Abstract
The mechanism for efficient nucleocapsid (NC) uptake into flavivirus particles which form by budding through the membranes of the endoplasmic reticulum (ER) was investigated by using Murray Valley encephalitis virus as a model. Budding of flavivirus membranes is driven by the viral transmembrane proteins prM and E independently of NC interaction. We show that control of signalase cleavage of the multimembrane-spanning flavivirus polyprotein by the catalytic function of the viral protease is critical for efficient virus morphogenesis. In wild-type virus, signalase cleavage of prM remains inefficient until cleavage of capsid at the cytosolic side of the signal sequence separating the two proteins has occurred. This obligatory sequence of cleavages was uncoupled in a mutant virus with the consequence of greatly reduced incorporation of NC into budding membranes and augmented release of NC-free virus-like particles. Efficient signalase cleavage of prM in the mutant virus resulted in partial inhibition of cleavage of capsid by the viral NS2B-3 protease. Our results support a model for flavivirus morphogenesis involving temporal and spatial coordination of NC assembly and envelopment by regulated cleavages of an ER membrane-spanning capsid-prM intermediate.
Many animal viruses acquire a lipid membrane by budding through cellular membranes. The membranes are modified with viral glycoproteins and envelop the nucleic acid-containing core particle or nucleocapsid (NC). Assembly of viral structural proteins on the cytosolic side, or within host membranes, into lattice structures is thought to provide the force needed for budding; this involves the induction of local membrane curvature required for the pinching off of the budding viral particle. Protein interactions that drive budding can involve (i) interaction of preassembled NC with membrane-associated viral glycoproteins, (ii) assembly of viral core proteins into core particles at the cytosolic side of membranes without involvement of viral transmembrane glycoproteins, or (iii) lateral interaction of viral transmembrane glycoproteins without involvement of core protein interactions (reviewed in references 8 and 10). A key question in the latter, NC-independent budding strategy is by what mechanism efficient uptake of NC into budding viral membranes is achieved. Here we report on such a mechanism for NC incorporation in flavivirus assembly.
The flaviviruses are medically important enveloped RNA viruses (reviewed in reference 4). Their plus-strand RNA genome (∼11 kb) encodes a single polyprotein which is cleaved into three structural proteins (capsid [C], precursor to membrane [prM], and envelope [E]) and at least seven nonstructural proteins. The polyprotein traverses the membranes of the endoplasmic reticulum (ER) multiple times and is proteolytically processed on the luminal side of the membrane by the host enzyme signalase and on the cytoplasmic side by a virally encoded (NS2B-3) protease. The ER is the putative assembly site for flavivirus particles. Virus particles are thought to form by budding into the lumen of the ER, followed by transport of virions out of the cell via the exocytic pathway (25). Given that recombinant expression of the flavivirus transmembrane proteins prM and E efficiently generates secreted virus-like particles (VLPs; these do not contain an NC [16, 36]) and that VLPs are also a significant by-product of virus replication in cell culture (37, 41), it is likely that flavivirus budding is NC independent. Furthermore, the majority of ultrastructural studies have failed to reveal assembly intermediates such as free or budding NC (reviewed in reference 22), indicating that flavivirus assembly and budding are rapid and tightly coordinated events. One exception to this may be the assembly of the Sarafend strain of West Nile virus, which is proposed to involve budding of preassembled NC at the plasma membrane (31, 32).
It has been proposed that the coordination of two cleavages at the junction of the flavivirus structural proteins C and prM is key in regulation of the assembly of the flaviviruses yellow fever virus (YFV) and Murray Valley encephalitis virus (MVE) (2, 21, 40). The C protein at the NH2-terminal end of the flavivirus polyprotein is separated from the prM protein by an internal signal sequence which directs translocation of prM, a type I transmembrane protein. The viral NS2B-3 protease catalyzes the cytoplasmic cleavage at the COOH terminus of C; interestingly, in the absence of C protein cleavage, ER luminal signalase cleavage of prM is inefficient (1, 23, 39, 44). This is the first example of regulation of cleavage by a signalase, a protease which generally cleaves cotranslationally (reviewed in references 11 and 27), by the catalytic function of a cytosolic protease. Conservation of the coordinated cleavages at the C-prM junction among members of the flavivirus genus suggested a crucial role in virus replication. This was confirmed by introducing a mutation into the cleavage region of the YFV and MVE prM signal sequences (PQAQA mutation) which uncoupled signalase cleavage of prM from its dependence on prior cleavage of C by the NS2B-3 protease (21, 40). The mutation did not markedly reduce viral macromolecular synthesis but prevented release of progeny virus, most likely due to a defect in virus assembly, when it was introduced into the genomic RNA of YFV and the virus was transfected into mammalian cells (21). Given the lethal phenotype of the prM signal sequence mutation, it was not possible to identify the event in flavivirus morphogenesis for which uncoupling of the coordinated cleavages at the C-prM junction was detrimental. Here we evaluate the effect of efficient signalase cleavage of prM on the morphogenesis of MVE. We show that the mutant virus is viable although restricted in growth in cell culture and demonstrate that the growth defect is due to a deficiency in NC incorporation into budding viral membranes.
MATERIALS AND METHODS
Cells and virus.
Vero (African green monkey kidney), BHK (baby hamster kidney), COS-7 (African green monkey kidney), and C6/36 (Aedes albopictus) cells were obtained from the American Type Culture Collection and grown in Eagle's minimal essential medium (MEM) supplemented with 5% fetal calf serum (FCS) or 8% FCS for C6/36 cells. The mammalian cells were grown at 37°C, and C6/36 cells were grown at 28°C.
The MVE-1-51 prototype strain was used; virus stocks were 10% infected-mouse-brain homogenates in Hanks' balanced salt solution containing 20 mM HEPES buffer (pH 8.0) and 0.2% bovine serum albumin or infected-C6/36-cell culture supernatants buffered at pH 8.0 and were stored in single-use aliquots at −70°C.
Anti-MVE and anti-C protein antisera.
Antisera against MVE and C protein were generated by using 107 PFU of MVE and 0.25 mg of a synthetic peptide with the sequence H-SKKPGGPGKPRVVNMLC-NH2 (corresponding to the NH2-terminal 16 amino acids of the MVE C protein) conjugated to diphtheria toxoid (Chiron, Clayton, Australia), respectively, for intradermal injection into rabbits in the presence of Freund's complete adjuvant. Three booster immunizations were given at 10-day intervals with the same amounts of antigen but in the presence of Freund's incomplete adjuvant. Sera were collected 7 days after the last booster immunization and frozen in aliquots at −20°C. One microliter of the sera was used for immunoprecipitations.
Plasmids and transfections.
Plasmid pM112.PQAQA is a full-length cDNA clone of MVE with the PQAQA mutation in the prM signal sequence. For its construction, a shuttle plasmid was generated by ligating an ApaI/XbaI fragment (1.7 kb) from the parent MVE cDNA clone, pM212 (20), into pcDNA3 (Invitrogen) digested with the same restriction enzymes. The region in this shuttle plasmid encoding the C-prM junction was replaced with a SpeI/HindIII fragment (0.3 kb) from plasmid pSTR.mutPQAQA (40). The PQAQA shuttle vector was digested with ApaI and XbaI to produce a 1.7-kb fragment containing the mutated prM signal sequence for ligation into ApaI/XbaI-digested pM112. Escherichia coli strains MC1061.1 and DH5α were used for transformations with pM212 and pcDNA3 derivatives, respectively. Transcription of RNA from full-length MVE cDNA clones and transfection of BHK cells with RNA by electroporation were as described previously (20).
To generate eukaryotic expression plasmids encoding the PQAQV and LQAQA reversions in the context of the MVE structural polyprotein, reverse transcription-PCR was performed on total RNA extracted from Vero cells infected with the PQAQV or LQAQA revertant (designated MVE.PQAQV or MVE.LQAQA, respectively) by using oligonucleotides flanking the C and E genes: 5′-GACCATTGATTAACGCGGTTT-3′ (sense) and 5′-CACACTTGAGCTCCCTCCT-3′ (antisense). The cDNA fragments (2 kb) were phosphorylated by treatment with T4 polynucleotide kinase and ligated into T-tailed pBluescript KS(+) (Stratagene) at the EcoRV restriction site. A shuttle plasmid comprising pBluescript digested with SalI and XbaI and a SalI/XbaI fragment (1.5 kb) from pcDNA-STR (23) was generated. The junction region between the C and prM genes from the LQAQA and PQAQV recombinant pBluescript plasmids was swapped as a 0.3-kb SpeI/HindIII fragment into the pBluescript-STR shuttle vector, which had been digested with SpeI and HindIII, dephosphorylated, and gel isolated as a 5-kb fragment. Recombinant shuttle vectors containing either the LQAQA or PQAQV sequence at the C-prM junction were digested with EcoRI, XmnI, and XbaI, and a 1.4-kb EcoRI/XmnI fragment was isolated for ligation into pcDNA-STR digested with EcoRI and XmnI. The ligation mixture was used to transformMC1061/P3 cells.
COS-7 cells were transfected with pcDNA.STR (23), p.STRmutPQAQA (40), p.STRrevPQAQV, or p.STRrevLQAQA by using 4 μg of plasmid DNA as previously described (24). Infectivity assays, virus growth assays, and infectivity titrations by Vero cell plaque assay or C6/36 cell focus-forming assay were as described previously (20).
Estimation of rates of RNA synthesis.
RNA synthesis kinetics were determined with modifications as described previously (30). BHK cell monolayers in 35-mm-diameter tissue culture dishes were infected at a multiplicity of 10 PFU/cell or left untreated. Prior to labeling, the monolayers were washed with phosphate-buffered saline (PBS) and incubated for 1 h with MEM containing 1% FCS and 5 μg of actinomycin D (Sigma)/ml to block cellular RNA synthesis. Labeling was for 3 h with 0.5 ml of MEM containing 1% FCS, 5 μg of actinomycin D/ml, and 5 μCi of [5,6-3H]uridine (Amersham) at the time intervals shown in Fig. 3. Subsequently, the monolayers were washed twice with PBS and dissociated by the addition of 0.2 ml of 1% sodium dodecyl sulfate (SDS) (wt/vol). Samples (25 μl) were dried, in duplicate, onto GF/A glass fiber disks (Whatman), washed with 5% trichloroacetic acid (three changes over 1 h) and then with ethanol (three changes over 10 min), and air dried. Radioactivity bound to the fiber disks was determined by liquid scintillation counting using 8 ml of Ready Flow III liquid scintillation cocktail (Beckman).
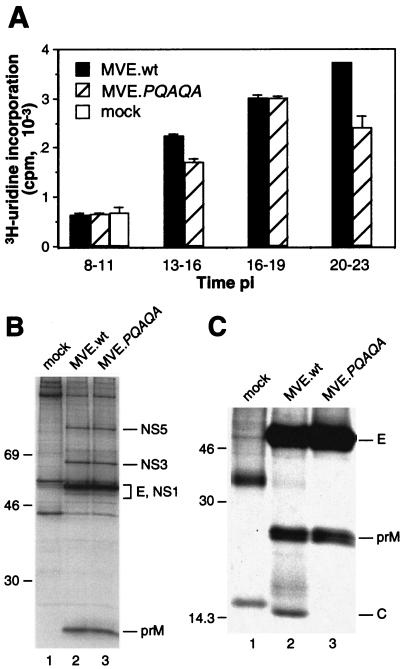
FIG. 3.
Virus-specific macromolecular synthesis in and protein secretion from cells infected with wt or mutant MVE. (A) Actinomycin D-resistant RNA synthesis in BHK cells infected with wt MVE (MVE.wt) or MVE.PQAQA or left uninfected (mock). Means ± standard errors of the means of results for two samples are presented. (B) Lysates of wt MVE-, MVE.PQAQA-, and mock-infected metabolically labeled BHK cells were subjected to immunoprecipitation with a polyclonal rabbit anti-MVE antiserum, and proteins were analyzed by SDS-PAGE (12% acrylamide). Sizes (in kilodaltons) of marker proteins are given on the left, and virus-specific proteins are labeled on the right. (C) Material secreted in the culture supernatants from mock-, wt MVE-, and MVE.PQAQA-infected BHK cells was precipitated with an E protein-specific monoclonal antibody (8E7) and analyzed by SDS-PAGE (15% acrylamide).
Sucrose gradient analyses.
For analysis of virions and VLPs secreted from infected cells, Vero cell monolayers (∼1 × 106 cells) in 60-mm-diameter plastic tissue culture dishes were infected with wild-type (wt) MVE, mutant MVE, or MVE revertants at multiplicities of 10 to 20 PFU/cell. At the times indicated in the legends to Fig. 4 and 5, the cells were starved for 0.5 h by replacement of the growth medium with 2 ml of methionine- and cysteine-free MEM followed by metabolic labeling for 6.5 h using 2 ml of methionine- and cysteine-free MEM containing 0.5% FCS, 2% MEM, 5 μg of actinomycin D/ml, and 100 μCi of Tran35S-label (ICN)/ml. At the end of the labeling period, the culture supernatants were clarified by centrifugation at 12,000 rpm for 15 min at 4°C in an Eppendorf microfuge and stored on ice overnight. Samples (0.5 to 1.0 ml) were loaded onto 10 to 30% continuous sucrose gradients (10.5 ml) in NTE-BSA buffer (120 mM NaCl, 12 mM Tris-HCl [pH 8.0], 1 mM Na2EDTA, 0.1% bovine serum albumin) and centrifuged at 35,000 rpm for 3 h at 4°C in an SW41 rotor (Beckman). Fractions of ∼0.4 ml each were collected from the bottoms of the gradients, 0.05 ml of each fraction was applied to a 96-well Luma plate (Packard), and radioactivity was measured in a TopCount NXT microplate scintillation counter (Packard).
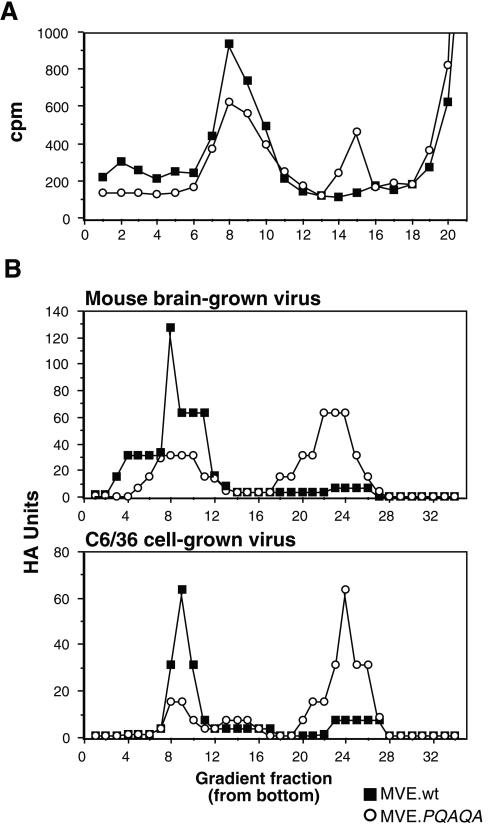
FIG. 4.
Sucrose density gradient analysis of particles secreted from MVE wt- and mutant-infected cells. (A) BHK cells were infected with wt or PQAQA mutant MVE and metabolically labeled from 40 to 47 h p.i. Infected culture supernatants were subjected, in parallel, to centrifugation on 10 to 30% sucrose gradients (10.5 ml), and radioactivity of fractions (∼0.4 ml) collected from the bottoms of the gradients was determined. (B) Comparable numbers of HA units from preparations of mouse brain- or C6/36 cell-grown wt MVE (MVE.wt) and MVE.PQAQA were subjected, in parallel, to centrifugation on 10 to 30% sucrose density gradients (33 ml), and the HA activity of fractions (∼1.1 ml) collected from the bottoms of the gradients was determined.
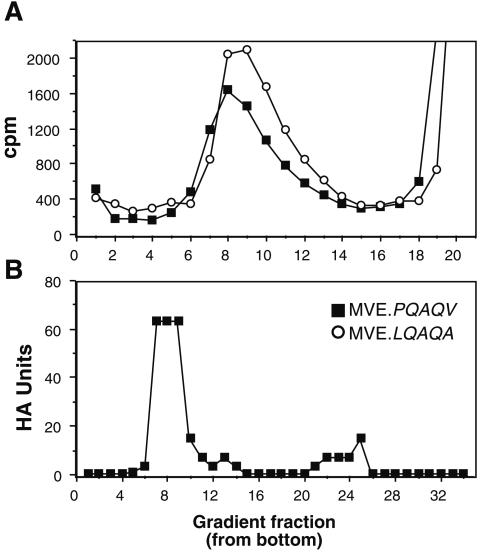
FIG. 5.
Characterization of particles secreted from mammalian and insect cells infected with revertants. (A) BHK cells were infected with MVE.LQAQA or MVE.PQAQV revertants and metabolically labeled from 20 to 26 h p.i. Infected culture supernatants were subjected to centrifugation on 10 to 30% sucrose gradients (10.5 ml), and radioactivity of fractions (∼0.4 ml) collected from the bottoms of the gradients was determined. (B) Culture fluid from C6/36 cells infected with MVE.PQAQV for 48 h was centrifuged on a 10 to 30% sucrose density gradient (33 ml), and the HA activity of fractions (∼1.1 ml) collected from the bottom of the gradient was determined.
For hemagglutination (HA) analysis of virions and VLPs in virus stocks, ∼12,800 HA units were applied to a 10 to 30% continuous sucrose gradient (33 ml) and centrifuged at 26,000 rpm for 3.25 h at 4°C in an SW28 rotor (Beckman). Fractions of ∼1.1 ml each were collected from the bottoms of the gradients and individually titrated for HA activity.
HA test.
HA tests were performed by a modification of the method of Clarke and Casals (5) in 96-well trays at pH 6.6 by using goose red blood cells. HA titers were calculated as the reciprocal of the last antigen dilution resulting in red blood cell agglutination.
Metabolic labeling, immunoprecipitation, and SDS-PAGE.
For analyzing virus-specific protein synthesis, BHK cell monolayers in 35-mm-diameter tissue culture dishes were infected at a multiplicity of 10 PFU/cell or left uninfected. At 16 h postinfection (p.i.), the cells were starved for 0.5 h by replacement of the growth medium with 1.0 ml of methionine- and cysteine-free MEM. For metabolic labeling, 0.5 ml of methionine- and cysteine-free MEM containing 100 μCi of Tran35S-label (ICN)/ml was added to the cell monolayers. For chases, the monolayers were washed with PBS, and MEM containing 5% FCS was added. Lysis, immunoprecipitation, SDS-polyacrylamide gel electrophoresis (PAGE), and fluorography were as described previously (40).
RESULTS
Enhanced signalase cleavage of prM reduces growth of MVE.
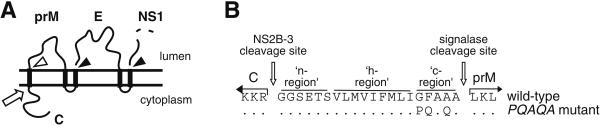
The flavivirus C and prM proteins are separated by an internal signal peptide which directs translocation of prM into the lumen of the ER (Fig. 1A). Signalase cleavage of prM remains inefficient until the cytosolic C protein is proteolytically removed by the viral NS2B-3 protease (1, 23, 39, 44). No requirement for prior cleavage of C in downstream signalase cleavages of E and NS1 proteins (Fig. 1A) has been found. The amino acid sequence of the prM signal peptide is depicted in Fig. 1. Consistent with classical signal peptides (33, 43), it contains basic residues in the NH2-terminal region (n-region), a hydrophobic core uninterrupted by charged or polar residues (h-region), and a −3, −1 amino acid motif in the COOH-terminal cleavage region (c-region) suitable for signalase recognition. An uncharacteristic feature of the prM signal peptide of MVE and other flaviviruses is the lack of polar residues in the c-region. Previously it has been shown that replacement of Gly, Phe, and Ala at positions −5, −4, and −2 with Pro, Gln, and Gln, respectively (PQAQA mutation), dramatically increases the extent of signalase cleavage of prM in vitro without requirement of prior cleavage of C (40).
FIG. 1.
(A) Schematic diagram showing the expected ER membrane topology of the flavivirus structural proteins C, prM, and E. Sites of efficient and inefficient cleavages by signal peptidase are indicated by closed and open arrowheads, respectively. The cleavage catalyzed by the viral NS2B-3 protease is denoted by an open arrow. (B) Amino acid sequence around the MVE C-prM junction, with residues that we have considered to be the n-, h-, and c-regions of the prM signal peptide indicated. The NS2B-3 protease and signalase cleavage sites are shown. Amino acid changes introduced to obtain the PQAQA mutation are shown below the wt MVE sequence with unchanged residues denoted as dots.
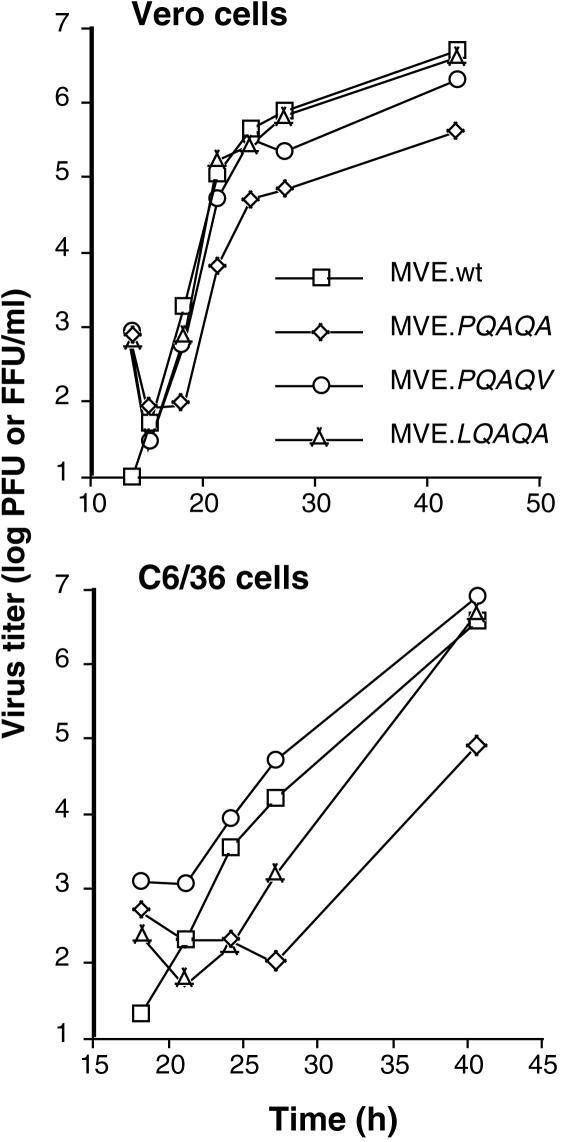
To investigate the biological role of the obligatory sequence of cleavage in the prM signal peptide, the PQAQA mutation was inserted into a full-length infectious cDNA clone of MVE (20). Transfection of BHK cells with comparable amounts of in vitro-synthesized wt or mutant RNA gave rise to detectable cytopathic effect on day 3 after transfection and yielded progeny virus in both cases, showing that the mutation in the prM signal sequence was not lethal for the growth of MVE. However, virus yield from transfected BHK cells was reduced for the mutant RNA relative to that for the wt RNA (2 × 104 and 3 × 103 PFU/ml of culture supernatant on day 3 after transfection). Reduced growth of the PQAQA mutant (designated MVE.PQAQA) relative to that of wt MVE was also found in Vero (monkey) and C6/36 (mosquito) cells in single-step growth experiments (Fig. 2). In the vertebrate cell line, the kinetics of PQAQA mutant virus release were marginally delayed and virus yield was >10-fold lower than that for wt MVE. In C6/36 cells infected with MVE.PQAQA, virus in the culture supernatant was detected only at a late time point p.i. and the titer was 100-fold lower than that of wt MVE. The growth phenotypes of revertant viruses depicted in Fig. 2 are discussed below. The recovery of viable but growth-impaired MVE.PQAQA offered the possibility to investigate the role of coordinated cleavages at the flavivirus C-prM junction in virus replication by characterizing the stage in the virus growth cycle detrimentally affected by the mutation.
FIG. 2.
Growth of wt MVE (MVE.wt), the PQAQA mutant, and revertants in mammalian and insect cells. Vero and C6/36 cells were infected at a multiplicity of 2 PFU and two focus-forming units (FFU)/cell, respectively, and growth samples were removed from the culture medium at the indicated time points. Virus titers in growth samples taken from Vero and C6/36 cells were determined by plaque assay on Vero cell monolayers and focus-forming assay on C6/36 cell monolayers, respectively.
Effect of PQAQA mutation on viral RNA and protein synthesis.
To investigate whether the reduced growth of PQAQA virus results from a defect in an early stage of virus replication, the kinetics of viral RNA synthesis and the efficiency of viral protein synthesis were compared to those in wt MVE in infected BHK cells. Virus-specific RNA synthesis was quantitated by measuring actinomycin D-resistant [3H]uridine incorporation into infected cells at time intervals shown in Fig. 3. Viral RNA synthesis was first apparent at 13 h p.i. for both viruses and increased during the subsequent labeling interval (16 to 19 h p.i.), suggesting that during the first cycle of growth the kinetics and magnitudes of RNA synthesis did not differ markedly in wt- and mutant virus-infected cells. Immunofluorescence staining for the viral E protein performed at 18 h p.i. on wt- and mutant-infected control monolayers showed that ∼50 and ∼30% of cells, respectively, were infected (data not shown). In the 20- to 23-h p.i. labeling interval, the two viruses differed in that virus-specific RNA synthesis increased further in wt-infected cells but decreased in mutant-infected cells, suggesting that second-round infection was more efficiently induced by the wt than the mutant virus.
Protein synthesis in BHK cells infected with wt or mutant MVE was evaluated by immunoprecipitation of virus-specific proteins from cell lysates metabolically labeled at 16 h p.i. for 0.5 h. Comparable amounts of NS5, NS3, NS1, E, and prM proteins were recovered with an anti-MVE antiserum for both viruses (Fig. 3B).
The PQAQA mutation prevents secretion of C but not of prM and E proteins from infected cells.
To examine the amounts of viral antigens secreted from wt- and PQAQA mutant-infected cells, metabolic labeling was performed at 16 h p.i. for 6.5 h and clarified culture fluids were subjected to immunoprecipitation with anti-E monoclonal antibody 8E7 (9). Secretion of E protein from MVE-infected cells is predominantly in the form of virion particles and VLPs which are both bound by the antibody in immunoprecipitations in the absence of detergent. Despite a >10-fold difference in the amounts of infectious virus released into the culture fluid during the labeling interval (3.5 × 107 and 2 × 106 PFU/ml for wt and mutant virus, respectively), comparable amounts of E and prM proteins were recovered (Fig. 3C). However, while the C protein was also precipitated from the culture fluid of wt virus-infected cells, it was detectable only following overexposure of the autoradiograph (data not shown) for the PQAQA mutant. This was consistent with the difference between the two viruses in amounts of infectious particles released and suggests that the mutation in the prM signal sequence impacts on the formation and/or secretion of infectious virus but not on the release of prM and E proteins, most likely in the form of VLPs.
The PQAQA mutation enhances the assembly and release of VLPs at the expense of virion particles.
Flavivirus VLPs differ from virion particles by lacking an NC, and this difference in densities between the two types of particles allows their separation by gradient centrifugation. When the supernatant from infected Vero cells, metabolically labeled from 40 to 47 h p.i., was analyzed by centrifugation through a 10 to 30% sucrose gradient, a single peak (fractions 7 to 10) corresponding to virion particles was found for wt MVE (Fig. 4A). In contrast, two peaks were prominent in the gradient profile for the PQAQA mutant, the virion peak and a slower-sedimenting peak (fractions 14 and 15) consistent with that for the less dense VLPs. The latter peak cosedimented with VLPs produced by recombinant expression of the MVE prM and E proteins in transfected COS-7 cells (data not shown).
The production of VLPs as a by-product of flaviviral infection was first noted because of the HA activity of gradient fractions that sedimented more slowly than infectious virus (37, 41). To compare the ratio of virions to VLPs present in wt and PQAQA virus stocks generated in infected suckling mouse brain or mosquito cells, infectivities and HA activities of the stocks were determined. The infectivities of the mouse brain- and C6/36 cell-grown wt virus stocks per HA unit exceeded those of the mutant virus stocks by 9- and 25-fold, respectively (8 × 104 and 9 × 103 PFU/HA unit and 5 × 104 and 2 × 103 PFU/HA unit, respectively). When comparable numbers of HA units were centrifuged on 10 to 30% sucrose gradients and the HA activities of the collected gradient fractions were determined, a distinct distribution of HA activity was found for the two viruses: the majority of wt virus HA activity was found in the virion peak whereas that of the PQAQA mutant was found in the slower-sedimenting VLP-containing peak (Fig. 4B).
Recovery of revertants from MVE.PQAQA-infected cells.
The growth kinetics of the PQAQA virus in C6/36 cells were markedly delayed and progeny virus yield was significantly reduced relative to that for wt MVE (Fig. 2). To test whether revertants or second-site mutants were produced following infection of C6/36 cells with MVE.PQAQA, the nucleotide sequence of the C-to-E gene region and phenotypic properties of plaque-purified progeny virus were analyzed. The great majority of progeny virus from infected C6/36 cells (9 out of 11 plaques analyzed) carried a reversion in the PQAQA sequence, one virus carried a second site mutation in the C protein, and one remained unchanged (Table 1). The dominant reversion was an Ala125→Val change (MVE.PQAQV) at the −1 position with respect to the signalase cleavage site in prM. Given that Val is rarely found at this position in eukaryotic signal peptides (33), it is likely that this change results in less-efficient signalase cleavage of prM. The LQAQA reversion was isolated only once; the Pro121→Leu change in this revertant shortens the length of the c-region of the prM signal peptide and removes the α-helix-breaking property of Pro, two factors which appear to influence the cleavage efficiency of the prM signal peptide. A third (pseudo)revertant was isolated with a Phe→Leu change at residue 54 in the C protein. This residue is in the center of a conserved internal hydrophobic domain which may promote membrane integration of the flaviviral C protein (26). No revertants were obtained from 12 plaque isolates and 3 plaque isolates of progeny virus released from BHK and Vero cells, respectively, following infection with MVE.PQAQA (Table 1).
TABLE 1.
Recovery of revertants after growth of MVE.PQAQA in insect or mammalian cells
| Host cell type | No. of plaques analyzed | Revertant | No. of times isolated | Amino acid changea (nucleotide change) |
|---|---|---|---|---|
| C6/36 | 11 | MVE.PQAQV | 8 | Ala125→Val (GCC)→(GUC) |
| MVE.LQAQA | 1 | Pro121→Leu (CCA)→(CUA) | ||
| Capsid F54b | 1 | Phe54→Leu (UUC)→(CUC) | ||
| BHK | 12 | |||
| Vero | 3 |
Amino acid numbering is from the first residue in the C protein (6).
Pseudorevertant with an amino acid change in the C protein.
Phenotypic properties of revertants.
The reversions significantly increased growth efficiency of MVE in mosquito and mammalian cells (Fig. 2). In C6/36 cells, virus yields for the two revertants were comparable to that for wt MVE at 40 h p.i., although the growth kinetics of the LQAQA revertant in the early phase of the growth curve were intermediate in comparison to those of wt virus and the PQAQA mutant. In Vero cells, the growth phenotypes of the two revertants were essentially restored relative to that of wt MVE (Fig. 2).
The main impact of the PQAQA mutation accounting for reduced growth in cell culture was at the stage of virion assembly and was reflected in the significantly reduced ratio of virions to VLPs released from infected cells. The PQAQV and LQAQA reversions repaired this defect: metabolically labeled particles released from MVE.PQAQV- and MVE.LQAQA-infected Vero cells sedimented as a single virion peak in 10 to 30% sucrose gradients in the absence of a detectable peak corresponding to VLPs; furthermore, the ratio of PFU to HA units in a mosquito cell-grown stock of MVE.PQAQV (2 × 104 Vero cell PFU/HA unit) was comparable to that for wt MVE and analysis of this stock by gradient centrifugation demonstrated that the HA activity was predominantly associated with the virion peak (fractions 7 to 9) (Fig. 5).
Effect of reversions on signalase cleavage of prM in vitro.
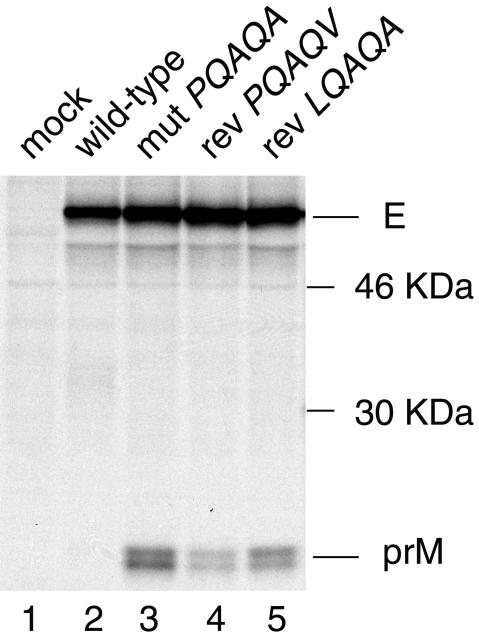
Dependence of efficient signalase cleavage of prM on prior cytosolic cleavage of the C protein by the viral NS2B-3 protease is detectable in recombinant expression systems but not in viral infections given the accumulation of the viral protease in infected cells. To investigate whether single amino acid reversions in the c-region of the prM signal sequence give rise to less-efficient signalase cleavage of prM relative to that in the PQAQA mutant, the corresponding structural polyprotein regions were transiently expressed in transfected COS-7 cells in the absence of the viral protease. Following metabolic labeling, the transfected cell lysates were subjected to immunoprecipitation with an E protein-specific monoclonal antibody which resulted in recovery of the E-prM heterodimer where the prM protein migrates as a double band due to a difference in the levels of carbohydrate maturation of the glycosylated protein (23).
Figure 6 shows that the PQAQV and LQAQA reversions reduced the efficiency of signalase cleavage of prM relative to that in the PQAQA mutant. However, the reversions did not reduce the efficiency of signalase cleavage of prM to the same level as that found for the wt construct in our in vitro expression system. Calculation of the E-to-prM ratio (made by subtracting background densities from average density measurements of the areas corresponding to the E and prM protein bands in Fig. 6, obtained by using the program NIH Image 1.62, and dividing by the number of cysteines and methionines in the respective proteins) gave values of 1.0, 2.8, and 1.6 for constructs mutPQAQA, revPQAQV, and revLQAQA, respectively. The ratio could not be calculated for the wt construct given that the density value for the area corresponding to the prM protein equaled that of background. Results comparable to those shown in Fig. 6 were obtained in two repeat experiments.
FIG. 6.
Effect of reversions on the efficiency of signalase cleavage of prM. COS-7 cells were transfected with eukaryotic expression plasmids encoding the wt MVE structural proteins (pcDNA.STR), the PQAQA mutation (pSTRmutPQAQA), or reversions in the prM signal sequence (pSTRrevPQAQV or pSTRrevLQAQA). At 2 days after transfection, the cells were metabolically labeled for 30 min and chased for 1 h, and the prM and E proteins were immunoprecipitated with monoclonal antibody 8E7 and analyzed by SDS-PAGE (12% acrylamide). Bands corresponding to prM and E are labeled, and positions of 46- and 30-kDa protein markers are shown.
Efficient signalase cleavage of prM results in inefficient cleavage of the C protein.
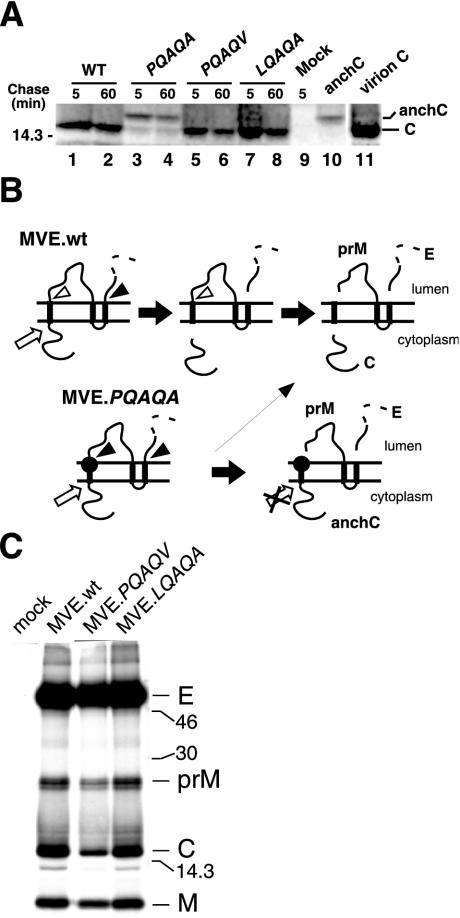
Our results, so far, suggest that the coordinated cleavages at the C-prM junction play a pivotal role in virus morphogenesis by preventing the loss of viral transmembrane proteins (in the form of VLPs) from the virus assembly site on the ER membrane. In Fig. 7, we show that a cryptic signalase cleavage site at the C-prM junction is also key for efficient cleavage of C by the viral NS2B-3 protease. Immunoprecipitation from lysates of MVE.PQAQA-infected BHK cells (by using an antiserum raised against a fusion peptide containing NH2-terminal residues of C) predominantly yielded a band with an electrophoretic mobility slower than that for authentic C recovered from lysates of wt MVE-infected cells (Fig. 7A). This band is referred to as anchored C (anchC) and corresponds to the C protein plus the prM signal peptide. Virion C and anchC immunoprecipitated from COS-7 cells transfected with plasmid pSTR.mutPQAQA (40) are shown as references for their respective electrophoretic mobilities in lanes 11 and 10, respectively (Fig. 7A). Bands corresponding to C or anchC were not apparent in lysates of uninfected BHK cells (lane 9). The identity of anchC has been confirmed elsewhere by using transient transfection experiments with eukaryotic expression plasmids encoding wt or mutant forms of C and anchC (38). In addition to anchC, a low-intensity band corresponding to authentic C was also recovered from MVE.PQAQA-infected cells. A precursor-product relationship between anchC and authentic C was not apparent following a 1-h chase of pulse-labeled MVE.PQAQA-infected cells (lanes 3 and 4 in Fig. 7A). Thus, it is not clear whether anchC is a substrate for cleavage by the viral protease. Alternatively, authentic C may be produced in MVE.PQAQA-infected cells when cytosolic cleavage by NS2B-3 precedes signalase cleavage of prM. A schematic diagram of the proteolytic processing events at the C-prM junction for wt and mutant MVE is given in Fig. 7B.
FIG. 7.
Effect of PQAQA mutation and reversions on the proteolytic processing of C protein. (A) BHK cells were infected with wt MVE (WT), MVE.PQAQA, or either the MVE.PQAQV or MVE.LQAQA revertant or left uninfected (Mock). At 16 h p.i., the cells were metabolically labeled for 1 h and chased as shown. Immunoprecipitation was with an anti-C antiserum, and proteins were subjected to SDS-PAGE (15% acrylamide). anchC and virion C proteins are shown in lanes 10 and 11, respectively, as references for their respective electrophoretic mobilities. Bands corresponding to C and anchC are labeled, and the position of a 14.3-kDa marker protein is shown on the left. (B) Schematic diagram showing the expected ER membrane topology of the MVE structural proteins and the putative sequence of proteolytic cleavage products for wt MVE (MVE.wt) and PQAQA mutant MVE. The PQAQA mutation in the prM signal sequence is represented by a closed circle, sites of efficient and inefficient cleavages by signal peptidase are indicated by closed and open arrowheads, respectively, and the cleavage catalyzed by the viral NS2B-3 protease is denoted by an open arrow. (C) BHK cells (106 cells) were infected with wt MVE, MVE.PQAQV, or MVE.LQAQA (multiplicity of infection, ∼10) or left uninfected and metabolically labeled from 16 to 22 h p.i. Products secreted in the culture supernatants were immunoprecipitated with an E protein-specific monoclonal antibody (8E7) and analyzed by SDS-PAGE (15% acrylamide). Sizes (in kilodaltons) of marker proteins are given, and viral protein bands are labeled.
The PQAQV and LQAQA reversions restored efficient cleavage of C; anchC was not detected in lysates of cells infected with either of the two revertants (Fig. 7A, lanes 5 to 8), and the ratios of virion-associated transmembrane proteins to virion C were comparable between revertants and wt virus (Fig. 7C). Given the saturating intensity of the E protein bands, the ratio of prM plus membrane (M) proteins to C proteins was calculated by using the program NIH Image 1.62, as described above, which gave values of 1.6, 1.8, and 1.7 for the wt, MVE.PQAQV, and MVE.LQAQA, respectively.
The apparently full restoration of NS2B-3 cleavage of C in the two revertants in vivo contrasts with the only partial restoration of inefficient signalase cleavage of prM in COS-7 cells transiently transfected with plasmids encoding the reversions in the context of the structural polyprotein region of MVE (Fig. 6). The most likely explanation for this discrepancy is that the in vitro expression model, in which the viral protease and the structural proteins are expressed from separate constructs in the absence of a number of nonstructural proteins, does not entirely mimic the efficiency of cleavages at the C-prM junction during viral infection, as noted by others (2).
DISCUSSION
We describe here a unique mechanism for efficient incorporation of NC into budding viral membranes. Central to this mechanism is the control of signalase cleavage of the flavivirus transmembrane protein prM by the catalytic activity of a viral protease at the cytosolic side of the signal sequence. The signalase cleavage site of prM is maintained in a predominantly cryptic conformation until cleavage of C by the viral protease has taken place. This regulation of cleavages at the C-prM junction prevents rapid (cotranslational) signalase cleavage of prM and hence is predicted to result in transient expression of a C-prM intermediate at the putative flavivirus assembly site on the ER membrane until cytosolic processing by the viral protease has taken place. We and others (28) have been unable to demonstrate a C-prM intermediate in flavivirus-infected mammalian cells, suggesting that, at the time of maximal viral protein synthesis, cleavage by NS2B-3 at the C-prM junction is too rapid for detection of C-prM by pulse-labeling and immunoprecipitation due to the accumulation of the viral protease in infected cells. However, the transient existence of a C-prM precursor has been shown in flavivirus-infected mosquito cells (28).
We propose that the coordination of cytosolic and luminal cleavages at the C-prM junction is key to efficient incorporation of NC during flavivirus assembly, given that budding of flavivirus membranes is driven by the two viral transmembrane proteins prM and E independently of C protein or assembled NC. We show that a mutant virus (MVE.PQAQA), which was constructed to uncouple efficient signalase cleavage of prM from its dependence on prior cytosolic cleavage of C, replicated >10-fold and >100-fold more poorly in mammalian and mosquito cells, respectively, than wt MVE as a result of a defect in NC incorporation into particles released from infected cells. Thus, the amount of virion particles released from MVE.PQAQA-infected cells was significantly reduced at the expense of augmented release of VLPs. The isolation of revertants with single amino acid substitutions in the PQAQA mutation showed that (partial) repair of the wt cleavage phenotype at the C-prM junction in vitro restored efficient virion production, lending further support for the critical role of inefficient signalase cleavage of prM in efficient NC uptake into budding flavivirus membranes.
We show that the signal sequence at the C-prM junction has a dual regulatory role in the proteolytic processing of C and prM. In addition to maintaining the signalase cleavage site of prM in a cryptic conformation, it also retains accessibility of the double-basic cleavage site at the COOH terminus of C for recognition by the viral protease. When the obligatory sequence of cleavages was altered by introduction of the PQAQA mutation such that signalase cleavage of prM became efficient, cytosolic cleavage of C was significantly reduced. Accordingly, the membrane-anchored form of C is a poor substrate for cleavage by the viral protease unless covalently attached to an ER luminal polypeptide. anchC was not detected in virions released from MVE.PQAQA-infected cells (data not shown), suggesting that it cannot assemble into NC. Infectious virus released from these cells contained authentic C which was visible as a minor cleavage product in lysates of cells infected with the mutant virus.
Currently, we cannot distinguish between whether (i) the loss of viral transmembrane proteins (in the form of VLPs) from the virus assembly site on the ER membrane or (ii) inefficient C protein production accounts for the detrimental effect of the PQAQA mutation on virus morphogenesis. A mutant virus in which both cleavages at the C-prM junction can occur efficiently and independently of each other is required to address this issue. Nevertheless, it is difficult to envisage why an interdependence of cytosolic and ER luminal cleavages at the C-prM junction should have evolved unless it orchestrates NC assembly and budding of flavivirus membranes to favor NC incorporation into budding particles. It should be noted that the assembly of infectious flavivirus particles is not exclusively dependent on the coordinated cleavages at the C-prM junction but that the mechanism enhances the efficiency of this process; this is shown by the viability of the MVE.PQAQA mutant described here and by others who demonstrated that expression of Kunjin virus C and prM/E proteins from separate coding units in cells stably transfected with subgenomic replicon RNA trans-complements replicon particle production, albeit inefficiently (13). It has been shown earlier that incorporation of the PQAQA mutation into the c-region of the prM signal sequence of YFV is lethal and that viability is restored by reversions at one of the mutagenized residues or by second-site mutations which cluster in the NH2-terminal part of the prM signal sequence (21). The latter YFV pseudorevertants were significantly attenuated for neurovirulence in mice and their growth in mammalian cells was reduced ∼10-fold. Given the finding in our present study demonstrating that enhanced signalase cleavage of prM is, in part, inhibitory for NS2B-3-mediated C protein cleavage, it appears that in the case of the YFV PQAQA mutant the lethal phenotype is a consequence of a deficiency of cleavage of anchC. It is most likely that in the pseudorevertants some authentic C protein production was restored but that growth and virulence phenotypes remained attenuated due to inefficient virus assembly analogous to that of the MVE PQAQA mutant described here.
Corona- and hepadnaviruses are examples of other virus families which employ an NC-independent budding strategy. The coronavirus M and E proteins drive budding of virus membranes and can assemble into VLPs in recombinant expression (42). During infection, the release of coronavirus NC-free particles is most likely kept to a minimum by the stable interaction of the cytosolic domain of the M protein with assembled NC (19, 29). VLPs are a major by-product of hepatitis B virus infection and are usually present in the sera from human carriers in a 104- to 106-fold excess over virion particles, suggesting an inefficient mechanism of virus morphogenesis (7). The small (S) envelope protein of hepatitis B virus drives the budding of ER membranes independently of other viral components. Incorporation of NC into budding particles is mediated by the large (L) envelope protein, which is translated in low abundance: the COOH-terminal half of the L protein oligomerizes with S proteins, inhibiting the release of the latter in the form of VLPs, while a cytosolic domain of the L protein binds to NCs, directing their assembly into virion particles (3, 34, 35).
The flaviviral prM and E proteins are devoid of a distinct NC-binding domain comparable to those of the coronavirus M and hepatitis B virus L proteins, given that in the flavivirus particle both transmembrane domains at the COOH termini of prM and E remain integrated in the viral membrane with the COOH-terminal residues on the exterior side (45). Flaviviral NCs are thought to interact only weakly with the virus membranes, which is reflected in a poorly ordered structure of the C proteins relative to the external, icosahedral scaffold of the transmembrane glycoproteins (17, 45). In the absence of a stable interaction between flaviviral C and transmembrane proteins, the uptake of NC into budding flavivirus membranes may be a stochastic process aided by (i) temporal and spatial coordination of NC assembly and formation of transmembrane protein lattices as a result of regulated cleavages at the C-prM junction, (ii) hydrophobic interaction of C proteins at the cytosolic surface of the ER membrane (12, 14, 15, 26), and (iii) a yet unknown contribution of the membrane-spanning NS2A protein, putatively associated with viral genomic RNA, as part of the NS2B-3 protease complex that cleaves at the cytosolic side of the prM signal peptide (18).
Acknowledgments
We thank Megan Pavy for excellent technical help and Alyssa Pyke for generously providing us with goose red blood cells.
This work was supported by a grant from the National Health and Medical Research Council of Australia.
REFERENCES
- 1.Amberg, S. M., A. Nestorowicz, D. W. McCourt, and C. M. Rice. 1994. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. J. Virol. 68:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amberg, S. M., and C. M. Rice. 1999. Mutagenesis of the NS2B-NS3-mediated cleavage site in the flavivirus capsid protein demonstrates a requirement for coordinated processing. J. Virol. 73:8083-8094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruss, V., X. Lu, R. Thomssen, and W. H. Gerlich. 1994. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 13:2273-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1126. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 5.Clarke, D. H., and J. Casals. 1958. Techniques for hemagglutination and hemagglutination-inhibition with arthropod-borne viruses. Am. J. Trop. Med. Hyg. 7:561-573. [DOI] [PubMed] [Google Scholar]
- 6.Dalgarno, L., D. W. Trent, J. H. Strauss, and C. M. Rice. 1986. Partial nucleotide sequence of the Murray Valley encephalitis virus genome. J. Mol. Biol. 187:309-323. [DOI] [PubMed] [Google Scholar]
- 7.Ganem, D., and R. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2970. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 8.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall, R. A., B. H. Kay, G. W. Burgess, P. Clancy, and I. D. Fanning. 1990. Epitope analysis of the envelope and non-structural glycoproteins of Murray Valley encephalitis virus. J. Gen. Virol. 71:2923-2930. [DOI] [PubMed] [Google Scholar]
- 10.Harrison, S. C. 2001. Principles of virus structure, p. 53-86. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 11.Johnson, A. E., and M. A. van Waes. 1999. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 15:799-842. [DOI] [PubMed] [Google Scholar]
- 12.Jones, C. T., L. Ma, J. W. Burgner, T. D. Groesch, C. B. Post, and R. J. Kuhn. 2003. Flavivirus capsid is a dimeric alpha-helical protein. J. Virol. 77:7143-7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khromykh, A. A., A. N. Varnavski, and E. G. Westaway. 1998. Encapsidation of the flavivirus Kunjin replicon RNA by using a complementation system providing Kunjin virus structural proteins in trans. J. Virol. 72:5967-5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kofler, R. M., F. X. Heinz, and C. W. Mandl. 2002. Capsid protein C of tick-borne encephalitis virus tolerates large internal deletions and is a favorable target for attenuation of virulence. J. Virol. 76:3534-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kofler, R. M., A. Leitner, G. O'Riordain, F. X. Heinz, and C. W. Mandl. 2003. Spontaneous mutations restore the viability of tick-borne encephalitis virus mutants with large deletions in protein C. J. Virol. 77:443-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konishi, E., S. Pincus, E. Paoletti, R. E. Shope, T. Burrage, and P. W. Mason. 1992. Mice immunized with a subviral particle containing the Japanese encephalitis virus prM/M and E proteins are protected from lethal JEV infection. Virology 188:714-720. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn, R. J., W. Zhang, M. G. Rossmann, S. V. Pletnev, J. Corver, E. Lenches, C. T. Jones, S. Mukhopadhyay, P. R. Chipman, E. G. Strauss, T. S. Baker, and J. H. Strauss. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kümmerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuo, L., and P. S. Masters. 2002. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 76:4987-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, E., and M. Lobigs. 2000. Substitutions at the putative receptor-binding site of an encephalitic flavivirus alter virulence and host cell tropism and reveal a role for glycosaminoglycans in entry. J. Virol. 74:8867-8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, E., C. E. Stocks, S. M. Amberg, C. M. Rice, and M. Lobigs. 2000. Mutagenesis of the signal sequence of yellow fever virus prM protein: enhancement of signalase cleavage in vitro is lethal for virus production. J. Virol. 74:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1042. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 23.Lobigs, M. 1993. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proc. Natl. Acad. Sci. USA 90:6218-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lobigs, M. 1992. Proteolytic processing of a Murray Valley encephalitis virus non-structural polyprotein segment containing the viral proteinase: accumulation of a NS3-4A precursor which requires mature NS3 for efficient processing. J. Gen. Virol. 73:2305-2312. [DOI] [PubMed] [Google Scholar]
- 25.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Markoff, L., B. Falgout, and A. Chang. 1997. A conserved internal hydrophobic domain mediates the stable membrane integration of the dengue virus capsid protein. Virology 233:105-117. [DOI] [PubMed] [Google Scholar]
- 27.Martoglio, B., and B. Dobberstein. 1998. Signal sequences: more than just greasy peptides. Trends Cell Biol. 8:410-415. [DOI] [PubMed] [Google Scholar]
- 28.Murray, J. M., J. G. Aaskov, and P. J. Wright. 1993. Processing of the dengue virus type 2 proteins prM and C-prM. J. Gen. Virol. 74:175-182. [DOI] [PubMed] [Google Scholar]
- 29.Narayanan, K., A. Maeda, J. Maeda, and S. Makino. 2000. Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol. 74:8127-8134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newton, S. E., N. J. Short, and L. Dalgarno. 1981. Bunyamwera virus replication in cultured Aedes albopictus (mosquito) cells: establishment of a persistent viral infection. J. Virol. 38:1015-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ng, M. L., J. Howe, V. Sreenivasan, and J. J. Mulders. 1994. Flavivirus West Nile (Sarafend) egress at the plasma membrane. Arch. Virol. 137:303-313. [DOI] [PubMed] [Google Scholar]
- 32.Ng, M. L., S. H. Tan, and J. J. Chu. 2001. Transport and budding at two distinct sites of visible nucleocapsids of West Nile (Sarafend) virus. J. Med. Virol. 65:758-764. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 34.Ostapchuk, P., P. Hearing, and D. Ganem. 1994. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 13:1048-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prange, R., and R. E. Streeck. 1995. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 14:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schalich, J., S. L. Allison, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1996. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 70:4549-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith, T. J., W. E. Brandt, J. L. Swanson, J. M. McCown, and E. L. Buescher. 1970. Physical and biological properties of dengue-2 virus and associated antigens. J. Virol. 5:524-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stocks, C. E. 2001. A study of coordinated cleavages by signal peptidase and NS2B-3 protease at the C-prM junction in the flavivirus polyprotein. Ph.D. thesis. Australian National University, Canberra, Australia.
- 39.Stocks, C. E., and M. Lobigs. 1995. Posttranslational signal peptidase cleavage of the flavivirus C-prM junction in vitro. J. Virol. 69:8123-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stocks, C. E., and M. Lobigs. 1998. Signal peptidase cleavage at the flavivirus C-prM junction: dependence on the viral NS2B-3 protease for efficient processing requires determinants in C, the signal peptide, and prM. J. Virol. 72:2141-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stollar, V. 1969. Studies on the nature of dengue viruses. IV. The structural proteins of type 2 dengue virus. Virology 39:426-438. [DOI] [PubMed] [Google Scholar]
- 42.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Heijne, G. 1986. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamshchikov, V. F., and R. W. Compans. 1993. Regulation of the late events in flavivirus protein processing and maturation. Virology 192:38-51. [DOI] [PubMed] [Google Scholar]
- 45.Zhang, Y., J. Corver, P. R. Chipman, W. Zhang, S. V. Pletnev, D. Sedlak, T. S. Baker, J. H. Strauss, R. J. Kuhn, and M. G. Rossmann. 2003. Structures of immature flavivirus particles. EMBO J. 22:2604-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]