Abstract
Emerging evidence suggests that mitochondrial (mt) DNA damage may be a trigger for apoptosis in oxidant challenged pulmonary artery endothelial cells (PAECs). Understanding the rate-limiting determinants of mtDNA repair may point to new targets for intervention in acute lung injury. The base excision repair (BER) pathway is the only pathway for oxidative damage repair in mtDNA. One of the key BER enzymes is Ogg1, which excises the base oxidation product, 8-oxoguanine. Previously we demonstrated that over-expression of mitochondrially-targeted Ogg1 in PAECs attenuated apoptosis induced by xanthine oxidase (XO) treatment. To test the idea that Ogg1 is a potentially rate-limiting BER determinant protecting cells from oxidant-mediated death, PAECs transfected with siRNA to Ogg1 were challenged with XO and the extent of mitochondrial and nuclear DNA damage was determined along with indices of apoptosis. Transfected cells demonstrated significantly reduced Ogg1 activity, which was accompanied by delayed repair of XO-induced mtDNA damage and linked to increased XO-mediated apoptosis. The nuclear genome was undamaged by XO in either control PAECs or cells depleted of Ogg1. These observations suggest that Ogg1 plays a critical and possibly rate-limiting role in defending PAECs from oxidant-induced apoptosis by limiting the persistence of oxidative damage in the mitochondrial genome.
Keywords: Reactive oxygen species, mitochondrial DNA, oxidative damage, Ogg1 depletion, DNA repair, apoptosis
Introduction
Oxidative stress can kill pulmonary vascular endothelial cells (ECs), but the sentinel molecules that trigger cell death pathways as a consequence of oxidative modifications are poorly understood. Emerging evidence, however, points to a conspicuous association between oxidative damage to mitochondrial (mt) DNA and induction of apoptosis. First, mtDNA is about 50 fold more sensitive to oxidative damage than the nuclear genome [1–3]. Global nuclear DNA damage cannot be detected at oxidant burdens far in excess of those nearly obliterating the mitochondrial genome, even using techniques capable of revealing damage at single nucleotide resolution applied to sequences with known sensitivity to oxidative stress [3]. Second, survival of pulmonary arterial (PA), microvascular (MV), and venous (PV) endothelial cells challenged with xanthine oxidase (XO)-generated reactive oxygen species (ROS) is inversely related to the rate by which mtDNA damage is repaired after oxidative damage [3]. MVECs repair oxidative mtDNA damage fastest and are least sensitive to oxidant-induced cell death, while PAECs and PVECs display intermediate and highest sensitivities, respectively. Finally, over-expression of a mitochondrially targeted DNA glycosylase, Ogg1, protects against mtDNA damage and apoptosis induced in PAECs by XO-generated ROS [4, 5]. Similar strategies for protecting against mtDNA damage also have been shown to suppress asbestos-induced apoptosis in bronchial epithelial and other cell lines [6, 7].
The ability of mitochondrially-targeted Ogg1 to protect against oxidative mtDNA damage in PAECs may be pharmacologically significant. Oxidative base damage to the mitochondrial genome is removed by the base excision pathway of DNA repair [8, 9] which appears to be generally similar to the four-step pathway operative in the nucleus. Oxidatively damaged bases are recognized by DNA glycosylases after which the repair intermediate is processed by Ape1/Ref-1, a new base is inserted by a DNA polymerase, and the incised strands are re-ligated by a DNA ligase. The enzymes governing these steps are encoded by nuclear genes and appear to be subject to regulated import into mitochondria. Which of these steps is functionally rate-limiting - that is, which determines cell survival in response to oxidant stress – is unknown. However, the above-mentioned findings that genetically increased mitochondrial Ogg1 activity suppresses oxidant-induced mtDNA damage and cell death suggests that the initial step in base excision repair may function in this capacity. To explore this hypothesis further, the present studies used siRNA to inhibit Ogg1 expression in PAECs and defined this intervention in terms of changes in XO-induced damage to the mitochondrial and nuclear genomes and the induction of apoptosis. Our results suggest that Ogg1 may indeed govern mtDNA repair rate and thereby serve as a rate-limiting step controlling PAEC fate in response to oxidant stress.
Materials and Methods
Rat pulmonary artery endothelial cell culture and treatment
Rat PAECs were isolated and cultured as described previously [3]. In brief, main pulmonary arteries were isolated from 250 – 300 g Sprague-Dawley male rats killed with an overdose of sodium pentobarbital. Isolated arteries were opened and the intimal lining was carefully scraped with a scalpel. The harvested cells were then placed into flasks (Corning Inc., Corning, NY) containing F12 Nutrient Mixture and Dulbecco’s modified Eagle’s medium (DMEM) mixture (1:1) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Invitrogen, Carlsbad, CA). Culture medium was changed once per week and, after reaching confluence, the cells were harvested using a 0.05% solution of trypsin (Invitrogen, Carlsbad, CA). The endothelial cell phenotype, confirmed by acetylated LDL uptake, Factor VIII-RAg immunostaining, and the lack of immunostaining with smooth muscle cell α-actin antibodies (Sigma, St. Louis, MO), persisted for at least 15 passages.
Enzymatic oxidation of hypoxanthine (HX) by XO was used as a source of reactive oxygen species for cell treatment. Confluent PAECs transfected with siRNA were challenged for 1 hour with the indicated concentrations of XO and 0.5 mM HX in Hanks’ Balanced Salt Solution (HBSS). After treatment, cells were either processed immediately or placed into cell culture media for recovery. Doses and durations of XO treatment and recovery were selected based on previous studies showing evolution of XO-mediated mtDNA damage and apoptosis over a 4–6 hour period [5].
Inhibition and assessment of PAEC Ogg1 expression
Cells were transfected with Ogg1-specific siRNA with the sense and antisense sequences of 5’-GCUUGAUGAUGUCACUUAUtt-3’ and 5’-AUAAGUGACAUCAUCAAGCtg-3’, respectively or with scrambled sequence siRNA (Ambion, Austin, TX) using DharmaFECT 1 transfection reagent (Dharmacon, Lafayette, CO) following the manufacturer’s transfection protocol. In brief, cells were seeded in 6 or 12-well plates 24 hours before transfection and grown to 50–70% confluence. At this point, cell medium was replaced by transfection medium containing 100 nM of siRNA. OptiMEM (Invitrogen, Carlsbad, CA) was used as a serum-free medium to prepare working solutions of transfection reagent and siRNA, which were mixed together, incubated at room temperature for 20 min and combined with 4 volumes of complete culture medium to obtain transfection medium. Ogg1 mRNA expression in transfected cells was determined 24 h and 48 h after transfection.
Ogg1 mRNA expression, normalized to the abundance of 28S RNA, was quantified using real time PCR. In brief, total RNA was isolated from cells using PrepEase RNA spin kit (USB, Cleveland, OH) according to the manufacturer’s protocol. Quantitative real time PCR was carried out on the iCycler iQ5 (Bio-Rad, Hercules, CA) by amplifying equal amounts of total RNA using the iScript One-Step RT-PCR Kit with SYBR Green (Bio-Rad, Hercules, CA), also according to the manufacturer’s protocol. PCR primers were designed with Beacon Designer 7 software (PREMIER Biosoft International, Palo Alto, CA) and were as follows: Ogg1, forward 5’-CTAAGAAGACAGAAGGCTAGGTAG-3’, reverse 5’-TGACTTTGATTTGGGATGTTTGC-3’, and 28S RNA, forward 5’-CAACCTATTCTCAAACTT-3’, reverse 5’-GTCTATATCAACCAACAC -3’.
Ogg1-like activity was determined in sham- and siRNA-transfected PAECs using an oligonucleotide cleavage assay [4] with modifications. In brief, cells were washed with cold PBS and harvested with ice-cold buffer containing 20 mM HEPES pH 7.6, 1 mM EDTA, 2 mM DTT, 100 mM KCl, 5% glycerol, 0.05% triton X-100 with 1% of Protease Inhibitor Cocktail (Sigma, St. Louis, MO). The cell suspension was incubated at 4°C for 30 min and cleared by centrifugation at 20,000 × g for 10 min. Samples were stored at −80°C. Ogg1-like DNA glycosylase activity was determined by incubating the above prepared cellular extract with a 24-mer, 32P-end-labeled, duplex oligonucleotide containing an 8-oxoguanine at the 10th position (Trevigen, Gaithersburg, MD). An identical oligonucleotide without the 8-oxoguanine was used in parallel reactions as a negative control. Enzyme assays were performed in 20 µl reaction volume, containing 10 µg of the cellular extract, 85 fmol of labeled duplex oligonucleotide and NTE reaction buffer (25 mM Tris pH 7.6, 2 mM EDTA, 70 mM NaCl) [10].The reaction was carried out at 37°C for 1 h and terminated by adding 10 µl of 3X denaturing loading dye (300 mM NaOH, 97% formamide, 0.02% bromophenol blue). Samples were heated to 95°C for 5 min and resolved on 20% acrylamide gels with 8 M urea in TBE buffer.
DNA isolation and damage assessment
Sham and siRNA transfected PAECs cultured on 6-well plates were challenged for 1 h with the indicated concentrations of XO and 0.5 mM HX in HBSS. To detect damage in mitochondrial genome, total DNA was isolated immediately after treatment or after indicated recovery period using a DNeasy Blood and Tissue Kit (Qiagen GmbH, Hamburg, Germany). Prior to isolation all buffers were purged with nitrogen to prevent DNA oxidation. Southern blot analysis was performed as published previously with minor modifications [5]. In brief, purified DNA was digested with BamHI (New England Biolabs, Beverly, MA) - 10 U/mg DNA, overnight at 37°C. Following restriction, samples were precipitated, dissolved in TE buffer and precise DNA concentrations were measured on the Hoefer DyNA Quant 200 fluorometer (Pharmacia Biotech, San Francisco, CA) using Hoechst 33258 dye. To reveal oxidative base modifications, DNA was treated with formamidopyrimidine glycosylase, Fpg (New England Biolabs, Beverly, MA), a bacterial DNA repair enzyme that cleaves DNA at sites of oxidized purines, thereby creating single-strand breaks detectable on alkaline Southern blot. Samples containing 500 ng DNA were treated with 8 units of Fpg in 20 µl of reaction volume at 37°C for 1 h. Subsequently, samples were treated with NaOH (final concentration 0.1 N) for 15 min at 37°C, mixed with loading dye and resolved in 0.6% agarose alkaline gel. After electrophoresis DNA was vacuum transferred to Hybond N+ membrane (Amersham Biosciences, Little Chalfont, UK) and hybridized with 32P-labeled probe overnight at 55°C. The probe to mtDNA was generated by PCR using rat mtDNA sequence as template and the following primers: 5’-GCTGGAACAGGATGAACAGTAT-3’ for the sense strand and 5’-GTATCATGAAGTACAATGTCAAGGGATGAG-3’ for the anti-sense strand. The 746-bp product was hybridized with a 10.8-kb fragment of rat mtDNA obtained after BamHI digestion. After hybridization, the membrane was exposed to XAR-5 X-ray film (Eastman Kodak Co., Rochester, NY) and autoradiographic images were scanned with a GelLogic 1500 Imaging System (Eastman Kodak Co., Rochester, NY). Changes in the equilibrium lesion density were calculated as negative ln of the quotient of hybridization intensities in treated and control bands.
To assess nuclear oxidative DNA damage, pulmonary artery endothelial cells transfected with scrambled or Ogg1-specific siRNA were subjected to Comet and Fpg FLARE assays, performed according to the manufacturer's protocol (Trevigen, Gaithersburg, MD), immediately after a 1 hour exposure to xanthine oxidase, or as a positive control, a 15 min exposure to 1 mM H2O2,. The combination of Fpg treatment with the Comet assay permits detection of oxidized purine base products.
Quantification of apoptotic PAECs
As a morphological assessment of XO-induced apoptosis in PAECs, we enumerated the proportion of cells displaying small, condensed nuclei coincident with increased activated caspase 3. PAECs were exposed to the indicated concentrations of XO and 0.5 mM HX for 1 hour and harvested 4 hours later. Cells were fixed with 2% paraformaldehyde in PBS (1 h at 4°C), washed 3 times with 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 0.1% Triton X-100 (TBS-T), and stained with Hoechst 33258 dye (Molecular Probes, Eugene, OR) to demarcate nuclear morphology. For immunocytochemical analysis of caspase 3, control and XO-treated PAECs were fixed and washed as just described, then blocked with 5% normal goat serum in TBS-T, and incubated with primary antibody against activated form of caspase 3 (Cell Signaling Technology, Beverly, MA), diluted 1:120 in TBS-T with 3% BSA, at 4°C overnight. Cells were incubated with Texas Red-labeled anti-rabbit IgG (Vector Laboratories, Burlingame, CA) diluted 1:100 in TBS-T with 3% BSA for 1 hour at room temperature. Slides were then mounted with fluorescent mounting medium (DAKO, Carpinteria, CA) and observed with Olympus IX70 fluorescence microscope. Photomicrographs were taken with a SPOT digital camera using SPOT 2 software (Diagnostic Instruments, Sterling Heights, MI).
Immunoblot analyses of activated caspase-3
As a biochemical index of apoptosis, the activated form of caspase 3 was detected in total cell lysate using Western immunoblot analyses applied 4 hours after termination of a 1 hour exposure to XO. For preparation of total cell lysate, cells were washed twice with PBS and lysed in 2% SDS electrophoresis loading buffer after which 15 – 25 µg protein was applied to an SDS/12% polyacrylamide gel. Following separation, samples were transferred to nitrocellulose filters (BioRad Laboratories, Hercules, CA). Membranes blocked in 5% nonfat dry milk in PBS with 0.1% Tween 20 (PBS-T) were incubated overnight at 4°C with primary rabbit polyclonal antibody (1:600 dilution) recognizing the activated form of caspase 3 (Cell Signaling Technology, Beverly, MA). After washing, the membranes were incubated with 1:7,500 diluted HRP-conjugated goat anti-rabbit IgG for 1 h at room temperature and then revealed by chemiluminescence with the SuperSignal West Dura detection substrate (Pierce, Rockford, IL).
Statistical analysis
Pooled data are presented as the mean +/− the standard error (SE). Group-dependent differences were sought using one-way ANOVAs in conjunction with Neumann-Kuels test where appropriate. Differences were considered significant when p < 0.05.
Results
Ogg1 mRNA expression and Ogg1-like activity in siRNA transfected PAECs
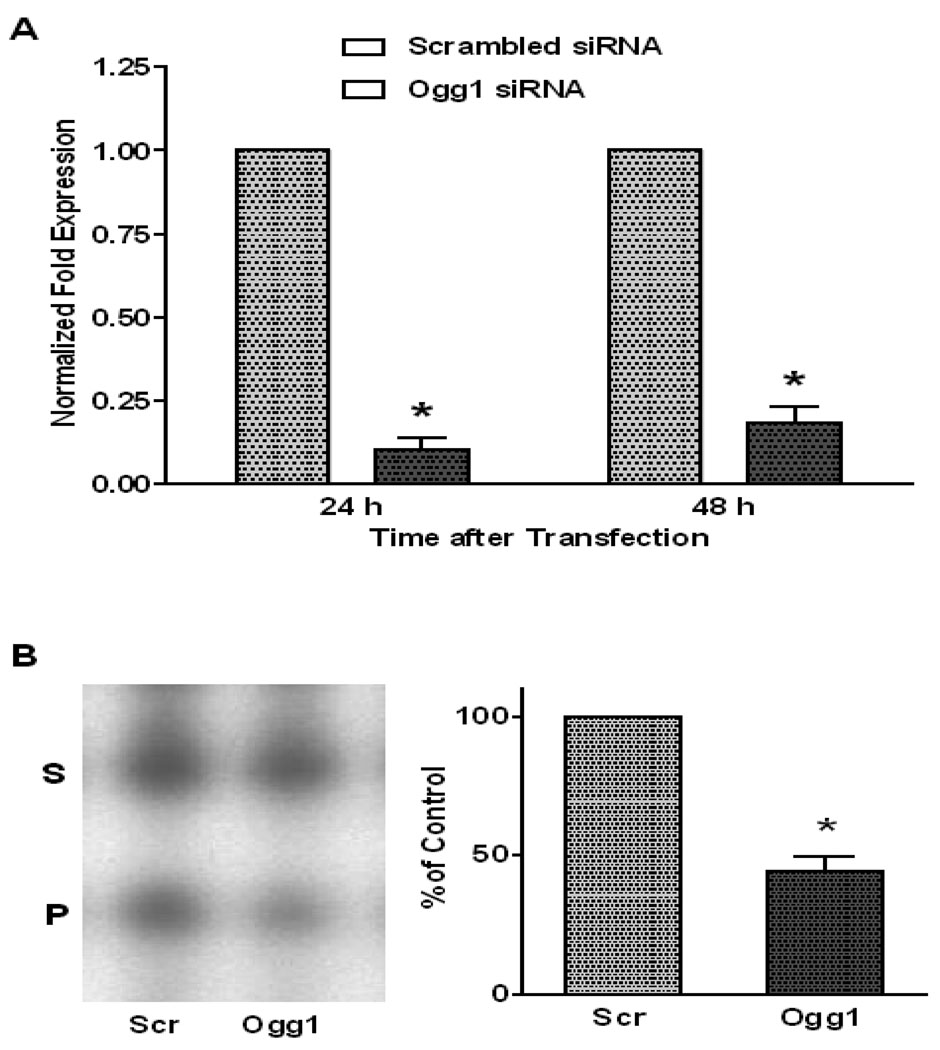
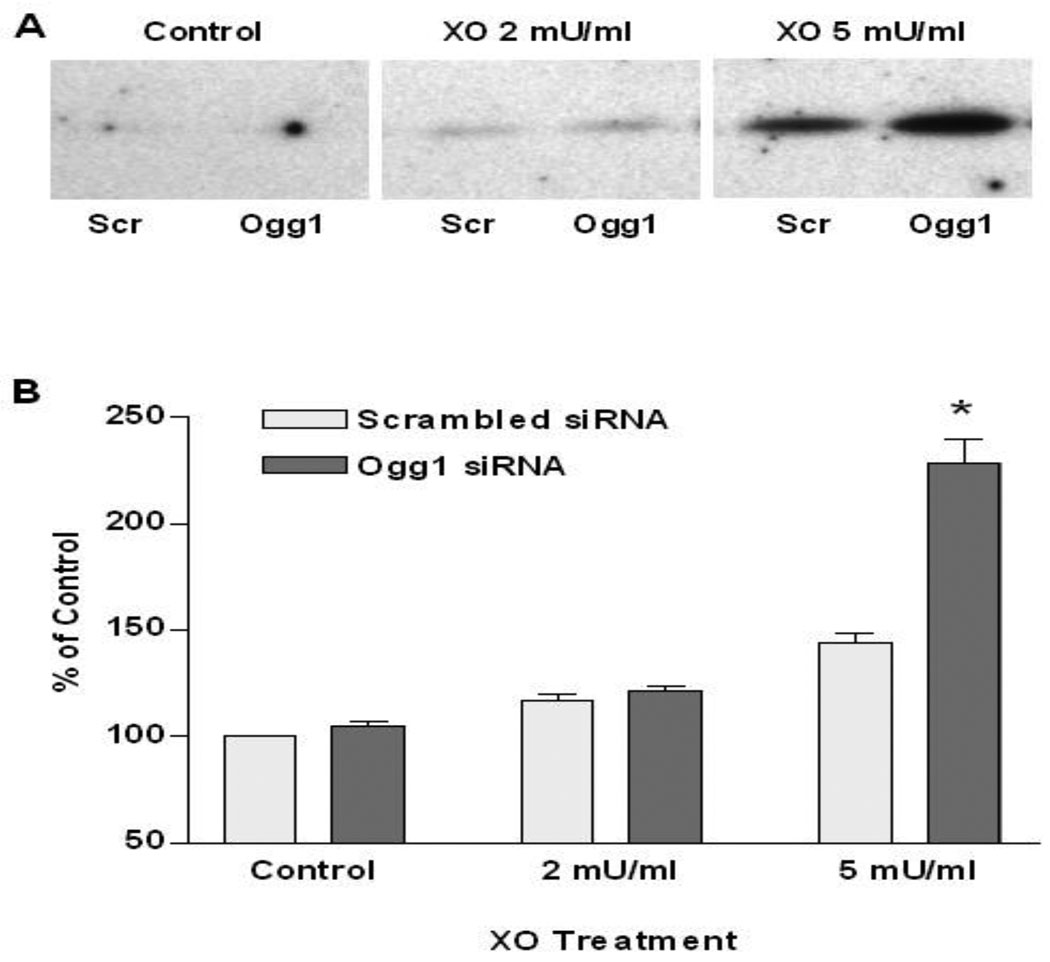
Ogg1 mRNA abundance and Ogg1-like activity in sham and siRNA-transfected PAECs are shown in FIGURE 1A and B, respectively. In comparison to sham treatment, siRNA transfection was associated with a substantial reduction in Ogg1 mRNA that persisted for at least 48h. Oligonucleotide cleavage assays revealed that siRNA transfection reduced DNA glycosylase activity by over 50%. It should be noted that this assay is not selective for Ogg1; rather, it reflects the entire constellation of glycosylases present in cultured PAECs, including Ogg1 and other enzymes with over-lapping specificities for 8-oxoguanine.
FIGURE 1.
Abundance of Ogg1 mRNA and Ogg1-like activity in pulmonary artery endothelial cells transfected with Ogg1-specific siRNA. (A) Decrease in Ogg1 mRNA accumulation normalized to the abundance of 28S RNA in PAECs transfected with Ogg1-specific siRNA compared with sham-transfected cells 24 and 48 hours after transfection. (Mean ± SE, N = 6, *p < 0.05). (B) Oligonucleotide cleavage assay for Ogg1-like activity in PAECs transfected with Ogg1-specific siRNA (Ogg1) and in cells transfected with a scrambled siRNA (Scr) 48 hours after transfection. LEFT: Representative autoradiograph of gel separation of 32P-labeled substrate (S) and product (P) of the Ogg1 enzymatic reaction. RIGHT: Results of oligonucleotide cleavage assays calculated as the ratio of product to substrate band intensities ratio and displayed as mean ± SE (N = 5, *p < 0.05).
Effect of Ogg1 knockdown on XO-induced nuclear and mitochondrial DNA damage
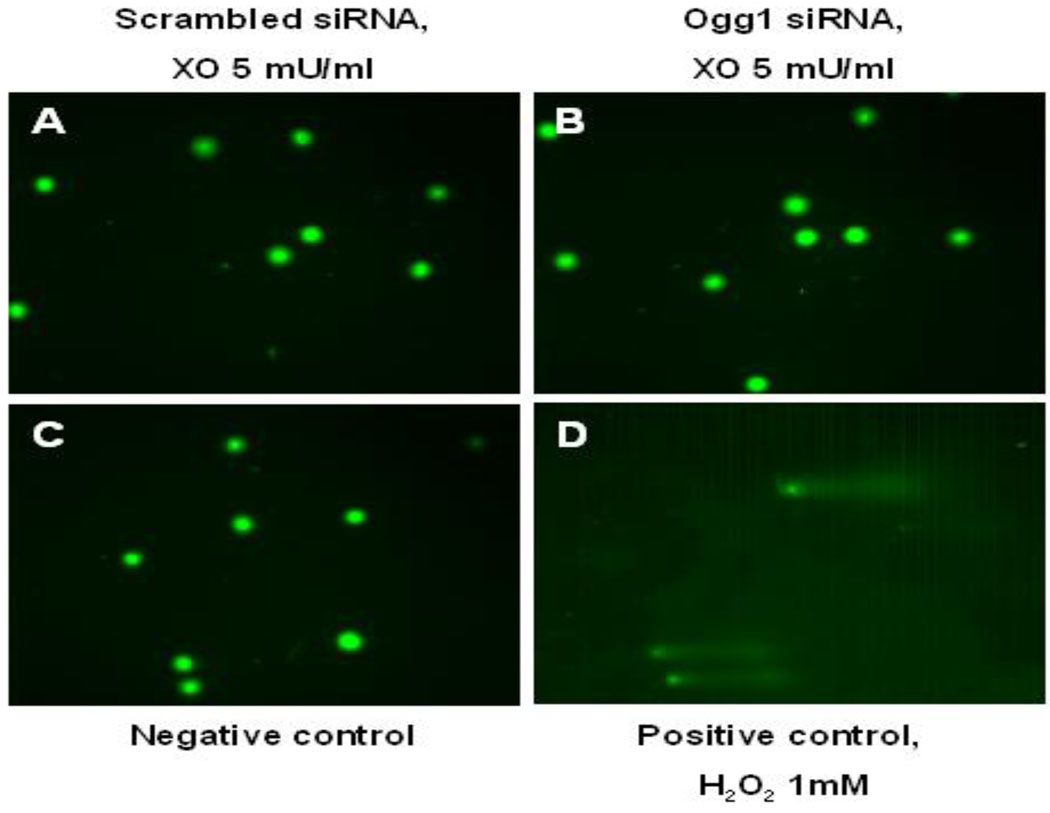
We applied the traditional Comet assay as well as a modification selective for 8-oxoguanine to search for oxidative nuclear DNA damage in PAECs immediately after XO + HX treatment. As shown in FIGURE 2, so-called Comet tails were not evident after XO-treatment in control cells, thus confirming previous reports that nuclear DNA is insensitive to ROS generation evoked by this concentration of XO + HX [3]. More interestingly, Comet tails also were not evident at any time in PAECs with siRNA-mediated reductions in cellular Ogg1 mRNA expression and Ogg1-like glycosylase activity.
FIGURE 2.
Lack of nuclear DNA fragmentation as detected by the Fpg-FLARE Comet assay in pulmonary artery endothelial cells transfected with either scrambled (A) or Ogg1-specific (B) siRNA and harvested immediately after 1 hour treatment with xanthine oxidase (5 mU/ml). Note lack of Comet “tails”. Negative results also were obtained in cells treated with 2 and 10 mU/ml of XO using either the conventional Comet assay or the Fpg-FLARE. (C) Pulmonary artery endothelial cells without any treatment were used as a negative control. (D) PAECs treated with 1 mM hydrogen peroxide for 15 min at 4°C were used as a positive control for the assay.
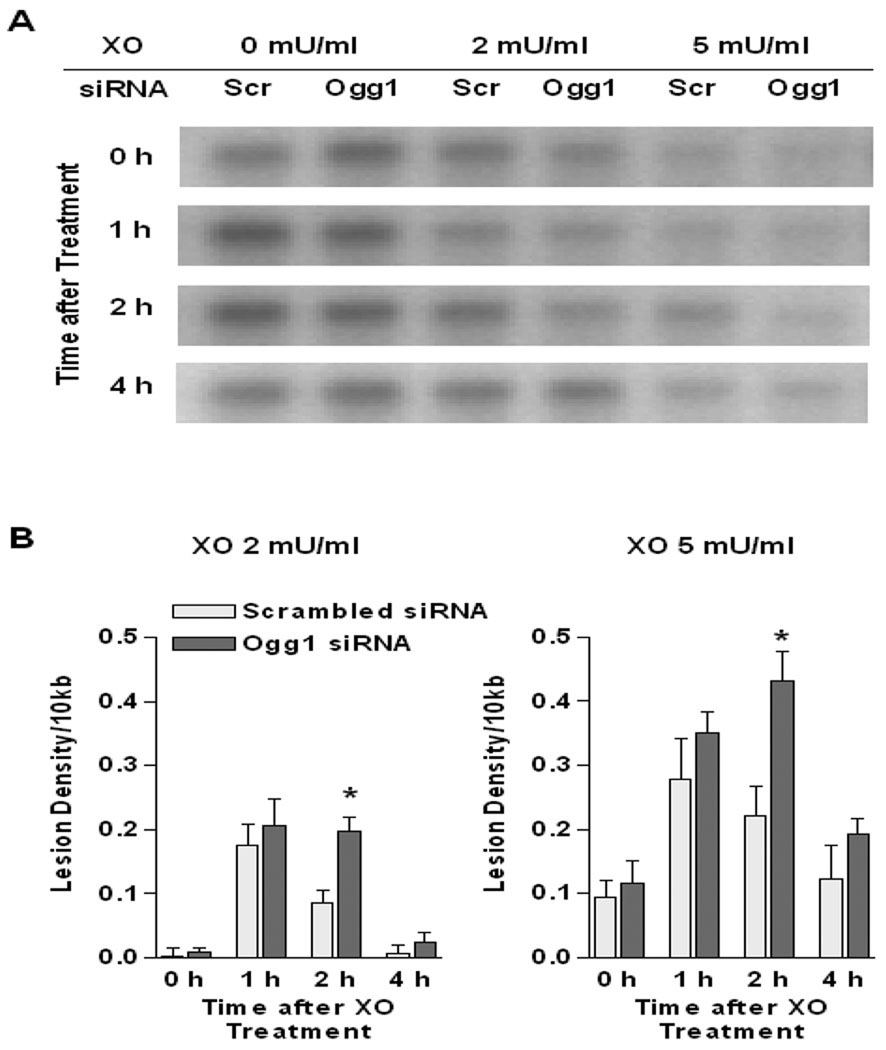
In contrast to the lack of impact of XO + HX on the nuclear genome, as shown in FIGURE 3 oxidative lesions in mtDNA were evident immediately after treatment with the ROS generating enzyme in both control and Ogg1 knock-down PAECs. In control cells, XO-induced ROS generation evoked transient accumulation of oxidative mtDNA damage, peaking at about 1 h after the 60 min exposure to XO + HX and declining to near baseline levels at 4 h. However, in cells with siRNA-mediated reductions in Ogg1, the application of XO + HX increased equilibrium mtDNA damage intensity to levels that exceed controls and peaked at 2 h after XO treatment, and which tended to remain elevated at 4h as well.
FIGURE 3.
Quantitative Southern blot analysis of Fpg-sensitive mtDNA damage in pulmonary artery endothelial cells. (A) Representative autoradiograph of Southern blot of mtDNA from PAECs transfected with scrambled (Scr) and Ogg1-specific (Ogg1) siRNA immediately after 1 h treatment with xanthine oxidase (XO, 2 or 5 mU/ml; 0 h) or without treatment (XO, 0 mU/ml; 0 h) and 1, 2 or 4 hours later. (B) Calculated changes in equilibrium lesion density in mtDNA normalized to 10 kb in PAECs immediately after 60 min treatment with XO, 2 or 5 mU/ml and at 1, 2 or 4 hours after removal of XO. Bars reflect means ± SE of 4 determinations. Note that lesions in mtDNA from Ogg1 siRNA transfected cells are significantly higher (*p < 0.05) at 2 h post XO treatment in comparison to sham-transfected cells.
Effect of Ogg1 knockdown on XO-induced PAECs apoptosis
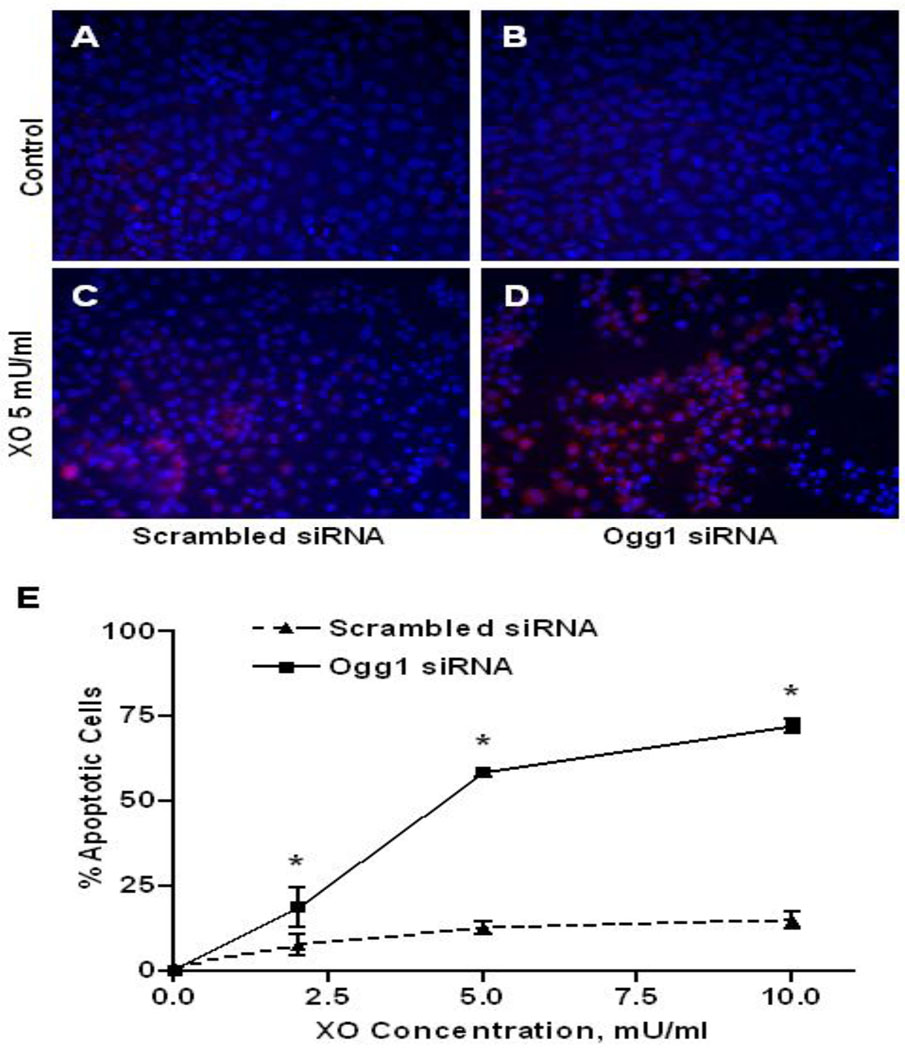
The time point for studying the apoptotic markers in XO-challenged PAECs was based on our earlier work [5] showing that markers of apoptosis (caspase 3 activation, DNA fragmentation) first appeared at 3 h and peaked at 4–6 h after exposure to XO generated ROS. Apoptosis in XO-challenged PAECs was assessed morphologically by enumerating the proportion of cells displaying small, condensed, apoptotic nuclei, in concert with activated caspase 3 immunostaining. As depicted in FIGURE 4, XO administered to control cells in concentrations of 2 – 10 mU/ml caused a modest induction of apoptosis, peaking at 10 – 15% of the total population. In contrast, the extent of morphologically detectable apoptotic PAECs was increased to about 75% in PAECs with reduced Ogg1 expression. Western immunoblot analysis of activated caspase 3 corroborated these findings (FIGURE 5); at a concentration of 5 mU/ml XO, activated caspase 3 was increased about 90% in PAECs with diminished Ogg1 expression.
FIGURE 4.
Immunocytochemical analysis of apoptotic markers in pulmonary artery endothelial cells transfected with scrambled and Ogg1-specific siRNA 4 h after cell treatment with xanthine oxidase. (A – D) Photomicrographs of PAECs transfected with scrambled ( A and C) or Ogg1-specific ( B and D) siRNA without treatment (A and B, Control) or after 1 h exposure to xanthine oxidase (C and D, XO 5 mU/ml) stained with Hoechst 33258 (blue) and antibodies against activated caspase 3 (red). (E) Pooled data displaying means ± SE of the percentage of apoptotic cells in cultures of siRNA transfected PAECs 4 h after exposure to xanthine oxidase (XO) in the indicated concentrations. (N = 4, *p < 0.05).
FIGURE 5.
Western blot analysis of pulmonary artery endothelial cells transfected with scrambled and Ogg1-specific siRNA for active caspase 3 after cell exposure to xanthine oxidase. (A) Representative Western blot analysis for active caspase 3 in PAECs transfected with scrambled (Scr) or Ogg1-specific (Ogg1) siRNA (Control) or harvested 4 hours after treatment with xanthine oxidase (2 or 5 mU/ml). (B) Pooled data for relative band intensities of activated caspase 3 hybridization displayed as mean ± SE (N = 3, *p < 0.05).
Discussion
Several lines of provocative evidence link oxidative damage to mtDNA to induction of apoptosis. In comparison to nuclear DNA, mtDNA is considerably more sensitive to oxidative damage [1–3]. Oxidative damage to the mitochondrial genome is repaired by the base excision pathway of DNA repair, and in a variety of cell types the rate of mtDNA repair is predictive of the cytotoxic response to exogenously applied or mitochondrially generated oxidants [3, 11]. Cells with high rates of mtDNA repair are relatively insensitive to ROS-mediated cytotoxicity, while those with low rates of repair are more sensitive. Finally, over-expression of the DNA glycosylase, Ogg1, targeted to mitochondria suppresses mtDNA damage and cytotoxicity in response to a variety of oxidant stresses in a variety of cells [4–6, 12, 13]. Despite these reports, many unanswered questions remain about how mtDNA integrity is maintained in response to oxidant stress and how oxidative damage to the mitochondrial genome triggers cell death pathways.
The findings that mitochondrially-targeted Ogg1 protects against ROS-induced mtDNA damage and cell death suggest that the initial, glycosylase-mediated step in base excision repair could be rate limiting in the cytotoxic response to oxidant stress. To address this idea, in the current study we used siRNA to decrease cellular Ogg1 expression in PAECs and defined its effect in terms of nuclear and mitochondrial DNA damage and cell death in response to ROS generated by XO + HX. Interestingly, while this inhibitory strategy nearly abolished Ogg1 mRNA expression, glycosylase activity directed against an oligonucleotide harboring 8-oxoguanine was reduced by only 50 – 60%. This disparity may reflect the presence of residual Ogg1 protein remaining even 48 h after persistent suppression of Ogg1 mRNA expression, but more likely represents the activities of the other glycosylases acting on 8-oxoguanine and known to be present in mammalian cells [14–16].
We found that the reduction in Ogg1 mRNA and diminution of Ogg1-like activity differentially impacted sensitivities of the nuclear and mitochondrial genomes to oxidant stress. Xanthine oxidase exposure failed to damage nuclear DNA not only in sham-transfected PAECs, but also in cells with reduced Ogg1, as detected by Comet and Fpg FLARE assays. These results are consistent with our previous findings; using the highly sensitive ligation-mediated PCR method to detect damage at single nucleotide resolution and quantitative Southern blot analysis to reveal base damage ≈4–10Kb sequences, we were unable to detect XO-mediated base damage at doses similar to that used in the current study [3, 17]. Indeed, damage to nuclear genes barely rose above the limit of detection at an XO concentration of 400 mU/ml, which produced 100% cytotoxicity and obliterated mtDNA integrity. Reports from other investigators also demonstrate that mtDNA is more prone to oxidative damage than nuclear genes. In Ogg1 knockout mice, for example, the mitochondrial genome contains almost 9 times more 8-oxoguanine than control animals, while in the nuclear DNA the level of 8-oxoguanine is increased only 2-fold [18]. There are multiple explanations for the divergent sensitivities of the two genomes to oxidant stress. For many years, the relative resistance of the nuclear genome to ROS damage, in comparison to mtDNA, was ascribed to the compact structure of chromatin and the presence of multiple, avid DNA repair pathways in the nucleus. By contrast, the mtDNA-protein nucleoid structure is believed to be more accessible to ROS and is protected principally by the base excision DNA repair (BER) pathway, with the presence of other repair mechanisms doubtful. In terms of the base excision repair pathway, there are various glycosylases present in the nucleus, many of which are also present in mitochondria. Our findings show that despite the reduction in Ogg1 mRNA expression, total cell glycosylase activity directed against the common base oxidation product, 8-oxoguanine, remained at about 45% of control. Thus, it is possible that the compact chromatin structure of nuclear DNA acting in concert with DNA glycosylases other than Ogg1 combined to retain nuclear DNA protection even in the face of limited Ogg1.
In marked contrast, Ogg1 knockdown exerted significant effects on the integrity of the mitochondrial genome. Reduction in Ogg1 expression did not affect the density of oxidative mtDNA damage in the baseline state, nor did it impact the initial increase in mtDNA lesion density evoked by 1h treatment with XO. Importantly, however, Ogg1 deficiency delayed mtDNA repair after XO-induced damage. The persistent oxidative mtDNA damage in cells with reduced Ogg1 activity was accompanied by a marked increase in the proportion of PAECs undergoing apoptosis. These observations are consistent with previous studies showing that over-expression of Ogg1 in mitochondria increases not only mtDNA repair rate after oxidative damage, but also enhances survival in multiple types of cells, including PAECs [4, 5, 7, 12, 13, 19]. Collectively, these findings support the view that Ogg1 plays a functionally significant role in governing repair of oxidative mtDNA damage and in initiation of apoptosis in response to exogenously generated oxidants. In addition, they point to the prospect that persistence of mtDNA damage after its initial introduction by an oxidant stress may interact with the magnitude of damage in terms of dictating cell survival.
The specific pathway linking mtDNA damage and repair to cell death may be more complex than previously surmised. Traditional concepts hold that mtDNA damage leads to a reduction in mtDNA encoded subunits of the electron transport chain, which in turn promotes a degenerative cycle of excessive ROS generation and increasing mtDNA damage ultimately culminating in mitochondrial-triggered, ROS-dependent apoptosis [20–22]. Thus, increasing or decreasing the persistence of mtDNA damage – for example, by increasing or decreasing the efficiency of mtDNA repair – would be predicted to exert coordinate effects on the propensity for oxidant-mediated apoptosis. In support of this idea, Chatterjee and co-authors demonstrated that targeting an R229Q mutant hOgg1 to mitochondria of HeLa cells significantly reduced mtDNA integrity and resulted in decreased cellular survival after exposure to oxidative stress [23]. Moreover, mitochondrially-targeted mutant hOgg1 caused more cell death than nuclear-targeted mutant hOgg1 upon exposure of cells to oxidative damage.
Challenging this traditional notion are recent findings by Kamp and coworkers showing that over-expression and mitochondrial targeting of either a wild type Ogg1 or a DNA glycosylase-deficient Ogg1 mutant protected against asbestos-induced apoptosis in A549 cells [6]. They also demonstrated that aconitase co-immunoprecipitated with over-expressed, mitochondrially targeted Ogg1, and on this basis proposed the intriguing hypothesis that the DNA repair enzyme functions to suppress oxidant mediated cell death not by contributing to mtDNA repair per se, but rather, by inhibiting the ROS-dependent mobilization of pro-apoptotic aconitase. Expanding on this concept of mtDNA-protein stability, several recent reports suggest that mtDNA-protein complexes – nucleoids - are considerably more dynamic than previously appreciated and are linked to the cell cytoskeleton by filaments spanning the inner and outer mitochondrial membranes [24]. It is thus tempting to speculate that persistent oxidative mtDNA damage may alter mtDNA-protein binding in the nucleoid complex which in turn results in release of pro-apoptotic proteins or abnormal interactions between mitochondria and cytoskeleton, either of which could trigger an apoptotic signal.
The concept that oxidative mtDNA damage interacts with an Ogg1-depedent step in mtDNA base excision repair pathway to regulate pulmonary vascular EC fate in response to oxidant stress could have interesting implications for identifying individuals at risk for oxidant mediated acute lung injury. In this regard, polymorphisms in the Ogg1 gene are known to be linked to an increased risk for various malignancies [25–27]. In light of the current finding that reduced Ogg1 expression and Ogg1-like activity sensitizes PAECs to ROS-induced apoptosis, it is tempting to speculate that such polymorphisms could confer an exaggerated risk for oxidant-mediated lung injury. Related to this concept, if Ogg1 is functionally rate-limiting in the survival response of pulmonary vascular ECs to oxidant stress, then pharmacologic strategies to enhance mitochondrial activity of the enzyme, perhaps using targeted fusion protein constructs, could be therapeutically advantageous. Obviously, considerable additional investigation will be required to address these interesting possibilities.
Acknowledgments
Dr. Glenn Wilson is acknowledged for his many helpful discussions concerning this work. We also appreciate the assistance of Ms. Gina Capley Bardwell.
This study was supported by National Heart, Lung and Blood Institute grants PO1 HL66299 and RO1 HL073244.
Abbreviations Used
- ANOVA
analysis of variance
- Ape1/Ref-1
apurinic/apyrimidinic endonuclease
- BER
base excision repair
- BSA
bovine serum albumin
- DTT
dithiothreitol
- ECs
endothelial cells
- FBS
fetal bovine serum
- Fpg
formamidopyrimidine DNA glycosylase
- HRP
horseradish peroxidase
- HX
hypoxanthine
- LDL
low density lipoprotein
- mtDNA
mitochondrial DNA
- MV
microvascular
- Ogg1
8-oxoguanine DNA glycosylase
- PA
pulmonary arterial
- PAECs
pulmonary artery endothelial cells
- PV
pulmonary venous
- ROS
reactive oxygen species
- RT
reverse transcription
- SE
standard error
- TBE
Tris/Borate/EDTA
- TE
Tris/EDTA
- XO
xanthine oxidase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ballinger SW, Patterson C, Yan CN, Doan R, Burow DL, Young CG, Yakes FM, Van Houten B, Ballinger CA, Freeman BA, Runge MS. Hydrogen peroxide- and peroxynitrite-induced mitochondrial DNA damage and dysfunction in vascular endothelial and smooth muscle cells. Circ Res. 2000;86:960–966. doi: 10.1161/01.res.86.9.960. [DOI] [PubMed] [Google Scholar]
- 2.Yakes FM, Van Houten B. Mitochondrial DNA damage is more extensive and persists longer than nuclear DNA damage in human cells following oxidative stress. Proc Natl Acad Sci U S A. 1997;94:514–519. doi: 10.1073/pnas.94.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grishko V, Solomon M, Wilson GL, LeDoux SP, Gillespie MN. Oxygen radical-induced mitochondrial DNA damage and repair in pulmonary vascular endothelial cell phenotypes. Am J Physiol Lung Cell Mol Physiol. 2001;280:L1300–L1308. doi: 10.1152/ajplung.2001.280.6.L1300. [DOI] [PubMed] [Google Scholar]
- 4.Dobson AW, Grishko V, LeDoux SP, Kelley MR, Wilson GL, Gillespie MN. Enhanced mtDNA repair capacity protects pulmonary artery endothelial cells from oxidant-mediated death. Am J Physiol Lung Cell Mol Physiol. 2002;283:L205–L210. doi: 10.1152/ajplung.00443.2001. [DOI] [PubMed] [Google Scholar]
- 5.Ruchko M, Gorodnya O, LeDoux SP, Alexeyev MF, Al-Mehdi AB, Gillespie MN. Mitochondrial DNA damage triggers mitochondrial dysfunction and apoptosis in oxidant-challenged lung endothelial cells. Am J Physiol Lung Cell Mol Physiol. 2005;288:L530–L535. doi: 10.1152/ajplung.00255.2004. [DOI] [PubMed] [Google Scholar]
- 6.Panduri V, Liu G, Surapureddi S, Kondapalli J, Soberanes S, de Souza-Pinto NC, Bohr VA, Budinger GR, Schumacker PT, Weitzman SA, Kamp DW. Role of mitochondrial hOGG1 and aconitase in oxidant-induced lung epithelial cell apoptosis. Free Radic Biol Med. 2009;47:750–759. doi: 10.1016/j.freeradbiomed.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shukla A, Jung M, Stern M, Fukagawa NK, Taatjes DJ, Sawyer D, Van Houten B, Mossman BT. Asbestos induces mitochondrial DNA damage and dysfunction linked to the development of apoptosis. Am J Physiol Lung Cell Mol Physiol. 2003;285:L1018–L1025. doi: 10.1152/ajplung.00038.2003. [DOI] [PubMed] [Google Scholar]
- 8.LeDoux SP, Wilson GL. Base excision repair of mitochondrial DNA damage in mammalian cells. Prog Nucleic Acid Res Mol Biol. 2001;68:273–284. doi: 10.1016/s0079-6603(01)68106-6. [DOI] [PubMed] [Google Scholar]
- 9.Bogenhagen DF. Repair of mtDNA in vertebrates. Am J Hum Genet. 1999;64:1276–1281. doi: 10.1086/302392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollenbach S, Dhenaut A, Eckert I, Radicella JP, Epe B. Overexpression of Ogg1 in mammalian cells: effects on induced and spontaneous oxidative DNA damage and mutagenesis. Carcinogenesis. 1999;20:1863–1868. doi: 10.1093/carcin/20.9.1863. [DOI] [PubMed] [Google Scholar]
- 11.Hollensworth SB, Shen C, Sim JE, Spitz DR, Wilson GL, LeDoux SP. Glial cell type-specific responses to menadione-induced oxidative stress. Free Radic Biol Med. 2000;28:1161–1174. doi: 10.1016/s0891-5849(00)00214-8. [DOI] [PubMed] [Google Scholar]
- 12.Rachek LI, Thornley NP, Grishko VI, LeDoux SP, Wilson GL. Protection of INS-1 cells from free fatty acid-induced apoptosis by targeting hOGG1 to mitochondria. Diabetes. 2006;55:1022–1028. doi: 10.2337/diabetes.55.04.06.db05-0865. [DOI] [PubMed] [Google Scholar]
- 13.Harrison JF, Rinne ML, Kelley MR, Druzhyna NM, Wilson GL, Ledoux SP. Altering DNA base excision repair: use of nuclear and mitochondrial-targeted N-methylpurine DNA glycosylase to sensitize astroglia to chemotherapeutic agents. Glia. 2007;55:1416–1425. doi: 10.1002/glia.20556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robertson AB, Klungland A, Rognes T, Leiros I. DNA repair in mammalian cells: Base excision repair: the long and short of it. Cell Mol Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meira LB, Burgis NE, Samson LD. Base excision repair. Adv Exp Med Biol. 2005;570:125–173. doi: 10.1007/1-4020-3764-3_5. [DOI] [PubMed] [Google Scholar]
- 16.Zharkov DO. Base excision DNA repair. Cell Mol Life Sci. 2008;65:1544–1565. doi: 10.1007/s00018-008-7543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grishko V, Solomon M, Breit JF, Killilea DW, Ledoux SP, Wilson GL, Gillespie MN. Hypoxia promotes oxidative base modifications in the pulmonary artery endothelial cell VEGF gene. FASEB J. 2001;15:1267–1269. doi: 10.1096/fj.00-0755fje. [DOI] [PubMed] [Google Scholar]
- 18.de Souza-Pinto NC, Eide L, Hogue BA, Thybo T, Stevnsner T, Seeberg E, Klungland A, Bohr VA. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. [PubMed] [Google Scholar]
- 19.Druzhyna NM, Hollensworth SB, Kelley MR, Wilson GL, Ledoux SP. Targeting human 8-oxoguanine glycosylase to mitochondria of oligodendrocytes protects against menadione-induced oxidative stress. Glia. 2003;42:370–378. doi: 10.1002/glia.10230. [DOI] [PubMed] [Google Scholar]
- 20.Mayer B, Oberbauer R. Mitochondrial regulation of apoptosis. News Physiol Sci. 2003;18:89–94. doi: 10.1152/nips.01433.2002. [DOI] [PubMed] [Google Scholar]
- 21.Ide T, Tsutsui H, Hayashidani S, Kang D, Suematsu N, Nakamura K, Utsumi H, Hamasaki N, Takeshita A. Mitochondrial DNA damage and dysfunction associated with oxidative stress in failing hearts after myocardial infarction. Circ Res. 2001;88:529–535. doi: 10.1161/01.res.88.5.529. [DOI] [PubMed] [Google Scholar]
- 22.Ricci C, Pastukh V, Leonard J, Turrens J, Wilson G, Schaffer D, Schaffer SW. Mitochondrial DNA damage triggers mitochondrial-superoxide generation and apoptosis. Am J Physiol Cell Physiol. 2008;294:C413–C422. doi: 10.1152/ajpcell.00362.2007. [DOI] [PubMed] [Google Scholar]
- 23.Chatterjee A, Mambo E, Zhang Y, Deweese T, Sidransky D. Targeting of mutant hogg1 in mammalian mitochondria and nucleus: effect on cellular survival upon oxidative stress. BMC Cancer. 2006;6:235. doi: 10.1186/1471-2407-6-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prachar J. Mouse and human mitochondrial nucleoid--detailed structure in relation to function. Gen Physiol Biophys. 2010;29:160–174. [PubMed] [Google Scholar]
- 25.Stanczyk M, Sliwinski T, Cuchra M, Zubowska M, Bielecka-Kowalska A, Kowalski M, Szemraj J, Mlynarski W, Majsterek I. The association of polymorphisms in DNA base excision repair genes XRCC1, OGG1 and MUTYH with the risk of childhood acute lymphoblastic leukemia. Mol Biol Rep. doi: 10.1007/s11033-010-0127-x. [DOI] [PubMed] [Google Scholar]
- 26.Li WQ, Zhang L, Ma JL, Zhang Y, Li JY, Pan KF, You WC. Association between genetic polymorphisms of DNA base excision repair genes and evolution of precancerous gastric lesions in a Chinese population. Carcinogenesis. 2009;30:500–505. doi: 10.1093/carcin/bgp018. [DOI] [PubMed] [Google Scholar]
- 27.Paz-Elizur T, Sevilya Z, Leitner-Dagan Y, Elinger D, Roisman LC, Livneh Z. DNA repair of oxidative DNA damage in human carcinogenesis: potential application for cancer risk assessment and prevention. Cancer Lett. 2008;266:60–72. doi: 10.1016/j.canlet.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]