Abstract
Phosphorylation is a ubiquitous protein post-translational modification that is intimately involved in most aspects of cellular regulation. Currently, most proteomic analyses are performed with phosphorylation searches for serine, threonine and tyrosine modifications, as the phosphorylated residues of histidine and aspartic acid are acid labile, and thus undetectable with most proteomic methodologies. Here, we present a novel buffer system to show histidine phosphorylation of NM23-H1, the product of the first identified putative human metastasis suppressor gene (NME1), which catalyzes the transfer of the γ-phosphate from nucleoside triphosphates to nucleoside diphosphates. Based on a pH titration of LC elution buffers and MS/MS identification, recombinant NM23-H1 subjected to auto-phosphorylation was shown to contain phosphorylated histidine at residue 118 at pH 5 and pH 6, with each level giving over 75% peptide coverage for identification. The solvent system presented permits the detection of all five possible phosphorylation moieties. Application of histidine and aspartic acid phosphorylation modifications to proteomic analyses will significantly advance the understanding of phosphorylation relay signaling in cellular regulation, including elucidation of the role of NM23-H1 in metastasis.
Introduction
Protein phosphorylation regulates many diverse processes in cells. Although phosphorylation of serine (Ser), threonine (Thr), and tyrosine (Tyr) are common among prokaryotes and eukaryotes, the phosphorylation of histidine (His) and aspartic acid (Asp) residues was historically considered a prokaryotic style of protein regulation, as exemplified in the bacterial chemotactic response1. In bacterial chemotaxis, two-component His kinase/regulator systems respond to environmental cues by initiating phospho-relay signaling via auto-phosphorylation of histidine. This phosphate group may then be transferred to other histidine or aspartate residues on interacting proteins, allowing the transmission of regulatory messages.
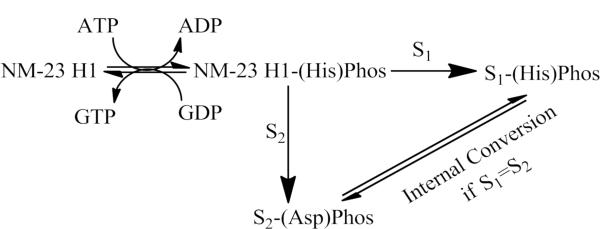
NM23-H1, the product of the first identified putative human metastasis suppressor gene2, was initially classified as a nucleoside diphosphate kinase (NDPK)3. This reaction catalyzes the transfer of the γ-phosphate from nucleoside triphosphates (NTPs), such as GTP and ATP, to nucleoside diphosphates (NDPs), including GDP, ADP, UDP and CDP4. It has been inferred thorough site-directed mutagenesis that the phosphorylation events occur via an auto-phosphorylation of His1185. Scheme 1 shows the mechanism of NM23-H1 phosphorylation. Initial auto-phosphorylation of histidine (pHis) may either form an alternate NTP or a meta-stable species capable of transferring its phosphate group to another histidine or an aspartic acid of an interacting protein6.
Scheme 1.
NM23-H1 as an NDPK and potential histidine and aspartic acid kinase. When acting as an NDPK, NM23-H1 converts NTPs to alternate NTPs through phosphorylation of NDPs. Alternatively, NM23-H1 can auto-phosphorylate His118 in an ATP-dependent manner. This phosphorylation can be transferred to either a histidine or aspartic acid residue on another protein (depicted as S1 or S2) in a `ping-pong' mechanism.
While NM23-H1 His kinase activity appears important in the regulation of G-Protein signaling pathways7, downstream recipients of phosphorylation transfer catalyzed by NM23-H1 remain largely uncharacterized, although several species have been identified, including ATP-citrate lyase8, Gβ9 and calcium-activated K+ channel (KCa3.1)10. NM23-H1 was first discovered as a cancer metastasis marker and it is currently the only described histidine kinase in mammals11. Histidine phosphorylation in mammals has been previously identified12, but these methodologies do not lend themselves to high-throughput schemes13. Elucidation of the NM23-H1 phosphorylation pathway may unravel its role in cancer and metastasis suppression.
The barrier to the observation of histidine and aspartic acid has been the acid labile nature of the phosphate group on histidine and aspartic acid residues. The inherent sensitivity of these modifications to low pH could render them undetected during MS/MS analysis in a typical proteomic run14. We report definitive mass spectrometric evidence using new solvent conditions to detect this regulatory moiety, the histidine phosphorylation of NM23-H1.
Materials and Methods
Methods
NME1 cloning and purification
Recombinant NM23-H1 was cloned and purified as the gene product of human NME1. Human NME1 cDNA was synthesized by GeneCopia (Rockville, MD) and a C-terminal hexaHistidine tag was engineered by amplification with primers NME1-for-NdeI (5'-CGCGCATATGGCCAACTGTGAGCGTA CC-3') and NME1-rev-HisBamHI (5'-CGCGGATCCTTAT CAATGATGATGATGATGATGTTCATAGATCCAGTTCT-3'). The resulting PCR product was cleaned with a Qiagen PCR cleanup kit, phosphorylated with T4 polynucleotide kinase and ligated via blunt ends into SmaI linearized plasmid puc118. The integrity of the resulting DNA was confirmed by sequencing. Next, NME1 cDNA was moved into the expression plasmid pET29a (Novagen) via flanking NdeI and BamHI sites. The resulting DNA was transformed into the inducible E. coli expression strain BL21(DE3). One liter of cells (O.D. 600 nm = ~1.0 AU) was induced with 1mM IPTG for 2hr at 37°C. Cells were harvested by centrifugation and stored in 20ml buffer A (50mM Tris-HCl pH 7.4, 300mM NaCl, 10mM imidazole, 10% (v/v) glycerol) at −80°C until purification. For purification, cells were thawed, lysozyme was added to 1mg/ml final concentration, and samples remained on ice for 60min. Protease inhibitors (PMSF, leupeptin, pepstatin, and chymostatin) were added from DMSO stocks at final concentrations of 1mM, 0.1mg/ml, 0.1mg/ml, and 0.025mg/ml respectively, and cells were lysed by 3 consecutive passages through a french press (SLM-Aminco) at 1000 psi. Residual cells and membranes were removed by centrifugation at 164,000×g for 1 hr at 4°C. Supernatant containing soluble proteins was then rocked at 4°C in a 50ml conical for 4 hr with 4ml slurry of Ni2+/NTA agarose beads (Qiagen), which had been pre-equilibrated with buffer A. Extract containing slurry was passed by gravity flow through a disposable column (BioRad), and beads were subsequently washed with an additional 50 mL buffer A, followed by 50 ml modified buffer A containing 35 mM imidazole. NM23-H1 was eluted with 20ml modified buffer A containing 500mM imidazole, and 1.5ml fractions were collected. Fractions containing protein, as determined by the Bradford method protein assay (BioRad), were analyzed by SDS-PAGE followed by Coomassie staining. Fractions containing peptide corresponding to the molecular weight of NM23-H1 were pooled and dialyzed against buffer B (50mM TrisHCl pH 8.0, 100mM NaCl). The concentration of NM23-H1 was determined using a Bradford protein assay (BioRad) with BSA as a standard. NM23-H1 was purified by anion exchange MonoQ 5/5 column (GE Life Sciences). MonoQ was equilibrated in buffer B, and sample was eluted with a linear NaCl gradient of 100mM to 2M over 15ml. Fractions containing NM23-H1 were quantitated by a Bradford protein assay (BioRad) and analyzed by SDS-PAGE followed by Coomassie staining.
Phosphorylation of NM23-H1
Auto-phosphorylation of recombinant NM23-H1 was performed using the addition of ATP as a co-factor for NM23-H1, as compared to a control lacking ATP. Five micrograms of NM23 H1-His6 was added to TMD buffer (20mM Tris-HCl (EMD), pH 8.0, 5mM MgCl2 (Sigma), 1mM DTT (Calbiochem) and 0.33μM ATP (Sigma)) as previously described15. One set of samples served as a control, with no ATP added to the buffer. After a 10-minute incubation at room temperature, the reacted proteins were buffer exchanged into 100mM ammonium bicarbonate, pH 8.0 using Zeba Desalting spin columns (Pierce) for enzymatic digestion.
Mass Spectrometry Analysis
Determination of histidine phosphorylation was performed using a novel solvent system for liquid chromatography (LC) elution and mass spectrometric identification. Phosphorylated and control NM23 H1 His-6 was digested with trypsin as previously described16, and 1 μg peptides were loaded onto a home-packed C18 column (6cm × 75um, Magic C18AQ 200Å 5μm - Michrom) via a pressure bomb. Solvent A consisted of LC/MS grade water (JT Baker) supplemented with 10mM ammonium bicarbonate (EMD), and the pH levels were adjusted to pH 8.0, 7.0, 6.0, 5.0, 4.0, 3.0 and 2.5 with formic acid (Pierce). Normal solvent conditions were LC/MS grade water supplemented with 0.1% formic acid alone. Solvent B for all runs was LC/MS grade methanol (JT Baker). Peptides were eluted with a gradient as follows: 5–15%B in 3 min., 15–50%B in 157 min., 50%B for 5 min., and finally return to baseline conditions of 5%B. A new column was prepared for each analysis. Mass spectrometry analysis was performed on an LCQ Deca XP Max 3-D ion trap mass spectrometer (ThermoFisher Scientific, San Jose, CA) in a data-dependent MS3 manner. A survey scan was performed followed by fragmentation of the three most abundant peaks. Additionally, if a neutral loss occurred, additional isolation was performed for MS3 analysis. Collision-induced dissociation (CID) was used as a means of fragmentation, with helium as the collision gas and normalized collision energy of 35%. Resultant RAW files were converted to mzxml files before being converted to mgf files (MassMatrix), which were then read into BioTools (Bruker Daltonics, Billerica, MA). The protein sequence of NM23 H1 His-6 was entered into SequenceEditor (Bruker Daltonics, Billerica, MA), theoretically digested, and the digest transferred to BioTools to match mass spectra with the peptide peaks from the samples.
Results and Discussion
The development of a unique buffered solvent system for characterization of phosphorylation sites at protein PTMs evolved from pH titration of assay buffer during chromatographic isolation of NM23-H1. As illustrated in Table 1, the single pH unit alterations performed caused substantial differences in peptide coverage and the number of successful peptide hits for both the control and auto-phosphorylated proteins. Only buffer systems at pH 5–6 successfully maintained the presence of the phosphate group on His118 for identification, though these pH levels did not provide optimum peptide coverage or the highest number of peptides mapped. Experiments at higher pH (pH 7.0 and pH 8.0) did not yield detection of peptide with the additional molecular weight of a phosphorylated histidine (866m/z (M+3H)), nor did they yield the non-phosphorylated ion (839m/z (M+3H)). The absence of a detected phosphorylation at the higher pH is likely due to the lower concentration of protons, and thus the inherent weaker ionization of this peptide species, rather than due to acid labile de-phosphorylation as expected at pH 3.0. A comparison of ions found at pH 3.0 (control), pH 5.0 (+ATP), and pH 6.0 (+ATP) are shown in Table 2.
Table 1.
Liquid chromatography conditions and mass spectrometry results. The pH of the aqueous phase was adjusted in intervals of 1 pH unit from pH 8.0 to pH 2.5, the latter being our normal operating condition. A control (no ATP) and experimental (with ATP) auto-kinase reaction were performed on NM23-H1. Shown are conditions, percent sequence coverage of the protein, number of unique peptides corresponding to NM23-H1 and presence of detectable histidine phosphorylation (indicated as detected or not-detected in the “pHis column”, with D indicating the absence of pHis and ND indicating the pHis was not detected).
| Sample | pH Solvent A | % Coverage | #Peptides | P-His |
|---|---|---|---|---|
| Control | 8 | 62.7 | 17 | ND |
| + ATP | 8 | 70.9 | 11 | ND |
| Control | 7 | 72.8 | 11 | ND |
| + ATP | 7 | 70.9 | 18 | ND |
| Control | 6 | 70.9 | 18 | ND |
| + ATP | 6 | 77.2 | 21 | D |
| Control | 5 | 91.8 | 23 | ND |
| + ATP | 5 | 76.6 | 20 | D |
| Control | 4 | 76.6 | 27 | ND |
| + ATP | 4 | 76.6 | 26 | ND |
| Control | 3 | 95.6 | 22 | ND |
| + ATP | 3 | 67.7 | 24 | ND |
| Control | 2.5 | 95.6 | 29 | ND |
| + ATP | 2.5 | 95.6 | 21 | ND |
Table 2.
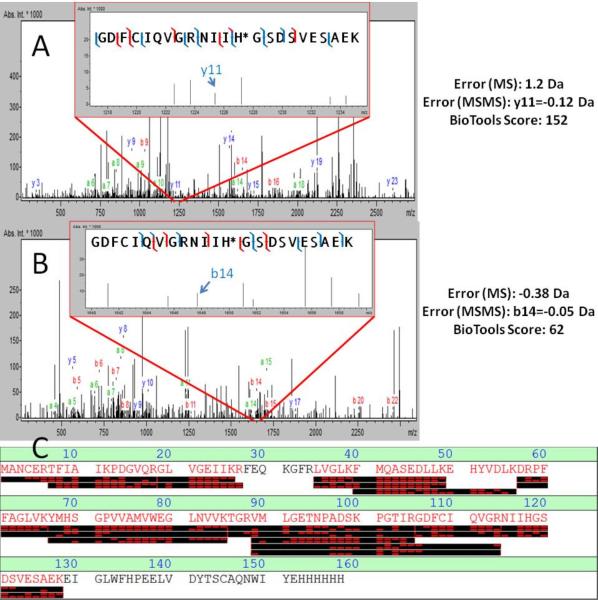
Corresponding b- and y-ions for phospho-peptide sequence. The peptide GDFC*IQVGRNIIH‡GSDSVESAEK, 866 m/z, is shown at pH 5.0 and pH 6.0. A corresponding nonphosphorylated peptide GDFC*IQVGRNIIHGSDSVESAEK, 839 m/z, from the pH 3.0 control was included as comparison. These are daughter ion data for the same peptides indicated in Figure 1.
| pH6.0 + ATP sample | pH5.0 + ATP sample | pH3.0 − ATP sample | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B-ion | Y-ion | B-ion | Y-ion | B-ion | Y-ion | |||||||||
| 1 | 58.029 | G | 2598.165 | 23 | 1 | 58.029 | G | 2598.165 | 23 | 1 | 58.029 | G | 2518.199 | 23 |
| 2 | 173.056 | D | 2541.144 | 22 | 2 | 173.056 | D | 2541.144 | 22 | 2 | 173.056 | D | 2461.178 | 22 |
| 3 | 320.124 | F | 2426.117 | 21 | 3 | 320.124 | F | 2426.117 | 21 | 3 | 320.124 | F | 2346.151 | 21 |
| 4 | 480.155 | C* | 2279.049 | 20 | 4 | 480.155 | C* | 2279.049 | 20 | 4 | 480.155 | C* | 2199.082 | 20 |
| 5 | 593.239 | I | 2119.018 | 19 | 5 | 593.239 | I | 2119.018 | 19 | 5 | 593.239 | I | 2039.052 | 19 |
| 6 | 721.297 | Q | 2005.934 | 18 | 6 | 721.297 | Q | 2005.934 | 18 | 6 | 721.371 | Q | 1925.968 | 18 |
| 7 | 820.366 | V | 1877.875 | 17 | 7 | 820.366 | V | 1877.875 | 17 | 7 | 820.366 | V | 1797.909 | 17 |
| 8 | 877.387 | G | 1778.807 | 16 | 8 | 877.387 | G | 1778.807 | 16 | 8 | 877.387 | G | 1698.841 | 16 |
| 9 | 1033.488 | R | 1721.785 | 15 | 9 | 1033.488 | R | 1721.785 | 15 | 9 | 1033.488 | R | 1641.819 | 15 |
| 10 | 1147.531 | N | 1565.684 | 14 | 10 | 1147.531 | N | 1565.684 | 14 | 10 | 1147.531 | N | 1485.718 | 14 |
| 11 | 1260.615 | I | 1451.641 | 13 | 11 | 1260.615 | I | 1451.641 | 13 | 11 | 1260.615 | I | 1371.675 | 13 |
| 12 | 1373.699 | I | 1338.557 | 12 | 12 | 1373.699 | I | 1338.557 | 12 | 12 | 1373.699 | I | 1258.591 | 12 |
| 13 | 1590.725 | H‡ | 1225.473 | 11 | 13 | 1590.725 | H‡ | 1225.473 | 11 | 13 | 1510.758 | H‡ | 1145.507 | 11 |
| 14 | 1647.746 | G | 1008.448 | 10 | 14 | 1647.746 | G | 1008.448 | 10 | 14 | 1567.78 | G | 1008.448 | 10 |
| 15 | 1734.778 | S | 951.427 | 9 | 15 | 1734.778 | S | 951.427 | 9 | 15 | 1654.812 | S | 951.427 | 9 |
| 16 | 1849.805 | D | 864.395 | 8 | 16 | 1849.805 | D | 864.395 | 8 | 16 | 1769.839 | D | 864.395 | 8 |
| 17 | 1936.837 | S | 749.368 | 7 | 17 | 1936.837 | S | 749.368 | 7 | 17 | 1856.871 | S | 749.368 | 7 |
| 18 | 2035.906 | V | 662.336 | 6 | 18 | 2035.906 | V | 662.336 | 6 | 18 | 1955.939 | V | 662.336 | 6 |
| 19 | 2164.948 | E | 563.267 | 5 | 19 | 2164.948 | E | 563.267 | 5 | 19 | 2084.982 | E | 563.267 | 5 |
| 20 | 2251.98 | S | 434.225 | 4 | 20 | 2251.98 | S | 434.225 | 4 | 20 | 2172.014 | S | 434.225 | 4 |
| 21 | 2323.017 | A | 347.193 | 3 | 21 | 2323.017 | A | 347.193 | 3 | 21 | 2243.051 | A | 347.193 | 3 |
| 22 | 2452.06 | E | 276.155 | 2 | 22 | 2452.06 | E | 276.155 | 2 | 22 | 2372.094 | E | 276.155 | 2 |
| 23 | 2580.155 | K | 147.113 | 1 | 23 | 2580.155 | K | 147.113 | 1 | 23 | 2500.189 | K | 147.113 | 1 |
on C indicates presence of carbamidomethylation, an artifact of the tryptic digest
on H indicates the presence of a phosphate group.
NM23-H1 is shown to be auto-phosphorylated at precisely His118 (Figure 1). Further, we verified phosphorylation of two other sites, Thr94 and Tyr53 (Supporting information). As these are present in both the samples containing ATP and those without ATP (Supporting information), they were likely phosphorylated in E. coli prior to purification. Additionally, we also found that NM23-H1 in E. coli acetylated Lys12 (Supporting information).
Figure 1.
Detection of histidine phosphorylation via LC/MS/MS. A and B) Fragmentation spectra of histidine phosphorylated peptide containing His118 from NM23-H1 in pH 5.0 and 6.0 solvents, respectively. Inlays represent zoomed regions showing appropriate fragment ions to indicate detection of histidine phosphorylation. Included with A and B are errors for the peptide and appropriate fragment, along with a map of detected ions and BioTools score. Blue hash marks show tolerances set to 1.5 Da for MS and 0.5 Da for MS/MS. Red hash marks indicate fragments detected when the tolerance for MS/MS was opened to 0.8 Da. C) Sequence coverage for NM23 H1-His6 from the pH 5.0 data-dependent MS3 run.
Histidine and aspartic acid phosphorylation events have not been well characterized due to their inherent pH sensitivity and the lack of phosphorylated His/Asp antibodies available commercially17 Whist others have employed a high pH solvent system to analyze pHis-containing peptides in histone H4, these data were obtained using milligram amounts of protein18, 19. In the present work, we show that pH 5–6 is best to observe this PTM, using microgram quantities of NM23-H1. Higher pH reduces protein coverage for identification, as demonstrated in Table 1. In addition, this methodology allows for analysis of phosphorylations from whole cell lysates in a high-throughput manner, allowing for rapid characterizations of large numbers of proteins. This is beneficial for screening studies where the target proteins or phosphorylation sites of interest are unknown.
Presently, the NM23-H1 phosphorylation relay signaling pathway remains largely uncharacterized. Although it has been thought that most regulatory phosphorylations in eukaryotes occur on threonine, serine and tyrosine residues, this could be the result of failed characterizations of histidine and aspartic acid residue phosphorylations. Without methods to identify the latter, their absence in prior works could be the result of the inability to see them, rather than their lack of regulatory function. Here we show that the histidine/aspartic acid axis for protein regulation may incorporate the use of histidine phosphorylation for message transmittance.
Using this method, the role of NM23-H1 as a metastasis suppressor can be further evaluated. The presence of high concentrations NM23-H1 has been shown to be an indicator of prognosis in many cancers17, 20–27, with its highest concentration in the non-tumorigenic state and a significantly reduced level in the metastatic state17. Over-expression of NM23-H1 can eradicate tumor cell motility and invasion28 while promoting cellular differentiation, and shuts down anchorage-independent growth29. The solvent system presented permits the detection of all five possible phosphorylation moieties (data not shown). Thus, these studies will significantly advance analysis of phosphorylation relay signaling in cellular regulation. Our goal is to employ this technique to define the phosphorylation relay pathway in cancer by mapping phosphorylation of all five possible phosphorylation residues, including histidine and aspartic acid residues. Based on the functional analysis of NM23-H1, these studies will provide unique insights into cellular regulation, cancer progression, and metastasis.
Synopsis.
Presented is a novel buffer system showing histidine phosphorylation of NM23-H1, a putative human metastasis suppressor protein and possibly part of a two-component regulation in humans. Based on a pH titration of LC elution buffers and MS/MS identification, recombinant NM23-H1 is auto-phosphorylated and shows pHis118 at pH 5.0 and pH 6.0. The solvent system presented permits the detection of five possible phosphorylation moieties: serine, threonine, tyrosine, histidine and aspartic acid.
Supplementary Material
Acknowledgement
We thank Michelle Friedman for insightful discussion and editing and the University of Rochester Proteomics Center for instrument time. AEF was funded by 1 UL1 RR024160-1 (NIH), FA9550-04-1-0430 (DOD), National Institute of Environmental Health Sciences Training Grant ES07026, and Center Grant ES01247 and U of R start-up funds (AEF).
Footnotes
Supporting Information Available: This information is available free of charge via the Internet at http://pubs.acs.org/.
Literature Cited
- 1.Klumpp S, Krieglstein J. Reversible phosphorylation of histidine residues in proteins from vertebrates. Sci Signal. 2009;2(61):pe13. doi: 10.1126/scisignal.261pe13. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS, Bevilacqua G, Kopper L, Thorgeirsson UP, Talmadge JE, Liotta LA, Sobel ME. Evidence for a novel gene associated with low tumor metastatic potential. J Natl Cancer Inst. 1988;80(3):200–4. doi: 10.1093/jnci/80.3.200. [DOI] [PubMed] [Google Scholar]
- 3.Roymans D, Willems R, Van Blockstaele DR, Slegers H. Nucleoside diphosphate kinase (NDPK/NM23) and the waltz with multiple partners: possible consequences in tumor metastasis. Clin Exp Metastasis. 2002;19(6):465–76. doi: 10.1023/a:1020396722860. [DOI] [PubMed] [Google Scholar]
- 4.Gounaris K, Thomas S, Najarro P, Selkirk ME. Secreted variant of nucleoside diphosphate kinase from the intracellular parasitic nematode Trichinella spiralis. Infect Immun. 2001;69(6):3658–62. doi: 10.1128/IAI.69.6.3658-3662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee HY, Lee H. Inhibitory activity of nm23-H1 on invasion and colonization of human prostate carcinoma cells is not mediated by its NDP kinase activity. Cancer Lett. 1999;145(1–2):93–9. doi: 10.1016/s0304-3835(99)00236-0. [DOI] [PubMed] [Google Scholar]
- 6.Wagner PD, Vu ND. Histidine to aspartate phosphotransferase activity of nm23 proteins: phosphorylation of aldolase C on Asp-319. Biochem J. 2000;346(Pt 3):623–30. [PMC free article] [PubMed] [Google Scholar]
- 7.Hippe HJ, Wolf NM, Abu-Taha I, Mehringer R, Just S, Lutz S, Niroomand F, Postel EH, Katus HA, Rottbauer W, Wieland T. The interaction of nucleoside diphosphate kinase B with Gbetagamma dimers controls heterotrimeric G protein function. Proc Natl Acad Sci U S A. 2009;106(38):16269–74. doi: 10.1073/pnas.0901679106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wagner PD, Vu ND. Phosphorylation of ATP-citrate lyase by nucleoside diphosphate kinase. J Biol Chem. 1995;270(37):21758–64. doi: 10.1074/jbc.270.37.21758. [DOI] [PubMed] [Google Scholar]
- 9.Cuello F, Schulze RA, Heemeyer F, Meyer HE, Lutz S, Jakobs KH, Niroomand F, Wieland T. Activation of heterotrimeric G proteins by a high energy phosphate transfer via nucleoside diphosphate kinase (NDPK) B and Gbeta subunits. Complex formation of NDPK B with Gbeta gamma dimers and phosphorylation of His-266 IN Gbeta. J Biol Chem. 2003;278(9):7220–6. doi: 10.1074/jbc.M210304200. [DOI] [PubMed] [Google Scholar]
- 10.Srivastava S, Li Z, Ko K, Choudhury P, Albaqumi M, Johnson AK, Yan Y, Backer JM, Unutmaz D, Coetzee WA, Skolnik EY. Histidine phosphorylation of the potassium channel KCa3.1 by nucleoside diphosphate kinase B is required for activation of KCa3.1 and CD4 T cells. Mol Cell. 2006;24(5):665–75. doi: 10.1016/j.molcel.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 11.Besant PG, Attwood PV. Mammalian histidine kinases. Biochim Biophys Acta. 2005;1754(1–2):281–90. doi: 10.1016/j.bbapap.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 12.Smtih DL, Bruegger BB, Halpern RM, Smith RA. New histone kinases in nuclei of rat tissues. Nature. 1973;246(5428):103–4. doi: 10.1038/246103a0. [DOI] [PubMed] [Google Scholar]
- 13.Besant PG, Attwood PV. Detection of a mammalian histone H4 kinase that has yeast histidine kinase-like enzymic activity. Int J Biochem Cell Biol. 2000;32(2):243–53. doi: 10.1016/s1357-2725(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 14.Wiener MC, Sachs JR, Deyanova EG, Yates NA. Differential mass spectrometry: a label-free LC-MS method for finding significant differences in complex peptide and protein mixtures. Anal Chem. 2004;76(20):6085–96. doi: 10.1021/ac0493875. [DOI] [PubMed] [Google Scholar]
- 15.Hartsough MT, Morrison DK, Salerno M, Palmieri D, Ouatas T, Mair M, Patrick J, Steeg PS. Nm23-H1 metastasis suppressor phosphorylation of kinase suppressor of Ras via a histidine protein kinase pathway. J Biol Chem. 2002;277(35):32389–99. doi: 10.1074/jbc.M203115200. [DOI] [PubMed] [Google Scholar]
- 16.Kim SC, Chen Y, Mirza S, Xu Y, Lee J, Liu P, Zhao Y. A clean, more efficient method for in-solution digestion of protein mixtures without detergent or urea. J Proteome Res. 2006;5(12):3446–52. doi: 10.1021/pr0603396. [DOI] [PubMed] [Google Scholar]
- 17.Steeg PS, Horak CE, Miller KD. Clinical-translational approaches to the Nm23-H1 metastasis suppressor. Clin Cancer Res. 2008;14(16):5006–12. doi: 10.1158/1078-0432.CCR-08-0238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zu XL, Besant PG, Imhof A, Attwood PV. Mass spectrometric analysis of protein histidine phosphorylation. Amino Acids. 2007;32(3):347–57. doi: 10.1007/s00726-007-0493-4. [DOI] [PubMed] [Google Scholar]
- 19.Besant PG, Attwood PV. Detection and analysis of protein histidine phosphorylation. Mol Cell Biochem. 2009;329(1–2):93–106. doi: 10.1007/s11010-009-0117-2. [DOI] [PubMed] [Google Scholar]
- 20.Kauffman EC, Robinson VL, Stadler WM, Sokoloff MH, Rinker-Schaeffer CW. Metastasis suppression: the evolving role of metastasis suppressor genes for regulating cancer cell growth at the secondary site. J Urol. 2003;169(3):1122–33. doi: 10.1097/01.ju.0000051580.89109.4b. [DOI] [PubMed] [Google Scholar]
- 21.Khaled HM, Bahnassy AA, Raafat AA, Zekri AR, Madboul MS, Mokhtar NM. Clinical significance of altered nm23-H1, EGFR, RB and p53 expression in bilharzial bladder cancer. BMC Cancer. 2009;9:32. doi: 10.1186/1471-2407-9-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki E, Ota T, Tsukuda K, Okita A, Matsuoka K, Murakami M, Doihara H, Shimizu N. nm23-H1 reduces in vitro cell migration and the liver metastatic potential of colon cancer cells by regulating myosin light chain phosphorylation. Int J Cancer. 2004;108(2):207–11. doi: 10.1002/ijc.11546. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, Lv F, Zhou L, Li X, Wu XX, Hoffman RM. Effect of verapamil on the expression of EGFR and NM23 in A549 human lung cancer cells. Anticancer Res. 2009;29(1):27–32. [PubMed] [Google Scholar]
- 24.Kapitanovic S, Cacev T, Berkovic M, Popovic-Hadzija M, Radosevic S, Seiwerth S, Spaventi S, Pavelic K, Spaventi R. nm23-H1 expression and loss of heterozygosity in colon adenocarcinoma. J Clin Pathol. 2004;57(12):1312–8. doi: 10.1136/jcp.2004.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seifert M, Welter C, Mehraein Y, Seitz G. Expression of the nm23 homologues nm23-H4, nm23-H6, and nm23-H7 in human gastric and colon cancer. J Pathol. 2005;205(5):623–32. doi: 10.1002/path.1724. [DOI] [PubMed] [Google Scholar]
- 26.Ismail NI, Kaur G, Hashim H, Hassan MS. Nuclear localization and intensity of staining of nm23 protein is useful marker for breast cancer progression. Cancer Cell Int. 2008;8:6. doi: 10.1186/1475-2867-8-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall JC, Lee JH, Steeg PS. Clinical-translational strategies for the elevation of Nm23-H1 metastasis suppressor gene expression. Mol Cell Biochem. 2009;329(1–2):115–20. doi: 10.1007/s11010-009-0116-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murakami M, Meneses PI, Lan K, Robertson ES. The suppressor of metastasis Nm23-H1 interacts with the Cdc42 Rho family member and the pleckstrin homology domain of oncoprotein Dbl-1 to suppress cell migration. Cancer Biol Ther. 2008;7(5):677–88. doi: 10.4161/cbt.7.5.5665. [DOI] [PubMed] [Google Scholar]
- 29.McDermott WG, Boissan M, Lacombe ML, Steeg PS, Horak CE. Nm23-H1 homologs suppress tumor cell motility and anchorage independent growth. Clin Exp Metastasis. 2008;25(2):131–8. doi: 10.1007/s10585-007-9128-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.