcasses from unfed females (U), females 6 h, 24 h, and 48 h post blood feeding and males were analysed individually. Bars show the mean of 20 samples for each condition. Error bars indicate the standard error of the mean.
(TIF)
Generation of transformants using plasmid pMinLuc. The 47 surviving adults from a total of 167 injected embryos were outcrossed with wild type A. stephensi in groups of same-sex individuals. The 24 females from the female group were allowed to lay eggs in isolation to determine the number of single founders. 4 female founders produced fluorescent individuals among their G1 progeny, indicative of a germline integration event. Transgenic progeny were interbred to achieve homozygosity of the transgene. Asterisks denote those lines that were assayed in detail for luciferase activity.
(DOCX)
Generation of transformants using pPB-G12EGFP. The 55 surviving adults from a total of 391 injected embryos were outcrossed with wild type A. stephensi in 4 groups of same-sex individuals. The 31 females from the female group were allowed to lay eggs in isolation (F1-F31) to determine the number of single founders. Segregation patterns of the transgene in subsequent outcrossing experiments revealed that many lines contained multiple transgene insertions at separate loci. Where possible lines were bred to achieve homozygosity at the transgenic loci.
(DOCX)
Quantitative real time PCR analysis of EGFP reporter expression driven by the G12 promoter in A. stephensi . Quantification was performed using TaqManTM primers and probes (ABI) specific for the EGFP gene and the S7 ribosomal gene as a reference control, as previously described [1]. The PCR cycle number (Ct) at which a threshold of amplification was calculated for each gene and the level of EGFP was calculated relative to the S7 internal control using the delta Ct method [2]. Supplementary References 1. Brown, A. E., Bugeon, L., Crisanti, A. & Catteruccia, F. Stable and heritable gene silencing in the malaria vector Anopheles stephensi. Nucleic Acids Res 31, e85 (2003). 2. Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101-8 (2008).
(DOCX)
Acknowledgments
We thank Elena Levashina for fluorescence microscopy and Nikolai Windbichler for help with preparation of figures. The KWA strain of A. gambiae was a kind gift by the late Chris Curtis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This project was supported by grants from the Wellcome Trust, BBSRC and from the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement N° 242095. FC received a bursary from the Fondazione Cenci Bolognetti. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO. Geneva; 2009. World Malaria Report 2009. [Google Scholar]
- 2.Abraham EG, Donnelly-Doman M, Fujioka H, Ghosh A, Moreira L, et al. Driving midgut-specific expression and secretion of a foreign protein in transgenic mosquitoes with AgAper1 regulatory elements. Insect Mol Biol. 2005;14:271–279. doi: 10.1111/j.1365-2583.2004.00557.x. [DOI] [PubMed] [Google Scholar]
- 3.Chen XG, Marinotti O, Whitman L, Jasinskiene N, James AA, et al. The Anopheles gambiae vitellogenin gene (VGT2) promoter directs persistent accumulation of a reporter gene product in transgenic Anopheles stephensi following multiple bloodmeals. Am J Trop Med Hyg. 2007;76:1118–1124. [PubMed] [Google Scholar]
- 4.Moreira LA, Edwards MJ, Adhami F, Jasinskiene N, James AA, et al. Robust gut-specific gene expression in transgenic Aedes aegypti mosquitoes. Proc Natl Acad Sci U S A. 2000;97:10895–10898. doi: 10.1073/pnas.97.20.10895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nirmala X, Marinotti O, Sandoval JM, Phin S, Gakhar S, et al. Functional characterization of the promoter of the vitellogenin gene, AsVg1, of the malaria vector, Anopheles stephensi. Insect Biochem Mol Biol. 2006;36:694–700. doi: 10.1016/j.ibmb.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 6.Yoshida S, Watanabe H. Robust salivary gland-specific transgene expression in Anopheles stephensi mosquito. Insect Mol Biol. 2006;15:403–410. doi: 10.1111/j.1365-2583.2006.00645.x. [DOI] [PubMed] [Google Scholar]
- 7.Sinden RE, Dawes EJ, Alavi Y, Waldock J, Finney O, et al. Progression of Plasmodium berghei through Anopheles stephensi is density-dependent. PLoS Pathog. 2007;3:e195. doi: 10.1371/journal.ppat.0030195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sinden RE, Winger L, Carter EH, Hartley RH, Tirawanchai N, et al. Ookinete antigens of Plasmodium berghei: a light and electron-microscope immunogold study of expression of the 21 kDa determinant recognized by a transmission-blocking antibody. Proc R Soc Lond B Biol Sci. 1987;230:443–458. doi: 10.1098/rspb.1987.0028. [DOI] [PubMed] [Google Scholar]
- 9.Collins FH, Sakai RK, Vernick KD, Paskewitz S, Seeley DC, et al. Genetic selection of a Plasmodium-refractory strain of the malaria vector Anopheles gambiae. Science. 1986;234:607–610. doi: 10.1126/science.3532325. [DOI] [PubMed] [Google Scholar]
- 10.Vernick KD, Barreau C, Seeley DC. Plasmodium: a quantitative molecular assay for detection of sporogonic-stage malaria parasites. Exp Parasitol. 1995;81:436–444. doi: 10.1006/expr.1995.1136. [DOI] [PubMed] [Google Scholar]
- 11.Franz AW, Sanchez-Vargas I, Adelman ZN, Blair CD, Beaty BJ, et al. Engineering RNA interference-based resistance to dengue virus type 2 in genetically modified Aedes aegypti. Proc Natl Acad Sci U S A. 2006;103:4198–4203. doi: 10.1073/pnas.0600479103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito J, Ghosh A, Moreira LA, Wimmer EA, Jacobs-Lorena M. Transgenic anopheline mosquitoes impaired in transmission of a malaria parasite. Nature. 2002;417:452–455. doi: 10.1038/417452a. [DOI] [PubMed] [Google Scholar]
- 13.Kim W, Koo H, Richman AM, Seeley D, Vizioli J, et al. Ectopic expression of a cecropin transgene in the human malaria vector mosquito Anopheles gambiae (Diptera: Culicidae): effects on susceptibility to Plasmodium. J Med Entomol. 2004;41:447–455. doi: 10.1603/0022-2585-41.3.447. [DOI] [PubMed] [Google Scholar]
- 14.Moreira LA, Ito J, Ghosh A, Devenport M, Zieler H, et al. Bee venom phospholipase inhibits malaria parasite development in transgenic mosquitoes. J Biol Chem. 2002;277:40839–40843. doi: 10.1074/jbc.M206647200. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida S, Matsuoka H, Luo E, Iwai K, Arai M, et al. A single-chain antibody fragment specific for the Plasmodium berghei ookinete protein Pbs21 confers transmission blockade in the mosquito midgut. Mol Biochem Parasitol. 1999;104:195–204. doi: 10.1016/s0166-6851(99)00158-9. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida S, Shimada Y, Kondoh D, Kouzuma Y, Ghosh AK, et al. Hemolytic C-type lectin CEL-III from sea cucumber expressed in transgenic mosquitoes impairs malaria parasite development. PLoS Pathog. 2007;3:e192. doi: 10.1371/journal.ppat.0030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller HM, Catteruccia F, Vizioli J, della Torre A, Crisanti A. Constitutive and blood meal-induced trypsin genes in Anopheles gambiae. Exp Parasitol. 1995;81:371–385. doi: 10.1006/expr.1995.1128. [DOI] [PubMed] [Google Scholar]
- 18.Muller HM, Crampton JM, della Torre A, Sinden R, Crisanti A. Members of a trypsin gene family in Anopheles gambiae are induced in the gut by blood meal. Embo J. 1993;12:2891–2900. doi: 10.1002/j.1460-2075.1993.tb05951.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skavdis G, Siden-Kiamos I, Muller HM, Crisanti A, Louis C. Conserved function of anopheles gambiae midgut-specific promoters in the fruitfly. Embo J. 1996;15:344–350. [PMC free article] [PubMed] [Google Scholar]
- 20.Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
- 21.Catteruccia F, Godfray HC, Crisanti A. Impact of genetic manipulation on the fitness of Anopheles stephensi mosquitoes. Science. 2003;299:1225–1227. doi: 10.1126/science.1081453. [DOI] [PubMed] [Google Scholar]
- 22.Giannoni F, Muller HM, Vizioli J, Catteruccia F, Kafatos FC, et al. Nuclear factors bind to a conserved DNA element that modulates transcription of Anopheles gambiae trypsin genes. J Biol Chem. 2001;276:700–707. doi: 10.1074/jbc.M005540200. [DOI] [PubMed] [Google Scholar]
- 23.Horn C, Schmid BG, Pogoda FS, Wimmer EA. Fluorescent transformation markers for insect transgenesis. Insect Biochem Mol Biol. 2002;32:1221–1235. doi: 10.1016/s0965-1748(02)00085-1. [DOI] [PubMed] [Google Scholar]
- 24.Kokoza V, Ahmed A, Wimmer EA, Raikhel AS. Efficient transformation of the yellow fever mosquito Aedes aegypti using the piggyBac transposable element vector pBac[3xP3-EGFP afm]. Insect Biochem Mol Biol. 2001;31:1137–1143. doi: 10.1016/s0965-1748(01)00120-5. [DOI] [PubMed] [Google Scholar]
- 25.Lombardo F, Lycett GJ, Lanfrancotti A, Coluzzi M, Arca B. Analysis of apyrase 5′ upstream region validates improved Anopheles gambiae transformation technique. BMC Res Notes. 2009;2:24. doi: 10.1186/1756-0500-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nolan T, Bower TM, Brown AE, Crisanti A, Catteruccia F. piggyBac-mediated germline transformation of the malaria mosquito Anopheles stephensi using the red fluorescent protein dsRED as a selectable marker. J Biol Chem. 2002;277:8759–8762. doi: 10.1074/jbc.C100766200. [DOI] [PubMed] [Google Scholar]
- 27.Tamura T, Thibert C, Royer C, Kanda T, Abraham E, et al. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat Biotechnol. 2000;18:81–84. doi: 10.1038/71978. [DOI] [PubMed] [Google Scholar]
- 28.Abraham EG, Islam S, Srinivasan P, Ghosh AK, Valenzuela JG, et al. Analysis of the Plasmodium and Anopheles transcriptional repertoire during ookinete development and midgut invasion. J Biol Chem. 2004;279:5573–5580. doi: 10.1074/jbc.M307582200. [DOI] [PubMed] [Google Scholar]
- 29.Catteruccia F, Nolan T, Loukeris TG, Blass C, Savakis C, et al. Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature. 2000;405:959–962. doi: 10.1038/35016096. [DOI] [PubMed] [Google Scholar]
- 30.de Wet JR, Wood KV, Helinski DR, DeLuca M. Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc Natl Acad Sci U S A. 1985;82:7870–7873. doi: 10.1073/pnas.82.23.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Handler AM, Harrell RA., 2nd Germline transformation of Drosophila melanogaster with the piggyBac transposon vector. Insect Mol Biol. 1999;8:449–457. doi: 10.1046/j.1365-2583.1999.00139.x. [DOI] [PubMed] [Google Scholar]
- 32.Klinakis AG, Loukeris TG, Pavlopoulos A, Savakis C. Mobility assays confirm the broad host-range activity of the Minos transposable element and validate new transformation tools. Insect Mol Biol. 2000;9:269–275. doi: 10.1046/j.1365-2583.2000.00183.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation and analysis of transgenic A.gambiae line AgPB1. A: map of transformation vector pPBLuc. pActin, D. melanogaster Actin5C promoter; HspT, D. melanogaster Hsp70 terminator sequence; pAntryp1, 1.1 kb of the upstream sequence of the Antryp1 gene; LUC, luciferase gene; SV40T, SV40 polyadenylation signal; pBL, piggyBac left arm; pBR, piggyBac right arm; E, EcoRI. Black bars represent the probe (PB) used in the Southern Blot analyses. B: Southern Blot analysis of genomic DNA from the A. gambiae line AgPB1, digested with EcoRI and hybridised with probe P (Figure 1A). Each single insertion is expected to give two hybridising bands, corresponding to each arm of the piggyBac element. C: Comparison of luciferase activity in A. gambiae line AgPB1 with the transgenic A. stephensi lines AsML10 and AsML12. Guts and carcasses from unfed females (U), females 6 h, 24 h, and 48 h post blood feeding and males were analysed individually. Bars show the mean of 20 samples for each condition. Error bars indicate the standard error of the mean.
(TIF)
Generation of transformants using plasmid pMinLuc. The 47 surviving adults from a total of 167 injected embryos were outcrossed with wild type A. stephensi in groups of same-sex individuals. The 24 females from the female group were allowed to lay eggs in isolation to determine the number of single founders. 4 female founders produced fluorescent individuals among their G1 progeny, indicative of a germline integration event. Transgenic progeny were interbred to achieve homozygosity of the transgene. Asterisks denote those lines that were assayed in detail for luciferase activity.
(DOCX)
Generation of transformants using pPB-G12EGFP. The 55 surviving adults from a total of 391 injected embryos were outcrossed with wild type A. stephensi in 4 groups of same-sex individuals. The 31 females from the female group were allowed to lay eggs in isolation (F1-F31) to determine the number of single founders. Segregation patterns of the transgene in subsequent outcrossing experiments revealed that many lines contained multiple transgene insertions at separate loci. Where possible lines were bred to achieve homozygosity at the transgenic loci.
(DOCX)
Quantitative real time PCR analysis of EGFP reporter expression driven by the G12 promoter in A. stephensi . Quantification was performed using TaqManTM primers and probes (ABI) specific for the EGFP gene and the S7 ribosomal gene as a reference control, as previously described [1]. The PCR cycle number (Ct) at which a threshold of amplification was calculated for each gene and the level of EGFP was calculated relative to the S7 internal control using the delta Ct method [2]. Supplementary References 1. Brown, A. E., Bugeon, L., Crisanti, A. & Catteruccia, F. Stable and heritable gene silencing in the malaria vector Anopheles stephensi. Nucleic Acids Res 31, e85 (2003). 2. Schmittgen, T. D. & Livak, K. J. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3, 1101-8 (2008).
(DOCX)