A quantitative RNA code for mRNA target selection by the germline fate determinant GLD-1
The Caenorhabditis elegans RNA-binding protein GLD-1 is a critical regulator of germline fate. Here, a quantitative approach to identifying GLD-1 target RNAs provides insight into the post-transcriptional regulatory networks underlying germ cell development.
Keywords: GLD-1, Quaking, RBP, ribonomics, STAR
Abstract
RNA-binding proteins (RBPs) are critical regulators of gene expression. To understand and predict the outcome of RBP-mediated regulation a comprehensive analysis of their interaction with RNA is necessary. The signal transduction and activation of RNA (STAR) family of RBPs includes developmental regulators and tumour suppressors such as Caenorhabditis elegans GLD-1, which is a key regulator of germ cell development. To obtain a comprehensive picture of GLD-1 interactions with the transcriptome, we identified GLD-1-associated mRNAs by RNA immunoprecipitation followed by microarray detection. Based on the computational analysis of these mRNAs we generated a predictive model, where GLD-1 association with mRNA is determined by the strength and number of 7-mer GLD-1-binding motifs (GBMs) within UTRs. We verified this quantitative model both in vitro, by competition GLD-1/GBM-binding experiments to determine relative affinity, and in vivo, by ‘transplantation' experiments, where ‘weak' and ‘strong' GBMs imposed translational repression of increasing strength on a non-target mRNA. This study demonstrates that transcriptome-wide identification of RBP mRNA targets combined with quantitative computational analysis can generate highly predictive models of post-transcriptional regulatory networks.
Introduction
Regulation of mRNA is crucial in both development and homoeostasis, and mutations affecting splicing, localization, turnover, or translation have been implicated in many human diseases ranging from neurological disorders to cancer (Lukong et al, 2008; Ule, 2008; Cooper et al, 2009; Kim et al, 2009; Sonenberg and Hinnebusch, 2009). Multiple aspects of mRNA metabolism are regulated by RNA-binding proteins (RBPs), which typically contain one or more domains determining their RNA-binding preferences (Lunde et al, 2007). Recent efforts from several laboratories using global approaches have begun to unravel the ‘RNA-binding code' for various RBPs, with the ultimate goal of understanding how individual RBPs and their combinations regulate expression of the transcriptome (Halbeisen et al, 2008). However, the majority of transcriptome-wide studies remain descriptive.
The signal transduction and activation of RNA (STAR) family of RBPs is characterized by a maxi-KH and two flanking QUA domains (Vernet and Artzt, 1997). The animal STAR proteins can be divided into three subfamilies: SAM68, SF-1, and Quaking related (QR). QR proteins are critical developmental regulators as well as tumour suppressors (Biedermann et al, 2010). The Caenorhabditis elegans QR protein GLD-1 is expressed in the germ line, and gld-1 mutants are defective in multiple germline events, such as sex determination, the mitosis/meiosis decision, or maintenance of germ cell identity (Francis et al, 1995a, 1995b; Jones et al, 1996; Ciosk et al, 2006; Biedermann et al, 2009) (Figure 1A). Several mutations that alter GLD-1 function are within the RNA-binding STAR domain (Jones and Schedl, 1995; Francis et al, 1995a). For example, substitution of a single amino acid (G227D), which is predicted to interact with RNA (Liu et al, 2001; Galarneau and Richard, 2009), phenocopies gld-1 deletion. This demonstrates that RNA binding is critical for GLD-1 function. To date, few GLD-1 targets have been reported (Jan et al, 1999; Lee and Schedl, 2001, 2004; Xu et al, 2001; Marin and Evans, 2003; Mootz et al, 2004; Schumacher et al, 2005; Biedermann et al, 2009). In vitro analysis of one GLD-1 target has suggested that GLD-1 binds a hexanucleotide RNA sequence termed STAR-binding element (SBE) (Ryder et al, 2004) (Supplementary Figure S1). However, the scale of mRNA interaction with GLD-1 and the general relevance of the SBE for GLD-1 binding and regulation remain unknown.
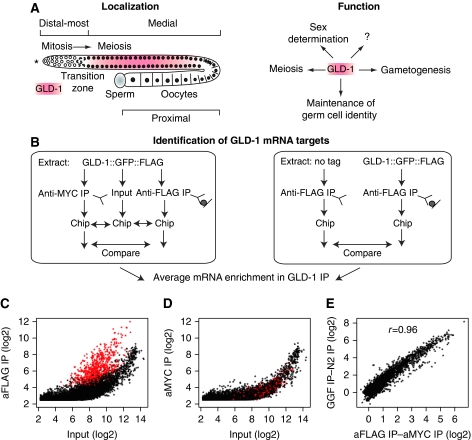
Figure 1.
Identification of mRNAs associated with GLD-1. (A) GLD-1 is expressed in the medial gonad and is a key regulator of germline development, yet the majority of its biological targets are unknown. ‘Distal-most', ‘medial', and ‘proximal' indicate parts of a wild-type adult worm gonad. The asterisk here and in subsequent figures indicates the distal end of the gonad. (B) Experimental outline of GLD-1 mRNA target identification. Two independent strategies were employed: (1) comparison of anti-FLAG IP (aFLAG) to anti-MYC IP (a MYC) on extract from worms expressing tagged GLD-1 (left panel); (2) comparison of anti-FLAG IP on extract from worms expressing tagged GLD-1 (GGF_IP) to anti-FLAG IP on extract from non-tagged (N2) worms (N2_IP) (right panel). To ensure that transcripts were detected with high confidence, an input aliquot (RNA purified from extract before IP) from strategy 1 was initially used to establish an input cutoff (>5.5). (C) A large set of mRNAs (red dots) are enriched greater than three-fold in a GLD 1 IP. Comparison of aFLAG to the corresponding input. (D) GLD-1-associated mRNAs (red dots, as in C) are not enriched in a control IP. Comparison of aMYC to the corresponding input. (E) The enrichment of associated mRNAs is reproducible between complementary GLD-1 IP approaches. Comparison of aFLAG—aMYC (C, D) on the x axis, to GGF_IP—N2_IP on the y axis gives a correlation coefficient of 0.96. All mRNAs detected on the array together with their average IP enrichment are given in Supplementary Dataset S1.
We present a comprehensive analysis of GLD-1 interaction with mRNA, providing a list of potential GLD-1 targets as well as a quantitative description of the determinants underlying mRNA recognition by GLD-1. These determinants are verified both in vitro and in live animals. Specifically, we find that GLD-1 associates with hundreds of mRNAs, and these interactions are mediated by 7-mer GLD-1-binding motifs (GBMs) that are distinct from SBEs. Furthermore, GBMs of varying strength can act in a multiplicative fashion to increase association with GLD-1. The value of this quantitative analysis is demonstrated by in vivo experiments, in which ‘weak' or ‘strong' GBMs are sufficient to impose GLD-1-mediated translational repression of increasing strength on an otherwise non-regulated mRNA.
Results
GLD-1 associates with a large number of mRNAs
To identify GLD-1 mRNA targets, we performed immunoprecipitation (IP) of GLD-1, followed by microarray analysis of the co-IPed mRNAs (RIP-chip) (Tenenbaum et al, 2000; Biedermann et al, 2009). Extracts from young adult transgenic worms expressing a rescuing FLAG and GFP-tagged GLD-1 (Schumacher et al, 2005), hereafter referred to as tagged GLD-1, were subjected to IP in triplicate with anti-FLAG (aFLAG IP) or anti-MYC (aMYC IP) antibodies as controls (Figure 1B). Comparison of aFLAG IP versus aMYC IP to input revealed a large population of GLD-1-associated transcripts (red dots in Figure 1C and D). We additionally performed complementary aFLAG IPs upon worms expressing either tagged GLD-1(GGF IP) or non-tagged GLD-1(N2 IP). Comparing transcript ‘IP-enrichment values' from both approaches revealed a correlation of 0.96, which indicated high reproducibility of GLD-1 association with specific mRNAs even on a quantitative level (Figure 1E). We calculated the mean IP enrichment from the two analyses and given a cutoff of three-fold, we identified 948 reproducibly enriched mRNAs (14.2%) from of a total of 6635 detected by the array (Supplementary Dataset S1). We suspect that this set is highly enriched for genuine targets because among similarly abundant mRNAs there is a strong bias against somatic transcripts (Supplementary Figure S2). To further validate the microarray analysis by an independent method, we performed deep sequencing (RNA-seq) after additional aFLAG IPs on the tagged (GGF) and non-tagged (N2) strains. Because of the high content of rRNA in the sequenced samples (Supplementary Table S1), only 2080 unique genes could be quantified (see Materials and methods). Despite this, comparison to the respective microarray data revealed a correlation of 0.705 demonstrating high reproducibility of GLD-1-associated mRNA detection and verifying the majority of mRNAs detected by RIP-chip (Supplementary Figure S3).
GLD-1-associated mRNAs are enriched for related sequences
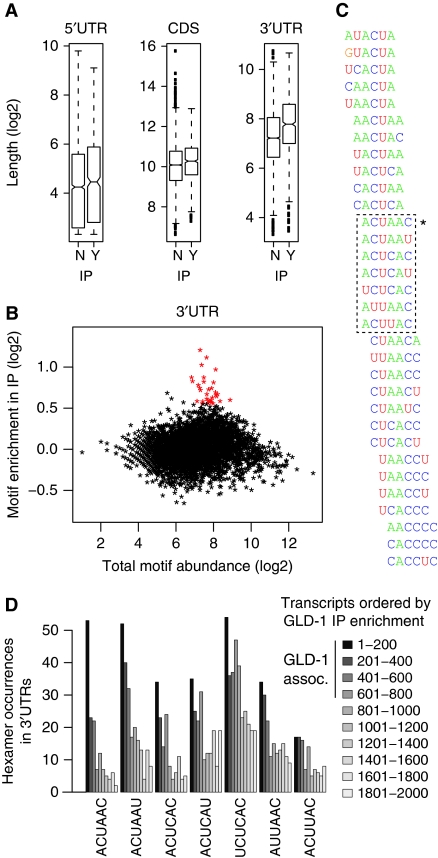
To test if GLD-1 associates with mRNA targets via a specific RNA motif, we searched for sequences enriched among GLD-1 targets. Initially, we found that GLD-1 targets contain substantially longer 3′UTRs when compared to non-targets (Figure 2A), suggesting that GLD-1 targets more likely contain regulatory motifs within the 3′UTR. To identify the potential binding motifs for GLD-1, we enumerated all hexanucleotides and looked to see if any were enriched among the 806 GLD-1-associated transcripts with annotated 3′UTRs (Supplementary Dataset S2). In total, 32 hexanucleotides (marked red in Figure 2B) were enriched and they showed high similarity when aligned, suggesting that they belong to a single degenerate motif, potentially longer than 6 nucleotides. (Figure 2C). We defined a motif core (Figure 2C) consisting of the highest scoring 6-mer (ACUAAC) and six non-overlapping variants. Given the highly reproducible IP-enrichment values (Figure 1E), we wondered whether they reflect GLD-1 affinity to mRNAs mediated by the enriched RNA motifs. Indeed, occurrences of the core 6-mers decreased progressively with IP enrichment, suggesting that this is the case (Figure 2D).
Figure 2.
GLD-1 targets are enriched for degenerate and overlapping sequences. (A) GLD-1-associated mRNAs have significantly longer 3′UTRs. The length distribution (log2) of all 3′UTRs, 5′UTRs, and CDSs is shown for both GLD-associated (Y) (greater than three-fold average IP enrichment) and non-associated (N) mRNAs. Only mRNAs with sufficient expression (>6.5) and with an annotated 5′UTR or 3′UTR were examined (Supplementary Dataset S2). (B) A subgroup of hexanucleotides is enriched in the 3′UTRs of GLD-1-associated mRNAs. For all possible hexamers (n=4096), the total number of occurrences in all 3′UTRs is depicted on the x axis, and the motif enrichment in the GLD-1-associated population on the y axis. Motif enrichment was computed for each hexanucleotide by counting the number of occurrences in GLD-1-associated 3′UTRs compared to all 3′UTRs, correcting for 3′UTR length differences. A pseudocount of 16 was added to limit enrichment values for hexanucleotides with very low number of occurrences. Red dots highlight 32 hexanucleotides enriched in the GLD-1 IP. (C) Alignment of the 32 enriched hexanucleotide sequences reveals overlapping motifs, which have sequence similarity. The asterisk indicates the highest scoring hexamer, ACUAAC and the dotted box demarcates the motif core containing other related non-overlapping variants. (D) Abundance of the core hexamers within 3′UTRs correlates with IP enrichment. The total number of occurrences of each core hexamer (indicated by the dotted box in C) was counted within sets of 200 transcripts ordered by IP enrichment. Sets corresponding to GLD-1-associated transcripts are indicated (GLD-1 assoc.).
GLD-1 binds a degenerate heptanucleotide RNA motif—GBM
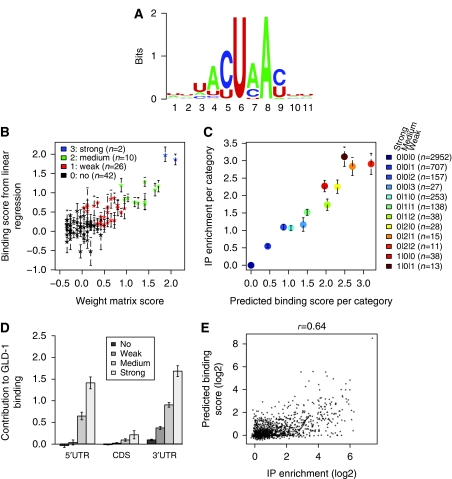
To identify the full motif and to obtain a quantitative weighting of the nucleotide preference in each position, we performed a unbiased search using the MEME program (Bailey and Elkan, 1994). The outcome of this search suggested that GLD-1 recognizes a 7-mer RNA motif containing nucleotides with various degrees of degeneracy, which can be quantitatively represented as a position specific weight matrix (Figure 3A). Importantly, such a weight matrix assumes independence between nucleotide positions and thus would not reveal if nucleotide changes at one position can impact upon the nucleotide preference at another position. To ask if this could be the case for GLD-1 binding, we took the 80 highest scoring 7-mers from the weight matrix and examined them by a linear regression to determine the contribution of each individual 7-mer to mRNA enrichment in the IP. This approach can account for any positional nucleotide dependency (Berger et al, 2006; Ray et al, 2009). The comparison between coefficients from linear regression and the weight matrix scores showed a good correlation (r=0.78), suggesting that positional dependencies have a minor role in GLD-1 binding to its motif (Figure 3B). The presence of a substantial number of 7-mers with a coefficient close to zero indicates that no relevant GLD-1-binding 7-mer was missed in the selection of the 80 7-mers using the weight matrix score. We computed the final binding score for each of the 80 7-mers using an average of both the weight matrix score and the coefficient from linear regression. Based on the predicted binding scores, we arbitrarily grouped the 80 7-mers into four classes: strong, medium, weak, or ‘no binding' to GLD-1 (Figure 3B, Table I). We refer to the 38 7-mers which we predict mediate strong, medium, or weak binding to GLD-1, as GBMs.
Figure 3.
GLD-1 binds degenerate 7-mer motifs, GBMs. (A) Identification of a GLD-1-binding motif (GBM). Weblogo of the position specific weight matrix (PSWM) detected within GLD-1 target 3′UTRs using MEME. (B) Positional nucleotide independence within the GBM. The scatter plot shows the score for the top 80 7-mers from the PSWM (A) on the x axis and the y axis indicates a coefficient of each 7-mer derived from a linear regression (which calculates the contribution of each 7-mer as a whole to GLD-1 IP enrichment). Motifs with the best average predicted binding score are shown in blue (strong), then green (medium), red (weak), and black (no predicted binding to GLD-1). For individual 7-mer scores, see Table I. (C) Multiple GBMs in a single 3′ UTR contribute to GLD-1 binding in a multiplicative fashion. All array detected transcripts with an annotated 3′UTR (n=806) were categorized depending on the number of strong, medium, and weak GBMs. For example, the ‘one medium together with two weak GBMs is denoted as (0¦1¦2). Only categories containing at least 10 3′UTRs were retained (legend). For each category the average GLD-1 IP enrichment from microarray (y axis) was compared to a predicted binding score per category (x axis). The later was computed by summing, in log space, the contribution of all GBMs present within a given 3′UTR, and subsequently by averaging the scores of all the transcripts that fall within a given category. (D) GLD-1 binds GBMs in both 5′ and 3′UTRs. Predicted relative GLD-1 binding contribution of strong, medium, and weak 7-mers in the 5′UTR, CDS, or 3′UTR was determined by linear regression (see text). (E) The sum of GBM scores is predictive of GLD-1 binding. A predicted binding score per transcript was calculated by summing, in log space, scores of any of the 80 7-mers (Table I) present in its 3′ and 5′UTR and this score compared to the actual GLD-1 IP enrichment (x axis). Comparison was limited to germline-detected transcripts (>4 tags) (Wang et al, 2009).
Table 1. Average predictive GLD-1-binding score of 80 7-mers, including GBMs.
| GBM motif | Score | Group | Other 7-mer | Score | Group | |
|---|---|---|---|---|---|---|
| 1 | UACUAAC | 1.981 | 3 | ACCUCAC | 0.222 | 0 |
| 2 | CACUAAC | 1.934 | 3 | UACUCAA | 0.215 | 0 |
| 3 | AACUAAC | 1.444 | 2 | GUCUAAC | 0.212 | 0 |
| 4 | UACUCAC | 1.362 | 2 | CGCUAAU | 0.208 | 0 |
| 5 | UACUAAU | 1.348 | 2 | GACUCAU | 0.206 | 0 |
| 6 | GACUAAC | 1.089 | 2 | AACUAAA | 0.206 | 0 |
| 7 | CACUCAC | 1.056 | 2 | CUCUCAC | 0.181 | 0 |
| 8 | UCCUAAC | 1.039 | 2 | CCCUCAC | 0.180 | 0 |
| 9 | AACUCAC | 1.000 | 2 | UAUUAAU | 0.177 | 0 |
| 10 | CACUAAU | 0.971 | 2 | AACUUAC | 0.176 | 0 |
| 11 | AACUAAU | 0.949 | 2 | UUCUAAU | 0.176 | 0 |
| 12 | UACUCAU | 0.890 | 2 | UCCUAAU | 0.171 | 0 |
| 13 | UACUAAA | 0.795 | 1 | AGCUAAC | 0.166 | 0 |
| 14 | GACUCAC | 0.777 | 1 | CAUUCAC | 0.145 | 0 |
| 15 | CCCUAAC | 0.743 | 1 | UAAUAAC | 0.078 | 0 |
| 16 | UAUUAAC | 0.743 | 1 | GACUAAA | 0.076 | 0 |
| 17 | UGCUAAC | 0.707 | 1 | CAUUAAU | 0.061 | 0 |
| 18 | UUCUAAC | 0.686 | 1 | UGCUAAU | 0.058 | 0 |
| 19 | CGCUAAC | 0.596 | 1 | AAUUCAC | 0.053 | 0 |
| 20 | UACUUAC | 0.549 | 1 | AUCUAAU | 0.042 | 0 |
| 21 | CAUUAAC | 0.537 | 1 | CUCUAAU | 0.041 | 0 |
| 22 | CACUCAU | 0.509 | 1 | CACUGAC | −0.021 | 0 |
| 23 | GACUAAU | 0.506 | 1 | GACUUAC | −0.022 | 0 |
| 24 | CUCUAAC | 0.470 | 1 | CCCUAAU | −0.023 | 0 |
| 25 | UACUGAC | 0.466 | 1 | AACUGAC | −0.026 | 0 |
| 26 | ACCUAAC | 0.434 | 1 | AAUUAAU | −0.037 | 0 |
| 27 | AAUUAAC | 0.401 | 1 | AGCUAAU | −0.056 | 0 |
| 28 | AACUCAU | 0.400 | 1 | CGCUCAC | −0.056 | 0 |
| 29 | GGCUAAC | 0.395 | 1 | CACUCAA | −0.070 | 0 |
| 30 | GAUUAAC | 0.373 | 1 | UACUUAU | −0.071 | 0 |
| 31 | CACUAAA | 0.339 | 1 | GAUUCAC | −0.087 | 0 |
| 32 | UUCUCAC | 0.336 | 1 | AACUCAA | −0.092 | 0 |
| 33 | AUCUCAC | 0.327 | 1 | ACCUAAU | −0.108 | 0 |
| 34 | CACUUAC | 0.324 | 1 | GUCUCAC | −0.119 | 0 |
| 35 | AUCUAAC | 0.320 | 1 | UUCUCAU | −0.128 | 0 |
| 36 | UCCUCAC | 0.300 | 1 | UAUUCAU | −0.130 | 0 |
| 37 | UAUUCAC | 0.279 | 1 | GCCUAAC | −0.162 | 0 |
| 38 | UGCUCAC | 0.270 | 1 | UCCUCAU | −0.165 | 0 |
| UGCUCAU | −0.209 | 0 | ||||
| GCCUCAC | −0.232 | 0 | ||||
| AGCUCAC | −0.276 | 0 | ||||
| GGCUCAC | −0.304 | 0 | ||||
| A final predictive binding score for the top 80 7-mers from the weight matrix was computed using an average of both the weight matrix score and the coefficient from linear regression. Using this predictive binding score the 7-mers were grouped into four classes: strong (3), medium (2), weak (1), or no (0) GLD-1 binding (Figure 3B). The 38 GBMs that mediate GLD-1 binding are indicated in bold. For GBMs tested in reporter assays, see Supplementary Table S3. Additionally the average IC50 and ΔΔG° values for GBMs, as calculated in Figure 4, are shown in Supplementary Dataset S3. | ||||||
| To identify the specific type, number, and location of GBMs within transcript(s) an R-based script is available on the FMI website http://www.fmi.ch/groups/gbioinfo. | ||||||
Multiple GBMs in a single 3′UTR contribute to GLD-1 binding in a multiplicative fashion
Next, we asked how GLD-1 interacts not only with a single target site but also with an entire mRNA, which can contain multiple binding sites with different strengths. We grouped transcripts into categories according to the number of strong, medium, and weak GBMs within the 3′UTR and compared their average IP enrichment per category to a binding score that we calculated assuming multiplicative motif contributions (Figure 3C). Comparison of the calculated score to the measured enrichment showed very high positive correlation (r=0.96) and all points lie close to a perfect diagonal. This shows that the affinity of GLD-1 to a 3′UTR can be predicted by multiplying (or adding in log space) the scores of individual GBMs. For example, 3′UTRs containing only one strong GBM have a similar affinity for GLD-1 as those which contain two medium GBMs or one medium and two weak GBMs. To exclude that our findings are biased by the linear regression approach that was used for motif scoring, we recalculated data shown in Figure 3C by cross-validation, where we infer 7-mer scores and calculate IP enrichments per category for independent sets of transcripts. This control leads to the same result confirming our analytical approach (Supplementary Figure S4).
Recognition of GBMs in 5′UTRs
Given that we were able to generate an accurate representation of GLD-1 binding to 3′UTRs, we wondered whether GBMs mediate GLD-1 binding to 5′UTRs or CDSs. To determine the relative importance of GBMs in the 5′UTR, CDS, or 3′UTR we performed a single linear regression using 7-mer classes (strong, medium, weak, or ‘no binding') as predictor variables and GLD-1 IP enrichment as response variables (12 predictor variables). The coefficients from the regression showed that GBMs located in 5′UTRs are bound almost as efficiently as motifs located in 3′UTRs (Figure 3D). In contrast, GBMs within the CDS are largely not recognized. However, though GBMs in the 3′ and 5′UTRs show similar contribution to GLD-1 binding, 3′UTRs are on average five times longer than 5′UTRs (Figure 2A), and therefore likely mediate the majority of GLD-1 interactions.
GLD-1-binding predictor
As GLD-1/mRNA interaction is influenced by the strength and number of GBMs within 5′ and 3′UTRs, we wondered if considering only this information alone is sufficient to predict which messages are GLD-1 targets. We therefore computed a predicted GLD-1-binding score for every mRNA based on summing (in log space) the score of each of the 7-mers present in either the 5′ or 3′UTRs (Supplementary Dataset S2). To assess the quality of our predictor, we compared it to the actual enrichment values of the GLD-1 IP. As GLD-1 is only found in the germ line, we restricted our analysis to germline-detected transcripts (Wang et al, 2009), as somatic mRNAs could potentially contain a GBM but would never ‘see' GLD-1. We found that the predicted and the actual association of mRNAs with GLD-1 showed a good correlation (R=0.64). We conclude that, based on the strength and number of GBMs only, GLD-1 binding to mRNA can be predicted with reasonable accuracy (Figure 3E).
GLD-1 targets and anti-targets
Top gene ontology terms among the 948 enriched GLD-1 mRNAs include cytokinesis, cell division, embryonic development, reproductive processes, cell cycle, and DNA replication (Supplementary Table S2). All of these terms are consistent with GLD-1 roles in meiotic cell cycle progression, sex determination, and cell fate specification (Francis et al, 1995b) (Figure 1A).
Additionally, we noticed that the most abundant mRNAs are not associated with GLD-1 (Figure 1C) and we wondered if this reflects actual avoidance of GBMs in these transcripts as was suggested for miRNA motifs (Stark et al, 2005). We inspected the deep sequencing data set in a similar way, and again the most abundant mRNAs (n=101) did not associate with GLD-1 (Supplementary Figure S5A) thus excluding that signal saturation on microarray could contribute to the observed lack of enrichment.
These highly expressed ‘non-targets', which contain many ribosomal genes, are highly depleted for GBMs (Supplementary Figure S5B), even when corrected for 3′UTR length. Such strong motif depletion suggests the possibility that highly expressed housekeeping mRNAs have evolved to avoid GBMs in their 3′UTR, as binding by GLD-1 might interfere with their translation.
Determining relative affinities of GLD-1 to GBMs in vitro
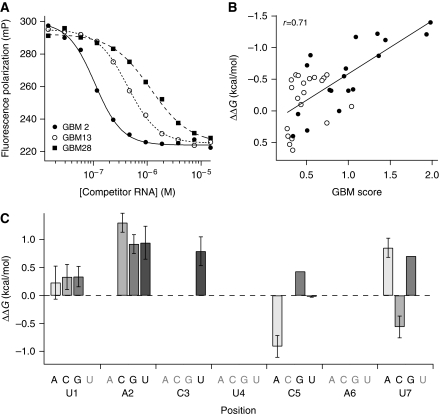
Computational analysis of the GLD-1 RIP chip predicts that the GBM score reflects GLD-1-binding affinity (Figure 2D). To determine relative affinities of all GBMs by an independent approach, we performed quantitative competition fluorescence polarization experiments that measure the ability of each GBM to inhibit binding of recombinant GLD-1 protein to a fluorescein-labelled TGE RNA sequence that mediates GLD-1 binding to one of its targets (Figure 4A) (Ryder et al, 2004). The GBM present within the TGE (UACUCAU) was used as a reference sequence. Remarkably, the change in standard free energy (ΔΔG°) values for all GBMs correlated well (r=0.71) with the GBM scores determined by the computational analysis of our RIP chip-enrichment values (Figure 4B; Supplementary Dataset S3).
Figure 4.
In vitro analysis of GLD-1 interaction with GBMs. (A) Representative example of determination of GLD-1/GBM affinities by the competition-binding assay. The relative affinity of each of the 38 GBMs was measured by competition-binding assay using purified recombinant protein. Fluorescence polarization is plotted as a function of unlabelled competitor RNA concentration for three competitor sequences containing GBMs: GBM2 (UUUCACUAACUUU, IC50,app=90±20 nM, filled circles), GBM13 (UUUUACUAAAUUU, IC50,app=400±30 nM, open circles), and GBM28 (UUUAACUCAUUUU, IC50,app=900±200 nM, filled squares). Data were fit to a sigmoidal dose response equation to determine the apparent IC50. (B) Correlation between GLD-1/GBM affinity in vitro (ΔΔG°) and in vivo (GBM score; Table I). The ΔΔG° for each of the 38 GBMs relative to the reference sequence (GBM12, UUUUACUCAUUUU, IC50,app=700±70 nM) was determined from the measured IC50 values using the following expression: ΔΔG°=−RT ln (IC50(variant)/IC50(reference)). The ΔΔG° measurement for each sequence was plotted as a function of its GBM score and fit to a line to determine the Pearson correlation coefficient (r=0.71). Filled circles represent GBMs which also score as SBEs, while open circles represent GBMs that are not identified as SBEs. (C) Context dependence of single nucleotide substitutions to GLD-1 affinity. The ΔΔG° for each sequence relative to every other sequence was determined, and the ΔΔG° for each occurrence of a single nucleotide change independent of sequence context was averaged. Each bar represents the average effect of the listed mutation at the listed position, and error bars represent context-dependent variance. Nucleotide changes in grey do not occur in the population of GBMs, and the C5G and U7G mutations are present only once and thus have no associated variance.
Consistent with the GBM prediction, the competition data reveal a few important differences from the previously reported SBE consensus. First, the data show a strong preference for a pyrimidine at the seventh position. A cytosine improves the binding over uridine by 0.5 kcal/mol, while purine substitutions reduce affinity by 0.5–0.7 kcal/mol. The contribution of this position was not explicitly tested in the previous work (Ryder et al, 2004). Second, an adenosine at the fifth position enhances the binding by five- to eight-fold relative to cytosine, whereas previous work revealed a two-fold enhancement (Ryder et al, 2004). Finally, an adenosine at the fifth position and/or cytosine at the seventh position provide sufficient favourable standard free energy to compensate for deleterious mutations at the second or third position, yielding GBMs that do not match the SBE that can bind to GLD-1 with affinity greater than or equal to the reference sequence.
To examine in vitro the effect of positional dependencies within the GBM, the ΔΔG° was determined for pairs of sequences differing by only one nucleotide (Figure 4C; Supplementary Dataset S4). The majority of single nucleotide mutations appear to be context independent agreeing with the comparison between weight matrix score and coefficient from linear regression (r=0.78) (Figure 3B), suggesting that positional dependencies have a minor role in GBM's affinity for GLD-1. Nucleotide substitutions at the first position, however, show some context dependence (Figure 4C), with a U contributing 0.5 kcal/mol in sequences with weak affinity but very little in sequences with high affinity. In summary, these experiments confirm that the GBM scores derived from IP enrichment reflect GLD-1 affinity for these motifs.
Validation of GBMs in vivo
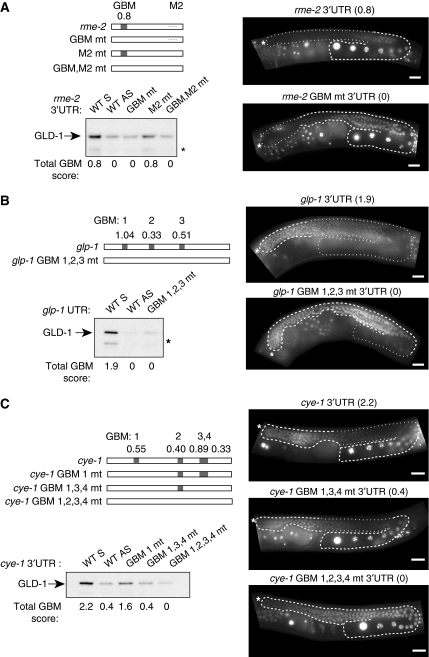
The 948 mRNAs that are enriched greater than three-fold in the GLD-1 IP contain 21 of 26 previously implicated GLD-1 targets (three targets were excluded due to input cutoff on the array data, one due to a probe detection problem; Supplementary Table S4). Several of these are repressed in the medial gonad and their repression was shown to be GLD-1 dependent (Grant and Hirsh, 1999; Lee and Schedl, 2001; Xu et al, 2001; Marin and Evans, 2003; Mootz et al, 2004; Schumacher et al, 2005; Biedermann et al, 2009). To test if their repression is mediated via GBMs, we mutated GBMs in the 3′UTRs and tested what effect it had on GLD-1 binding and mRNA repression. GLD-1 binding was assayed in vitro by incubating biotinylated 3′UTRs in a C. elegans extract and examining if they interact with GLD-1. Beads alone and a non-target 3′UTR were used as controls to establish the background signal (Supplementary Figure S6). To test the requirement of the GBM for mRNA repression in vivo, we generated transgenic lines expressing in the germ line a GFP:H2B-tagged reporter fused either to wild-type or GBM-mutated 3′UTRs (Merritt et al, 2008). We started with a simple case, the rme-2 3′UTR, which contains only one weak GBM (Figure 5A) and another unrelated sequence, M2, previously suggested to bind GLD-1 (Lee and Schedl, 2001). Mutating the GBM reduced GLD-1 binding to background levels, whereas mutating the M2 sequence did not (Figure 5A). This suggests that GLD-1 binds the rme-2 3′UTR predominantly via the GBM. Furthermore, transgenic lines expressing a GFP:H2B-tagged reporter fused to the wild-type rme-2 3′UTR caused repression in the medial gonad, where GLD-1 is expressed (Figure 5A). Importantly, this repression was eliminated when the GBM was mutated (Figure 5A). We additionally studied the glp-1 3′UTR that contains one medium and two weak GBMs. Mutating all three GBMs inhibited GLD-1 binding to RNA and caused reporter de-repression in the medial gonad (Figure 5B).
Figure 5.
GBMs bind GLD-1 in vitro and mediate mRNA repression in vivo. (A) The rme-2 3′UTR interacts with GLD-1 via a GBM, which mediates regulation of a linked reporter. The top left panel shows putative GLD-1 regulatory elements within the rme-2 3′UTR and versions where they are mutated. Boxes within the 3′UTR indicate GBMs and their predictive score is above. All of the 3′UTRs depicted were tested by biotin-RNA pulldown assay for their ability to pull down tagged GLD-1 from a worm extract (bottom left panel). A GFP∷H2B reporter is repressed in the medial gonad via the GBM in the rme-2 3′UTR (right panel). Images show adult gonads expressing GFP:H2B fused to either wild-type or GBM-mutated rme-2 3′UTR. (B) GLD-1 associates with the glp-1 3′UTR via multiple GBMs. The wild-type or GBM1,2,3 mutated glp-1 3′UTRs (upper left panel) were tested for their interaction with tagged GLD-1 from a worm extract (lower left panel). Images of adult gonads expressing GFP∷H2B fused to either wild-type or GBM1,2,3 mutated glp-1 3′UTR (right panels). (C) Multiple GBMs in the cye-1 3′UTRS act in concert to increase association with and repression by GLD-1. The wild-type or GBM-mutated cye-1 3′UTRs (upper left panel) were tested in biotin-RNA pulldown assay for interaction with tagged GLD-1 (lower left panel). Right panels show images of adult gonads expressing GFP∷H2B fused to either wild-type or mutated versions of the cye-1 3′UTR, as indicated. The gonads in A, B, and C are outlined with dashed lines highlighting areas of expression and dotted lines indicating repression. The total GBM score for each 3′UTR is shown below the relevant gel lanes and also above gonad images in parentheses. *Degradation product of GLD-1. Scale bars are 20 μm.
The computational analysis indicated that GLD-1 can recognize multiple GBMs within a 3′UTR leading to increased GLD-1 association (Figure 3). To test this, we sequentially mutated GBMs within the cye-1 3′UTR, which contains two separate GLD-1-binding regions (Biedermann et al, 2009). We predicted that the cye-1 3′UTR contains one medium (third) and three weak (first, second, fourth) GBMs (Figure 5C). Mutating GBM 1 reduced GLD-1 interaction but did not prevent it (Figure 5C). Mutating GBMs 3 and 4 together with GBM 1 further reduced GLD-1 binding. However, this 3′UTR was still able to repress the GFP:H2B reporter in vivo (Figure 5C). An additional mutation in the remaining GBM 2 was needed both to reduce GLD-1 binding to background levels and to cause de-repression of the reporter in vivo (Figure 5C). Therefore, a single GBM can mediate GLD-1 regulation but having multiple GBMs within a 3′UTR increases association with GLD-1.
As at least one QR protein, HOW, was reported not to bind dsRNA (Israeli et al, 2007; Volk et al, 2008) we wondered whether the ability of GLD-1 to recognize a GBM may similarly depend on GBM accessibility. To verify this, we focussed on the short rme-2 3′UTR, as secondary structure predictions on short sequences are generally more reliable (Figure 5A). Two different mutations of rme-2 3′UTR were predicted to place the GBM within either a ssRNA loop or a dsRNA stem. In contrast to the GBM within the ssRNA loop, the GBM within the dsRNA stem did not associate with GLD-1 and was unable to mediate GLD-1 repression in vivo (Supplementary Figure S7). We then tested if predicting GBM accessibility on a global scale can improve target prediction. We observed only little increase in the predictive power (Supplementary Figure S7D), suggesting either that current tools cannot sufficiently predict mRNA structure in vivo or that the majority of GBMs are accessible for GLD-1 binding.
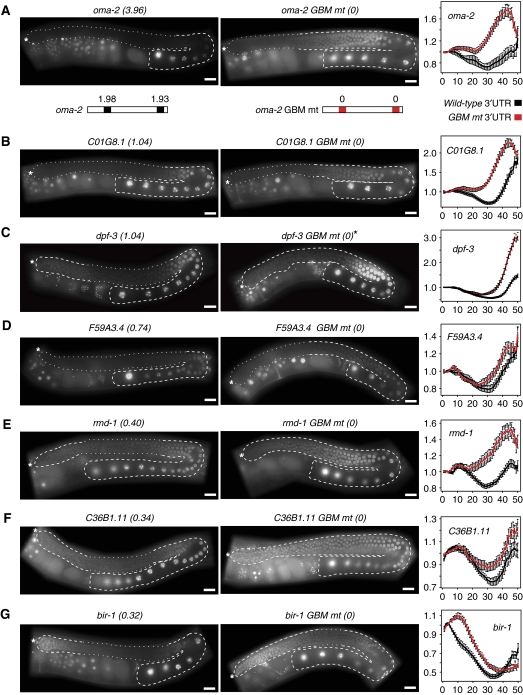
Validation of several additional GBMs within novel GLD-1 targets in vivo
To confirm that some of the novel GLD-1 targets we identified are regulated in vivo, and at the same time stringently test our GBM predictions, we tested several transcripts from among those enriched in the GLD-1 IP. Going top-down through the list we selected those that contained only one GBM, distinct from the SBE, within their 3′UTR (Supplementary Table S3). Tested targets matching these criteria were bir-1, rmd-1, dpf-3, C01G8.1, C36B1.11, and F59A3.4 (Figure 6B–G). Four of these contain one weak, and two have one medium scoring GBM. As a control, we also included the known GLD-1 target oma-2 (Lee and Schedl, 2004) that has two strong GBMs within its 3′UTR. GBM regulation was monitored by creating transgenic lines containing either wild-type or GBM-mutated 3′UTRs by Mos1 mediated single copy gene insertion (MosSCI) (Frokjaer-Jensen et al, 2008). Importantly, in every case, wild-type 3′UTRs caused repression of a linked GFP:H2B reporter in the medial gonad (Figure 6A–G, left panels), suggesting that all of these mRNAs are regulated by GLD-1 in vivo. Consistently, mutating the GBM caused de-repression of the reporter in the medial gonad in every case (Figure 6A–G, central panels).
Figure 6.
GBMs mediate GLD-1-dependent regulation in vivo in several novel GLD-1 targets. GBMs mediate repression in the medial (GLD-1 expressing) part of the gonad (A–G). Images of gonads expressing a GFP∷H2B reporter under the control of either wild-type 3′UTRs (left panels) or GBM-mutated 3′UTRs (central panels). Gonads are outlined: dashed lines highlighting expression and dotted lines repression. The 3′UTR total GBM score is in parentheses. Scale bars are 20 μm. Quantification of GFP intensity comparing wild-type and GBM-mutated 3′UTRs is shown in right most panels. Each line shows the average relative GFP intensity±s.e.m. X axis shows the relative distance across the gonad from most distal to the bend. Y axis indicates the normalized expression mean. Numbers of gonads quantified are as follows: (A) wt n=10, GBM1,2 mut n=25; (B) wt n=20, GBM mut n=27; (C) wt n=33, GBM mut n=32; (D) wt n=22, GBM mut n=22; (E) wt n=26, GBM mut n=25; (F) wt n=22, GBM mut n=20; and (G) wt n=22, GBM mut n=25. *The dpf-3 3′UTR permits expression in the most distal and proximal parts of the gonad and repression in the medial gonad. The dpf-3 GBM-mutated version is expressed more evenly across the distal and medial parts of the gonad as expected. However, there is a strong up-regulation of expression in the most proximal part, which we cannot explain.
By creating single-copy-integrated reporters, we were able to estimate the strength of GBM-mediated repression by quantifying GFP intensity dependent on GBMs in the 3′UTR (Figure 6A–G, right panels). Generally, higher scoring targets were regulated more strongly, but among weaker scoring targets the correlation was weaker. This is consistent with our observation that the strength prediction for weak GBMs is less certain (Figure 3B). It is also possible that the structural context of the GBM, or additional RBPs, may influence the GLD-1 binding or regulation of a particular 3′UTR. Therefore, to test the correlation between GBM strength (GLD-1 binding) and mRNA regulation, we tested GBMs of different strengths in the same RNA context.
GBMs induce strength-dependent translational repression
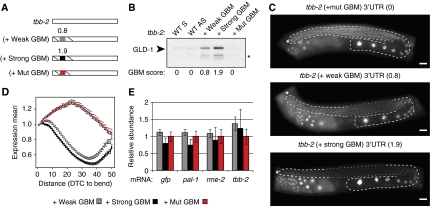
We introduced GBMs of differing strength into a 3′UTR that is normally not regulated by GLD-1. We chose the tbb-2 3′UTR as it permits GFP:H2B reporter expression throughout the entire germ line (Merritt et al, 2008) and contains no GBMs (Figure 7A). Addition of a weak GBM to the tbb-2 3′UTR resulted in efficient GLD-1 binding in vitro and repression in vivo (Figure 7B and C). As expected, the GFP repression was suppressed upon gld-1 (RNAi) (Supplementary Figure S8).
Figure 7.
A GBM inserted into a non-regulated 3′UTR confers strength-dependent GLD-1 binding and translational repression in vivo. (A) Schematic of the tbb-2 3′UTR or versions where the first 28 nts were replaced with 28 nts of the rme-2 3′UTR, containing either a weak, strong, or mutated GBM. (B) A strong GBM inserted into a non-target RNA sequence associates with more GLD-1 than a weak GBM. The in vitro transcribed RNAs in A were assayed for their ability to interact with tagged GLD-1. The total GBM score of each construct is shown. *GLD-1 degradation product. (C) Inserting GBMs of differing strength into the tbb-2 3′UTR causes strength-dependent repression of a GFP:H2B reporter in the medial gonad. Representative images of transgenic lines expressing a GFP:H2B reporter fused to tbb-2 (+mutated GBM), tbb-2 (+weak GBM), or tbb-2 (+strong GBM). Gonads are outlined: dashed lines highlighting expression and dotted lines repression. The total GBM score for each 3′UTR is in parentheses. Scale bars are 20 μm. (D) Quantification of GFP intensity observed in transgenic lines shown in C. The relative GFP intensity was plotted for transgenic lines where the GFP:H2B reporter is fused to the tbb-2 (+mutated GBM) 3′UTR (90 gonads, 2 strains of n=45), the tbb-2 (+weak GBM) 3′UTR (90 gonads, 2 strains of n=45), and the tbb-2 (+strong GBM) 3′UTR (90 gonads, 2 strains of n=45). Each line shows the average relative intensity across the gonad from the distal end indicated by an asterisk in C, to the gonad bend±s.e.m. (E) Quantification of mRNA levels for the transgenic lines quantified in D. The graph shows relative amounts of indicated mRNAs, determined by quantitative RT–PCR. Each bar represents an average of three independent biological replicates and error bars reflect the s.e.m. Primers for GFP were used to detect the transgenic GFP:H2B reporter in every line and three endogenous genes were also detected as controls.
To test if a stronger GBM induces stronger repression, we mutated the last nucleotide of the weak GBM in the tbb-2 (+weak GBM) 3′UTR from A to C. According to our prediction, this converts it from a weak (score=0.8) to a strong GBM (score=1.9). Indeed, the tbb-2 (+strong GBM) 3′UTR bound more GLD-1 than the tbb-2 (+weak GBM) 3′UTR (Figure 7B) and, importantly, induced stronger GFP repression (Figure 7D). Consistent with GLD-1 functioning as a translational repressor, the GBM-mediated repression affected protein, but not mRNA levels, of the reporters (Figure 7E). We conclude that the strength of a GBM is relevant not only for GLD-1 binding to RNA but also for the extent of mRNA target regulation in vivo.
Discussion
GLD-1 association with mRNA
Our approach, combining ‘quantitative ribonomics' with in vitro and in vivo studies, has uncovered several criteria that determine mRNA target selection by GLD-1. Firstly, GLD-1's affinity for RNA depends on a 7-mer GBM of differing strength that is distinct from previous motifs proposed for the QR proteins (Ryder et al, 2004; Galarneau and Richard, 2005; Hafner et al, 2010). It has also been suggested that Quaking and GLD-1 recognize a shorter ‘half-site', in addition to their core RNA-binding motif (Ryder et al, 2004; Galarneau and Richard, 2005, 2009). However, this does not seem to be a general feature of RNA recognition by GLD-1, as we found no evidence for any particular predictive trinucleotide sequence 20 bp upstream or downstream of the GBM (Supplementary Figure S9). Also, 24 of the 43 RNAs in which the ‘half-site' was reported contain multiple GBMs, suggesting that at least in some cases a half-site is part of a degenerate full-length-binding motif (our unpublished observation). Secondly, we find that although a single GBM is sufficient to ensure GLD-1 regulation, multiple GBMs work in concert to increase affinity for GLD-1. Thus, the strength and number of GBMs present within UTRs will largely determine the GLD-1 interaction with mRNA. Given that there is an absolute conservation of the amino acids involved in RNA binding within the QR subfamily (Ryder et al, 2004; Galarneau and Richard, 2009), we expect that our ‘GBM predictor' (http://www.fmi.ch/groups/gbioinfo) can be used to predict interactions between other QR, and possibly STAR, proteins and their mRNA targets.
GLD-1 regulation of mRNA expression
GLD-1 has been shown to translationally repress many important mRNAs in the medial gonad (Grant and Hirsh, 1999; Lee and Schedl, 2001; Xu et al, 2001; Marin and Evans, 2003; Mootz et al, 2004; Schumacher et al, 2005; Biedermann et al, 2009). Consistently, every GLD-1 target we analysed by GFP reporter was repressed in the medial gonad in a GBM-dependent way. To see if repression in the medial gonad is a general feature of identified GLD-1 targets, we mined the literature for their protein expression patterns. Of the 948 mRNAs enriched over three-fold in the GLD-1 IP, 128 had an antibody available and for 53 the germline expression pattern was available. The majority (34/53) had an expression pattern that was compatible with GLD-1 regulation. For the 19 that did not, this may indicate that their expression is activated by other RBPs, or that GLD-1 may regulate a different aspect of their RNA metabolism (Supplementary Table S5).
Some RBPs have been suggested to co-regulate mRNAs encoding functionally related proteins (Tenenbaum et al, 2000; Ule et al, 2003; Gerber et al, 2004; Keene, 2007; Licatalosi et al, 2008). Somewhat consistent with this idea, we see several members of functional groups enriched among GLD-1 targets such as cytokinesis, cell division, embryonic development, reproductive processes, cell cycle, and DNA replication (Supplementary Table S2). Our recent study has shown that GLD-1 function in tumour suppression and cell fate decision involves translational repression of cyclin E mRNA and perhaps B-type cyclins (Biedermann et al, 2009). This global analysis suggests that apart from cyclins, GLD-1 may also regulate cyclin ‘effectors' such as DNA replication factors. Thus, it is possible that at least some biological roles of GLD-1 reflect the regulation of protein networks, which may enable effective buffering against occasional mis-expression of individual targets due to physiological or other insults.
Much of the control of gene expression in the germ line lies with RBPs. Consistently, genes encoding RBPs are expressed four-fold higher in the germ line than in the soma (Wang et al, 2009). Remarkably still, our analysis suggests that GLD-1 associates with approximately 10% of germline mRNAs detected to date (Wang et al, 2009) (Supplementary Figure S2). Interestingly, the function of miRNAs, which can regulate large numbers of transcripts, is globally suppressed in mouse oocytes (Ma et al, 2010; Suh et al, 2010). Whether this suppression is conserved in the worm germ line is presently unknown. However, it is possible that in the absence of miRNA function the regulation could be mediated by RBPs such as GLD-1, which through a degenerate RNA motif interacts with, and potentially regulates, a large part of the germline transcriptome.
Quantitative prediction of RBP association with and regulation of mRNAs
Considering both the affinities that individual 7-mer GBMs have for GLD-1 and their multiplicative effect on GLD-1 binding, we can largely predict the strength of GLD-1 interactions with its targets. Although this prediction may be influenced by other RBPs, RNA structure, and/or other regulatory motifs, our predictor is a good representation of GLD-1 mRNA interactions. Remarkably, addition of a weak GBM is sufficient to impose GLD-1 regulation on an otherwise unregulated mRNA and converting this motif by a single nucleotide change to a strong GBM increased the association with, and translational repression by, GLD-1. Therefore, the extent of mRNA regulation by GLD-1 may, at least in some cases, be estimated based on the combined ‘strength' of GBMs, though regulation of many targets will likely be influenced by additional factors. Thus, this study is an important step towards predicting the outcome of GLD-1-mediated regulation, and we envisage that similar analysis may be useful for modelling post-transcriptional regulatory networks mediated by other RBPs and their combinations.
Materials and methods
GLD-1 IP and isolation of co-immunoprecipitated RNA
GLD-1 IP was performed as described (Biedermann et al, 2009). Wild-type (N2) or gld-1(q485); ozIs2 [gld-1∷gfp∷flag] (Schumacher et al, 2005) worms were synchronized and harvested as young adults. To identify GLD-1-associated transcripts, two independent IP strategies were performed: (1) comparison of anti-FLAG IP to anti-MYC IP on the gld-1(q485); ozIs2 [gld-1∷gfp∷flag] worm extract, (2) comparison of anti-FLAG IP on the gld-1(q485); ozIs2 [gld-1∷gfp∷flag] worm extract to anti-FLAG IP on wild-type extract. All IPs were performed in triplicate (total=12 IPs). Antibodies: mouse anti-FLAG M2 (Sigma) or mouse anti-Myc (9E10). RNA was eluted from beads using TRIzol (Invitrogen) following the manufacturer's instructions. Precipitation efficiency was enhanced by addition of 10 μg total RNA from mouse brain (Stratagene) to each IP sample. RNA was also isolated from an aliquot of each input. For IPs analysed by deep sequencing, anti-FLAG M2 agarose (Sigma) was used on both N2 and gld-1∷gfp∷flag extracts. Elution of RNP complexes was performed using FLAG peptide (Sigma) followed by RNA extraction with TRIzol (mouse RNA was not added). All RNA pellets were re-suspended in 20 μl DEPC-treated water.
Hybridization of co-immunoprecipitated RNA to 3′ gene expression analysis arrays
The samples (5 μg of each input and one-third of each IP) were amplified once, subsequently labelled, and hybridized according to the ‘GeneChip Expression Analysis Technical Manual' (Affymetrix). In total, 15 μg of labelled aRNA (amplified RNA) was fragmented and used for hybridization to the Affymetrix C. elegans 3′ gene expression array. Microarray sample preparation, hybridization, and scanning were performed in FMI genomics facility.
Analysis of 3′ gene expression array data
All the CEL files were processed in R (http://www.r-project.org) (Gentleman and Ihaka, 1996) using bioconductor (Gentleman et al, 2004). The data were background corrected with gcRMA (Wu et al, 2004) but not quantile normalized. Array annotation information was downloaded from wormbase (affy_oligo_mapping.gz) and gene sequence names were used to relate probe sets to genes. In cases where multiple probe sets existed for one gene, we selected the one with the highest expression in the input sample. The resulting table was used to create all the contrasts by averaging the three replicates and subtracting respective control IP from GLD-1 IP. Only sufficiently detected transcripts (input >5.5) were retained (Supplementary Dataset S1). Sequence data for 5′UTRs, CDSs, and 3′UTRs were downloaded from wormmart (WS195) and for every gene, the transcript with the longest annotated 3′UTR, of at least 9 nts, was used as a representative. This set was used for all downstream motif analyses (Supplementary Dataset S2). Searches for k-mer occurrences were performed with various perl scripts. MEME (Bailey and Elkan, 1994) motif finder was performed on the average IP-enriched (greater than three-fold) transcripts using the parameters ‘-w 7 –nsites 700' and an order 4 Markov background which was created with fasta_get_markov using all 3′UTRs. For the weblogo, the resulting motifs from the motif finder were extended by two nucleotides on each side. All linear regressions throughout the manuscript were performed in R using the function lm.
Analysis of co-immunoprecipitated RNA by Solexa next generation sequencing
In total, 100 ηg of RNA from each IP was used for the generation of the cDNA library, prepared essentially following the ‘preparing samples for mRNA sequencing' protocol (Illumina), but omitting the oligo dT purification. Sequencing was performed on an Illumina GA2 instrument. Annotation and quantification of the deep sequencing data were performed as described (Emmerth et al, 2010) using the ce6 C. elegans genome assembly (www.genome.ucsc.edu) and an annotation database (tRNA, rRNA, snRNA, snoRNA, miRNA, and mRNA) including wormbase transcript sequences. To robustly estimate IP enrichments (comparing IP and control), we required at least 10 tags in both samples resulting in 2080 transcripts used for the comparison with the array data.
Competitive-binding assays
Purification of recombinant GLD-1 was performed as described in Ryder et al (2004). Competitions were performed as described in Ryder et al (2004) with the following modifications. Binding experiments were performed in 50 mM Tris pH 8.0, 100 mM NaCl, 0.1 mg/ml tRNA, and 0.1% IGEPAL CA-630. Following equilibration, displacement of the labelled TGE RNA was measured by fluorescence polarization. Each sample was measured five times, and the average of each measurement was plotted as a function of competitor RNA concentration. The data were then fit to a sigmoidal dose response function to determine the IC50,apparent. Reported IC50,apparent values are the average±s.d. of at least three independent replicates.
Biotin-RNA pulldown assay
Biotin-RNA pulldown assays were performed as described (Lee and Schedl, 2001; Biedermann et al, 2009). For details of RNA sequences used as well as GBMs present in each 3′UTR and their mutations, see Supplementary Tables S6 and S7.
Transgenic constructs and strains created by microparticle bombardment
Plasmids carrying the pie-1 promoter (pCG142), the GFP:H2B fusion (pCM1.35), the tbb-2 3′UTR (pCM1.36), the unc-119 (+) gene (pCG150), and the transgenic line JM2252 containing the glp-1 wild-type 3′UTR, were kindly provided by Geraldine Seydoux (Merritt et al, 2008). Various additional wild-type and mutant 3′UTR fusions were constructed using the MultiSite Gateway Three Fragment Vector Construction Kit (Invitrogen). PCR products containing the relevant 3′UTR and flanking attB3 and attB2 sequences were cloned into pDONRP2RP3 entry vector. The resulting 3′ entry clones were recombined (LR reaction) with pCG142, pCM1.35, and pCG150 to create the final pie-1 promoter∷GFP∷H2B∷3′UTR fusion. All 3′UTRs contained sequences from the STOP codon up to 30–80 bp downstream from the poly A site. GBMs present in each 3′UTR and their corresponding mutations are shown in Supplementary Table S6. Each destination construct was transformed into unc-119(ed3) worms by microparticle bombardment (Praitis et al, 2001). Two or more independent lines were obtained for all constructs (Supplementary Table S8). Lines showed either stable (most worms maintain expression for many generations) or transient (in which germline expression got silenced over time) expression. All transgenic lines were grown at 25°C to maintain transgene expression.
Transgenic constructs and strains created by MosSCI
Constructs were generated using the Multisite Gateway cloning system (Invitrogen). The middle entry clone pCM1.35 was modified by mutating the XbaI site within the GFP, and the second and third codons of GFP were mutated to form an XbaI site. The MODC PEST sequence from pd1EGFP-N1 was cloned into the new XbaI site in GFP forming an N terminal PEST fusion (pBMF2.7). All of the wild-type and mutated 3′UTR fusions as well as the mex-5 promoter plasmid were amplified from genomic DNA (for oligos, see Supplementary Table S10) and inserted into entry vectors pDONRP4P1R for the mex-5 promoter and pDONRP2RP3 for the various 3′UTRs. All 3′UTRs contained sequences from the STOP codon up to 20–40 bp downstream from the poly A site. The resulting 3′ entry clones were recombined (LR reaction) with plasmids carrying the mex-5 promoter (pCS210), the PEST:GFP:H2B fusion (pBMF2.7), and the chromosome II integration sites together with the unc-119 (+) gene (pCFJ150) to create the final mex-5 pro∷PEST∷GFP∷H2B∷3′UTR fusion. Transgenic strains were then created by injecting these plasmids into EG4322 (ttTi5605 II; unc-119(ed3) III) worms http://sites.google.com/site/jorgensenmossci/protocols (Frokjaer-Jensen et al, 2008). For details of lines, see Supplementary Tables S6 and S9.
Microscopy
A Zeiss AxioImager Z1 microscope equipped with an Axiocam MRm REV 2 CCD camera was used to capture all images. If possible adult gonad images were obtained in one section, and if not, in two parts which were later merged. Images were acquired and normalized in the linear mode of the Axiovision software (Zeiss) and then exported into Photoshop CS3 and processed in an identical manner.
Relative quantification of GFP intensity in images
GFP intensity was plotted over a segmented line (thickness 35) from the distal tip to the bend of each gonad arm using ImageJ. The background mean grey value of each image was also recorded. For analysis and normalization of all expression values an R-based script was used, which is available upon request. Briefly, values from each gonad were binned into 50 groups, the background value subtracted from each 50, a ‘distal mean' calculated for the first 5% of values and this distal mean used to normalize the GFP intensity values across each gonad. Average values were then calculated for all gonads belonging to the same genotype. Error bars in figures show s.e.m.
Relative quantification of mRNA abundance by RT–PCR
Worms were synchronized by bleaching, harvested as young adults, and worm pellets frozen in liquid N2. Pellets were re-suspended in 10 volumes TRIzol (Invitrogen) and grinded in a mortar and pestle under liquid N2. RT reactions were performed with ImProm II Reverse transcription system (Promega) using 850 ng of isolated RNA and oligo dT. In all, 1/20th of cDNA was used in real-time PCR with SYBR GREENER mix (Invitrogen) and the ABI 7500 Fast sequence detection system (Applied Biosystems). At least one primer in each pair was specific for an exon–exon junction. Standard curves for every primer pair were generated using a serial dilution of cDNA obtained from pooled RT reactions using 1 μg RNA from the tbb-2 (+mut GBM) lines, and were used to derive the amount of each transcript in the different lines. All values for a particular transcript were normalized first to elt-2 mRNA expression and then the mean of the tbb-2+ mut GBM values. Error bars show the s.e.m. for three biological replicates from two independent lines per genotype. Primer sequences are available upon request.
Accession numbers
Array data are deposited at the Gene Expression omnibus (GEO). Accession number GSE21591.
Supplementary Material
Acknowledgments
We are grateful to Iskra Katic and Sandra Pauli for technical assistance, Laurent Gelman for discussion on GFP intensity quantification and CGC for strains. We also thank Helder Ferreira, Michael Stadler, and Dirk Schubeler for comments on the manuscript. JEW was supported by a long-term EMBO Postdoctoral Fellowship and by a National Ataxia Foundation fellowship award. Work in the laboratory of SPR is supported by National Institutes of Health Grant GM081422. This work was partly sponsored by a Swiss National Science Foundation (SNF) grant to RC. The Friedrich Miescher Institute is supported by the Novartis Research Foundation.
Author contributions: JEW designed and performed most experiments, analysed the data, and co-wrote the paper; DG designed and performed computational experiments, analysed the data, and co-wrote the paper; MS performed biotin-RNA pulldown assays and some GLD-1IPs; BMF and SPR designed, performed, and analysed the competition binding experiments; EW designed the secondary structure constructs; RC designed and supervised experiments and co-wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2: 28–36 [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW III, Bulyk ML (2006) Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol 24: 1429–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedermann B, Hotz HR, Ciosk R (2010) The quaking family of RNA-binding proteins: coordinators of the cell cycle and differentiation. Cell Cycle 9: 1929–1933 [DOI] [PubMed] [Google Scholar]
- Biedermann B, Wright J, Senften M, Kalchhauser I, Sarathy G, Lee MH, Ciosk R (2009) Translational repression of cyclin E prevents precocious mitosis and embryonic gene activation during C. elegans meiosis. Dev Cell 17: 355–364 [DOI] [PubMed] [Google Scholar]
- Ciosk R, DePalma M, Priess JR (2006) Translational regulators maintain totipotency in the Caenorhabditis elegans germline. Science (New York, NY) 311: 851–853 [DOI] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G (2009) RNA and disease. Cell 136: 777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerth S, Schober H, Gaidatzis D, Roloff T, Jacobeit K, Buhler M (2010) Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev Cell 18: 102–113 [DOI] [PubMed] [Google Scholar]
- Francis R, Barton MK, Kimble J, Schedl T (1995a) gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139: 579–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis R, Maine E, Schedl T (1995b) Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139: 607–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM (2008) Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40: 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galarneau A, Richard S (2005) Target RNA motif and target mRNAs of the Quaking STAR protein. Nat Struct Mol Biol 12: 691–698 [DOI] [PubMed] [Google Scholar]
- Galarneau A, Richard S (2009) The STAR RNA binding proteins GLD-1, QKI, SAM68 and SLM-2 bind bipartite RNA motifs. BMC Mol Biol 10: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman R, Ihaka R (1996) R: a language for data analysis and graphics. J Comput Graph Stat 5: 299–314 [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G et al. (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5: R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber AP, Herschlag D, Brown PO (2004) Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol 2: E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B, Hirsh D (1999) Receptor-mediated endocytosis in the Caenorhabditis elegans oocyte. Mol Biol Cell 10: 4311–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbeisen RE, Galgano A, Scherrer T, Gerber AP (2008) Post-transcriptional gene regulation: from genome-wide studies to principles. Cell Mol Life Sci 65: 798–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israeli D, Nir R, Volk T (2007) Dissection of the target specificity of the RNA-binding protein HOW reveals dpp mRNA as a novel HOW target. Development 134: 2107–2114 [DOI] [PubMed] [Google Scholar]
- Jan E, Motzny CK, Graves LE, Goodwin EB (1999) The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J 18: 258–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Francis R, Schedl T (1996) GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev Biol 180: 165–183 [DOI] [PubMed] [Google Scholar]
- Jones AR, Schedl T (1995) Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev 9: 1491–1504 [DOI] [PubMed] [Google Scholar]
- Keene JD (2007) RNA regulons: coordination of post-transcriptional events. Nat Rev Genet 8: 533–543 [DOI] [PubMed] [Google Scholar]
- Kim MY, Hur J, Jeong S (2009) Emerging roles of RNA and RNA-binding protein network in cancer cells. BMB Rep 42: 125–130 [DOI] [PubMed] [Google Scholar]
- Lee MH, Schedl T (2001) Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev 15: 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MH, Schedl T (2004) Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev 18: 1047–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB (2008) HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Luyten I, Bottomley MJ, Messias AC, Houngninou-Molango S, Sprangers R, Zanier K, Kramer A, Sattler M (2001) Structural basis for recognition of the intron branch site RNA by splicing factor 1. Science (New York, NY) 294: 1098–1102 [DOI] [PubMed] [Google Scholar]
- Lukong KE, Chang KW, Khandjian EW, Richard S (2008) RNA-binding proteins in human genetic disease. Trends Genet 24: 416–425 [DOI] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G (2007) RNA-binding proteins: modular design for efficient function. Nat Rev 8: 479–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Flemr M, Stein P, Berninger P, Malik R, Zavolan M, Svoboda P, Schultz RM (2010) MicroRNA activity is suppressed in mouse oocytes. Curr Biol 20: 265–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin VA, Evans TC (2003) Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development 130: 2623–2632 [DOI] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G (2008) 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol 18: 1476–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootz D, Ho DM, Hunter CP (2004) The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development 131: 3263–3272 [DOI] [PubMed] [Google Scholar]
- Praitis V, Casey E, Collar D, Austin J (2001) Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Chan ET, Pena Castillo L, Chaudhry S, Talukder S, Blencowe BJ, Morris Q, Hughes TR (2009) Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol 27: 667–670 [DOI] [PubMed] [Google Scholar]
- Ryder SP, Frater LA, Abramovitz DL, Goodwin EB, Williamson JR (2004) RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat Struct Mol Biol 11: 20–28 [DOI] [PubMed] [Google Scholar]
- Schumacher B, Hanazawa M, Lee MH, Nayak S, Volkmann K, Hofmann R, Hengartner M, Schedl T, Gartner A (2005) Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell 120: 357–368 [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG (2009) Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136: 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM (2005) Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123: 1133–1146 [DOI] [PubMed] [Google Scholar]
- Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R (2010) MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol 20: 271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum SA, Carson CC, Lager PJ, Keene JD (2000) Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci USA 97: 14085–14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J (2008) Ribonucleoprotein complexes in neurologic diseases. Curr Opin Neurobiol 18: 516–523 [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB (2003) CLIP identifies Nova-regulated RNA networks in the brain. Science (New York, NY) 302: 1212–1215 [DOI] [PubMed] [Google Scholar]
- Vernet C, Artzt K (1997) STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet 13: 479–484 [DOI] [PubMed] [Google Scholar]
- Volk T, Israeli D, Nir R, Toledano-Katchalski H (2008) Tissue development and RNA control: ‘HOW' is it coordinated? Trends Genet 24: 94–101 [DOI] [PubMed] [Google Scholar]
- Wang X, Zhao Y, Wong K, Ehlers P, Kohara Y, Jones SJ, Marra MA, Holt RA, Moerman DG, Hansen D (2009) Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics 10: 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Irizarry RA, Gentleman R, Martinez-Murillo F, Spencer F (2004) A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc 99: 909 [Google Scholar]
- Xu L, Paulsen J, Yoo Y, Goodwin EB, Strome S (2001) Caenorhabditis elegans MES-3 is a target of GLD-1 and functions epigenetically in germline development. Genetics 159: 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.