Abstract
A highly efficient lambda phage recombination system previously utilized for studies of bacterial artificial chromosome (BAC)-maintained mouse chromosomal DNA was adapted for the study of the role of human cytomegalovirus (HCMV)-encoded pp28 (UL99) in virus replication. Incorporating a two-step mutagenesis strategy with blue/white selection in Escherichia coli containing a HCMV AD169 BAC, we have shown that we can rapidly introduce point mutations into the HCMV BAC using linear PCR fragments. All manipulations were carried out in bacteria, which greatly accelerated the introduction and analysis of mutations in the viral genome. Our results indicated that HCMV pp28 was essential for the production of infectious virus and that introduction of a single base change that resulted in loss of the myristylation site on pp28 was also associated with the lack of production of infectious virus. Although the block in the viral morphogenesis cannot be determined from these studies, the latter finding suggested that authentic intracellular localization of pp28, not only the expression of the protein, is required for virus assembly.
The role of individual herpesvirus genes in virus replication and the pathogenesis of virus-induced disease have been best understood from studies using viral genetics. The propagation of infectious clones of herpesviruses as bacterial artificial chromosomes (BACs) has revolutionized the experimental manipulation of viral genomes. Importantly, this technology has allowed application of tools of prokaryotic molecular biology to the study of viral genetics, thereby facilitating the genetic analysis of many herpesviruses (4-6, 16, 23, 24, 27). Previously, mutagenic approaches based on homologous recombination in eukaryotic cells were limited to genetic manipulation of only the most rapidly replicating and promiscuous herpesviruses, such as herpes simplex virus (HSV) and pseudorabies virus (PRV) (19). In this study, we have adapted a recombination strategy originally developed for mutagenesis of BACs containing mouse chromosomes to mutagenize the human cytomegalovirus (HCMV) genome (8, 10, 25, 26). This methodology takes advantage of a lambda phage recombination system (RED locus) expressed from a temperature-sensitive promoter that enables the use of linear single-stranded DNA to target genes of interest carried on BACs maintained in Escherichia coli (8, 26). We have used this recombination system together with a blue/white lacZ-based selection approach to define the importance of HCMV tegument protein pp28 (UL99) for HCMV replication. The product of the HCMV UL99 open reading frame (ORF) is a 190-amino-acid (aa) phosphorylated tegument protein, which is modified by myristylation (17, 18, 21). Homologues of pp28 are found in all herpesviruses. Studies with the HSV homologue UL11 have indicated that this protein is also myristylated, traffics in the cytoplasm, and is essential for wild-type levels of virus replication in vitro (2, 3, 11-13). Furthermore, more recent studies of the PRV UL11 have demonstrated that it also was required for virus replication and that viruses with deletions in UL11 were defective in secondary cytoplasmic envelopment of tegumented capsids (9). The HCMV pp28 is a true late protein that localizes in a cytoplasmic assembly compartment, suggesting that the protein may be involved in late steps of viral morphogenesis such as final tegumentation or envelopment (20). To date, it is unknown whether pp28 is essential for the production of infectious HCMV.
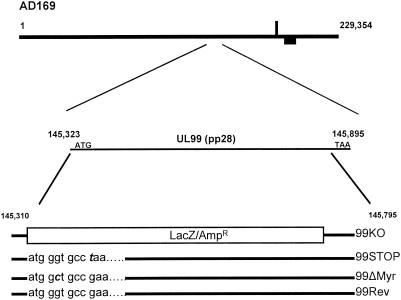
Our strategy for site-specific mutagenesis of the UL99 ORF involved a two-step procedure. In the first step, the UL99 ORF was deleted from a BAC-maintained HCMV genome by replacement with an Ampr lacZ cassette. Growth in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside)-X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) enables identification of HCMV recombinants by visual inspection for blue bacterial colonies. The mutagenesis was performed with RED locus-mediated recombination to delete sequences between positions 145310 and 145795 of the AD169 genome in the HB5 BAC (kindly provided by Martin Messerle and Ulrich Kozsinowski, University of Munich) (4). A primer set amplifying sequences from positions 145210 (forward primer) and 146025 (reverse primer) was used to produce a recombination cassette of approximately 2.0 kbp that carried the Ampr lacZ cassette together with approximately 100 to 200 bp of AD169 sequence flanking the site of deletion within the HCMV genome (Fig. 1). The cassette was recombined into the AD169 BAC by RED locus-mediated recombination using the protocol described by Lee et al., resulting in production of greater than 200 Ampr blue colonies (10). Insertion of the cassette resulted in the deletion of the entire UL99 ORF except for 100 bp of the 3′ end (recombinant designated 99KO BAC; Fig. 1). To ensure correct targeting of the recombination cassette into the desired genomic location, recombinant BAC DNA was digested with HindIII, electrophoresed, and transferred to nitrocellulose membranes, followed by hybridization using a 32P-labeled probe directed against the Ampr lacZ cassette. Analysis of three independent clones revealed a single 8.0-kbp fragment that hybridized to the probe. This fragment corresponds to the 6-kbp UL99 ORF containing a HindIII R fragment that was increased in size by 2.0 kbp following insertion of the Ampr lacZ cassette into this region (Fig. 2A). Nucleotide sequence analysis of PCR products amplified from these recombinant BACs confirmed correct insertion of the Ampr lacZ cassette and replacement of the UL99 ORF (data not shown).
FIG. 1.
Construction of UL99 recombinant BACs. The genome of HCMV AD169 is shown at the top with the UL99 ORF and its coordinates listed below. This AD169 BAC was transferred to E. coli strain DY380 for these experiments (25). The position of insertion of the Ampr lacZ cassette in the UL99 deletion BAC (99KO) is shown. The UL99 deletion BAC, 99KO, was then repaired with 800-bp linear PCR products encoding either wild-type sequences (99Rev) or mutant sequences (99STOP and 99ΔMyr) by previously described methods (10, 25, 26). The sequences containing the point mutations in BACs 99STOP and 99ΔMyr along with the corresponding sequence of the wild-type repaired BAC, 99Rev, are shown at the bottom, with changes in boldface italics.
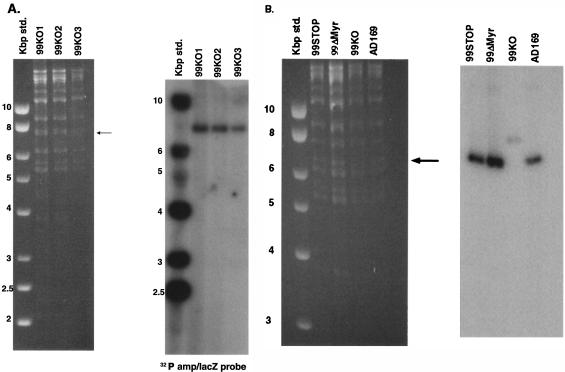
FIG. 2.
Analysis of 99KO, 99STOP, and 99ΔMyr BAC by Southern hybridization. (A) DNA prepared from three independent blue, ampicillin-resistant colonies of BAC-containing E. coli that had been recombined with the linear Ampr lacZ cassette were digested with HindIII, subjected to electrophoresis in agarose gel, and transferred to nitrocellulose membranes. Hybridization was carried out with a 32P-labeled probe generated by nick translation of DNA containing the Ampr lacZ cassette. Arrow, migration of the approximately 8-kbp HindIII fragment containing the Ampr lacZ cassette. Note that a single fragment hybridized with the probe, indicating a single site of insertion. (B) DNA prepared from E. coli containing the wild-type AD169 BAC (AD169), 99KO BAC, mutant 99STOP BAC, and mutant 99ΔMyr BACs was digested with HindIII and analyzed as described for panel A by Southern hybridization with a 32P-labeled probe generated by nick translation of DNA containing the intact UL99 ORF. Arrow, migration of the HindIII R fragment at approximately 6 kbp. The weak signal in the lane containing the 99KO DNA is secondary to the approximate 100 bp of the 3′ end of the UL99 ORF that remains following this insertion and deletion.
In the second step, RED recombination was used to replace the UL99 deletion in 99KO BAC with either the wild-type UL99 ORF sequence or the UL99 sequence containing desired mutations by using a linear DNA fragment. Recombination removes the Ampr lacZ cassette from the 99KO BAC, and repaired or mutagenized recombinants can be simply and rapidly identified by visual inspection for white colonies. The same primer set described above was used to prepare linear DNA fragments containing the desired mutations by PCR from a UL99 template as previously described (10, 25). The first mutation consisted of a single nucleotide substitution at position 145333 that altered a glutamic acid codon (GAA) to a stop codon (TAA; recombinant designated 99STOP BAC; Fig. 1). The second mutation was positioned at 145328 and changed a glycine codon (GGT) to an alanine codon (GCT; recombinant designated 99ΔMyr BAC; Fig. 1). This second mutation was introduced to delete the myristylation site that we have previously shown to be critical for intracellular localization of the UL99-encoded pp28 protein (21). A revertant containing the wild-type UL99 ORF sequence (designated 99Rev BAC) was also prepared to ensure that any observed phenotype was a result of the UL99 ORF mutation. Gel-purified PCR products were recombined into the 99KO BAC, and E. coli cells carrying BACs with the desired UL99 ORF mutations were readily recovered after overnight incubation at 30°C on X-Gal plates by selection of white colonies. In multiple experiments the frequency of white colonies varied from 0.8 to 0.1%.
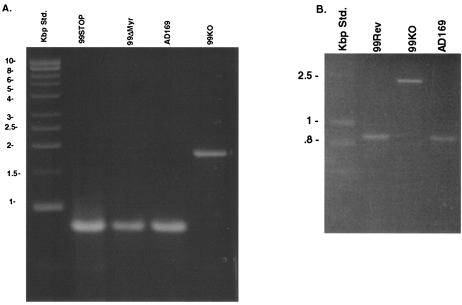
Analysis of the mutagenized BACs demonstrated that the UL99 deletion BAC (99KO BAC) had been repaired. DNA prepared from the mutagenized BACs was digested with HindIII and analyzed by agarose gel electrophoresis, followed by Southern transfer to nitrocellulose membranes and hybridization. The membrane was probed with a 32P-labeled probe generated by nick translation of the entire UL99 ORF. Both the 99STOP and 99ΔMyr mutagenized BACs had the same restriction fragment pattern as that observed for the parental wild type, AD169, including the 6-kbp HindIII fragment, which was absent from the 99KO BAC (Fig. 2B). The UL99 32P-labeled probe hybridized with the 6-kbp fragment in both mutant BACs and the wild-type AD169 BAC (Fig. 2B). A weak UL99-specific signal was also observed for the 8.0-kbp fragment in the 99KO BAC, presumably derived from hybridization to the 100 bp of the UL99 ORF that remains within this BAC (Fig. 2B). The same primer set was also used to amplify DNA from the BACs and readily amplified the expected 800-bp product from the parental AD169 BAC as well as those from the two mutagenized BACs, 99STOP and 99ΔMyr BAC (Fig. 3A). A 2.0-kb fragment was amplified from the 99KO BAC as predicted due to the presence of the Ampr lacZ cassette (Fig. 3A). Sequence analysis of PCR products from 99STOP and 99ΔMyr BAC confirmed the presence of the expected mutation and complete fidelity of the UL99 sequence following comparison with the published sequence of AD169 (with the exception of the introduced point mutations) (Fig. 1). Similarly, PCR amplification of the 99Rev BAC produced the expected 800-bp fragment (Fig. 3B), and sequence analysis revealed that this PCR product was identical to the parental AD169 nucleotide sequence (7).
FIG. 3.
PCR analysis of 99KO, 99STOP, 99ΔMyr, and the wild-type repaired (99Rev) BACs. (A) DNA from 99KO, 99STOP, or 99ΔMyr BACs was amplified by primer pairs used to generate the recombination cassette, and the resulting products were analyzed by agarose gel electrophoresis. The expected product from the wild-type parent BAC (AD169) was approximately 800 bp. The amplimers from the 99STOP and 99ΔMyr mutant BACs migrated similarly, whereas the amplimer of the 99KO BAC was approximately 1.2 kbp larger. (B) DNA from the wild-type parent BAC (AD169), the 99KO BAC, and the wild-type repaired BAC, 99Rev, was amplified as described for panel A, and the amplimers were analyzed by agarose gel electrophoresis. The amplimers of the wild-type parent BAC and the wild-type repaired BAC migrated at approximately 800 bp.
To determine whether the UL99 gene was essential for virus infectivity in normal human dermal fibroblasts (NHDF), DNA was purified from the BAC-containing E. coli cells and electroporated into NHDF. Infectivity was monitored by observing the production of visible plaques and by assaying cells from NHDF monolayers for expression of HCMV-encoded proteins with HCMV monoclonal antibodies (MAbs) reactive with IE-1, pp28, and gB. Infectious virus was consistently recovered from both the wild-type parental AD169 BAC DNA and from the 99Rev wild-type revertant BAC DNA (Table 1). Similarly, MAb reactivity for IE-1, pp28, and gB was observed in cells that had been electroporated with the parental AD169 BAC and 99Rev BAC (Table 1). In contrast, in multiple experiments (at least four independent experiments) infectious virus was never recovered from NHDF electroporated with DNA from BACs containing either the 99STOP or the 99ΔMyr mutation, suggesting that the pp28 gene is essential for the production of infectious HCMV (Table 1). Importantly, the inability of 99ΔMyr to produce infectious virus suggests that that correct localization of pp28 due to the presence of the myristic acid modification may be critical for virus infectivity.
TABLE 1.
Recovery of infectious virus from HCMV BAC DNAa
| BAC | Recovery of infectious virusb | Expression of HCMV-encoded proteinsc |
|---|---|---|
| AD169 | 5/5 | 5/5 |
| 99KO | 0/4 | 0/3 |
| 99Rev | 2/2 | 2/2 |
| 99STOP | 0/4 | 0/2 |
| 99ΔMyr | 0/4 | 0/2 |
Approximately 3 to 6 μg of BAC DNA plus 1 μg of a plasmid DNA encoding pp71 were electroporated into human fibroblasts under conditions previously described (14). The efficiency of electroporation was monitored by including 0.5 μg of an expression plasmid encoding enhanced green flurescent protein in each electroporation. Typical HCMV plaques were observed between 12 and 16 days following electroporation, and cells were cultured for approximately 3 weeks prior to assaying for infectivity.
Infectious virus was determined by the appearance of plaques in monolayers of human fibroblasts and the capacity of a freeze-thaw lysate of cells to transfer infectious virus to an uninfected monolayer of NHDF cells. Values are the numbers of times that virus was recovered/the numbers of experiments.
Expression of virus-encoded proteins in electroporated cells was determined by reactivity of fixed cells harvested 3 weeks following electroporation with MAbs reactive with IE-1, gB, and pp28 as determined in a immunofluorescence assay (20). Values are numbers of times reactivity with all MAbs was observed/the total number of experiments.
The recombination strategy we have described in this report provides a simple and rapid method to introduce site-specific mutations into the HCMV genome. Once the viral sequence of interest has been replaced by the Ampr lacZ cassette, repair of this BAC with either wild-type sequences or sequences containing specific mutation can be accomplished within 2 days. Importantly, as opposed to other approaches, our approach enables insertion of only the specific mutations within the herpesvirus genome without leaving residual foreign sequences (e.g., frt sites). In our experience, this recombination procedure has consistently allowed recombination into a predicted site and unexpected recombination or deletion in the parental BAC has been observed only infrequently. We have used this approach to introduce stop codons at various positions in the UL99 sequence as well as to introduce mutations into the UL55, UL100, UL73, and UL32 ORFs (data not shown). In each case, the targeting of the Ampr lacZ cassette and the subsequent repair of the deletion mutation has been confirmed by both Southern hybridization and sequence analysis. Thus, we believe that this adaptation of the powerful technology originally developed by Court et.al. offers a rapid and reproducible method to introduce specific mutations into the HCMV genome (10, 25, 26).
Our finding that the tegument protein, pp28, was an essential protein for in vitro replication of HCMV was not unexpected. Previous studies using HSV have demonstrated that the UL99 homologue, UL11, was not essential for virus replication, but UL11 deletion mutants produced several orders of magnitude less virus than wild-type HSV or PRV (3, 9). The 99STOP mutation resulted in a translational stop after the second codon of the pp28 gene. This mutation did not alter the coding sequence of the overlapping UL98 ORF, a gene encoding the alkaline nuclease that is likely required for HCMV replication (1). The mutation present in 99ΔMyr BAC changed aa 567 of ppUL98 from a valine to a leucine. This is a conservative change, and, more importantly, this residue lies outside of any conserved domains or motifs that have been assigned to herpesvirus alkaline nucleases based on amino acid alignment (15, 22). In addition, alignment of the reported amino acid sequence of alkaline nucleases from other alpha-, beta-, and gammaherpesviruses revealed that the region surrounding HCMV UL98 aa 567 was highly variable and in only one instance was the valine conserved at this position. Consequently, we believe that this mutation introduced into the ppUL98 amino acid sequence does not account for the null phenotype of the 99ΔMyr mutant. The more probable explanation is that the 99ΔMyr mutant encodes a pp28 that lacks a myristic acid modification and is mislocalized compared to the myristylated protein of the wild-type virus. This suggests that authentic intracellular localization of pp28 is required for HCMV morphogenesis.
We have not discerned the step in virus replication that is blocked by mutations in pp28. However, the block is presumably at a late step in virion morphogenesis, since pp28 is a true late protein that is produced following expression of virion glycoproteins and other structural proteins such as capsid proteins (20). Furthermore, the assembly compartment in virus-infected cells has been shown to contain HCMV structural proteins prior to the detection of pp28 or localization of this protein within the assembly compartment (20). Consistent with this interpretation were findings that early-late and late HCMV proteins (pp65/UL38, pp150/UL32, MCP/UL86, pp52/UL44, and gB/UL55) could be detected in single cells from cultures electroporated 7 to 10 days earlier with DNA from either the 99ΔMyr or 99STOP mutant BAC (data not shown). These results suggested that the block in virus replication was late in the replicative cycle of the virus, possibly in the assembly of the infectious particle. Because pp28 cannot be detected in the nucleus at any time during its expression in infected cells, our findings also provide strong support that envelopment of infectious virions takes place in the cytoplasm, likely in proximity to the assembly compartment, and that pp28 plays an integral role in the latter stages of this process. Our recent observations have demonstrated that pp28 interacts with HCMV glycoproteins including gB (unpublished data) and raise the possibility that pp28 could serve either a targeting or adaptor function in the final steps of virion assembly. Importantly, with the availability of a rapid HCMV mutagenesis system, the functions of specific domains of the pp28 molecule that have been defined in transient expression assays should now be possible within the context of replicating virus.
Acknowledgments
We thank Don Court (National Cancer Institute, Frederick, Md.), Martin Messerle and Ulrich Kozsinowski (University of Munich), and Pat Higgins (University of Alabama) for kindly providing critical reagents and advice for this study.
This work was supported through funding from the PHS to W.J.B. (AI35602 and AI50189) and to M.J. and J.N. (AI21640 and AI10418).
REFERENCES
- 1.Anders, D. G., and L. A. McCue. 1996. The human cytomegalovirus genes and proteins required for DNA synthesis. Intervirology 39:378-388. [DOI] [PubMed] [Google Scholar]
- 2.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borst, E. M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britt, W. J. 2000. Infectious clones of herpesviruses: a new approach for understanding viral gene function. Trends Microbiol. 8:262-265. [DOI] [PubMed] [Google Scholar]
- 6.Brune, W., C. Menard, U. Hobom, S. Odenbreit, M. Messerle, and U. H. Koszinowski. 1999. Rapid identification of essential and nonessential herpesvirus genes by direct transposon mutagenesis. Nat. Biotechnol. 17:60-64. [DOI] [PubMed] [Google Scholar]
- 7.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-170. [DOI] [PubMed] [Google Scholar]
- 8.Copeland, N. G., N. A. Jenkins, and D. L. Court. 2001. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2:769-779. [DOI] [PubMed] [Google Scholar]
- 9.Kopp, M., H. Granzow, W. Fuchs, B. G. Klupp, E. Mundt, A. Karger, and T. C. Mettenleiter. 2003. The pseudorabies virus UL11 protein is a virion component involved in secondary envelopment in the cytoplasm. J. Virol. 77:5339-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, E. C., D. Yu, J. Martinez de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56-65. [DOI] [PubMed] [Google Scholar]
- 11.Loomis, J. S., J. B. Bowzard, R. J. Courtney, and J. W. Wills. 2001. Intracellular trafficking of the UL11 tegument protein of herpes simplex virus type 1. J. Virol. 75:12209-12219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.MacLean, C. A., B. Clark, and D. J. McGeoch. 1989. Gene UL11 of herpes simplex virus type 1 encodes a virion protein which is myristylated. J. Gen. Virol. 70:3147-3157. [DOI] [PubMed] [Google Scholar]
- 13.MacLean, C. A., A. Dolan, F. E. Jamieson, and D. J. McGeoch. 1992. The myristylated virion proteins of herpes simplex virus type 1: investigation of their role in the virus life cycle. J. Gen. Virol. 73:539-547. [DOI] [PubMed] [Google Scholar]
- 14.Marchini, A., H. Liu, and H. Zhu. 2001. Human cytomegalovirus with IE-2 (UL122) deleted fails to express early lytic genes. J. Virol. 75:1870-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez, R., L. Shao, J. C. Bronstein, P. C. Weber, and S. K. Weller. 1996. The product of a 1.9-kb mRNA which overlaps the HSV-1 alkaline nuclease gene (UL12) cannot relieve the growth defects of a null mutant. Virology 215:152-164. [DOI] [PubMed] [Google Scholar]
- 16.Messerle, M., I. Crnkovic, W. Hammerschmidt, H. Ziegler, and U. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer, H., A. T. Bankier, M. P. Landini, C. M. Brown, B. G. Barrell, B. Ruger, and M. Mach. 1988. Identification and procaryotic expression of the gene coding for the highly immunogenic 28-kilodalton structural phosphoprotein (pp28) of human cytomegalovirus. J. Virol. 62:2243-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pande, H., K. Campo, B. Tanamachi, and J. A. Zaia. 1991. Human cytomegalovirus strain Towne pp28 gene: sequence comparison to pp28 of HCMV AD169 and stable expression in Chinese hamster ovary cells. Virology 184:762-767. [DOI] [PubMed] [Google Scholar]
- 19.Post, L. E., and B. Roizman. 1981. A generalized technique for deletion of specific genes in large genomes: α gene 22 of herpes simplex virus 1 is not essential for growth. Cell 25:227-232. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez, V., K. D. Greis, E. Sztul, and W. J. Britt. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanchez, V., E. Sztul, and W. J. Britt. 2000. Human cytomegalovirus pp28 (UL99) localizes to a cytoplasmic compartment which overlaps the endoplasmic reticulum-Golgi-intermediate compartment. J. Virol. 74:3842-3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheaffer, A. K., S. P. Weinheimer, and D. J. Tenney. 1997. The human cytomegalovirus UL98 gene encodes the conserved herpesvirus alkaline nuclease. J. Gen. Virol. 78:2953-2961. [DOI] [PubMed] [Google Scholar]
- 23.Smith, G. A., and L. W. Enquist. 1999. Construction and transposon mutagenesis in Escherichia coli of a full-length infectious clone of pseudorabies virus, an alphaherpesvirus. J. Virol. 73:6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith, G. A., and L. W. Enquist. 2000. A self-recombining bacterial artificial chromosome and its application for analysis of herpesvirus pathogenesis. Proc. Natl. Acad. Sci. USA 97:4873-4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Swaminathan, S., H. M. Ellis, L. S. Waters, D. Yu, E. C. Lee, D. L. Court, and S. K. Sharan. 2001. Rapid engineering of bacterial artificial chromosomes using oligonucleotides. Genes. J. Genet. Dev. 29:14-21. [DOI] [PubMed] [Google Scholar]
- 26.Yu, D., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu, D., G. A. Smith, L. W. Enquist, and T. Shenk. 2002. Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76:2316-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]