Abstract
Nitric oxide (NO) is a cell-signaling molecule involved in a number of physiological and pathophysiological processes. Modification of cysteine residues by NO (or NO metabolites), i.e. S-nitrosation, changes the function of a broad spectrum of proteins. This reaction represents an important post-translational modification that transduces NO dependent signals. However, the detection and quantification of S-nitrosation in biological samples remains a challenge mainly because of the lability of S-nitrosation products: S-nitrosothiols (SNO). In this review we summarize recent developments of the methods to detect S-nitrosation. Our focus is the methods which can be used to directly conjugate the site(s) of S-nitrosation.
Introduction
As an important signaling molecule, NO has received a great deal of attention in the past several decades. The cellular response to NO is transduced via multiple chemical reactions. In particular, the reaction of NO or NO metabolites with protein cysteine residues that results in S-nitrosation is believed to be critical for redox-sensitive cell signaling. This biological reaction has been also suggested as a mechanism by which NO can transmit signals both within and between cells and tissues. To date, a large group of proteins have been characterized as targets for S-nitrosation and in many cases S-nitrosation is believed to regulate protein activity and function. However, most of the protein targets of S-nitrosation were originally identified using exogenous NO donors. It has not always been established whether S-nitrosation of these proteins is associated with endogenous NO activity. This is primarily due to the technical limitations in detecting S-nitrosation. The products of S-nitrosation, i.e. S-nitrosothiols (SNO), are known as unstable species that can easily undergo decomposition catalyzed by light, metal ions, enzymes, or other conditions. Therefore, the detection of SNO remains a challenge.
To date, five major SNO detection methods have been developed: chemiluminescence, colorimetry/fluorimetry, electrochemical/amperometric, anti-S-nitroso-cysteine antibody, and mass spectrometric (MS) based assays (Figure 1). However, these methods applications in analyzing cell extracts or tissue samples are still limited. Chemiluminescence, colorimetry/fluorimetry, and electrochemical methods are indirect methods that detect SNO decomposition products (NO radical or NO+). Therefore, other NO metabolites in biological systems, such as nitrite, may interfere with the results. In addition, these methods can only determine the overall concentration of SNO moieties. They can not identify the site(s) of S-nitrosation. While anti-S-nitroso-cysteine antibody and MS-based assays are direct methods, they are not very useful in identifying SNO proteins in a pool of unnitrosated proteins. It is worth noting that SNO formation in vitro or using purified proteins often does not truly reflect SNO formation in cells. Since SNO groups are unstable species, modifications that are made between the time that the protein is being extracted from cells and the time that SNO is measured can dramatically affect results.
Figure 1.
Common methods for SNO detection
Even with all of the aforementioned methods available, the detection of SNO is still artifact-prone. The analytical deficiencies become evident when it is observed that reported values of the analysis of the same tissue or biological fluid by different research groups cover several orders of magnitude [1]. The detection methods for SNO have been reviewed multiple times previously [for example, see references 2–7]. In this review, we will only focus on recent developments of chemical methods for SNO detection; specifically methods which can be used (or potentially used) to label SNO in proteins.
Biotin switch technique and recent improvements
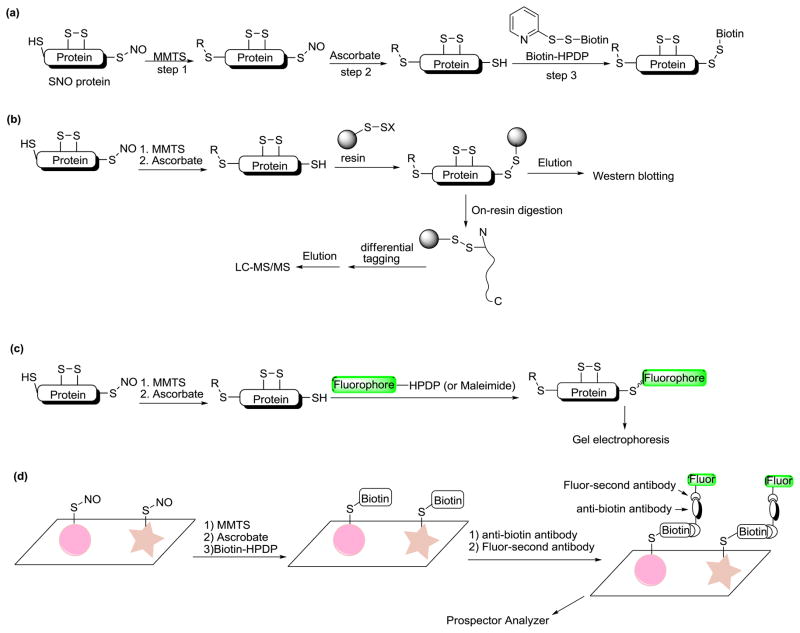
The biotin switch method is the most popular method so far which can be used to detect and isolate SNO proteins from cell extracts [8, 9]. As shown in Figure 2(a), this method involves three chemical transformation steps that selectively target and convert unstable SNO to stable biotin conjugates: 1) free thiols in a protein or a protein mixture are blocked by thiol-specific reagents like MMTS (methyl methanethiosulfonate) or NEM (N-ethylmaleimide). In this step proteins usually need to be denatured to ensure access of blocking reagents to all thiols. 2) SNO groups are selectively reduced by ascorbate, resulting in the formation of free thiols. However, the ascorbate reduction mechanism is still not completely clear [10, 11]. 3) The newly formed thiols are treated with biotin-HPDP (N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)-propionamide), a thiol specific biotinylating reagent, to form the desired biotin conjugates.
Figure 2.
Biotin switch based methods for SNO detection. (a) Original biotin switch. (b) Resin assisted capture. (c) Fluorescence switch. (d) Microarray based assay. R shown here indicates thiol blocking groups.
Using this method, SNO proteins can be fished out from complex biological samples and subjected to mass spectrometric analysis or Western blot analysis. The biotin switch method has been applied in many protein systems and has made a significant contribution towards SNO research. Recently researchers have made some important improvements on the biotin switch technique to facilitate the identification of SNO proteins. These include resin-assisted capture [12], fluorescence labeling [13–15], and microarray-based assay [16].
Resin-assisted capture (SNO-RAC)
Protein free thiol residues can be easily conjugated on solid supports and this strategy has long been used for the enrichment of thiol-containing proteins. Stamler and co-workers creatively combined the biotin switch strategy and solid phase separation. This technique, named SNO-RAC (Figure 2-b), is an effective way for quantitative and on-site identification of SNO [12]. Compared to the classic biotin switch, the labeling and pulldown steps are shortened into one step by the formation of covalent disulfide bonds between the SNO site and resin. This is very suitable for on-site trypsinization and peptide labeling/sequencing. One major advantage of SNO-RAC is that it has better sensitivity for high mass proteins (>100 KDa). Moreover, when combined with the iTRAQ labeling technique, SNO-RAC can be used to detect the kinetics of S-nitrosation/denitrosation and confirm the global occurrence of nitrosation.
Fluorescence switch
Based on the biotin-switch principle, fluorescent dyes can also be conjugated to SNO residues to facilitate the detection of SNO proteins. Different fluorophores such as 7-amino-4-methylcoumarin-3-acetic acid (AMCA) [13], Cy-dyes (Cy3 and Cy5) [14], and fluorescein [15] have been applied in studies (Figure 2-c). This fluorescence switch strategy eliminates the need for Western blot or avidin enrichment protocols. It allows direct comparison of the S-nitrosation state between two samples in the same gel lane or on the same 2D gel with a low background and a high signal to noise ratio.
Protein microarray-based analysis
Due to sensitivity problems, the original biotin switch method is biased toward the identification of abundant proteins, and therefore is believed to be limited as a proteomic tool. To address such limitations, Stamler et al. recently developed a protein microarray-based biotin switch technique [16] to study determinants of S-nitrosation. This method uses an anti-biotin antibody and fluorescently labeled secondary antibody to label and analyze SNO-proteins on microarrays (Figure 2-d). This has proven advantageous for not only performing relative quantification of SNO-reactivity across a proteome, but also the ability to assess the effects of multiple S-nitrosation reagents and cofactors.
Potential problems of biotin switch
Although biotin switch has been applied in many systems, this method is still not ideal and has three major drawbacks: 1) being subtractive in nature, the efficiency/sensitivity of this assay relies on blocking 100% of free thiols in the sample. However, due to the low concentration of SNO in real biological systems, this is not always achievable. 2) The specificity of biotin switch has been questioned. The reduction of SNO by ascorbate seems very substrate dependent. Some SNO moieties cannot be reduced by ascorbate efficiently [4, 17–19], while under some circumstances ascorbate has been shown to not only reduce SNO groups, but also reduce disulfides [20–22]. 3) A third issue of the biotin switch is the potential for disulfide exchange after reduction of cysteine SNO. This could lead to incorrect identification of the nitrosated cysteine of a protein. In addition, recent studies by Snyder et al. even found that biotin switch also generated signals from S-sulfurhydration (-SSH) [23]. Many studies have been undertaken in order to understand these inconsistencies. For example, Stamler et al. revealed that indirect sunlight from a laboratory window is the likely cause of the false-positive signal (i.e. the reduction of disulfides) [19]. Hogg et al. suggested that copper should be used in the experiments to accelerate SNO reduction while maintaining specificity [17]. Mutus and coworkers suggested that sinapinic acid could be another effective and selective reductant of SNO for use in biotin switch [24]. Nevertheless, current results suggest that additional control experiments are necessary in order to positively identify SNO in complex protein mixtures by the biotin switch method [1, 25].
New bioorthogonal reactions of SNO
Given the problems and limitations of current methods there is a clear need of more effective and reliable methods for SNO detection. From a chemistry point-of-view, SNO is a unique functional group that should have distinct reactivity from other biological functional groups such as thiols (-SH), disulfides (-S-S-), sulfenic acids (-SOH), etc. If new reactions can be developed to specifically target SNO moieties and convert unstable SNO to stable/detectable products, such reactions would hold considerable promise in applications for SNO detection. Ideally, a useful bioorthogonal reaction of SNO should meet the following criteria: 1) the reaction should be selective for SNO. Other biological functionalities, especially disulfides or other oxidized thiol derivatives such as sulfenic and sulfinic acids, should not be affected; 2) the reaction should proceed rapidly under physiological conditions; 3) the reaction product should be stable for analysis.
Reductive ligation of SNO
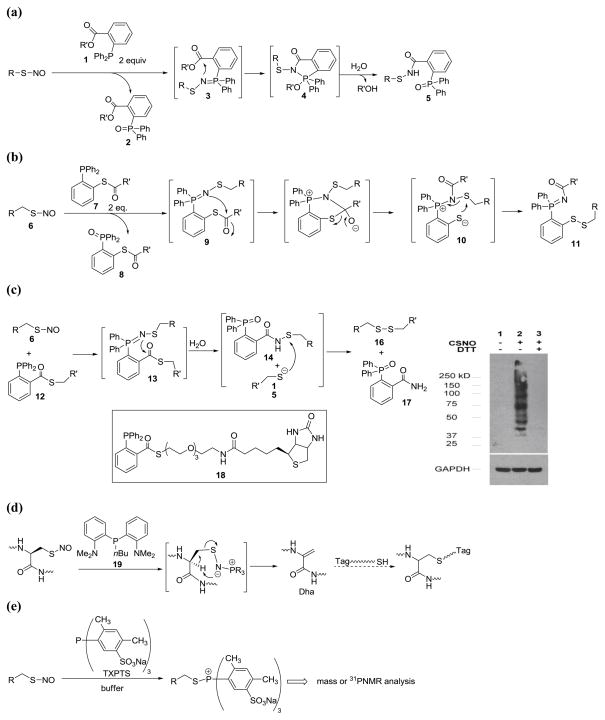
With the criteria for bioorthogonality of SNO reactions in mind, our group initiated a program to study phosphine-mediated reactions of SNO. We noticed that triaryl-substituted phosphines react with SNO rapidly to form azaylide intermediates under very mild conditions. It is known that azaylides are potential nucleophilic species. If a suitable electrophilic group to trap azaylides is attached to the phosphine, a spontaneous intramolecular reaction might proceed, which could lead to stable products. Such a reaction should not interfere with other biological functionalities on proteins. In 2008, a reductive ligation of SNO was developed using phosphine-ester conjugates [26]. As shown in Figure 3-a, these type of phosphine compounds have been used in the well-known Staudinger ligation to label azides. When SNO was treated with substrates like 1, stable sulfenamide products were obtained in good yields. In addition, this reductive ligation was proven to be a rapid process. The products typically formed in minutes. The reaction mechanism is believed to be similar to the Staudinger ligation: the azaylide 3 first forms upon treatment of SNO with 1 (in this step, two equivalents of phosphine are consumed). Azaylide 3 then undergoes an intramolecular reaction with the ester functionality to provide phosphorane 4. Finally, hydrolysis of 4 in the presence of water leads to the final product 5. As proven in the applications of Staudinger ligation, substrates like 1 are expected to be bioorthogonal reagents for SNO. Therefore, this reaction can potentially be used for SNO detection.
Figure 3.
Phosphine based methods for SNO detection. (a) Reductive ligation. (b) Bis-ligation. (c) One-step disulfide formation. (d) Reductive elimination. (e) S-alkylphosphonium formation. R shown here indicates different S-nitrosothiol substrates, mainly cysteine derivatives. R′ indicates aryl or alkyl substituent groups.
Bis-ligation of SNO
In an effort to develop a “traceless” version of the reductive ligation [27], we discovered a unique bis-ligation of SNO (Figure 3-b) [28]. When biologically relevant primary SNO were treated with phosphine-thioester substrates like 7, stable disulfide-iminophosphorane products 11 were obtained in good yield. The reaction mechanism was proposed as follows: the reaction first leads to the formation of an azaylide intermediate 9. Then, acyl transfer from the thioester to the nitrogen atom provides intermediate 10. Finally, the nucleophilic phenylthiolate attacks the pseudo-sulfenamide linkage via a fast intramolecular process to furnish the disulfide-iminophosphorane product 11. As the disulfide linkage in 11 is more stable than the sulfenamide linkage in biological systems, the bis-ligation holds considerable promise in applications for SNO detection.
One-step disulfide formation of SNO
Inspired by the high reactivity of the sulfenamide towards thiolate, as observed in bis-ligation, we recently developed a reductive ligation mediated one-step disulfide formation with SNO (Figure 3-c) [29]. With substrates like 12, a regular reductive ligation should occur first to generate sulfenamide product 14 and thiolate 15. Then, an intermolecular reaction between 14 and 15 will proceed spontaneously to provide stable disulfide 16 and liberate phosphine oxide 17. The formation of simple disulfide products, without the bulky phosphine adducts, would have attractive applications in protein systems. Control experiments proved this reaction is selective for SNO and disulfides are unaffected. Most importantly, this strategy has been used in SNO protein labeling using substrate 18. Briefly, COS-7 cells were first treated with S-nitrosocysteine (CSNO). The resulting SNO proteins were found to form biotin conjugates via disulfides upon incubation with 18 (lane 2). In the control experiment, non-CSNO treated cells were carried through the same process but no labeled proteins were observed (lane 1).
Reductive elimination of SNO
In the study of phosphine-based ligation reactions of SNO, we envisioned that the azaylide intermediate could serve as a base in some cases. If an acidic proton is present, such as the α-proton in cysteine derivatives, azaylide may promote an intramolecular elimination to provide dehydroalanine (Dha) products. With this idea in mind, a series of trialkyl- and triaryl-phosphines were tested with S-nitrosocysteine derivatives and some effective phosphines like 19 were found to promote the formation of Dha in good yields (Figure 3-d) [30]. This fast reaction (<1 min) was proven to proceed through an intramolecular elimination process and to be selective for SNO cysteine groups [30]. No Dha was observed when disulfides were the substrates. Dha formation is potentially useful for the detection of protein S-nitrosation, as Dha derivatives would allow a straightforward chemoselective incorporation of various sulfide-containing molecules through a Michael addition. Although this strategy requires a two-step operation, the simplicity of both the phosphine substrates and thiol-based tags, as well as the excellent stability of the final conjugates make it very attractive.
Direct labeling of SNO by S-alkylphosphonium adducts
King’s group recently reported an interesting method for directly labeling SNO using the phosphine reagent [31]. They found that a water-soluble phosphine tris(4,6-dimethyl-3-sulfonatophenyl)-phosphine trisodium salt hydrate (TXPTS) could react with SNO to form covalent S-alkylphosphonium adducts (Figure 3-e). Both small molecule SNO and protein SNO work in this reaction. However, their experiments also showed that TXPTS could react with certain disulfides (GSSG and cystine) to small extents (3%~15%). Therefore, careful controls are needed when using this method for SNO detection in complex systems. This method is amenable to mass detection and 31PNMR spectroscopy. It is not clear if protein S-alkylphosphonium adducts are stable for other common protein analysis methods like Western blotting.
Conclusion
Recent technological developments have made it possible to study the S-nitrosoproteome. Currently the biotin switch method and its modifications are the mainstream methods in the field. Being subtractive in nature, these methods suffer some problems/limitations in selectivity and reproducibility issues. In our opinion, future method development in this area should focus on direct methods which only target SNO moieties. To this end, the phosphine-based methods are promising as they only react with S-nitroso groups. However, further evaluations using complex protein samples are required for further development of phosphine-based reagents as reliable SNO labeling tools.
Acknowledgments
We apologize to those authors whose work could not be cited due to space limitations. We thank the NSF CAREER Award (0844931), NIH (R01GM08226), and the American Heart Association Scientist Development Grant #0932120N to MX for the support of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1••.Gladwin MT, Wang X, Hogg N. Methodological vexation about thiol oxidation versus S-nitrosation-A commentary on “An ascorbate-dependent artifact that interferes with the interpretation of the biotin-switch assay”. Free Radic Biol Med. 561;2006:41–557. doi: 10.1016/j.freeradbiomed.2006.05.025. The authors point out possible problems of the biotin switch method and suggest that careful control experiments be necessary when biotin switch is used to analyze protein S-nitrosation. [DOI] [PubMed] [Google Scholar]
- 2.Gow A, Doctor A, Mannick J, Gaston B. S-Nitrosothiol measurement in biological systems. J Chromatogr B Analyt Technol biomed Life Sci. 2007;851:140–151. doi: 10.1016/j.jchromb.2007.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giustarini D, Milzani A, Dale-Donne I, Rossi R. Detection of s-nitrosothiol in biological fluids: a comparison among the most widely applied methodologies. J Chromatogr B Analyt Technol biomed Life Sci. 2007;851:124–139. doi: 10.1016/j.jchromb.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 4.Kettenhofen NJ, Broniowska KA, Keszler A, Zhang Y, Hogg N. Proteomic methods for analysis of S-nitrosation. J Chromatogr B Analyt Technol biomed Life Sci. 2007;851:152–159. doi: 10.1016/j.jchromb.2007.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacArthur PH, Shiva S, Galdwin MT. Measurement of circulating nitrite and S-nitrosothiols by reductive chemiluminescence. J Chromatogr B Analyt Technol biomed Life Sci. 2007;851:93–105. doi: 10.1016/j.jchromb.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Hausladen A, Rafikov R, Angelo M, Singel D, Nudler E, Stamler JS. Assessment of nitric oxide signals by triiodide chemiluminescence. Proc Natl Acad Sci U S A. 2007;104:2157–2162. doi: 10.1073/pnas.0611191104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torta F, Usuelli V, Malgaroli A, Bachi A. Proteomic analysis of protein S-nitrosylation. Proteomics. 2008;8:4484–4494. doi: 10.1002/pmic.200800089. [DOI] [PubMed] [Google Scholar]
- 8.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 9.Jaffrey SR. Detection and characterization of protein nitrosothiols. Methods Enzymol. 2005;396:105–118. doi: 10.1016/S0076-6879(05)96011-4. [DOI] [PubMed] [Google Scholar]
- 10.Kirsch M, Büscher AM, Aker S, Schulz R, de Groot H. New insights into the S-nitrosothiol-ascorbate reaction. The formation of nitroxyl. Org Biomol Chem. 2009;7:1954–1962. doi: 10.1039/b901046g. [DOI] [PubMed] [Google Scholar]
- 11.Holmes AJ, Williams DLH. Reaction of ascorbic acid with S-nitrosothiols: clear evidence for two distinct reaction pathways. J Chem Soc Perkin Trans 2. 2000:1639–1644. [Google Scholar]
- 12•.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. The authors use resin-assisted capture coupled with biotin switch to study SNO proteomics. The use of resin capture significantly enhanced the sensitivity of the method. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han P, Zhou X, Huang B, Zhang X, Chen C. On-gel fluorescent visualization and the site identification of S-nitrosylated proteins. Anal Biochem. 2008;377:150–155. doi: 10.1016/j.ab.2008.03.023. [DOI] [PubMed] [Google Scholar]
- 14.Santhanam L, Gucek M, Brown TR, Mansharamani M, Ryoo S, Lemmon CA, Romer L, Shoukas AA, Berkowitz DE, Cole RN. Selective fluorescent labeling of S-nitrosothiols (S-FLOS): A novel method for studying S-nitrosation. Nitric Oxide. 2008;19:295–302. doi: 10.1016/j.niox.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tello D, Tarín C, Ahicart P, Bretón-Romero R, Lamas S, Martínez-Ruiz A. A “fluorescence switch” technique increases the sensitivity of proteomic detection and identification of S-nitrosylated proteins. Proteomics. 2009;9:5359–5370. doi: 10.1002/pmic.200900070. [DOI] [PubMed] [Google Scholar]
- 16.Foster MW, Forrester MT, Stamler JS. A protein microarray-based analysis of S-nitrosylation. Proc Natl Acad Sci U S A. 2009;106:18948–18953. doi: 10.1073/pnas.0900729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Kettenhofen NJ, Shiva S, Hogg N, Gladwin MT. Copper dependence of biotin switch assay: Modified assay for measuring cellular and blood nitrosated proteins. Free Radic Biol Med. 2008;44:1362–1372. doi: 10.1016/j.freeradbiomed.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Keszler A, Broniowska KA, Hogg N. Characterization and application of the biotin-switch assay for the identification of S-nitrosated proteins. Free Radic Biol Med. 2005;38:874–881. doi: 10.1016/j.freeradbiomed.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Forrester MT, Foster MW, Stamler JS. Assessment and application of the biotin switch technique for examining protein s-nitrosylation under conditions of pharmacologically induced oxidative stress. J Biol Chem. 2007;282:13977–13983. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- 20.Landino LM, Koumas MT, Mason CE, Alston JA. Ascorbic acid reduction of microtubule protein disulfides and its relevance to protein S-nitrosylation assays. Biochem Biophys Res Commun. 2006;340:347–352. doi: 10.1016/j.bbrc.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Giustarini D, Dalle-Donne I, Colombo R, Milzani A, Rossi R. Is ascorbate able to reduce disulfide bridges? A cautionary note. Nitric Oxide. 2008;19:252–258. doi: 10.1016/j.niox.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Huang B, Chen C. An ascorbate-dependent artifact that interferes with the interpretation of the biotin switch assay. Free Radic Biol Med. 2006;41:562–567. doi: 10.1016/j.freeradbiomed.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 23.Mustafa AK, Gadalla MM, Sen N, Kim S, Mu W, Gazi SK, Barrow RK, Yang G, Wang R, Snyder SH. H2S Signals through protein S-sulfhydration. Sci Signal. 2009;2:ra72. doi: 10.1126/scisignal.2000464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kallakunta VM, Staruch A, Mutus B. Sinapinic acid can replace ascorbate in the biotin switch assay. Biochim Biophys Acta. 2010;1800:23–30. doi: 10.1016/j.bbagen.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Forrester MT, Foster MW, Benhar M, Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic Biol Med. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Wang H, Xian M. Fast reductive ligation of S-nitrosothiols. Angew Chem Int Ed Engl. 2008;47:6598–6601. doi: 10.1002/anie.200801654. This paper reports the first phosphine-mediated reaction which can selectively convert unstable S-nitrosothiols to stable products. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Wang H, Xian M. Exploration of the “traceless” reductive ligation of S-nitrosothiols. Org Lett. 2009;11:477–480. doi: 10.1021/ol802663q. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Wang H, Xian M. An unexpected bis-ligation of S-nitrosothiols. J Am Chem Soc. 2009;131:3854–3855. doi: 10.1021/ja900370y. [DOI] [PubMed] [Google Scholar]
- 29•.Zhang J, Li S, Zhang D, Wang H, Whorton AR, Xian M. Reductive ligation mediated one-step disulfide formation of S-nitrosothiols. Org Lett. 2010;12:4208–4211. doi: 10.1021/ol101863s. This paper demonstrates the phosphine-mediated disulfide formation can be used to conjugate SNO proteins in cell extracts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Zhang J, Xian M. Facile formation of dehydroalanine from S-nitrosocysteines. J Am Chem Soc. 2009;131:13238–13239. doi: 10.1021/ja905558w. [DOI] [PubMed] [Google Scholar]
- 31.Bechtold E, Reisz JA, Klomsiri C, Tsang AW, Wright MW, Poole LB, Furdui CM, King SB. Water-soluble triarylphosphines as biomarkers for protein S-nitrosation. ACS Chem Biol. 2010;5:405–414. doi: 10.1021/cb900302u. [DOI] [PMC free article] [PubMed] [Google Scholar]