Abstract
As also found for other retroviruses, the Rous sarcoma virus structural protein Gag is necessary and sufficient for formation of virus-like particles (VLPs). Purified polypeptide fragments comprising most of Gag spontaneously assemble in vitro at pH 6.5 into VLPs lacking a membrane, a process that requires nucleic acid. We showed previously that the minimum length of a DNA oligonucleotide that can support efficient assembly is 16 nucleotides (nt), twice the protein's binding site size. This observation suggests that the essential role of nucleic acid in assembly is to promote the formation of Gag dimers. In order to gain further insight into the role of dimerization, we have studied the assembly properties of two proteins, a nearly full-length Gag (ΔMBDΔPR) capable of proper in vitro assembly and a smaller Gag fragment (CTD-NC) capable of forming only irregular aggregates but with the same pH and oligonucleotide length requirements as for assembly with the larger protein. In analyses by sedimentation velocity and by cross-linking, both proteins remained monomeric in the absence of oligonucleotides or in the presence of an oligonucleotide of length 8 nt (GT8). At pH 8, which does not support assembly, binding to GT16 induced the formation of dimers of ΔMBDΔPR but not of CTD-NC, implying that dimerization requires the N-terminal domain of the capsid moiety of Gag. Assembly of VLPs was induced by shifting the pH of dimeric complexes of ΔMBDΔPR and GT16 from 8 to 6.5. An analogue of GT16 with a ribonucleotide linkage in the middle also supported dimer formation at pH 8. Even after quantitative cleavage of the oligonucleotide by treatment of the complex with RNase, these dimers could be triggered to undergo assembly by pH change. This result implies that protein-protein interactions stabilize the dimer. We propose that binding of two adjacent Gag molecules on a stretch of nucleic acid leads to protein-protein interactions that create a Gag dimer and that this species has an exposed surface not present in monomers which allows polymerization of the dimers into a spherical shell.
The retrovirus polyprotein Gag is the structural protein that assembles into virus particles that bud from the plasma membrane. Late in this process, the nascent particles undergo a maturation step in which Gag is cleaved by the viral protease (PR) to generate the mature proteins matrix (MA), capsid (CA), and nucleocapsid (NC) as well as several small peptides or proteins. NC is a small basic domain of Gag that binds to genomic or other RNA during assembly. By a mechanism that remains to be elucidated, it promotes protein-protein interactions that lead to assembly, probably indirectly by virtue of binding to RNA (5, 7, 13, 15, 44, 45, 55). CA is the domain of Gag that is probably the major locus of protein-protein interactions during assembly (51). After liberation as a mature protein, CA forms the shell of the viral core (19, 35).
In vitro systems have been developed to study assembly for several retroviruses, including Mason-Pfizer monkey virus (28, 43), Rous sarcoma virus (RSV) (10, 11, 37, 54), and human immunodeficiency virus type 1 (HIV-1) (8, 9, 19-22, 36, 41, 46, 47, 50). The Gag protein purified from Escherichia coli or translated in vitro spontaneously forms spherical or tubular virus-like particles (VLPs). Under physiological conditions, both the NC domain and nucleic acid are required for assembly of spherical particles, which closely resemble authentic immature virions lacking a membrane. In a previous report, we demonstrated that DNA oligonucleotides support efficient assembly (37). The minimum size of a DNA oligonucleotide required, 16 nucleotides (nt), was determined to be twice the binding site size of Gag. Based on this observation, we hypothesized that the role of nucleic acid in assembly is to promote formation of Gag dimers, which is the species that polymerizes into immature VLPs. This hypothesis is consistent with the ability of a heterologous protein dimerization domain, a leucine zipper, to functionally replace NC (2, 24, 56).
Little is known about the Gag sequences involved in protein dimerization or multimerization. HIV-1 CA, which is composed of two domains like all retroviral CA proteins (4, 12, 17, 23, 26, 27, 40, 53), dimerizes in vitro through its C-terminal domain (CTD) (17). Based on their similar folding, the CA proteins of other retroviruses have been assumed to form dimers in a similar fashion. However, neither RSV CA (27) nor other CA proteins (26) have been observed to dimerize in solution. One model that provides a framework for picturing Gag-Gag interaction derives from the cryo-electron microscopy (cryo-EM) reconstruction of tubes assembled in vitro with HIV-1 CA (35). This structure presumably represents the CA-CA interaction in a mature viral core. In the reconstruction the only types of protein-protein interactions are homotypic. Hexagonal CA rings are held together by contacts between the N-terminal domains (NTDs), while each CA molecule is linked to a CA in an adjoining hexagonal ring by interactions between the CTDs. The same organization probably applies to retroviruses in general, as inferred from two-dimensional crystals (18, 38, 39). More recent data in the HIV system imply that heterotypic NTD-CTD interactions also occur in the mature capsid shell (31, 32). It remains unclear to what extent any of these interactions are relevant for assembly of immature particles. From an extensive mutagenesis analysis of the CA domain of Gag in HIV-1 it can be inferred that surfaces important for mature assembly overlap only partially with surfaces of CA important for immature assembly (51; B. K. Ganser, U. K. von Schwedler, K. M. Stray, C. Aiken, and W. I. Sundquist, submitted for publication).
In this report we describe the oligomeric state of RSV Gag in the presence of two DNA oligonucleotides, GT8 and GT16, which are long enough to allow the binding of one or two Gag molecules, respectively. At pH 8.0, a pH that does not allow in vitro assembly, Gag remains monomeric in the presence of GT8 but forms dimers in the presence of GT16. These dimers are held together not only by the oligonucleotide but by protein-protein interactions. The NTDs are necessary for the formation of the stable Gag dimers. Rapid in vitro assembly of the dimers into VLPs can be triggered by reduction of the pH to 6.5. The acidic pH appears to be required for inducing conformational changes in CTD, which are crucial for the protein multimerization step leading to VLP formation.
MATERIALS AND METHODS
DNA constructs and protein purification.
The plasmids pET-ΔMBDΔPR and pET-CTD-NC were constructed by using common subcloning techniques based on the pET3XC vector and propagated in the DH5α strain of E. coli. Construction of pET-ΔMBDΔPR has been described previously (11). pET-CTD-NC was created by replacing an XhoI-SacII fragment of pET-ΔMBDΔPR with a PCR product containing the CTD region with the same restriction sites. E. coli BL21 DE3(pLysS) cells were grown and induced for protein expression as previously described (11, 37, 54).
The ΔMBDΔPR protein was purified as described previously (37). Briefly, frozen cell pellets were resuspended on ice in buffer A (20 mM Tris · Cl [pH 8.0], 0.5 M NaCl, 0.1% Nonidet P-40, 10% glycerol, 50 μM ZnCl2, 10 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]) at 25 ml/liter of cell culture, and the cells were broken by sonication. Insoluble debris, ribosomes, and nucleic acids were removed by addition of 0.3% polyethyleneimine followed by ultracentrifugation at 45,000 rpm for 3 h using a Ti50.2 rotor. Soluble protein was first precipitated by addition of ammonium sulfate to 25% saturation and then resuspended in buffer B (20 mM Tris · Cl [pH 8.0], 50 μM ZnCl2, 10 mM DTT, 1 mM PMSF) plus 0.1 M NaCl at 5 ml/liter of cell culture. Insoluble material was removed by centrifugation, and the supernatant was applied to a DEAE-cellulose (DE-52) column. The resin was washed with buffer B plus 0.1 M NaCl. The flowthrough and wash fractions were pooled and loaded onto a phosphocellulose (Whatman P11) column. The resin with bound protein was first washed with buffer B plus 0.1 M NaCl and then with buffer B plus 0.3 M NaCl, and then the protein was eluted with buffer B plus 0.5 M NaCl. The final purified protein was first dialyzed against buffer B plus 0.1 M NaCl and 200 μM ZnCl2 overnight and then dialyzed against buffer B plus 0.1 M NaCl and 20 μM ZnCl2 for 4 h to remove the bulk of the free zinc. Protein aliquots were stored in liquid nitrogen for no more than 2 months. CTD-NC was purified using the same strategy, except that 35% saturated ammonium sulfate was used to precipitate protein.
Protein concentration was determined by spectrophotometry. The A260/A280 ratio of purified protein was determined to be about 0.7, indicating lack of significant nucleic acid contamination. The final protein was about 90% pure as judged by Coomassie blue staining after sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE). The protein used in the experiments described here was frozen and thawed only once.
Analysis of in vitro assembly.
A direct dilution method was used for in vitro assembly as described previously (37). In brief, protein (typically 5 mg/ml) in buffer B plus 0.1 M NaCl was first diluted with 4 volumes of buffer D (50 mM morpholineethanesulfonic acid [MES] [pH 6.5], 0.1 M NaCl) followed by addition of DNA oligonucleotide (10% [wt/wt] ratio of nucleic acid to protein). Assembly reactions were incubated at room temperature for 30 min. Particles formed under these conditions were negatively stained with 2% uranyl acetate (pH 5) on Formvar-carbon coated grids. In some experiments, particles first were collected by centrifugation in an Eppendorf microcentrifuge for 30 min at 14,000 rpm.
The hybrid oligonucleotide GT16rC (dGdTdGdTdGdTdGdTrCrCdGdTdGdTdGdTdGdT) was end labeled with [32P]ATP and polynucleotide kinase by standard procedures. When labeled GT16rC was used to promote assembly, the protein and oligonucleotide first were mixed in buffer B with 0.1 M NaCl. RNase then was added to the mixture at a 1% (wt/wt) RNase to protein ratio, followed by incubation at room temperature for 20 min. To allow assembly, the reaction was then adjusted to 50 mM MES (pH 6.5) using 1 M MES, followed by another 30-min incubation at room temperature. Prior to pH adjustment, a portion of the protein-oligonucleotide mixture was loaded directly into the well of a 25% polyacrylamide-8 M urea gel and electrophoresed, in order to verify complete cleavage of the oligonucleotide by RNase. Brief diethyl pyrocarbonate treatment of the mixture after the RNase digestion was used to eliminate the possibility that cleavage occurred after the sample loading step. End-labeled GT16 and GT8 were also included as size markers. After electrophoresis in standard Tris-acetate buffer, the gel was dried and exposed to film.
Sedimentation and cross-linking.
For rate zonal sedimentation assays, a 4-ml sucrose gradient was made in buffer E (0.1 M NaCl, 50 μM ZnCl2, 10 mM DTT, 1 mM PMSF) with 20 mM Tris · Cl (pH 8.0) or with 20 mM MES (pH 6.5), using eight steps of sucrose between 10 and 30% (wt/vol). The sucrose was allowed to diffuse overnight at 4°C to form a smooth gradient. In a typical reaction mixture (200 μl), proteins (1 mg/ml) and DNA oligonucleotides (100 μg/ml) were mixed in buffer E with 20 mM Tris · Cl (pH 8.0) or with 20 mM MES (pH 6.5), followed by incubation at room temperature for 30 min. The mixture or protein size markers (typically containing myosin [220 kDa], phosphorylase B [97 kDa], bovine serum albumin [66 kDa], and ovalbumin [46 kDa]) were then loaded on top of the sucrose gradient, followed by centrifugation at 50,000 rpm for 24 h using an SW60 rotor. Seventeen fractions (200 μl each) were collected from the top of the gradient and analyzed by SDS-PAGE.
For cross-linking assays, in a typical reaction mixture (40 μl) proteins (1.5 μM) and DNA oligonucleotides (10% [wt/wt] ratio of nucleic acid to protein) were mixed with buffer F (50 mM triethanolamine [pH 8.0] and 0.1 M NaCl). Dimethylsuberimidate (DMS) was added to the mixture to a final concentration of 2 mM, and the reaction was incubated at room temperature for 2 h before quenching with 0.1 M glycine and analysis by SDS-PAGE.
RESULTS
GT16 induces the formation of ΔMBDΔPR dimers.
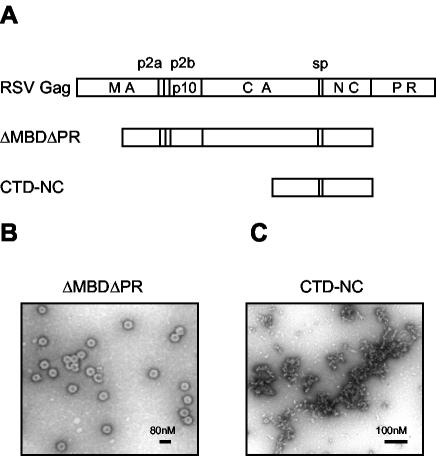
To study assembly in vitro, we have used a truncated version of full-length Gag called ΔMBDΔPR. This protein, which lacks the N-terminal half of MA as well as the PR domain, can be purified readily as a soluble protein from E. coli (Fig. 1A). At a pH between 6.0 and 7.0 and in the presence of nucleic acid, ΔMBDΔPR assembles spontaneously into spherical particles (Fig. 1B). In ultrastructure these VLPs are indistinguishable from immature virions without a membrane, as demonstrated by negative-stain and thin-section electron microscopy (EM) and scanning transmission EM, as well as cryo-EM (54). With DNA oligonucleotides composed of the dinucleotide repeat GT (GT-mers), the binding site size of ΔMBDΔPR in the presence of Zn2+ ions is about 8 nt, while the minimum length of oligonucleotide required for efficient assembly is 16 nt (37).
FIG. 1.
Schematic representation of proteins and electron micrographs of assembly products. (A) The structure of RSV Gag is shown as the uppermost rectangle, with vertical lines denoting proteolytic cleavage sites. The proteins ΔMBDΔPR and CTD-NC are the focus of the present study. (B) Negative-stain EM of particles formed by ΔMBDΔPR at pH 6.5 and 0.1 M NaCl in the presence of the oligonucleotide GT50. (C) Negative-stain EM of aggregates formed by CTD-NC at pH 6.5 and 0.1 M NaCl in the presence of GT50.
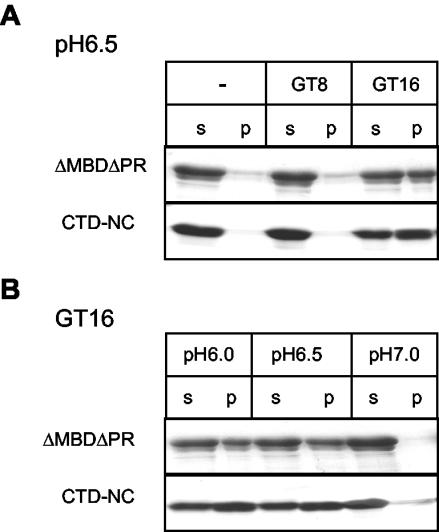
In the present study, we first used a simple centrifugation assay to directly compare the effect of oligonucleotide length and pH on ΔMBDΔPR assembly. VLPs were collected by centrifugation for 30 min in a microcentrifuge, while the unassembled protein remained in the supernatant. At pH 6.5, about one-half of the total ΔMBDΔPR protein was found in the pellet fraction in the presence of GT16, indicating efficient assembly under these conditions (Fig. 2A). Very little protein could be spun down in the absence of nucleic acid or in the presence of GT8, even though this oligonucleotide binds to the protein (37), implying that Gag-nucleic acid binding is necessary but not sufficient for assembly. In the presence of GT16, assembly was most efficient at pH 6.0 and 6.5 and greatly reduced at pH 7.0 (Fig. 2B). The pH dependence of assembly became less stringent with longer nucleic acids, since with GT50 a few assembled particles could be detected even at pH 8.0 (data not shown).
FIG. 2.
Effect of oligonucleotide length and pH on assembly and aggregation. (A) ΔMBDΔPR or CTD-NC were mixed with no oligonucleotide or GT8 or GT16 at pH 6.5 and incubated for 30 min at room temperature. The particles or aggregates were collected by centrifugation for 30 min in a microcentrifuge. The supernatant (s) and the pellet (p) fractions were analyzed by SDS-PAGE and Coomassie blue staining. (B) A similar experiment was carried out with GT16 at pH 6.0, 6.5, or 7.0.
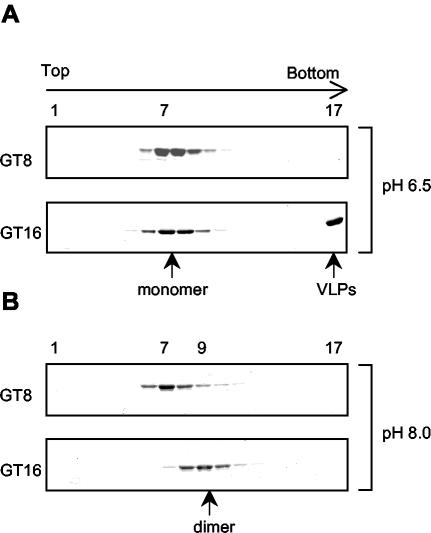
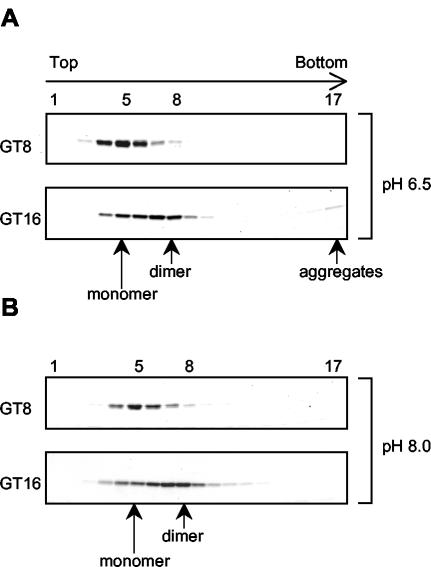
We utilized velocity sedimentation analysis to search for the Gag dimers as well as other possible assembly intermediates. The oligomeric state of ΔMBDΔPR was determined in the absence of nucleic acid or in the presence of GT8 or GT16. The sedimentation assays first were performed under assembly conditions, pH 6.5 and 0.1 M NaCl. ΔMBDΔPR sedimented as a monomer in the absence of nucleic acid (data not shown), consistent with the previous report that RSV CA does not dimerize even at very high protein concentration (27). In the presence of GT8, ΔMBDΔPR also remained monomeric (Fig. 3A). That binding of GT8 to ΔMBDΔPR occurred was demonstrated by nitrocellulose filter binding (37) and by the observation that the GT8 peak (measured by A260) shifted in the presence of Gag to overlap with the protein peak (measured by A280) (data not shown). In some experiments the protein peak with GT8 shifted by one fraction towards the bottom of the gradient, suggesting that a conformational change might occur in the NC domain of ΔMBDΔPR upon binding. Structural studies have shown that NC becomes more compact when it binds to nucleic acid (3, 16, 34), which could explain the slight increase in sedimentation rate. In the presence of GT16, the VLPs formed under these conditions sedimented to the pellet in the bottom fraction of the gradient, while the unassembled ΔMBDΔPR remained as monomers that sedimented to the middle of the gradient (Fig. 3A). There was little evidence for the presence of Gag dimers or other assembly intermediates, which we interpret to mean that the Gag dimers are transient intermediates and are rapidly incorporated into the growing particle at this protein concentration.
FIG. 3.
Sedimentation analysis of ΔMBDΔPR in the presence of GT8 or GT16. The protein-oligonucleotide mixtures were layered on a 10-to-30% (wt/vol) sucrose gradient and centrifuged for 24 h at 50,000 rpm in an SW60 rotor. The gradients were divided into 17 fractions and analyzed by SDS-PAGE and Coomassie blue staining. (A) Reaction and gradient at pH 6.5; (B) reaction and gradient at pH 8.0.
Reasoning that blocking assembly might lead to accumulation of intermediates, we attempted to gain evidence for such intermediates by sedimentation analysis under nonassembly conditions. In vitro assembly can be arrested under at least two conditions: high salt concentration and basic pH. High salt abolished protein-nucleic acid binding, which resulted in monomeric Gag (data not shown). At pH 8.0 and 0.1 M NaCl, ΔMBDΔPR remained monomeric in the absence of nucleic acid or in the presence of GT8 (data not shown and Fig. 3B). In the presence of GT16, no protein sedimented to the bottom fraction of the gradient, where VLPs would be pelleted, confirming the EM result that assembly was inhibited at pH 8.0. However, the ΔMBDΔPR peak shifted two fractions towards the bottom of the gradient (fraction 9) compared with the monomeric protein peak (fraction 7). This is the same position as that of the phosphorylase B (97-kDa) marker, indicating the formation of a Gag dimer, which would have a molecular mass of 98 kDa. The Gag dimerization promoted by binding of GT16 could be due simply to Gag-DNA oligonucleotide binding, or additional Gag-Gag interaction could be induced when the protein molecules are brought into close proximity (see below). The ΔMBDΔPR dimer appeared to represent a true assembly intermediate, rather than a dead end product, because adjusting the pH from 8.0 to 6.5 led to efficient particle formation which could be visualized readily by EM (data not shown). When longer oligonucleotides were used in the sedimentation assays, broader protein peaks were observed as expected, since longer strands of nucleic acid have the ability to link more Gag molecules together (data not shown).
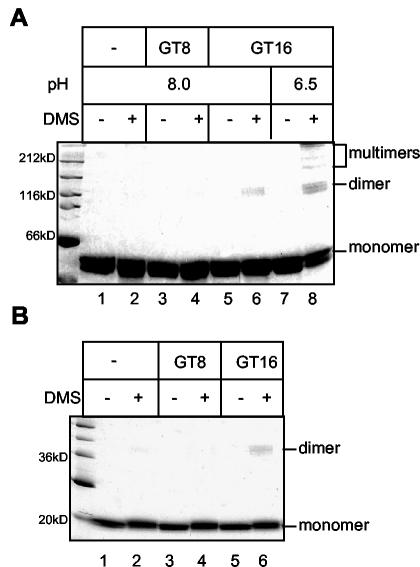
The oligonucleotide length dependence of Gag dimerization was confirmed by protein cross-linking with the bifunctional, amino group-reactive reagent DMS. DMS was added to the mixture of ΔMBDΔPR and GT oligonucleotides at pH 8.0. A low protein concentration (1 μM) was used to minimize the possibility of nonspecific cross-linking from transient interactions. Dimeric ΔMBDΔPR species were observed by SDS-PAGE in the presence of DMS and GT16 (Fig. 4A, lane 6), but not in the presence of GT8 or with protein alone. No higher-order cross-linked species were detected with GT16, even after longer exposure, suggesting that the Gag dimer was the final product at pH 8.0, consistent with the sedimentation result. Cross-linking also was carried out with fully formed ΔMBDΔPR VLPs, after resuspension of the particles in triethanolamine buffer (pH 8.0) followed by addition of DMS. The preformed particles were not disrupted at an alkaline pH, even though they do not form under this condition. Extensive cross-linking was observed for the VLPs, including high-molecular-weight species in addition to Gag dimers (Fig. 4A, lane 8). Multiple bands corresponding to the Gag dimers could be detected, in contrast to a single dimeric band at pH 8, indicating the presence of more than one Gag-Gag interface in the VLPs.
FIG. 4.
Cross-linking analysis. Protein-oligonucleotide mixtures were incubated with or without 2 mM DMS at pH 8.0 for 2 h at room temperature and then were analyzed by SDS-PAGE and Coomassie blue staining. (A) ΔMBDΔPR. For lanes 7 and 8 assembly reactions were carried out at pH 6.5 and, subsequently, the pH was changed to 8.0 with or without the addition of DMS. (B) CTD-NC.
Acidic pH triggers conformational changes in the CTD.
We hypothesize that the RSV CA domain is crucial for GT16-induced ΔMBDΔPR dimerization, since under physiological conditions CA-NC is the minimum construct that can assemble into regular structures in vitro, while the primary role of NC is nucleic acid binding. Mature HIV-1 CA forms dimers in solution through its CTD. Despite the similarity in protein folding, RSV CA remains as a monomer even at very high protein concentration (20 mg/ml). According to the dimerization model, binding to nucleic acid is necessary to bring two Gag molecules into close proximity, which then triggers conformational changes to allow extensive CA-CA interaction.
To analyze the sequences that are necessary for Gag dimerization upon nucleic acid binding, we constructed the protein CTD-NC, comprising only the CTD and the NC domains of Gag (Fig. 1A). At acidic pH and in the presence of nucleic acid, CTD-NC formed irregular aggregates that could be visualized by negative-stain EM (Fig. 1C). This observation suggests that the NTD of CA participates in protein-protein interactions that underlie formation of a regular higher-order structure. We studied the effect of pH and oligonucleotide length on CTD-NC aggregation for comparison with ΔMBDΔPR assembly. As evidenced by a simple centrifugation assay, CTD-NC aggregates were formed in the presence of GT16 but not in the presence of GT8 or without nucleic acid (Fig. 2A). The aggregates were produced most efficiently at an acidic pH, and not at pH 7.0 (Fig. 2B). Thus, CTD-NC aggregation and ΔMBDΔPR assembly share the same requirements for oligonucleotide length and pH. These findings suggest that the NC and CTDs alone are responsible for the oligonucleotide length requirement in assembly and that low pH triggers conformational changes in the CTD. These changes somehow create surfaces involved in protein-protein interactions that ultimately lead to higher-order structures, which remain irregular in the absence of the NTD.
We carried out 24-h rate zonal sedimentation assays to determine the oligomeric state of CTD-NC in the presence of GT oligonucleotides under different pH conditions. At both pH 6.5 and 8, CTD-NC remained monomeric by itself or in the presence of GT8 (Fig. 5 and data not shown). The binding between GT8 and CTD-NC was confirmed by the overlapping position of the A260 and A280 peaks in the gradient (data not shown). At pH 6.5 but not at pH 8, CTD-NC formed aggregates in the presence of GT16, as evidenced by appearance of the protein in the bottom fraction of the gradient. However, in repeated assays the amount of protein fraction 17 was always significantly less than that in the pellet fraction of a simple centrifugation assay, indicating that the aggregates probably dissociated during the 24-h centrifugation (compare Fig. 5A with 2A). The majority of the CTD-NC protein formed a broad peak covering the location of both a monomer and a dimer, with little evidence of species larger than dimers. When the material from fraction 8 of the gradient was adjusted to pH 6.5, massive aggregation occurred immediately, as visualized by EM (data not shown).
FIG. 5.
Sedimentation analysis of CTD-NC in the presence of GT8 or GT16. Protein-oligonucleotide mixtures were analyzed by sucrose gradient centrifugation as described in the legend for Fig. 3. (A) Reaction and gradient at pH 6.5; (B) reaction and gradient at pH 8.0.
GT16-dependent CTD-NC dimerization was confirmed by DMS cross-linking. A dimeric CTD-NC species was observed only in the presence of DMS and GT16, but not with GT8 or protein only (Fig. 4B). However, we were not able to study the protein-protein interaction in the CTD-NC aggregates by the same approach as for VLPs, since the aggregates were rapidly disrupted when resuspended in a buffer at pH 8.0, the condition for cross-linking with bifunctional imidates like DMS.
As for ΔMBDΔPR, dimerization of CTD-NC might be due simply to protein-nucleic acid binding. Alternatively, additional protein-protein interaction might be involved to stabilize the dimers. We assume that the binding affinities of ΔMBDΔPR and CTD-NC for DNA oligonucleotides were the same, since both proteins contain the same NC domain that is the locus for interaction with nucleic acid. However, the sedimentation profiles of ΔMBDΔPR and CTD-NC in the presence of GT16 differed significantly. While binding to GT16 at pH 8.0 resulted in a shift of the ΔMBDΔPR protein peak to the position of a dimer, it led only to a broadening of the CTD-NC peak. One explanation for this observation is that additional protein-protein contacts in the ΔMBDΔPR dimers stabilize them.
NTD is required for the formation of particles and stable Gag dimers.
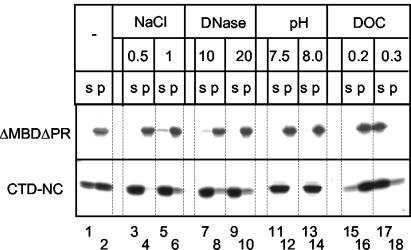
Although the formation of ΔMBDΔPR particles and formation of CTD-NC aggregates share the same oligonucleotide length and pH dependence, we observed significant differences in their stability (Fig. 6). VLPs and aggregates were first collected by centrifugation, followed by resuspension in various buffers, incubation, and a second centrifugation. ΔMBDΔPR particles remained intact after high-salt or DNase treatment, both of which should abolish protein-nucleic acid interactions. Thus, even though nucleic acid binding is necessary initially for triggering Gag dimerization and particle formation, it becomes dispensable after assembly is complete, contributing little to the stability of the VLPs. Neither did an alkaline pH disrupt the particles, suggesting that the putative conformational change in CTD responsible for protein multimerization is not reversible on this time scale. We hypothesize that additional protein interactions are present in the VLPs, such that abolition of the CTD-CTD interaction is not sufficient to disrupt the particles. In contrast to ΔMBDΔPR VLPs, CTD-NC aggregates were extremely unstable. One half of the protein already became soluble after reincubation in the original buffer, suggesting an equilibrium between protein in aggregates and free protein molecules (Fig. 6). High-salt, DNase treatment, or pH 8.0 rapidly and nearly completely solubilized the CTD-NC pellet. These results imply that protein-nucleic acid interaction is crucial for stability of the aggregates and that the putative low pH-triggered conformational change in CTD is reversible. Thus, it is likely that the NTD in ΔMBDΔPR contributes substantially to the stability of the higher-order protein structures. However, although stability of CTD-NC aggregates and of ΔMBDΔPR particles could be distinguished by these several treatments, under some nondenaturing conditions both types of structures were readily disrupted. For example, both particles and aggregates were solubilized by deoxycholate (DOC) above a concentration of 0.3% (wt/wt) (Fig. 6).
FIG. 6.
Disassembly of ΔMBDΔPR VLPs or CTD-NC aggregates. Particles or aggregates were formed with GT16, collected by centrifugation, and then resuspended in various conditions. After 1 h of incubation at room temperature a second centrifugation was carried out, and the supernatant (s) and pellet (p) fractions were analyzed by SDS-PAGE and Coomassie blue staining. Lanes 1 and 2, incubation in reference buffer (pH 6.5; 0.1 M NaCl); lanes 3 to 6, 0.5 or 1.0 M NaCl; lanes 7 to 10, 10 or 20 μg of DNase/ml; lanes 11 to 14, pH 7.5 or 8.0; lanes 15 to 18, 0.2 or 0.3% deoxycholate.
In order to provide further evidence for NTD-NTD interactions in the formation of ΔMBDΔPR dimers, we created the protein NTD-NC, which is composed of the NTD fused to the 12-amino-acid spacer between CTD and NC, followed by the NC domain. In rate zonal sedimentation, the position of the NTD-NC protein shifted from that of a monomer to that of a dimer in the presence of GT16 (data not shown), implying some form of dimerization. Dimerization occurred at both pH 6.5 and pH 8.0, without further oligomerization, as evidenced by EM and by sedimentation analysis (data not shown). These results corroborate the conclusion that the CTD is the locus of the low pH-triggered conformational changes that apparently are critical for formation of higher-order structures, whether these be aggregates or VLPs.
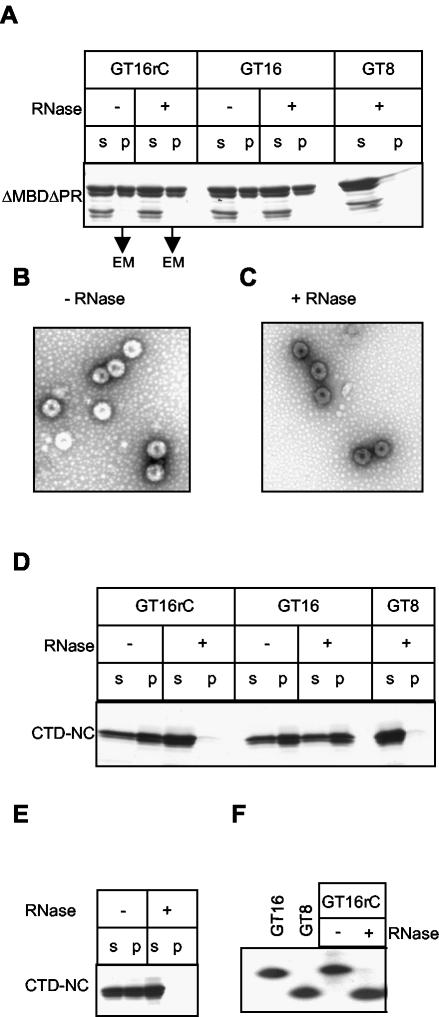
The results described above strongly support the notion that not only NC-nucleic acid interactions but also protein-protein interactions stabilize the dimeric Gag species that is capable of polymerizing into VLPs. To provide more direct evidence for this hypothesis, we designed an oligonucleotide that could be severed experimentally after it had been incorporated into a dimeric complex. GT16rC is a hybrid that contains two ribonucleotides inserted into the middle of the oligodeoxyribonucleotide GT16, allowing it to be cleaved by RNase A. GT16rC was first mixed with ΔMBDΔPR at pH 8.0 to promote protein dimerization but not assembly, followed by RNase treatment. We reasoned that if extensive protein-protein interaction occurred after binding to the oligonucleotide, the Gag dimers would remain intact even after cleavage. These dimers then should proceed to form particles when conditions are adjusted to an acidic pH.
Consistent with the prediction, after RNase treatment and pH adjustment efficient assembly occurred with GT16rC, as shown by a centrifugation assay (Fig. 7A). Particles assembled after RNase treatment were morphologically indistinguishable by EM, compared with VLPs formed in the absence of RNase (Fig. 7C, compare with B). RNase itself did not have an apparent effect on assembly when GT16 was used. As a negative control, no VLPs were detected when GT8 was used. However, after RNase treatment of GT16rC, the ΔMBDΔPR dimers formed at pH 8.0 appeared as monomers in the 24-h sedimentation assays (data not shown), suggesting that in the absence of a contiguous oligonucleotide the dimeric species was not stable enough to survive for long periods. Complete cleavage of GT16rC by RNase was confirmed by a urea gel analysis performed in parallel with the assembly and sedimentation assays. 32P-end-labeled GT16rC migrated to a position similar to that of GT16 or GT8, in the absence or presence of RNase, respectively (Fig. 7F).
FIG. 7.
ΔMBDΔPR but not CTD-NC forms stable dimers at pH 8.0. Protein was mixed at pH 8.0 with GT16 or GT8 or the hybrid oligonucleotide GT16rC. The latter has two ribonucleotide linkages in the middle and hence can be cleaved by RNase. After 20 min of incubation at room temperature in the presence or absence of RNase A at a 1% (wt/wt) ratio with the Gag protein, the pH was adjusted to 6.5 and the reactions were incubated for another 30 min to allow assembly or aggregation. Formation of VLPs or aggregates was monitored by analysis of the supernatant (s) or pellet (p) fractions as described in the legend for Fig. 2 or by negative-stain EM. (A) Assembly of ΔMBDΔPR monitored by pelleting assay. The top line shows the oligonucleotides used, and the next line indicates the presence or absence of RNase treatment. EM, pellets analyzed by EM. (B and C) Representative pictures of particles formed in these reactions. (D) Aggregation of CTD-NC monitored by pelleting assay. The symbols are the same as in panel A. (E) Preformed CTD-NC aggregates were treated with RNase and then analyzed by the same pelleting assay. (F) Cleavage of the end-labeled hybrid oligonucleotide was monitored by PAGE. GT16 and GT8 are marker lanes, and the other lanes show the presence or absence of incubation of the ΔMBDΔPR/GT16rC complexes with RNase.
We then exploited the same approach to determine whether stable protein-protein interactions occurred in the CTD-NC dimers. Complexes of CTD-NC bound to GT16rC at pH 8.0 were treated with control buffer or RNase, followed by adjustment of pH to 6.5. The aggregation of CTD-NC was completely abolished by RNase, as if the protein had bound to GT8 (Fig. 7D). This result suggests that the interaction between CTD domains is weak at best, and thus after the cleavage of GT16rC the two CTD-NC molecules do not remain together and thus are unable to form any higher-order structure. Consistent with this model, RNase digestion of preformed CTD-NC/GT16rC aggregates led to their dissolution (Fig. 7E), similar to the effect of high-salt and DNase treatment of the aggregates shown in Fig. 6.
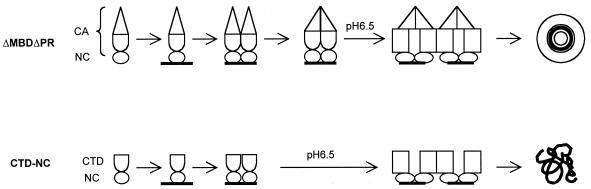
We draw the following overall conclusions about the dimerization of ΔMBDΔPR and CTD-NC, as summarized in the model in Fig. 8: first, binding to GT16 is required to bring two Gag molecules together; second, NTD-NTD interaction helps to lock the ΔMBDΔPR dimers together; third, low-pH-triggered conformational changes in CTD are required for further dimer-dimer interaction.
FIG. 8.
Dimerization model for RSV assembly. Only the CA and NC portions of Gag are shown, with the two domains of CA, NTD and CTD, having distinct shapes. Binding of GT16 induces Gag dimerization, mediated primarily by the NTDs, as indicated by the juxtaposed triangles. Acidic pH triggers conformational changes in the CTD, as indicated by the change of shape to a rectangle. Dimer-dimer contacts are mediated primarily between CTDs, as indicated by the juxtaposition of rectangles of adjacent dimers. In the presence of the NTD (and perhaps immediately upstream sequences [data not shown]), dimer-dimer contacts lead ultimately to the formation of regular spherical particles. In absence of the NTD, similar dimer-dimer contacts lead to the formation of irregular aggregates.
DISCUSSION
We previously proposed a dimerization model to explain the role of nucleic acid in retrovirus assembly (37). In this model, adjacent Gag molecules bound to nucleic acid form dimers, which are the immediate building blocks in assembly. For the dimerization step to occur in vitro, a DNA oligonucleotide must be long enough, about 16 nt, to accommodate two Gag molecules simultaneously. The results presented in the present study show that after oligonucleotide binding, both the NTD and the CTD of RSV CA are required for further assembly steps (Fig. 8). We envision that the proximity of the two Gag molecules promotes conformational changes that lock the Gag dimer together and also create new protein-protein interaction surfaces, ultimately leading to polymerization of dimers to yield the immature protein shell of the virus. The data suggest that the NTD is necessary for the stable Gag dimerization upon nucleic acid binding and that the CTD as well as an acidic pH are necessary for the further interactions among dimers.
According to this model, Gag-Gag interactions are intrinsically weak and are triggered only when the CA domains and immediately adjoining sequences are in close proximity. This explanation is congruent with our present observations as well as published data on Gag multimerization. As shown above, RSV ΔMBDΔPR remains as a monomer in the absence of nucleic acid, or in the presence of a DNA oligonucleotide that is not long enough to allow simultaneous binding of two Gag proteins. RSV CA (27) also remains monomeric in solution even at high concentrations, in contrast to HIV-1 CA, which dimerizes through its CTD with a Kd in the range of 20 μM (17). The RSV MA domain appears not to play an essential role in multimerization, since ΔMBDΔPR lacks most of MA, and the remainder also can be deleted without loss of ability to assemble in vitro (25). MA-MA interactions also appear to be dispensable for assembly in vivo, since MA deletion mutant proteins can be readily incorporated into particles when wild-type Gag is coexpressed (52). From nuclear magnetic resonance studies, the mature NC protein of HIV-1 is monomeric in solution, both as a free protein and when bound to RNA (3, 16, 34); presumably, RSV NC shares these properties, but relevant data have not been published. HIV-1 NC-NC interactions have been reported from yeast two-hybrid analysis and glutathione S-transferase pull-down assays, but those studies were performed in the presence of nucleic acid and probably reflect RNA-mediated interactions (7, 55). Finally, dimerization may be a principle for building other viral capsid structures, as has been inferred for alphaviruses (48, 49) and generalized for other viruses (57).
The locus and nature of interactions in the inferred RSV Gag dimer remain unknown, as do the putative dimer-dimer interactions that lead to Gag polymerization. However, several studies provide clues to these questions. In HIV-1, the structure of the mature core, made up only of CA protein, has been deduced by cryo-EM reconstruction of tubular particles assembled in vitro (19, 35). In that structure, hexagonal rings of CA are held together by contacts between NTDs, which include interactions involving helices 1 and 2. At the same time, each CA is linked to a molecule in an adjoining hexagonal ring by interactions between CTDs, which project inward from the surface of the tube. Heterotypic NTD-CTD contacts between helices 3 and 4 on the NTD and helix 8 on the CTD, which were not envisioned in the original cryo-EM reconstruction, apparently also occur in tubes formed in vitro (31, 32), as well as in vivo in the RSV system by inference from genetic studies (6). However, overall it is unclear to what extent the structure of the mature core can be applied to immature Gag particles. Results from alanine scanning mutagenesis of the HIV-1 CA domain suggest the existence of essential interaction surfaces in the NTD, including parts of helices 4, 5, and 6, which are dispensable for mature core formation (51; Ganser et al., submitted). Similarly, the contacts between helices 2 that are necessary for efficient or proper mature core formation appear not to be essential for immature assembly in vivo.
Another clue to the nature of RSV Gag-Gag interactions stems from the crystal structure of a protein termed 25-NTD, which comprises the NTD of CA plus the upstream 25 residues of the Gag protein p10 (42). In RSV these 25 residues play a key shape-determining role in particle formation, both in vitro (25) and in vivo in budding of VLPs in the baculovirus expression system (24). That is, Gag forms immature spherical particles when these residues are present but forms tubular particles when they are deleted. Since tubular particles are considered to represent a mature form of assembly, these 25 residues somehow function to promote immature polymerization. In the crystal structure, the 25-NTD protein is a dimer with an extensive dimer interface comprising portions of the 25 residues as well as helices 4 and 6, approximately matching the interaction surface implied by the mutagenesis studies on HIV-1 CA (51; Ganser et al., submitted). Our working hypothesis is that the NTD-NTD contacts inferred from the in vitro assembly experiments presented here map to the 25-NTD dimer interface observed in the crystals.
The present study suggests that in the RSV system a CTD-CTD interaction, promoted by a conformational change triggered by acidic pH, underlies the polymerization of dimers. The locus of this interaction probably includes the spacer peptide between CTD and NC, since in both HIV-1 (1, 22, 29) and RSV (14, 30; data not shown) this short sequence is essential for proper immature assembly. In addition, the NC domain itself, or at least the N-terminal portion of NC that is inferred to be critical for assembly (44, 45, 54, 55), also may be directly involved in the nucleic acid-dependent aggregation of RSV CTD-NC at acid pH. This possibility is suggested by the ability of the mature HIV-1 NC to form aggregates of defined size in the presence of poly(A) (33). It remains to be determined what titratable group is responsible for this pH effect, but apparently it is not a histidine residue. The RSV CTD is devoid of His residues, and although NC has two such residues in the zinc-binding motifs, these are dispensable for assembly in vitro (25; data not shown). The pH dependence of assembly in vitro is less stringent when nucleic acid longer than 16 nt is used and is quite different for HIV-1 than for RSV. Thus, its relevance for assembly in vivo is uncertain. Conceivably, the implied conformational change in the CTD can be brought about by other factors in living cells.
Acknowledgments
This work was supported by U.S. Public Health Service grant CA20081.
REFERENCES
- 1.Accola, M. A., S. Höglund, and H. G. Göttlinger. 1998. A putative α-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Accola, M. A., B. Strack, and H. G. Göttlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amarasinghe, G. K., R. N. De Guzman, R. B. Turner, K. J. Chancellor, Z. R. Wu, and M. F. Summers. 2000. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J. Mol. Biol. 301:491-511. [DOI] [PubMed] [Google Scholar]
- 4.Berthet-Colominas, C., S. Monaco, A. Novelli, G. Sibaï, F. Mallet, and S. Cusack. 1999. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 18:1124-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 72:9034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowzard, J. B., J. W. Wills, and R. C. Craven. 2001. Second-site suppressors of Rous sarcoma virus CA mutations: evidence for interdomain interactions. J. Virol. 75:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, S., R. J. Fisher, E. M. Towler, S. Fox, H. J. Issaq, T. Wolfe, L. R. Phillips, and A. Rein. 2001. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc. Natl. Acad. Sci. USA 98:10875-10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell, S., and V. M. Vogt. 1997. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 71:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campos-Olivas, R., J. L. Newman, and M. F. Summers. 2000. Solution structure of the Rous sarcoma virus capsid protein and comparison with capsid proteins of other retroviruses. J. Mol. Biol. 296:633-649. [DOI] [PubMed] [Google Scholar]
- 13.Cimarelli, A., S. Sandin, S. Höglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type-1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Craven, R. C., A. E. Leure-duPree, C. R. Erdie, C. B. Wilson, and J. W. Wills. 1993. Necessity of the spacer peptide between CA and NC in the Rous sarcoma virus Gag protein. J. Virol. 67:6246-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dawson, L., and X.-F. Yu. 1998. The role of nucleocapsid of HIV-1 in virus assembly. Virology 251:141-157. [DOI] [PubMed] [Google Scholar]
- 16.De Guzman, R. N., Z, R. Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384-388. [DOI] [PubMed] [Google Scholar]
- 17.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 18.Ganser, B. K., A. Cheng, W. I. Sundquist, and M. Yeager. 2003. Three-dimensional structure of the M-MuLV CA protein on a lipid monolayer: a general model for retroviral capsid assembly. EMBO J. 22:2886-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 20.Gross, I., H. Hohenberg, and H.-G. Kräusslich. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249:592-600. [DOI] [PubMed] [Google Scholar]
- 21.Gross, I., H. Hohenberg, C. Huckhagel, and H.-G. Kräusslich. 1998. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J. Virol. 72:4798-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross, I., H. Hohenberg, T. Wilk, K. Wiegers, M. Grättinger, B. Müller, S. Fuller, and H.-G. Kräusslich. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin, Z., L. Jin, D. L. Peterson, and C. L. Lawson. 1999. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J. Mol. Biol. 286:83-93. [DOI] [PubMed] [Google Scholar]
- 24.Johnson, M. C., H. M. Scobie, Y. M. Ma, and V. M. Vogt. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 76:11177-11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi, S. M., and V. M. Vogt. 2000. Role of the Rous sarcoma virus p10 domain in shape determination of Gag virus-like particles assembled in vitro and within Escherichia coli. J. Virol. 74:10260-10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khorasanizadeh, S., R. Campos-Olivas, and M. F. Summers. 1999. Solution structure of the capsid protein from the human T-cell leukemia virus type-1. J. Mol. Biol. 291:491-505. [DOI] [PubMed] [Google Scholar]
- 27.Kingston, R., E. Z. Eisenmesser, T. Fitzon-Ostendorp, G. W. Schatz, V. M. Vogt, C. B. Post, and M. G. Rossmann. 2000. Structure and self-association of the Rous sarcoma virus capsid protein. Structure 8:617-628. [DOI] [PubMed] [Google Scholar]
- 28.Klikova, M., S. S. Rhee, E. Hunter, and T. Ruml. 1995. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J. Virol. 69:1093-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kräusslich, H.-G., M. Fäcke, A.-M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krishna, N. K., S. Campbell, V. M. Vogt, and J. W. Wills. 1998. Genetic determinants of Rous sarcoma virus particle size. J. Virol. 72:564-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lanman, J., T. T. Lam, S. Barnes, M. Sakalian, M. R. Emmett, A. G. Marshall, and P. E. Prevelige, Jr. 2003. Identification of novel interactions in HIV-1 capsid protein assembly by high-resolution mass spectrometry. J. Mol. Biol. 325:759-772. [DOI] [PubMed] [Google Scholar]
- 32.Lanman, J., J. Sexton, M. Sakalian, and P. E. Prevelige, Jr. 2002. Kinetic analysis of the role of intersubunit interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro. J. Virol. 76:6900-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Cam, E., D. Coulaud, E. Delain, P. Petitjean, B. P. Roques, D. Gerard, E. Stoylova, C. Vuilleumier, S. P. Stoylov, and Y. Mely. 1998. Properties and growth mechanism of the ordered aggregation and growth of a model RNA by the HIV-1 nucleocapsid protein: an electron microscopy investigation. Biopolymers 45:217-229. [DOI] [PubMed] [Google Scholar]
- 34.Lee, B. M., R. N. De Guzman, B. G. Turner, N. Tjandra, and M. F. Summers. 1998. Dynamical behavior of the HIV-1 nucleocapsid protein. J. Mol. Biol. 279:633-649. [DOI] [PubMed] [Google Scholar]
- 35.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 36.Lingappa, J. R., R. L. Hill, M. L. Wong, and R. S. Hegde. 1997. A multistep ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J. Cell Biol. 136:567-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma, Y. M., and V. M. Vogt. 2002. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J. Virol. 76:5452-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mayo, K., M. L. Vana, J. McDermott, D. Huseby, J. Leis, and E. Barklis. 2003. Analysis of Rous sarcoma virus capsid protein variants assembled on lipid monolayers. J. Mol. Biol. 316:667-678. [DOI] [PubMed] [Google Scholar]
- 39.Mayo, K., D. Huseby, J. McDermott, B. Arvidson, L. Finlay, and E. Barklis. 2003. Retrovirus capsid protein assembly arrangements. J. Mol. Biol. 325:225-237. [DOI] [PubMed] [Google Scholar]
- 40.Momany, C., K. C. Kovari, A. J. Prongay, W. Keller, R. K. Gitti, B. M. Lee, A. E. Gorbalenya, L. Tong, J. McClure, L. S. Ehrlich, M. F. Summers, C. Carter, and M. G. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 41.Morikawa, Y., T. Toshiyuki, and K. Sano. 1999. In vitro assembly of human immunodeficiency virus type 1 Gag protein. J. Biol. Chem. 274:27997-28002. [DOI] [PubMed] [Google Scholar]
- 42.Nandhagopal, N., A. Simpson, M. C. Johnson, A. B. Francisco, G. W. Schatz, V. M. Vogt, and M. G. Rossmann. Dimeric Rous sarcoma virus capsid protein structure relevant to immature Gag assembly. J. Mol. Biol., in press. [DOI] [PubMed]
- 43.Sakalian, M., S. D. Parker, R. A. Weldon, Jr., and E. Hunter. 1996. Synthesis and assembly of retrovirus Gag precursors into immature capsids in vitro. J. Virol. 70:3706-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandefur, S., R. M. Smith, V. Varthakavi, and P. Spearman. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 74:7238-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 72:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh, A. R., R. L. Hill, and J. R. Lingappa. 2001. Effect of mutations in Gag on assembly of immature human immunodeficiency virus type 1 capsids in a cell-free system. Virology 279:257-270. [DOI] [PubMed] [Google Scholar]
- 47.Spearman, P., and L. Ratner. 1996. Human immunodeficiency virus type 1 capsid formation in reticulocyte lysates. J. Virol. 70:187-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tellinghuisen, T. L., and R. J. Kuhn. 2000. Nucleic acid-dependent cross-linking of the nucleocapsid protein of Sindbis virus. J. Virol. 74:4302-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tellinghuisen, T. L., R. Perera, and R. J. Kuhn. 2001. In vitro assembly of Sindbis virus core-like particles from cross-linked dimers of truncated and mutant capsid proteins. J. Virol. 75:2810-2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Schwedler, U. K., K. M. Stray, J. E. Garrus, and W. I. Sundquist. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77:5439-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wills, J. W., R. C. Craven, R. A. Weldon, Jr., T. D. Nelle, and C. R. Erdie. 1991. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J. Virol. 65:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Worthylake, D. K., H. Wang, S. Yoo, W. I. Sundquist, and C. P. Hill. 1999. Structures of the HIV-1 capsid protein dimerization domain at 2.6 Å resolution. Acta Crystallogr. D 55:85-92. [DOI] [PubMed] [Google Scholar]
- 54.Yu, F., S. M. Joshi, Y. M. Ma, R. L. Kingston, M. N. Simon, and V. M. Vogt. 2001. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J. Virol. 75:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zabransky, A., E. Hunter, and M. Sakalian. 2002. Identification of a minimal HIV-1 Gag domain sufficient for self-association. Virology 294:141-150. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zlotnick, A. 2003. Are weak protein-protein interactions the general rule in capsid assembly? Virology 315:269-274. [DOI] [PubMed]