Abstract
Many of the products of the ca. 80 genes encoded by alphaherpesviruses have already been identified and, at least tentatively, functionally characterized. Among the least characterized proteins are the products of the genes homologous to herpes simplex virus UL3, which are present only in the subfamily Alphaherpesvirinae. To identify the UL3 protein of the porcine alphaherpesvirus pseudorabies virus (PrV), the complete PrV UL3 open reading frame was cloned, expressed in Escherichia coli as a glutathione S-transferase fusion protein, and used for immunization of a rabbit. In Western blots, the generated antiserum specifically detected a 34-kDa protein in PrV-infected cells, which was absent from purified virus preparations, indicating that PrV UL3 encodes a nonstructural protein. In indirect immunofluorescence analysis, the anti-UL3 serum produced predominantly nuclear staining in transfected as well as in infected cells, which was not altered in the absence of other virus-encoded nuclear proteins such as the UL31 and UL34 gene products. To investigate UL3 function, a deletion mutant, PrV-ΔUL3F2, was constructed and characterized. This mutant replicated and formed plaques on noncomplementing cells indistinguishable from wild-type PrV, demonstrating that PrV UL3 is not required for virus propagation in cultured cells. Moreover, ultrastructural examinations revealed no impairment of capsid formation in the nucleus, nuclear egress of capsids, virion maturation in the cytoplasm, or virus release. Thus, the overall properties of PrV UL3 are similar to those described for the homologous herpes simplex virus proteins which may be indicative of a common, yet hitherto unknown, function in alphaherpesvirus replication. However, based on our studies, an involvement of the UL3 homologs in virion formation appears unlikely.
Alphaherpesviruses encode ca. 80 different proteins, and approximately half of them are not essential for viral replication in cultured cells (41, 42). Although most of the alphaherpesvirus genes and gene products have been characterized in some detail, among the less well characterized proteins are the products of genes homologous to herpes simplex virus (HSV) UL3, which are conserved only in the alphaherpesvirus subfamily. In HSV type 1 (HSV-1) and HSV-2, the UL3 proteins are nonglycosylated but phosphorylated proteins which are located in the nuclei of infected cells (18, 32, 46, 47). Moreover, HSV-1 UL3 has been shown to be dispensable for viral replication in cell culture (2).
For pseudorabies virus (PrV), a porcine alphaherpesvirus causing Aujeszky's disease, only the sequence of the UL3 open reading frame (ORF) has so far been determined (10). The PrV UL3 gene is located in a gene cluster containing genes UL1 (coding for gL), UL2 (encoding uracil DNA glycosylase), and UL3.5 (Fig. 1). All four genes are transcribed with early-late kinetics into 3′-coterminal mRNAs that use a common polyadenylation signal located downstream of UL3.5 (10). The PrV UL3 ORF has the capacity to code for a protein of 237 amino acids with a predicted molecular mass of 26 kDa. However, the ORF contains two additional in-frame ATG codons, which, if used, would yield protein products of 231 and 230 amino acids (10). The deduced amino acid sequence shows a high degree of homology to the corresponding gene products of other members of the Alphaherpesvirinae, which is most prominent in the C-terminal half of the protein (32). Although the HSV-1 UL3 protein has initially been proposed to represent a viral membrane-associated protein (33), primary and secondary structure predictions yielded no evidence for hydrophobic regions in the PrV UL3 protein which could serve as signal sequences or transmembrane domains nor for consensus sequences for N glycosylation. This indicated that the PrV UL3 protein may not be a membrane constituent. In contrast, a nuclear localization signal (NLS) which is conserved in all UL3 homologs is present between amino acids 195 and 198 (10, 32).
FIG. 1.
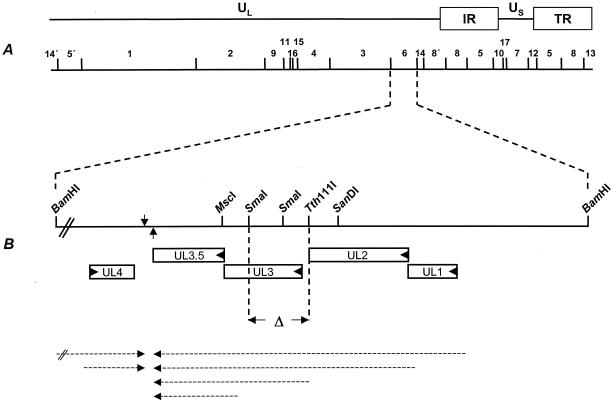
Diagram of the UL3 coding region and construction of PrV-ΔUL3F2. (A) A schematic map of the PrV genome shows the unique long (UL) and the unique short (US) regions, the inverted repeat regions (IR and TR), and the positions of BamHI restriction sites. Numbers indicate BamHI restriction fragments. (B) Enlargement of the UL3 coding region on BamHI fragment 6. Genes coding for UL1, UL2, UL3, and UL3.5 are transcribed into 3′-coterminal mRNAs (dashed arrows pointing leftward) which share a common polyadenylation signal (arrow pointing upward). UL4 and UL5 (not shown) are transcribed from the opposite strand into mRNAs (dashed arrows pointing rightward) that coterminate at a common polyadenylation signal (arrow pointing downward). The left end of BamHI fragment 6 is not drawn to scale. For generation of PrV-ΔUL3F2, sequences between the Tth111I and SmaI sites were deleted. After recombination only approximately 100 nt of foreign sequences encompassing the Flp recombinase recognition target (FRT) site remained (data not shown).
One of our main interests is the elucidation of the mechanisms of capsid transit from the nucleus to the cytosol at the molecular level (reviewed in reference 36). Capsids leave the nucleus by budding at the inner nuclear membrane, thereby acquiring a primary envelope (36, 41). This primary envelope then fuses with the outer nuclear membrane, resulting in the release of capsids into the cytosol (36). It was previously shown that the PrV UL31 and UL34 gene products, which physically interact, are important for nuclear egress (15, 26). Both proteins are conserved within the family Herpesviridae, indicating that they play fundamental roles in herpesvirus replication. Indeed, the HSV-1 homologs have also been shown to interact and to be important for nuclear egress (5, 39, 40, 43). Whereas the UL31 and UL34 proteins are important for primary envelopment, the US3 products, which are conserved only in the alphaherpesviruses, modulate de-envelopment. In their absence, enveloped primary virions accumulate in the perinuclear space (25, 45). So far, no function has been assigned to any of the UL3 gene products, but the nuclear localization of the HSV-1 and HSV-2 homologs suggests that the protein might be involved in DNA replication, capsid assembly, or nuclear egress of capsids.
We wanted to identify the PrV UL3 gene product and investigate its functional role in the viral replication cycle. To this end, a UL3-specific antiserum was generated and a UL3 deletion and a corresponding rescue mutant were isolated and characterized.
MATERIALS AND METHODS
Cells and viruses.
All PrV mutants were derived from the laboratory strain Kaplan (PrV-Ka) (22). Viruses were grown in rabbit kidney (RK13) or porcine kidney (PSEK) cells in minimum essential medium supplemented with 10 or 5% fetal calf serum, respectively. RK13-UL3 cells, which constitutively express UL3 under control of the human cytomegalovirus immediate early promoter-enhancer complex, were established after transfection of RK13 cells with plasmid pcDNA-UL3 (see below) followed by selection with 0.5 mg of Geneticin (Invitrogen, Karlsruhe, Germany)/ml and immunofluorescence screening with the monospecific anti-UL3 serum (see below). Cloning of PrV as a bacterial artificial chromosome, pPrV-ΔgB, has been described elsewhere (30). The mutants PrV-ΔUL28 (PrV-332-31) (37), PrV-ΔUL31 (15), and PrV-ΔUL34 (25) were propagated on transcomplementing cells as described previously, and mutants PrV-ΔUS3 (25) and PrV-ΔUL46-49 (16) were grown on RK13 cells.
Preparation of monospecific anti-UL3 serum.
For generation of a UL3-specific antiserum, the complete UL3 ORF of PrV-Ka was amplified by PCR with platinum Pfx DNA polymerase (Invitrogen), primers UL3FOR (5′-CACAGAATTCGGGATGGACGGAGGAGAGCG-3′, nucleotides [nt] 2658 to 2677, GenBank accession no. L13855) and UL3REV (5′-CACACTCGAGCTCATGGCGCCGCAAACAC-3′, nt 3375 to 3357, GenBank accession no. L13855), and cloned genomic BamHI fragment 6 as a template. UL3 start and stop codons are shown in bold, and EcoRI and XhoI restriction sites introduced for convenient cloning are printed in italics. The resulting 0.7-kb PCR product was digested with EcoRI and XhoI and cloned into the appropriately cleaved vector pcDNA3 (Invitrogen), giving rise to pcDNA-UL3, and into vector pGEX-4T-1 (Amersham Biosciences, Freiburg, Germany), yielding plasmid pGEX-UL3. Plasmid pcDNA-UL3 was used for generation of a complementing cell line (RK13-UL3, see above) as well as for coupled in vitro transcription and translation (TNT; Promega). Plasmid pGEX-UL3 was used for UL3 expression in Escherichia coli. The 52-kDa glutathione S-transferase (GST)-UL3 fusion protein was isolated from sodium dodecyl sulfate (SDS)-8% polyacrylamide gels and used for immunization of a rabbit as described previously (27). Serum obtained after the fourth immunization was used in this study.
Construction of PrV-ΔUL3F2.
For deletion of the PrV UL3 ORF, the genomic BamHI fragment 6 was cloned into pUC19 and subsequently shortened to 1,065 bp by double digestion with HindIII and SanDI as well as EcoRI and MscI (Fig. 1) (HindIII and EcoRI sites are located in the multiple cloning site of vector pUC19). To delete major parts of the UL3 coding sequences without interfering with transcription of the neighboring genes, a 543-bp Tth111I/SmaI fragment representing codons 1 to 164 of UL3 as well as the intergenic region between UL2 and UL3 was removed (Fig. 1) and replaced with a 1.3-kb EcoRI fragment containing a kanamycin resistance gene flanked by FRT sites (30), giving rise to plasmid pUC19-ΔUL3IIKF. The 1.7-kb insert of this plasmid was amplified by PCR with vector-specific M13 and M13reverse primers (New England Biolabs, Frankfurt, Germany), and the PCR product was excised after gel electrophoresis and used for mutagenesis of pPrV-ΔgB (30) in E. coli, utilizing the Red recombinase of bacteriophage λ (8). After deletion of the kanamycin resistance gene by Flp recombinase (7) and cotransfection with calcium phosphate (19) with plasmid pUC-B1BclI, which contains the authentic gB gene of PrV-Ka (30) and restores gB expression, the virus mutant PrV-ΔUL3F2 could be isolated. For generation of a UL3 rescuant, genomic DNA of PrV-ΔUL3F2 was cotransfected with a plasmid containing the complete BamHI fragment 6. Transfection progeny was tested for expression of UL3 by dot blot analysis with the monospecific serum and plaque purified to homogeneity. One single plaque isolate, PrV-ΔUL3FR, was further used. The deletion and rescue mutants were characterized by restriction typing and Southern blot analysis, and the introduction of the correct deletion in PrV-ΔUL3F2 was confirmed by PCR and DNA sequencing (data not shown).
Western blot analysis.
Virus purification and preparation of infected cell lysates was done as described recently (30). Samples of infected cell lysates and purified virions were separated by SDS-10% polyacrylamide gel electrophoresis and electrotransferred onto nitrocellulose membranes (Schleicher & Schuell, Dassel, Germany). Blots were blocked with 6% skim milk in phosphate-buffered saline (PBS) and incubated with monospecific sera against the UL3 (1:100,000) (this study), UL3.5 (1:10,000) (14), UL31 (1:100,000) (15), UL34 (1:100,000) (26), UL37 (1:100,000) (27), UL49 (1:100,000) (4), gH (1:50,000) (24), and gL (1:1,000) (28) proteins or a monoclonal antibody against gB (b43-b5) (38). Bound antibody was detected with peroxidase-conjugated secondary antibodies (Dianova, Hamburg, Germany) and visualized by chemiluminescence (Super Signal; Pierce) recorded on X-ray films.
In vitro radiolabeling.
For in vitro coupled transcription and translation, RNA was transcribed from plasmid pcDNA-UL3 and translated in the presence of [35S]methionine/cysteine. The translation product was separated in an SDS-10% polyacrylamide gel and visualized after drying by exposure to an X-ray film (X-Omat; Kodak). For metabolic labeling, cells were infected at a multiplicity of infection (MOI) of 10 with either wild-type PrV-Ka or PrV-ΔUL3F2 in medium containing either [35S]methionine/cysteine (ICN) or [32P]orthophosphate (ICN). At 16 h after infection, cells were lysed and radioactively labeled proteins were precipitated as described previously (25). Proteins were visualized after electrophoresis in an SDS-10% polyacrylamide gel by phosphorimaging.
Indirect immunofluorescence and confocal laser scan microscopy.
RK13 cells grown on coverslips were infected for approximately 18 h with wild-type PrV-Ka or respective recombinant viruses with approximately 300 PFU per well of a six-well culture dish. Thereafter, infected cells were fixed for 20 min with 3% paraformaldehyde followed by 10 min of incubation in paraformaldehyde plus 0.3% Triton X-100. They were subsequently incubated for 1 h with antisera against UL3 (1:500) (this study), UL31 (1:500) (15), UL34 (1:500) (26), or US3 (1:500) (25) and Alexa 488-conjugated secondary antibodies (Molecular Probes, Leiden, The Netherlands). Coverslips were washed repeatedly with PBS after each step, and fluorescence was preserved with a 9:1 mixture of glycerol and PBS containing 2.5 mg of 1,4-diazabicyclooctane per ml and 1 μg of propidium iodide per ml for chromatin counterstaining. The slides were analyzed with a confocal laser scan microscope (LSM 510; Zeiss, Jena, Germany).
In vitro replication properties.
RK13 cells were infected at an MOI of 10 with PrV-Ka, PrV-ΔUL3F2, or PrV-ΔUL3FR and incubated on ice for 1 h. Cells were then overlaid with prewarmed medium and incubated at 37°C for 1 h. Remaining extracellular virus was inactivated by low-pH treatment (35), and cells were scraped into the medium immediately as well as after 4, 8, 12, 24, and 36 h and lysed by freezing (−70°C) and thawing (37°C). Titers of progeny virus were determined by titration on RK13 cells. Mean values of three independent experiments were calculated and drawn into a graph with corresponding standard deviations. Plaque sizes were determined as described previously (30).
Electron microscopy.
For ultrathin sectioning, RK13 cells were infected at a MOI of 1 with PrV-ΔUL3F2 and fixed at 10 h postinfection (p.i.). Fixation, dehydration, and embedding for routine microscopy were performed as described previously (20). The counterstained ultrathin sections were analyzed with an electron microscope (Tecnai 12; Philips, Eindhoven, The Netherlands).
RESULTS
Identification of the PrV UL3 gene product.
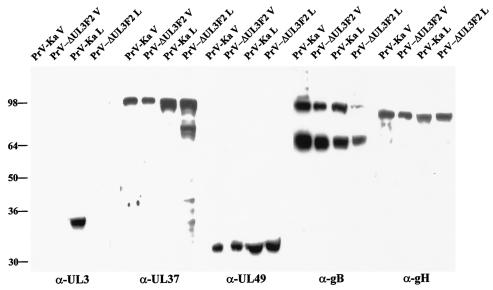
For identification of the PrV UL3 protein, the complete UL3 ORF was expressed as a GST fusion protein in E. coli and used for immunization of a rabbit. In Western blot experiments with PrV-Ka-infected cells, a single 34-kDa protein was detectable (Fig. 2) which was absent in noninfected cells (Fig. 3B) or in cells infected with the UL3 deletion mutant PrV-ΔUL3F2 (Fig. 2). No UL3-specific signal was detectable in lysates of purified virions, indicating that UL3 is not a structural component of virus particles (Fig. 2). As controls, tegument proteins UL37 and UL49 as well as envelope glycoprotein H (gH) and gB were readily detectable in infected lysates and purified virions (Fig. 2). Moreover, the UL1 gene encoding gL, which is located upstream but in the same gene cluster, was also transcribed and translated similarly in PrV-Ka- and PrV-ΔUL3F2-infected cells (Fig. 4), indicating that the introduced deletion of UL3 did not eliminate expression of upstream genes within the same cluster.
FIG. 2.
Identification of the PrV UL3 protein. (A) Purified PrV-Ka and PrV-ΔUL3F2 virions (V) and the respective infected cell lysates (L) were analyzed by Western blotting with monospecific sera against the UL3 (α-UL3), UL37 (α-UL37), and UL49 (α-UL49) proteins and gH (α-gH) or a monoclonal antibody against gB (α-gB). Proteins were separated on SDS-10% polyacrylamide gels, and signals were visualized by chemiluminescence. Locations of molecular mass (in kilodaltons) markers are shown on the left.
FIG. 3.
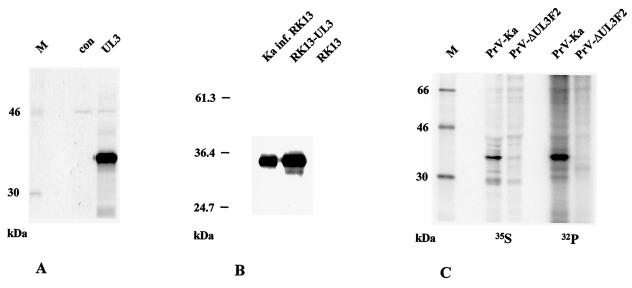
Characterization of the PrV UL3 protein. (A) Plasmid pcDNA-UL3 was used for coupled in vitro transcription and translation, and products were visualized by autoradiography. The lane labeled Con contains products without the addition of exogenous plasmid. (B) Western blot analysis of lysates of RK13-UL3 and RK13 cells in comparison with PrV-Ka-infected (inf.) RK13 cells. (C) Metabolic labeling. Cells were infected with PrV-Ka or PrV-ΔUL3F2 and radiolabeled with [35S]methionine/cysteine or [32P]orthophosphate. Proteins were precipitated with the anti-UL3 serum, separated by SDS-10% polyacrylamide gel electrophoresis, and visualized by phosphorimaging. Locations of molecular mass markers (lanes M) are shown to the left of each panel.
FIG. 4.
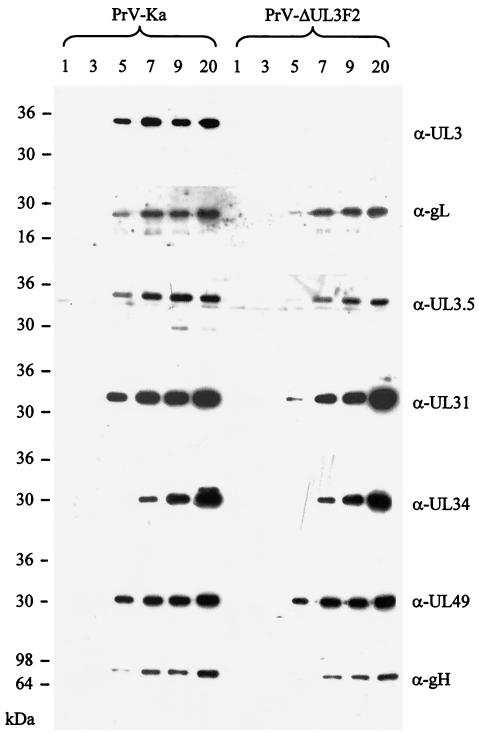
UL3 expression kinetics during infection. RK13 cells were infected with PrV-Ka or PrV-ΔUL3F2 at an MOI of 10 and harvested at the indicated times (in hours) thereafter. Proteins were separated on SDS-10% polyacrylamide gels and analyzed on Western blots with monospecific sera against UL3 (α-UL3), gL (α-gL), UL3.5 (α-UL3.5), UL31 (α-UL31), UL34 (α-UL34), UL49 (α-UL49), and gH (α-gH). Locations of molecular mass markers are indicated on the left.
The apparent molecular mass of PrV UL3 is greater than the prediction of 26 kDa (10). To investigate whether the size difference is due to protein modifications, plasmid pcDNA-UL3 was used for coupled in vitro transcription and translation. Again, a single 34-kDa product arose, as shown in Fig. 3A, and a protein of the same size was also detected in lysates of cells constitutively expressing UL3 (RK13-UL3) (Fig. 3B).
To analyze whether the PrV UL3 protein is phosphorylated, as has been shown for the HSV-1 and HSV-2 homologs (18, 32, 46), cells infected with PrV-Ka or PrV-ΔUL3F2 were radioactively labeled with either [35S]methionine/cysteine or [32P]orthophosphate and radiolabeled proteins were precipitated with the anti-UL3 antiserum. As shown in Fig. 3C, the antiserum specifically precipitated a [35S]methionine/cysteine-labeled 34-kDa protein from PrV-Ka-infected cells which is not present in cells infected with PrV-ΔUL3F2. A protein of identical molecular mass was also observed after precipitation of 32P-labeled proteins from PrV-Ka-infected cells but not PrV-ΔUL3F2-infected cells. These data demonstrated that the PrV UL3 protein is indeed phosphorylated.
To investigate the time course of UL3 expression, Western blot experiments with cell lysates harvested at different time points after infection were performed. As shown in Fig. 4, the UL3 protein was first detectable at 5 h p.i. and is expressed with kinetics similar to those of gL and UL3.5. This finding is in agreement with previous studies demonstrating that UL1, UL2, UL3, and UL3.5 genes are transcribed into 3′-coterminal early-late mRNAs (10). UL49- and UL31-specific signals are also found starting from 5 h p.i., while gH- and UL34-specific signals were first detectable at 7 h p.i. (Fig. 4).
Localization of PrV UL3 in infected cells.
To investigate the subcellular localization of the PrV UL3 protein, RK13, cells were infected with PrV-Ka and investigated by confocal laser scan microscopy at approximately 18 h p.i. No specific fluorescence was found in uninfected cells, but cells infected with PrV-Ka exhibited a homogeneous nuclear staining (Fig. 5). Diffuse nuclear staining was also predominant in cells transfected with plasmid pcDNA-UL3 or RK13-UL3 cells (data not shown), indicating that UL3 is located in the nucleus in the absence of other viral gene products. No specific granular localization of PrV UL3 was obvious either in transfected or infected cells.
FIG. 5.
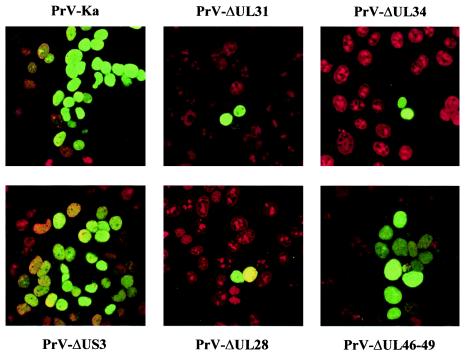
Intracellular localization of the PrV UL3 protein. RK13 cells were infected under plaque assay conditions with PrV-Ka, PrV-ΔUL31, PrV-ΔUL34, PrV-ΔUS3, PrV-ΔUL28, and PrV-ΔUL46-49, and immunofluorescence analysis was performed by confocal laser scan microscopy with the monospecific anti-UL3 serum and Alexa 488-conjugated secondary antibodies (green). Chromatin was counterstained with propidium iodide (red).
Since PrV UL3 has been identified as a nuclear protein, we were interested in whether its localization may be influenced by other viral nuclear proteins. To this end, RK13 cells were infected with mutants lacking UL31 (15), UL34 (26), US3 (25), or UL28 (37). In addition, a virus mutant lacking genes UL46, UL47, UL48, and UL49 (PrV-ΔUL46-49) (16), which encode tegument proteins that, in the case of the UL47 and UL48 proteins, have also been shown to be present in the nucleus (17, 29), was also analyzed. No difference in nuclear localization of UL3 was evident in any of the mutants investigated (Fig. 5), indicating that those proteins do not influence UL3 localization.
PrV UL3 is not essential for viral replication in cell culture.
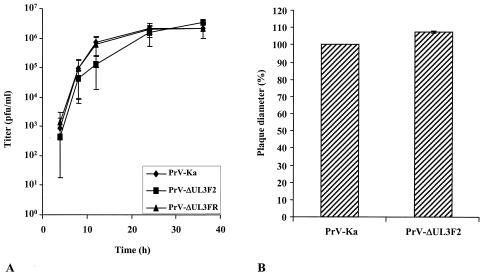
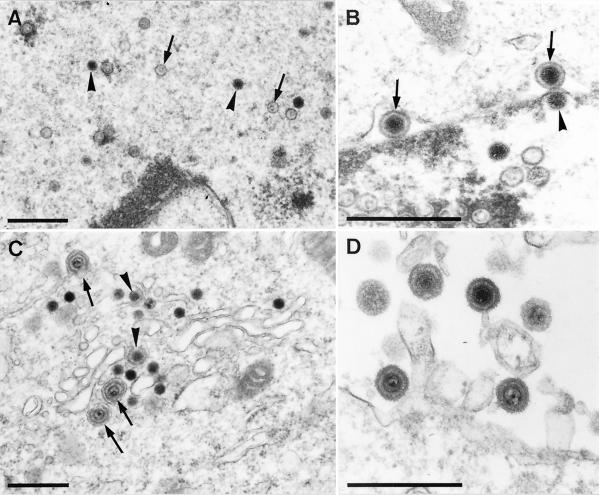
The fact that the mutant PrV-ΔUL3F2 could be isolated on noncomplementing cells already demonstrated that the PrV UL3 protein is not required for viral replication in cell culture. For a more thorough analysis, one-step growth kinetics were performed (Fig. 6A) and plaque sizes were determined (Fig. 6B). Neither assay revealed any significant difference between cells infected with PrV-ΔUL3F2 and cells infected with PrV-Ka or PrV-ΔUL3FR (Fig. 6), besides a minimal delay in replication of mutant PrV-ΔUL3F2 between 8 and 24 h p.i. Moreover, electron microscopic examinations of RK13 cells infected with PrV-ΔUL3F2 were performed 10 h p.i. (Fig. 7). In PrV-ΔUL3F2-infected cells, all the maturation stages typical for herpesvirus morphogenesis (20, 21, 36) were observed, including intranuclear capsid formation (Fig. 7A), nuclear egress (Fig. 7B), secondary envelopment in the cytosol (Fig. 7C), and formation of cell-free virions (Fig. 7D). Thus, absence of the UL3 protein apparently does not impair intranuclear assembly of capsids, nuclear egress, secondary envelopment, or viral egress.
FIG. 6.
Growth properties of PrV-ΔUL3F2. (A) RK13 cells were infected with PrV-Ka, PrV-ΔUL3F2, or PrV-ΔUL3FR at an MOI of 10 and harvested immediately or 4, 8, 12, 24, and 36 h p.i. Virus progeny was titrated on RK13 cells. Mean virus titers and standard deviations of the results from three independent experiments are shown. (B) RK13 cells were infected with PrV-Ka or PrV-ΔUL3F2 under plaque assay conditions, and plaque diameters were measured microscopically at approximately 48 h p.i. The mean value of PrV-Ka was set at 100%, and average relative plaque sizes of PrV-ΔUL3F2 as well as standard deviations from the results of two independent experiments were calculated.
FIG. 7.
Electron microscopy of PrV-ΔUL3F2-infected RK13 cells. RK13 cells were infected with PrV-ΔUL3F2 at an MOI of 1 and analyzed 10 h after infection. Panel A shows the presence of intranuclear empty (arrows) and DNA-filled (arrowheads) nucleocapsids. In panel B, two primary enveloped virions in the perinuclear space (arrows) can be observed as well as one nucleocapsid that starts to bud at the inner nuclear membrane (arrowhead). Panel C shows intracytosolic capsids during early (arrowheads) and late (arrows) stages of secondary envelopment, whereas panel D demonstrates the presence of extracellular mature virions. Bar, 500 nm.
Absence of UL3 does not alter localization of other viral nucleus-localizing proteins.
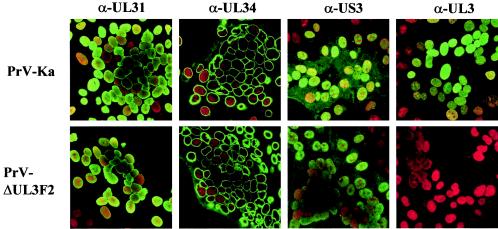
Since UL3 was shown to locate to the nucleus of infected cells, it might function in targeting other viral proteins to this cell compartment. Therefore, we investigated whether in the absence of UL3 localization of several other viral nuclear proteins is different. To this end, cells were infected with PrV-Ka or PrV-ΔUL3F2 and processed for confocal laser scan microscopy at approximately 18 h p.i. Besides the absence of UL3-specific staining in PrV-ΔUL3F2-infected cells, no difference was detectable in the intracellular localization of the UL31, UL34, or US3 protein between PrV-Ka- and PrV-ΔUL3F2-infected cells (Fig. 8).
FIG. 8.
Subcellular distribution of viral nuclear localizing proteins in the absence of PrV UL3. RK13 cells were infected with PrV-Ka or PrV-ΔUL3F2 under plaque assay conditions and processed for immunofluorescence at 18 h p.i. with sera against the UL31 (α-UL31), UL34 (α-UL34), US3 (α-US3), and UL3 (α-UL3) gene products. Green, protein-specific fluorescence; red, propidium iodide staining.
DISCUSSION
UL3 homologs are found only in the Alphaherpesvirinae subfamily of the Herpesviridae (1, 6, 9, 10, 11, 13, 23, 33, 44, 48). However, within the alphaherpesvirus UL3 proteins, a strikingly high degree of amino acid conservation is found, which is most pronounced in the C-terminal half of the protein (32). Conservation includes a probable NLS, whereas an N glycosylation signal sequence is found in several but not in all UL3 homologs. No function has been assigned to any of the UL3 proteins up to now. The HSV-1 UL3 protein had first been proposed to represent a membrane glycoprotein (33). Recent studies demonstrated that the HSV-1 UL3 protein is not glycosylated and is localized in the nucleus of infected cells (18, 31, 46). Since this could indicate that the UL3 proteins play a role in nuclear egress, one of our main topics of research, we were interested in identifying the UL3 protein of PrV, analyzing its subcellular localization and functional role, and comparing its properties with those of the HSV-1 and HSV-2 homologs.
By using a rabbit antiserum raised against the complete UL3 protein expressed as a GST fusion protein in E. coli, we identified the PrV UL3 protein as a 34-kDa polypeptide in PrV-Ka-infected cell lysates. No reactivity was found in purified virion preparations, demonstrating that the UL3 protein is not a structural component of virions. To our knowledge, until now nobody has tested whether HSV-1 UL3 is a virion component. For HSV-1 UL3, six different protein isoforms were described, varying in size between 34 and 42 kDa (32). They originate from a translational start at two alternative start codons and from at least two posttranslational modifications, including phosphorylation by the virus-encoded UL13 protein kinase (32). In contrast, Western blot analysis of PrV-infected cells or cells stably expressing UL3 showed only a single UL3-specific protein, although three alternative in-frame methionine codons are present (10). However, since the second and third ATG codons are only 18 and 21 nt downstream from the first one, our gel system might not be able to resolve the slight differences in protein migration patterns. Amino acid analysis of PrV UL3 with the program NetPhos, version 2.0 (3) (http://www.cbs.dtu.dk/services/NetPhos), predicts 11 possible sites for phosphorylation, 8 on threonine and 3 on serine residues, and like the homologous HSV-1 UL3, the PrV UL3 protein is also phosphorylated. This might be the cause for the observed size difference between the predicted molecular mass of 26 kDa and the apparent molecular mass of 34 kDa, since it has been shown that phosphorylated proteins migrate differently in SDS-polyacrylamide gels based on the extent of phosphorylation (12). Since the in vitro-translated UL3 protein and the UL3 protein expressed in transfected RK13 cells exhibit the same molecular mass, it is unlikely that phosphorylation is performed exclusively or primarily by virus-encoded kinases.
In infected cells, PrV UL3-specific labeling was predominantly found diffusely distributed in the nucleus. The same fluorescence pattern was apparent in transfected cells, indicating that UL3 has the intrinsic property for nuclear targeting and that the predicted NLS is functional. Nuclear localization was also described for the HSV-1 and HSV-2 homologs (31, 46, 47). However, in addition to a diffuse nuclear staining, these proteins were found in large speckles which were later identified as nucleoli in HSV-2 (47) or small dense nuclear bodies in HSV-1 (31). The nucleolus is a highly structured and specialized organelle that functions as the site of both the synthesis of rRNA and the assembly and processing of preribosomal ribonucleoprotein particles (34). For nucleolar targeting, a second NLS seems to be important (47) which is not conserved in all UL3 homologues (32). It is also absent from the PrV UL3 protein, which might account for the different staining patterns. The function of the small dense nuclear bodies is unknown. In HSV-1, late in infection, UL3 and UL4 colocalize in these structures together with and dependent on US1 (ICP22), suggesting that these proteins play a role late in the viral replicative cycle (31). In contrast to HSV-1, where UL3 is expressed with late kinetics, PrV UL3 seems to be expressed as a delayed early gene (10). In the absence of ICP22, which has been shown to be necessary for upregulation of a subset of true late genes (31), HSV-1 UL3 also shows a more diffuse nuclear staining pattern, comparable to the pattern found in PrV (31). Therefore, temporal regulation may be different in PrV, resulting in the differences in subcellular distribution.
The diffuse nuclear staining pattern of PrV UL3 did not change in the absence of several other viral proteins which are found in the nucleus during infection, in particular those involved in nuclear egress such as the nuclear membrane protein UL34 (26), its complex partner UL31 (15), or the protein kinase US3, which is necessary for efficient release of primary virions from the perinuclear space (25, 45). Absence of the UL28 protein, which is involved in DNA replication and nucleocapsid maturation (37), also did not alter intracellular distribution of the PrV UL3 protein. Correlating with these results, intracellular distribution of the UL31, UL34, or US3 protein was not affected by the absence of UL3, indicating that these proteins are not interdependent for nuclear localization. Whereas the HSV-1 UL3 protein has been proposed to represent a putative membrane protein (2, 33), and a perinuclear distribution has been observed early after HSV-1 infection (46), we did not observe any UL3-specific perinuclear immunofluorescence in PrV-infected cells.
No thorough investigation of UL3 function has been undertaken up to now. An HSV-1 deletion mutant which was unable to express the UL3 ORF was replication competent in noncomplementing cells, which indicated that UL3 is not essential for HSV-1 replication in cell culture (2). We isolated a virus mutant lacking PrV UL3, PrV-ΔUL3F2, which replicated on RK13 cells almost as efficiently as PrV-Ka. Moreover, the capacity for direct cell-to-cell spread as measured by plaque sizes appears unaffected. Electron microscopic examinations of cells infected with the deletion mutant gave no indication for a replication defect, and all stages of virus assembly in the nucleus and cytoplasm, such as primary envelopment, de-envelopment, tegumentation, and secondary envelopment in the cytoplasm, and viral egress seem to be undisturbed. Even after intranasal infection of mice with PrV-ΔUL3F2, no significant differences compared to wild-type PrV-Ka infection could be observed (R. Klopfleisch, J. Teifke, and T. C. Mettenleiter, unpublished data). A sequence homology search of the aligned UL3 sequences gave a weak homology to DNA binding proteins (data not shown), which might suggest that UL3 is involved in DNA replication or transcription regulation, but at least HSV-2 UL3 does not appear to directly interact with DNA or RNA (46).
In summary, we identified PrV UL3 as a nucleus-localizing phosphoprotein which is dispensable for viral replication in vitro. These properties are shared by the PrV UL3 protein and its homologs in HSV-1 and HSV-2. Since there is no indication that PrV UL3 is involved in virion formation, it is likely that homologous proteins of other alphaherpesviruses also do not play a role in this process.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (grant ME 854/4-2).
We thank Uta Hartwig, Petra Meyer, and Nadine Müller for expert technical assistance and E. Zorn for photographic help.
REFERENCES
- 1.Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. F. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature (London) 310:207-211. [DOI] [PubMed] [Google Scholar]
- 2.Baines, J. D., and B. Roizman. 1991. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J. Virol. 65:938-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blom, N., S. Gammeltoft, and S. Brunak. 1999. Sequence- and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 294:1351-1362. [DOI] [PubMed] [Google Scholar]
- 4.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, Y. E., and B. Roizman. 1993. The product of the UL31 gene of herpes simplex virus 1 is a nuclear phosphoprotein, which partitions with the nuclear matrix. J. Virol. 67:6348-6356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchinson, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 7.Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 10.Dean, H. J., and A. K. Cheung. 1993. A 3′coterminal gene cluster in pseudorabies virus contains herpes simplex virus UL1, UL2, and UL3 gene homologs and a unique UL3.5 open reading frame. J. Virol. 67:5955-5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Draetta, G., and D. Beach. 1988. Activation of cdc2 protein kinase during mitosis in human cells: cell cycle dependent phosphorylation and subunit rearrangement. Cell 54:17-26. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs, W., and T. C. Mettenleiter. 1996. DNA sequence and transcriptional analysis of the UL1 to UL5 gene cluster of infectious laryngotracheitis virus. J. Gen. Virol. 77:2221-2229. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs, W., B. G. Klupp, H. Granzow, H.-J. Rziha, and T. C. Mettenleiter. 1996. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J. Virol. 70:3517-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs, W., H. Granzow, and T. C. Mettenleiter. 2003. A pseudorabies virus recombinant simultaneously lacking the major tegument proteins encoded by the UL46, UL47, UL48, and UL49 genes is viable in cultured cells. J. Virol. 77:12891-12900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghiasi, H., G. Perng, S. Cai, A. B. Nesburn, and S. L. Wechsler. 1996. The UL3 open reading frame of herpes simplex virus type 1 codes for a phosphoprotein. Virus Res. 44:137-142. [DOI] [PubMed] [Google Scholar]
- 19.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 20.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Granzow, H., F. Weiland, A. Jöns, B. G. Klupp, A. Karger, and T. C. Mettenleiter. 1997. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J. Virol. 71:2072-2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaplan, A. S., and A. E. Vatter. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 23.Khattar, S. K., S. van Drunen Littel-van den Hurk, L. A. Babiuk, and S. K. Tikoo. 1995. Identification and transcriptional analysis of a 3′-coterminal gene cluster containing UL1, UL2, UL3, and UL3.5 open reading frames of bovine herpesvirus-1. Virology 213:28-37. [DOI] [PubMed] [Google Scholar]
- 24.Klupp, B. G., and T. C. Mettenleiter. 1999. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J. Virol. 73:3014-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 26.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klupp, B. G., W. Fuchs, E. Weiland, and T. C. Mettenleiter. 1997. Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J. Virol. 71:7687-7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Markovitz, N. S., and B. Roizman. 2000. Small dense nuclear bodies are the site of localization of herpes simplex virus UL3 and UL4 proteins and of ICP22 only when the latter protein is present. J. Virol. 74:523-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Markovitz, N. S., F. Filatov, and B. Roizman. 1999. The UL3 protein of herpes simplex virus 1 is translated predominantly from the second in-frame start methionine codon and is subjected to at least two posttranslational modifications. J. Virol. 73:8010-8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGeoch, D. J., M. B. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, and D. McNab. 1988. The complete DNA sequence of the unique long region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 34.Melese, T., and Z. Xue. 1995. The nucleolus: an organelle formed by the act of building a ribosome. Curr. Opin. Cell Biol. 7:319-324. [DOI] [PubMed] [Google Scholar]
- 35.Mettenleiter, T. C. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623-625. [DOI] [PubMed] [Google Scholar]
- 36.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mettenleiter, T. C., A. Saalmüller, and F. Weiland. 1993. Pseudorabies virus protein homologous to herpes simplex virus type 1 ICP18.5 is necessary for capsid maturation. J. Virol. 67:1236-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nixdorf, R., B. G. Klupp, and T. C. Mettenleiter. 2001. Restoration of function of carboxy-terminally truncated pseudorabies virus glycoprotein B by point mutations in the ectodomain. J. Virol. 75:11526-11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roizman, B., and D. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 42.Roizman, B., and P. Pellet. 2001. The family Herpesviridae: a brief introduction, p. 2381-2397. In D. M. Knipe and P. M. Howley (ed.), Virology, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 43.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Telford, E. A. R., M. S. Watson, D. McBirde, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus 1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 45.Wagenaar, F., J. M. Pol, B. Peeters, A. L. Gielkens, N. de Wind, and T. G. Kimman. 1995. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J. Gen. Virol. 76:1851-1859. [DOI] [PubMed] [Google Scholar]
- 46.Worrad, D. M., and S. Caradonna. 1993. The herpes simplex virus type 2 UL3 open reading frame encodes a nuclear localizing phosphoprotein. Virology 195:364-376. [DOI] [PubMed] [Google Scholar]
- 47.Yamada, H., Y. Jiang, H. Zhu, K. Inagaki-Ohara, and Y. Nishiyama. 1999. Nucleolar localization of the UL3 protein of herpes simplex virus type 2. J. Gen. Virol. 80:2157-2164. [DOI] [PubMed] [Google Scholar]
- 48.Yoshida, S., L. F. Lee, N. Yanagida, and K. Nazerian. 1994. Identification and characterization of a Marek's disease virus gene homologous to glycoprotein L of herpes simplex virus. Virology 204:414-419. [DOI] [PubMed] [Google Scholar]