miR-605 joins p53 network to form a p53:miR-605:Mdm2 positive feedback loop in response to stress
Mdm2, a transcriptional target of p53, promotes p53 degradation in a well-established negative feedback loop. Here, microRNA miR-605 is found to break this loop: being induced by p53 in response to stress, and downregulating Mdm2 to promote robust p53 expression.
Keywords: apoptosis, gene expression, Mdm2, miRNA, p53
Abstract
In cancers with wild-type (WT) p53 status, the function of p53 is inhibited through direct interaction with Mdm2 oncoprotein, a negative feedback loop to limit the function of p53. In response to cellular stress, p53 escapes the p53:Mdm2 negative feedback to accumulate rapidly to induce cell cycle arrest and apoptosis. We demonstrate herein that an microRNA miR-605 is a new component in the p53 gene network, being transcriptionally activated by p53 and post-transcriptionally repressing Mdm2. Activation of p53 upregulated miR-605 via interacting with the promoter region of the gene. Overexpression of miR-605 directly decreased Mdm2 expression at the post-transcriptional level but indirectly increased the transcriptional activity of p53 on miR-34a via downregulating Mdm2; knockdown of miR-605 did the opposite. Mdm2 inhibitor upregulated expression of both miR-34a and miR-605, which was mitigated by p53 inhibitor. miR-605 preferentially induced apoptosis in WT p53-expressing cells, an effect abolished by p53 inhibition. These results indicate that miR-605 acts to interrupt p53:Mdm2 interaction to create a positive feedback loop aiding rapid accumulation of p53 to facilitate its function in response to stress.
Introduction
In ∼50% of human cancers, the gene (TP53) encoding p53 protein, a DNA damage responsive transcription factor that affects diverse cellular processes including transcription, DNA synthesis and repair, cell cycle arrest, senescence and apoptosis, is mutated rendering its tumour suppressor activity inactivated. In the remaining cancers with wild-type (WT) p53 status, its function is inhibited through direct interaction with the human murine double minute 2 (Mdm2) oncoprotein; p53 activates the transcription of Mdm2, which in turns targets p53 for degradation and transcription repression, therefore creating a negative feedback loop to limit the function of p53. Mdm2 is often present at high levels in tumours with WT p53. This autoregulatory feedback loop constitutes the core module of a network of regulatory interactions activated under cellular stresses including DNA damage, hypoxia and oncogene activation. In normal cells, the activity of p53 proteins is kept low by Mdm2, that is Mdm2 negatively regulates the stability of p53 and the transcription of TP53 (Momand et al, 1992; Haupt et al, 1997; Kubbutat et al, 1997; Eischen and Lozano, 2009). In the case of DNA damage, p53:Mdm2 interaction is weakened that allows rapid activation of the p53-mediated pathways leading to cell cycle arrest or apoptosis in case of excessive damage (Barak et al, 1993; Harris and Levine, 2005). Blocking the p53:Mdm2 interaction to reactivate the p53 function is a promising anti-cancer strategy. However, recent findings have shown this network to be more complex than previously envisioned and our current understanding of this network is incomplete. In particular, it remained vague how p53 can escape the p53:Mdm2 negative feedback loop to accumulate rapidly in the cell in response to stresses. There may be a ‘third' player that can operate to break the p53:Mdm2 negative feedback loop. This study was designed to exploit this issue.
Results and discussion
Mdm2 as a target of miR-605 for post-transcriptional repression
Recent studies have identified an miRNA miR-34 as a direct transcriptional target of p53 (He et al, 2007; Raver-Shapira et al, 2007; Tarasov et al, 2007), indicating important participation of miRNAs in the p53 gene network. In an initial effort to explore the potential role of miRNAs in regulating the p53:Mdm2 negative feedback loop, we studied miR-34 (including miR-34a, miR-34b and miR-34c). However, our computational analysis and experimental results excluded the role of miR-34 in targeting p53 and Mdm2, consistent with the view that miR-34 acts as a downstream component mediating the function of p53 (He et al, 2007; Raver-Shapira et al, 2007; Tarasov et al, 2007).
With theoretical prediction of miRNA target genes and of putative cis-acting elements for transcription regulation of miRNA genes (Supplementary data), we identified miR-605 as a top-priority miRNA for our study for it has multiple potential binding sites (12) in the 3′UTR of Mdm2 mRNA (Supplementary Figure S1) and the promoter region of its host gene PRKG1 contains two high-scored p53 consensus binding sites (half-sites) within 1125 bp upstream its transcription start site (Supplementary Figure S2; The precursor sequence of miR-605 is located within the intron 2 of PRKG1 at the genomic location of 52 729 339–52 729 421[+] in the chromosome 10) (Lee et al, 2009). These analyses suggest a possibility of miR-605 to regulate Mdm2 expression at the post-transcriptional level on one hand and to be regulated in its own expression at the transcriptional level by p53 on the other hand.
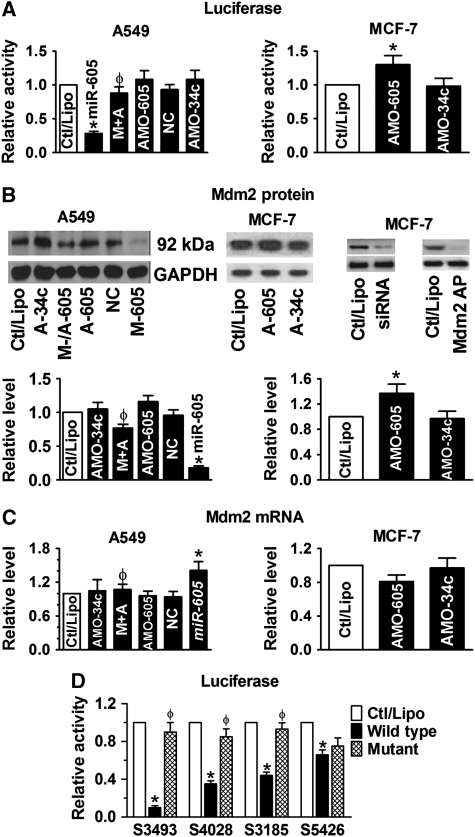
We set up to examine our hypothesis using the following approaches. We first studied the interactions between miR-605 and Mdm2 by luciferase reporter gene assay. Transfection of synthetic mature miR-605 produced ∼75% reduction of the luciferase activity of the pMIR-REPORT™ luciferase vector carrying the 3′UTR of Mdm2 to replace that of the luciferase gene in A549 human lung cancer cells that express a low level of endogenous miR-605 (Figure 1A; Supplementary Figure S3). And this effect was effectively reversed by co-transfection with its antisense oligoribonucleotides (AMO-605). Additionally, application of AMO-605 alone in MCF-7 human breast cancer cells that express a higher level of authentic miR-605 (MCF-7:A549=14:1; Supplementary Figure S3) enhanced the luciferase activity, indicating the tonic repression of the luciferase gene expression by endogenous miR-605. The repressive effect of miR-605 was then confirmed at the protein level using western blot analysis; miR-605 decreased the protein level of Mdm2 by ∼80% in A549 cells (Figure 1B). Similar results were observed in MCF-7 cells (Supplementary Figure S4). Moreover, AMO-605 alone was able to increase the protein level of Mdm2 in MCF-7 cells, presumably by knocking down the endogenous miR-605 in these cells. Quantitative real-time RT–PCR (qPCR) analyses revealed that Mdm2 mRNA level changed in the opposite direction: transfection of miR-605 caused ∼30% upregulation of Mdm2 mRNA (Figure 1C), which was abolished by co-transfection with AMO-605. This mRNA-increasing effect is likely ascribed to the net outcome between the enhanced p53 activity by miR-605 (see below) and an miR-605-induced degradation process as miR-605 has a high percentage of complementarity to Mdm2 (Supplementary Figure S1). As negative controls, neither miR-34a nor a scrambled miRNA caused any significant changes on luciferase activity, and protein and mRNA levels of Mdm2 (Figure 1A–C). Important to note is that in A549 cells that expressed a low level of endogenous miR-605, AMO-605 alone failed to alter Mdm2 expression determined by luciferase assay, western blot and qPCR (Figure 1A–C), whereas in MCF-7 cells that express a higher level of miR-605, AMO-605 was able to produce derepression of Mdm2.
Figure 1.
Mdm2 as a target of miR-605 for post-transcriptional repression. (A) Role of miR-605 in repressing Mdm2, determined by luciferase activity assays with the pMIR-REPORT luciferase miRNA expression reporter vector carrying the 3′UTR of miR-605 target gene Mdm2 in A549 (left) and MCF-7 (right) cells. AMO-605 and AMO-34a: antisense nucleotides to miR-605 and miR-34a (used as a control), respectively; M+A: co-transfection of miR-605 and AMO-605; NC: negative control with scrambled miRNAs. Control cells were mock treated with lipofectamine 2000 (Ctl/Lipo). *P<0.05 versus Ctl/Lipo; φP<0.05 versus miR-605 alone; n=6 for each group. (B) Western blot analysis revealing repression of Mdm2 protein by miR-605 in A549 cells (left) and in MCF-7 cells (right). M-/A-605: co-transfection of miR-605 and AMO-605; A-605: AMO-605; A-34a: AMO-34a; M-605: miR-605; NC: negative control with scrambled miRNAs; Mdm2 AP: antigenic peptide; siRNA: small interference RNA to Mdm2. *P<0.05 versus Ctl/Lipo; φP<0.05 versus miR-605 alone; n=4 for each group. (C) Effect of miR-605 on Mdm2 mRNA level in A549 (upper) and MCF-7 (lower). M+A: co-transfection of miR-605 and AMO-605; NC: negative control with scrambled miRNAs. *P<0.05 versus Ctl/Lipo; φP<0.05 versus miR-605 alone; n=4 for each group. (D) Comparison of the effects of miR-605 on luciferase activities in A549 cells generated by the pMIR-REPORT vectors each carrying a wild-type or mutated fragment of Mdm2 3′UTR that spans a single predicted binding site for miR-605. Note the differential strengths of inhibition luciferase activities by miR-605 among the different fragments and the abolishment or abrogation of the effects with the mutant fragments. S3493 indicates the fragment spanning the predicted binding sites starting at location of 3493 of Mdm2 mRNA (Supplementary Figure S1); the same meaning applies to other three labels. *P<0.05 versus Ctl/Lipo; φP<0.05 wild-type versus mutant; n=3 for each group.
To obtain confirmative evidence for that the effects observed above are ascribed to the specific interaction between miR-605 and the binding sites for miR-605 in the 3′UTR of Mdm2 mRNA, we introduced nucleotide-substitution mutations to 4 out of 12 predicted binding motifs corresponding to the seed site of miR-605 to disrupt base paring between miR-605:Mdm2. The binding site S3493 has the highest degree of complementarity among the 12 predicted binding motifs (Supplementary Figure S1), S4028 has a medium degree of complementarity, and S3185 and S5426 have the lowest degrees of complementarity. As expected, among the four sites, the site with the highest degree of complementarity (S3493) conferred the greatest decrease in luciferase activities and mutation of the site (MT S3493) nearly abolished the effect of miR-605 (Figure 1D). By comparison, the other three sites demonstrated weaker effects.
miR-605 as a target of p53 for transcriptional activation
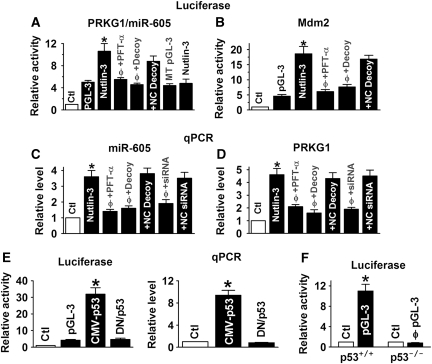
An intronic miRNA is expected to be co-expressed with its host gene. Our experiments indeed confirmed this relationship between miR-605 and its host gene PRKG1. In the first set of experiments, we constructed a pGL-3 luciferase vector by inserting a 2.5-kb fragment spanning the two putative half-sites for p53 upstream the host gene PRKG1 into the promoter region of the luciferase gene for studying transcription regulation. Treatment with the Mdm2 inhibitor nutlin-3 (1 μM) to activate p53 caused a significant increase in luciferase activity in MCF-7 cells and this effect was abolished by pretreatment with the p53 inhibitor pifithrin-α (PFT-α; 30 μM; Figure 2A). Co-transfection with either p53 decoy oligodeoxyribonucleotide (ODN) to sequestrate p53 (Morishita et al, 1997; Gao et al, 2006) prevented the nutlin-3-induced luciferase activity, but their respective negative control constructs failed to do so. On the other hand, when the two p53-binding sites were mutated, nutlin-3 failed to affect the luciferase activity. Similar stimulation and silencing of luciferase activities by nutlin-3 and p53 inhibition, respectively, were observed with a bona fide p53-responsive promoter—the first intron of the Mdm2 gene (Figure 2B). Consistent with the above results, nutlin-3 elevated the level of miR-605 (Figure 2C), along with the upregulation of PRKG1 mRNA (Figure 2D), which was reversed when co-treated with PFT-α, p53 decoy or siRNA to knockdown p53.
Figure 2.
miR-605 as a target of p53 for transcriptional activation. (A) Role of p53 in transactivating the expression of the miR-605-host gene PRKG1, assessed by luciferase activity assays with the pGL-3 luciferase reporter vector carrying the promoter region of PRKG1 in MCF-7 cells. Except for control and pGL-3 Base alone, all experiments were preformed in the presence of nutlin-3. Nutlin-3 (1 μM): the Mdm2 inhibitor to activate p53; PFT-α: the p53 inhibitor pifithrin-α (30 μM); Decoy: the decoy oligodeoxyribonucleotide fragment containing a perfect p53-binding site to sequestrate endogenous p53; NC decoy: negative control decoy containing mutated nucleotides in the core region for p53 binding; MT pGL-3: the luciferase reporter vector carrying the mutated p53-binding sites in the promoter region of PRKG1. *P<0.05 versus pGL-3 base; φP<0.05 versus nutlin-3 alone; n=4 for each group. (B) Role of p53 in transactivating the expression of the Mdm2 gene, assessed by luciferase activity assays with the pGL-3 luciferase reporter vector carrying the promoter region (the first intron) of Mdm2 in MCF-7 cells. *P<0.05 versus pGL-3 base; φP<0.05 versus nutlin-3 alone; n=4 for each group. (C, D) Role of p53 in regulating miR-605 and PRKG1 levels, respectively, evaluated by real-time qRT–PCR in MCF-7 cells. siRNA, small interference RNA to p53; NC siRNA, negative control siRNA. *P<0.05 versus pGL-3 base; φP<0.05 versus nutlin-3 alone; n=4 for each group. (E) Luciferase (left) and qPCR (right) evaluation of the effects of p53 overexpression by transient transfection of CMV plasmid carrying wild-type p53 (CMV-p53) in p53-null H1299 cells. The construct carrying the dominant-negative p53 (DN/p53) was used as a control. *P<0.05 versus pGL-3 base; n=4 for each group. (F) Comparison of luciferase activities generated by the pGL-3 luciferase reporter vector carrying the promoter region of PRKG1 between isogenic colorectal carcinoma HCT116 cells (p53+/+ and p53−/−). *P<0.05 versus Ctl; φP<0.05 versus p53+/+; n=3 for each group.
In the second set of experiments, we assessed the ability of p53 overexpression by the CMV plasmid carrying p53 cDNA in the p53-null human lung adenocarcinoma cancer cell line H1299. As shown in Figure 2E, transfection of p53 vector induced a robust increase in PRKG1/miR-605 promoter activity (left) and miR-605 level (right), which was not seen with the plasmid carrying the dominant-negative p53.
Next, we compared the luciferase activities between isogenic colorectal carcinoma HCT116 cells (p53+/+ and p53−/−), and we found substantially higher luciferase activities in p53+/+ than in p53−/− HCT116 cells (Figure 2F).
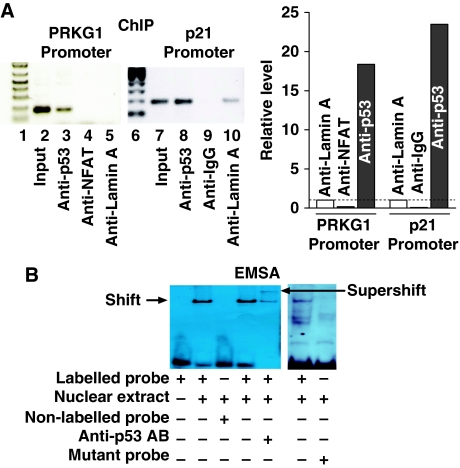
Finally, we verified the physical binding of p53 to the cis-elements in the promoter region of the miR-605 host gene PRKG1 using chromatin immunoprecipitation (ChIP) in conjunction with qPCR semi-quantification (Figure 3A) and electrophoresis mobility shift assay (EMSA) with supershift (Figure 3B), using anti-p53 antibody to indicate the specificity of interactions.
Figure 3.
Binding of p53 to the promoter region of the miR-605 host gene. (A) Chromatin immunoprecipitation (ChIP) assay for the presence of p53 on its cis-acting elements in the PRKG1 promoter region in MCF-7 cells. p21 promoter region was used as a positive control, NFAT, IgG and lamin A were sued as negative controls. The recovered DNA by anti-p53 to indicate the relative level of p53:PRKG1 or p53:p21 binding was measured by qPCR following ChIP and is expressed as fold changes over anti-lamin A band. Lanes 1 and 6: DNA marker; lane 2: the PCR-amplified band using PRKG1 primer pairs and human genomic DNA without immunoprecipitation; lane 3: the PCR-amplified band using PRKG1 primer pairs and human genomic DNA after p53 antibody immunoprecipitation as a template; lane 4: the PCR-amplified band using PRKG1 primer pairs and human genomic DNA after NFAT antibody immunoprecipitation as a template for negative control; lane 5: the PCR-amplified band using PRKG1 primer pairs and human genomic DNA after lamin-A antibody immunoprecipitation as a template for negative control; lane 7: the PCR-amplified band using p21 primer pairs and human genomic DNA without immunoprecipitation; lane 8: the PCR-amplified band using p21 primer pairs and human genomic DNA after p53 antibody immunoprecipitation as a template for positive control; lane 9: the PCR-amplified band using p21 primer pairs and human genomic DNA after IgG antibody immunoprecipitation as a template for negative control; lane 10: the PCR-amplified band using p21 primer pairs and human genomic DNA after lamin-A antibody immunoprecipitation as a template for negative control. (B) Electrophoresis mobility shift assay (EMSA) to indicate the in vitro physical binding of p53 to the cis-elements in the PRKG1 promoter region using the nuclear extract from MCF-7 cells. A positive control experiment was performed using the p53 decoy containing the canonical p53-binding sites (right panel). Note the shift band representing protein–DNA binding that is abolished with excessive non-labelled probes or with the mutant fragment. Also note the supershift band picked up by the p53 antibody (anti-p53 AB) representing p53–cis-elements binding. The probe was a digoxigenin (DIG)-labelled oligonucleotides fragment containing p53-binding site.
While these above data clearly indicate the transcriptional control of miR-605 by p53, we sought to clarify whether miR-605 can produce post-transcriptional influence on p53 gene (TP53). Our experiments excluded this possibility by showing that miR-605 failed to alter the luciferase activity elicited by the pMIR-REPORT vector carrying the 3′UTR of TP53 (Supplementary Figure S5). This is consistent with the lack of binding sites for miR-605 in the TP53 gene. By comparison, miR-214, which has its binding site in the 3′UTR of TP53, robustly decreased the luciferase activity as expected.
These findings prompted us to deduce that miR-605 might serve as a regulatory molecule of p53:Mdm2 interaction, being transactivated by p53 on one hand and repressing Mdm2 on the other hand. And joining of miR-605 is able to switch the p53:Mdm2 negative feedback loop to a positive feedback loop favouring p53 function. If this is true, then we should be able to see the following. First, there is an increase in p53 transcriptional activity while miR-605 is overexpressed to repress Mdm2. Second, disruption of p53:Mdm2 interaction should be able to upregulate miR-605 expression. The results from our following experiments indeed support these notions.
miR-605 enhances transactivation activity of p53
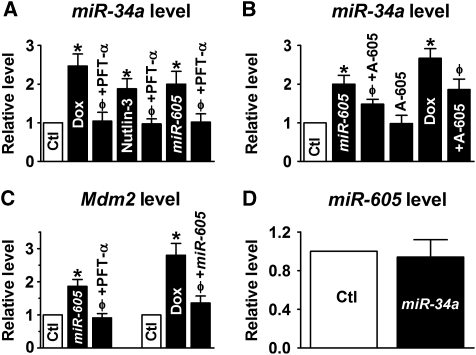
Both MCF-7 and A549 cells are known to express WT p53. It was anticipated that by repressing Mdm2, miR-605 should be able to enhance p53 transcriptional activity. To test this notion, we assessed the effect of p53 activation by doxorubicin (Dox), transfection of miR-605, and by inhibition of Mdm2 by nutlin-3 (Vassilev, 2007; Shangary and Wang, 2009) on the expression level of miR-34a, an established transcriptional target of p53 (Chang et al, 2007; He et al, 2007; Raver-Shapira et al, 2007; Tarasov et al, 2007; Welch et al, 2007). As shown in Figure 4A, these manoeuvers all increased miR34a transcript and strikingly, their effects were all antagonized by the inhibitor of p53 (PFT-α). Further, the effect of miR-605 was also reversed by AMO-605 (Figure 4B). Moreover, application of AMO-605 alone did not significantly alter miR-34a level, but in the presence of Dox this treatment reduced miR-34a level (Figure 4B). Increase in p53 transcriptional activity by miR-605 was further evidenced by elevated mRNA levels of p53 target gene p21/CDKN1A (Supplementary Figure S6A). The results suggest that the upregulation of miR-34a induced by miR-605, as well as by p53 activation strategies (by DNA-damaging agent and Mdm2 inhibition), was mediated by p53. In other words, miR-605 acts through enhancing p53 activity; without p53 activation, miR-605 was unable to influence miR-34a expression. miR-605 did not significantly change p53 protein level, neither did nutlin-3 in MCF-7 cells 48 h after the treatments (Supplementary Figure S6B). However, when measured 24 h after treatment with nutlin-3 or miR-605 upregulation of p53 proton levels were observed in p53+/+ HCT116 (Supplementary Figure S6C), possibly due to reduced E3 ubiquitin ligase activity of Mdm2 (Haupt et al, 1997; Honda et al, 1997).
Figure 4.
miR-605 enhances transactivation activity of p53. (A, B) Regulation of miR-34a expression by p53 in MCF-7 cells, determined by real-time RT–PCR. Ctl/Lipo: cells mock-treated with lipofectamine 2000 for control; miR-605: cells transfected with miR-605 (10 nM); +AMO-605 (10 nM): co-transfection of AMO-605 (10 nM) with miR-605 (10 nM); AMO-605: treatment with AMO-605 alone; A-605: AMO-605; Dox: p53 activator doxorubicin (0.5 μM); PFT-α: p53 inhibitor pifithrin-α (30 μM); Nutlin-3 (1 μM): Mdm2 inhibitor. *P<0.05 versus Ctl; φP<0.05 versus Dox, Nutlin-3 or miR-605 alone; n=5 for each group. (C) Regulation of Mdm2 mRNA level in MCF-7 cells. *P<0.05 versus Ctl; φP<0.05 versus miR-605 or Dox; n=4 for each group. (D) Effects of miR-34a on miR-605 expression in MCF-7 cells (n=3 for each group).
We have shown in Figure 1C that miR-605 increased the transcript level of Mdm2. To see whether this was attributable to the enhancement of p53 activity, we pretreated MCF-7 cells with PFT-α before miR-605 transfection. As depicted in Figure 4C, without PFT-α, miR-605 was able to elevate Mdm2 mRNA level as expected, but in the presence of PFT-α, miR-605 lost the ability to do so. On the other hand, cells incubated with Dox demonstrated increased transcription of Mdm2, but pretreatment with AMO-605 abrogated the effect (Figure 4C).
While we have shown that miR-605 was able to upregulate miR-34a (Figure 4A), we found that miR-34a failed to alter expression of miR-605 (Figure 4D), consistent with the view that miR-34a does not target p53 (Chang et al, 2007; He et al, 2007; Raver-Shapira et al, 2007; Tarasov et al, 2007; Welch et al, 2007) nor Mdm2.
Implication of miR-605 participation in the p53:Mdm2 signalling pathway
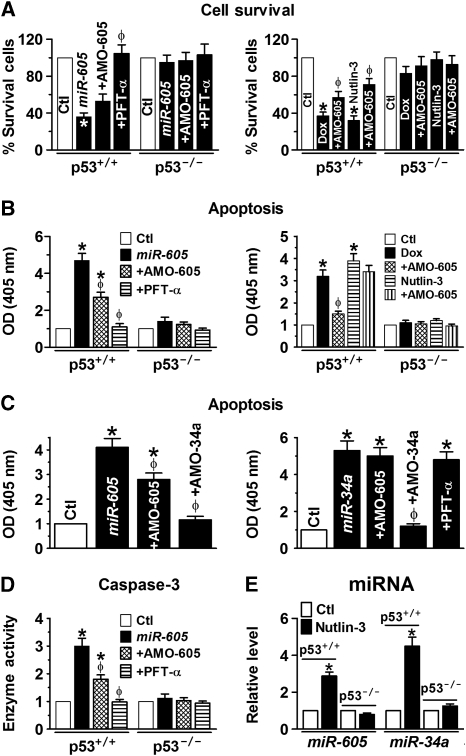
In order to see whether the participation of miR-605 in the p53 gene network can be translated into corresponding cellular functions, we carried out the following experiments. We first demonstrated that like Dox or nutlin-3, miR-605 preferentially induced apoptotic cell death in p53+/+ over p53−/− HCT116 cells (Figure 5A and B). Intriguingly, co-transfection of AMO-605 only partially reversed, whereas treatment of PFT-α was able to abolish, the apoptosis promoting of miR-605 (Figure 5A and B, left panels) in p53+/+ HCT116 cells, despite that AMO-605 was able to completely wipe out miR-605 (Supplementary Figure S7). Further, pretreatment with AMO-605 alone to knockdown endogenous miR-605 rendered a significantly less extent of apoptosis induced by either Dox or nutlin-3 (Figure 5A and B, right panels). Strikingly, AMO-34a prevented the apoptosis-promoting effect of miR-605 more efficiently than AMO-605 though it had no effect on miR-605 level (Figure 5C; Supplementary Figure S7). By comparison, p53−/− HCT116 cells did not respond to miR-605. On the other hand, AMO-605 failed to affect the apoptotic effect of miR-34a, neither did PFT-α (Figure 5C). The apoptotic cell death was further verified by measurement of caspase-3 activities (Figure 5D). We subsequently confirmed that in response to p53 activation by nutlin-3, miR-605 level was significantly upregulated only in p53+/+ HCT116 cells, and so was miR-34a level (Figure 5E), further verifying that both of these two miRNAs are target genes of p53 for transcriptional regulation.
Figure 5.
Role of miR-605 in regulating cell survival and apoptosis. (A) Effects of miR-605 on cell viability in isogenic colorectal carcinoma HCT116 cells (p53+/+ and p53−/−), as determined by MTT methods. Note that AMO-605 only partially reversed the inhibitory effects of miR-605 (left panel) and of Dox (0.5 μM) or Nutlin-3 (3 μM; right panel) on cell survival; whereas PFT-α completely reversed the effects. miR-605 and Dox failed to produce any significant effect in p53−/− HCT116 cells. Ctl: cells mock-treated with lipofectamine 2000; +AMO-605: co-transfection of AMO-605 and miR-605; AMO-605: transfection of AMO-605 alone; +PFT-α: application of PFT-α after transfection with miR-605. *P<0.05 versus Ctl; φP<0.05 versus miR-605 or Dox or Nutlin-3 alone; n=4 for each group. (B) Effects of miR-605 on apoptotic cell death in p53+/+ and p53−/− HCT116 cells, as determined by optical density (OD at 405 nm) obtained by ELISA. *P<0.05 versus Ctl; φP<0.05 versus miR-605 or Dox or Nutlin-3 alone; n=4 for each group. (C) Effects of AMO-605 and AMO-34a on apoptosis induced by miR-605 (left panel) and miR-34a (right panel) in p53+/+ HCT116 cells. *P<0.05 versus Ctl; φP<0.05 versus miR-605 or miR-34a alone; n=4 for each group. (D) Mean OD values (n=4) indicating caspase-3 activities, determined by cellular caspase-3 activity assay. *P<0.05 versus Ctl; φP<0.05 versus miR-605 alone; n=5 for each group. (E) Effects of nutlin-3 (3 μM) on miR-605 level in p53+/+ HCT116 cells as compared with those in p53−/− HCT116 cells. Note that nutlin-3 is able to upregulate both miR-605 and miR-34a only in p53+/+ HCT116 cells.*P<0.05 versus Ctl; φP<0.05 versus miR-605 alone; n=3 for each group.
Qualitatively, the same effects of miR-605 on apoptosis were also observed in WT p53-expressing MCF-7 cells than of p53-null human breast cancer MDA-MB-436 cells (Lacroix and Leclercq, 2004; Keimling and Wiesmüller, 2009) (Supplementary Figure S8).
In summary, we have demonstrated here that (1) miR-605 post-transcriptionally represses Mdm2; (2) miR-605 is transcriptionally activated by p53; (3) activation of p53 stimulates miR-605 expression and vice versa overexpression of miR-605 increases the transactivation activity of p53 resulting in upregulation of miR-34a; (4) inhibition of Mdm2 function upregulates both miR-605 and miR-34a expression; and (5) miR-605 preferentially induces cell death in WT p53-expressing cells.
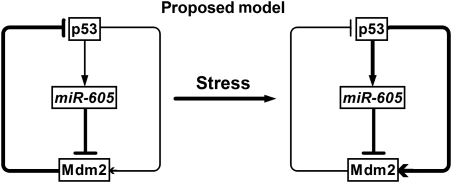
Together with all these results, it appears that miR-605 is able to disrupt p53:Mdm2 interaction similar to, but distinct from, a DNA-damaging agent or an Mdm2 inhibitor. DNA-damaging drugs activate p53 that in turn transcriptionally promotes Mdm2 expression. Mdm2 inhibitors bind to Mdm2 without changing Mdm2 level directly but increasing Mdm2 expression indirectly through releasing p53 from p53:Mdm2 interaction. By comparison, miR-605 represses Mdm2 expression without directly affecting p53, and this repression relieves the inhibitory effect of Mdm2 on p53 leading to an indirect increase in p53 activity and expression that then further enhances miR-605 expression. In this way, the participation of miR-605 creates a positive feedback loop that helps p53 escape the p53:Mdm2 negative feedback loop (Figure 6).
Figure 6.
A proposed model describing the participation of miR-605 to create a positive feedback loop that helps p53 escape the p53:Mdm2 negative feedback loop. The thickness of the lines indicates the strength of actions. In response to cellular stress, miR-605 expression is enhanced due to initial p53 activation and this miR-605 upregulation then feeds back to increase the transactivation activity of p53 by repressing Mdm2 to relieve the inhibitory effects of Mdm2 on p53.
p53 is at the centre of a complex molecular network regulating diverse physiological responses to cancer-related stresses. Activated p53 in response to DNA damage or oncogene activation induces cell cycle arrest, which can be transient or permanent (senescence), or promotes apoptosis in cases where the damage is too severe. The Mdm2 oncoprotein is a primary regulator of p53, mediating p53 control via ubiquitin-dependent proteasomal degradation. Activation of p53 in tumour cells by inhibiting its physical interaction with Mdm2 has been in the focus of cancer drug discovery (Dickens et al, 2010; Wade et al, 2010; Weber, 2010). Our study herein revealed that miR-605 is a new component in the p53 gene network, serving as an on-and-off switch between the p53:Mdm2 negative feedback loop and the p53:miR-605:Mdm2 positive feedback loop. In response to cellular stresses, miR-605 expression is enhanced due to initial p53 activation and this elevated miR-605 level subsequently feeds back to increase the transactivation activity of p53 by repressing Mdm2 to relieve the inhibitory effects of Mdm2 on p53. Through this positive feedback loop, miR-605 is able to aid rapid accumulation of p53 to induce cell cycle arrest or apoptosis. It appears that miR-605 participates in the p53 signalling network differently from miR-34a though both are downstream transcriptional targets of p53. miR-34a is known to mediate the cellular function of p53 to regulate cell cycle progression, cellular senescence and apoptosis by targeting its downstream genes (Chang et al, 2007; Raver-Shapira et al, 2007; Welch et al, 2007; Rokhlin et al, 2008; Hermeking, 2010). By comparison, miR-605 seems not to be directly involved in the cellular function of p53; rather it only acts to facilitate the function of p53 by enhancing the transactivation activity through inhibiting Mdm2. The data that AMO-34a is able to mitigate the apoptosis-promoting effects of miR-605 but AMO-605 failed to affect the proapoptotic action of miR-34a (Figure 5C), plus the partial reversal of effect of miR-605 by AMO-605, provides a strong evidence. In other words, miR-605 itself is unable to induce apoptosis in the absence of p53 activation; this is supported by the results shown in Figure 5A and B that miR-605 failed to induce apoptosis when p53 function was inhibited. Our findings therefore revealed that in addition to mediate the cellular function of p53 like the miR-34 family, miRNAs, like miR-605, could also act to fine tune or amplify the activity of p53 to enhance its cellular function.
It should be noted, however, that we could not draw a conclusion from the present study as to whether the p53–miR-605–Mdm2 feedback signalling loop exists in other cell types neither could we draw a notion on what physiological role this pathway may have under in vivo conditions. Nonetheless, this study unraveled a novel molecular mechanism for fine tuning the dynamic shift of the p53 signalling network to favour or disfavour the function of p53 on demand, laying the groundwork for future studies on the implications of this type p53–miRNA interaction.
Materials and methods
Cell culture
Human breast cancer cells line MCF-7 with functional p53 status and MDA-MB-436 with non-functional mutant p53 (Lacroix and Leclercq, 2004; Keimling and Wiesmüller, 2009), human lung cancer cell line A549 (WT p53), the p53-null human lung adenocarcinoma cancer cell line H1299 used in this study was purchased from American Type Culture Collection (ATCC, Manassas, VA) and cultured in Dulbecco's Modified Eagle Medium or in RPMI 1640 supplemented with 10% FCS. Isogenic colorectal carcinoma HCT116 cells with or without p53 expression (p53+/+ and p53−/−, respectively) were gracefully provided by Dr Bert Vogelstein (The Howard Hughes Medical Institute and The Johns Hopkins Oncology Center, Baltimore, MD) (Bunz et al, 1998; Ericson et al, 2010).
Quantitative real-time RT–PCR analysis
The mirVana™ qRT–PCR miRNA Detection Kit (Ambion) was used in conjunction with TaqMan real-time PCR for quantification of miRNAs in our study, as previously described in detail (Xiao et al, 2007; Yang et al, 2007; Luo et al, 2008). Total RNA samples were isolated from various cell lines with Ambion's mirVana miRNA Isolation Kit and mRNA samples from various human tissues were purchased from Ambion. Reactions contained mirVana qRT–PCR primer sets specific for human, rat and mouse miR-605, and for TP53 and Mdm2 were supplied by ABI. qRT–PCR was performed on a thermocycler ABI Prism® 7500 fast (Applied Biosystems) for 40 cycles. Fold variations in expression of an mRNA between RNA samples were calculated.
Western blot analysis
The cytosolic protein samples were extracted 48 h after treatments with the procedures essentially the same as described in detail elsewhere (Xiao et al, 2007; Yang et al, 2007; Luo et al, 2008). The protein content was determined by BCA Protein Assay Kit using bovine serum albumin as the standard. Protein sample (∼50 μg) was fractionated by SDS–PAGE (12% polyacrylamide gels) and transferred to PVDF membrane (Millipore, Bedford, MA). The sample was incubated overnight at 4°C with the primary antibodies in 1:200. Affinity purified rabbit polyclonal anti-p53 (mAb421; Enzo Life Sciences) and anti-Mdm2 antibodies (Cell Signaling) were used as the primary antibodies. Other primary antibodies (the antibodies to NFAT, IgG and lamin A) were all purchased from Santa Cruz Biotech. Next day, the membrane was incubated with secondary antibodies (Molecular Probes) diluted in PBS for 2 h at room temperature. Finally, the membrane was rinsed with PBS before scanning using the Infrared Imaging System (LI-COR Biosciences). Anti-GAPDH and anti-α-acting antibodies were used as internal controls for equal input of protein samples from cancer cell lines and from cardiac preparations, respectively. Western blot bands were quantified using QuantityOne software by measuring the band intensity (Area × OD) for each group and normalizing to GAPDH or α-acting. The final results are expressed as fold changes by normalizing the data to the control values.
Synthesis of miRNAs and anti-miRNA antisense inhibitors
miR-605 (5′-UAAAUCCCAUGGUGCCUUCUCCU-3′) and its antisense oligoribonucleotide fragment (AMO-605: 3′-AUUUAGGGUACCACGGAAGAGGA-5′) were synthesized by Integrated DNA Technologies Inc (IDT, Coralville, IA). Five nucleotides or deoxynucleotides at both ends of the antisense molecules were locked (the ribose ring is constrained by a methylene bridge between the 2′-O- and the 4′-C atoms). Additionally, a scrambled RNA was used as negative control; sense: 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense: 5′-ACGGACACGUUCGGAGAATT-3′ (Xiao et al, 2007; Yang et al, 2007; Luo et al, 2008).
Construction of luciferase—miRNA-target site fusion plasmids
To construct reporter vectors bearing miRNA-target sites, we synthesized the 3′UTR of miR-605 target gene Mdm2, or the 3′UTR of TP53, by PCR amplification. The sense and antisense strands of the oligonucleotides were annealed by adding 2 μg of each oligonucleotides to 46 μl of annealing solution (100 mM K-acetate, 30 mM HEPES-KOH, pH 7.4 and 2 mM Mg-acetate) and incubated at 90°C for 5 min and then at 37°C for 1 h. The annealed oligonucleotides were digested with HindIII and SpeI. These inserts were ligated into HindIII and SpeI sites in the pMIR-REPORT luciferase miRNA expression reporter vector (Ambion) (Lin et al, 2007; Xiao et al, 2007; Yang et al, 2007; Luo et al, 2008).
Construction of PRKG1 promoter—luciferase fusion plasmids
A fragment (2861-AAAGTTCTTTCAGCTGTAATATACTATGACATGTTTTTTAATAATTCAGTGTTTATTCTTCTACCAGCATGTCTTTTCTCAAGCTTTCCTTGTGGTCTAA-2960) containing the three putative p53-binding motifs upstream the cGMP-dependent protein kinase 1, β-isozyme (PRKG1, the host gene of the intronic miR-605) was synthesized by Invitrogen. The fragment was then used as a template for PCR amplification with 5′-GGGGTACCAAAGTTCTTTCAGCTGTA-3′ and 5′-CCGCTCGAGTTAGACCACAAGGAAAGC-3′ as forward and reverse primers, respectively, using PCR Advantage and Advantage-GC genomic polymerase mixes (Clontech). The PCR product was subcloned into luciferase-containing pGL-3-Basic (Promega) vector, as described elsewhere (Lin et al, 2007). The integrity and orientation of all constructs were confirmed by restriction endonuclease analysis and DNA sequencing.
Preparation of decoy ODNs
Single-stranded phosphorothioate decoy ODNs were synthesized by IDT (Gao et al, 2006). The ODN was washed in 70% ethanol, dried and dissolved in sterilized Tris–EDTA buffer (10 mM Tris+1 mM EDTA). The supernatant was purified using Micro Bio-spin30 columns (Bio-Rad, Hercules, CA) and quantified by spectrophotometry. The double-stranded decoy ODN was then prepared by annealing complementary single-stranded ODN by heating to 95°C for 10 min followed by cooling to room temperature slowly over 2 h. The decoy ODN was prepared at a concentration of 50 μM in saline. The sequence of p53 decoy ODN is sense 5′-AGACATGCCTAGACATGCCT-3′ and antisense 5′-TCTGTACGGATCTGTACGGA-3′ (The consensus core sequence is bold and underlined). For negative control, a mutant decoy ODN was also used: sense 5′-AGAtgcaaaTAGAtgcaaaT-3′ and antisense 5′-TCTgacgtgATCTacgtggA-3′. Nucleotide-substitution mutations to p53 decoy ODN were made using PCR-based methods.
Electrophoresis mobility shift assay
EMSA was performed with the digoxigenin (DIG) Gel Shift kit (Roche, Mannheim, Germany), as described previously (Gao et al, 2006; Lin et al, 2007). Varying amounts of nuclear protein extracts from MCF-7 cells were incubated with DIG-labelled double-stranded oligonucleotides containing the putative p53 cis-element from the PRGK1 promoter. For competition experiments, 100-fold excess of unlabelled double-stranded p53 consensus oligonucleotides, and for supershift experiments, 1 μg of p53 antibody (Cell Signaling), were added to the reaction. The generated chemiluminescent signals were recorded on the X-ray film. The sequences of the p53 cis-element examined are GTTTATTCTTCTACCAGCATGTCTTTTCTC; and the mutant p53 cis-element are GTTTATTCTTCTACCAGtcgcTCTTTTCTC (the replaced nucleotides are indicated by lower case letters.
Chromatin immunoprecipitation assay
ChIP assays were conducted with the EZ ChIP kit according to the manufacturer's instructions (Upstate Cell Signaling Solutions, Lake Placid, NY) (Lin et al, 2007). Briefly, MCF-7 cells were grown to subconfluency, washed and fixed in 1% formaldehyde for 10 min to crosslink nucleoprotein complexes and scraped in phosphate-buffered saline containing protease inhibitor cocktail. Pelleted cells were then lysed and sonicated in detergent lysis buffer. Sheared DNA–protein complexes were immunoprecipitated by incubating overnight the lysates with 2 μg antibodies against p53 or lamin A (as a control). Protein A/G Plus beads (Santa Cruz) were used, and after extensive washing, crosslinks were removed at 65oC over overnight in an elution buffer (1% SDS, 0.1 M NaHCO3). The DNA was isolated using the QIAquick PCR purification kit (Qiagen) and the presence of the PRKG1 (the host gene of the intronic miR-605) promoter or of the p21 promoter as a positive control was analysed by PCR amplification using 10% of purified DNA. The primers used for the PRKG1 promoter sequence containing p53 cis-elements were 5′-GAGACCCTCACTCAGACGCA-3′ (forward) and 5′-CAGCACTAGGCTTCGGGTGG-3′ (reverse). The primers used for the p21 promoter sequence containing p53 cis-elements were from 5′-GAGCCTCCCTCCATCCCTATG-3′ (forward) and 5′-CTCCCAGCACACACTCACAC-3′ (reverse). The PCR products were analysed by gel electrophoreses on an 8% non-denaturing polyacrylamide gel and subsequent ethidium bromide staining.
p53 overexpression and knockdown
To overexpress p53 or dominant-negative p53, the plasmids contained in the p53 Dominant-Negative Vector Set (Clontech) were transiently transfected into H1299 cells using lipofectamine 2000 reagent. p53 siRNA and Mdm2 siRNA, used to knockdown endogenous p53 and Mdm2, respectively, were purchased from Santa Cruz Biotech.
Transfection procedures
Cell lines or neonatal rat ventricular myocytes were transfected with a final concentration of 10 nM of miR-605 and/or 5 nM of AMO-605, and negative control miRNAs or AMOs with lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Forty-eight hours after transfection, cells were used for luciferase assay or were collected for total RNA or protein purification.
For decoy ODNs, cells were washed with serum-free medium once and then incubated with 50 μl fresh fetal bovine serum (FBS)-free medium. Decoy ODNs and lipofectamine 2000 (0.25 μl, Invitrogen, Carlsbad, CA) were separately mixed with 25 μl of Opti-MEM® I Reduced Serum Medium (Gibco, Grand Island, NY) for 5 min. Then, the two mixtures were combined and incubated for 20 min at RT. The lipofectamine:ODNs mixture was added dropwise to the cells and incubated at 37°C for 5 h. Subsequently, 25 μl fresh medium containing 30% FBS was added to the well and the cells were maintained in the culture until use.
Luciferase activity assay
For luciferase assay involving miRNA function, cells were transfected with the pMIR-REPORT luciferase miRNA expression reporter vector carrying the 3′UTR of miR-605 target genes (Lin et al, 2007; Xiao et al, 2007; Yang et al, 2007; Luo et al, 2008).
For luciferase assay involving analysis of promoter activities of the miR-605 host gene PRKG1, MCF-7 cells were simultaneously transfected with 10 nM miR-605, 1 μg PGL-3-target DNA (firefly luciferase vector) and 0.1 μg PRL-TK (TK-driven Renilla luciferase expression vector) with lipofectamine 2000. Following transfection (48 h), luciferase activities were measured with a dual luciferase reporter assay kit (Promega) on a luminometer (Lumat LB9507). For all experiments, transfection took place 24 h after starvation of cells in serum-free medium.
MTT assay for cell viability
Cell Proliferation Kit I (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT); Roche Molecular Biochemicals, Laval, PQ, Canada) was used to quantify surviving cells from oxidative stress, as previously described in detail (Xu et al, 2007; Lu et al, 2009).
Enzyme-linked immunosorbent assay (ELISA)
The Cell Death Detection ELISA kit (Roche Molecular Biochemicals) was employed to quantify DNA fragmentation on the basis of antibody detection of free histone and fragmented DNA, as detailed elsewhere (Gao et al, 2006; Xu et al, 2007; Lu et al, 2009).
Assays for caspase-3
The procedures for the assay for cellular caspase-3 activities have been previously described in detail elsewhere (Han et al, 2001). Briefly, after experimental treatment, cells were centrifuged (1000 g, 4°C, 10 min) and lysed. Sample was centrifuged at 10 000 g (4°C, 10 min), and the supernatant (10 μl) was incubated with 10 μl of substrate (2 mM Ac-DEVDpNA) in 80 μl of assay buffer at 37°C. Absorbance at 405 nm was read at several time points until readings reached plateau levels. Optical density changes relative to zero-time values were obtained as a measure of enzyme activity.
Terminal deoxyribonucleotide transferase-mediated dUTP nick end labelling (TUNEL)
DNA fragmentation of individual cells was detected with the In Situ Cell Death Detection kit, Fluorescein (Roche Molecular Biochemicals), as detailed previously (Han et al, 2001; Wang et al, 2002). TUNEL staining was analysed with a confocal microscope (Zeiss).
Data analysis
Group data are expressed as mean±s.e.m. Statistical comparisons among multiple groups were performed by analysis of variance (ANOVA). If significant effects were indicated by ANOVA, a t-test with Bonferroni correction or a Dunnett's test was used to evaluate the significance of differences between individual means. Otherwise, baseline and drug data were compared by paired Student's t-test and age-matched comparisons between control and treatment were done by unpaired Student's t-test. Group comparisons for AF incidence were performed using χ2-test. A two-tailed P<0.05 was taken to indicate a statistically significant difference.
miRNA (miR-605 and miR-214) target gene was predicted using miRBase and cis-elements for transcription factor binding sites were analysed with MatInspector V2.2 (Genomatix).
Supplementary Material
Acknowledgments
We thank XiaoFan Yang and Huizhen Wang for their excellent technical supports. This work was supported by the Canadian Institute of Health Research and Fonds de la Recherche de l'Institut de Cardiologie de Montreal.
Author contributions: Zhiguo Wang generated the hypothesis for this study, provided overall direction and supervision on the project, and wrote the manuscript; Jiening Xiao is the primary investigator who coordinated and designed the study and performed experiments and data analyses on luciferase, western blot and cell death; Huixian Lin is the second primary investigator involving every aspect of the study being responsible for characterizing the promoter region of miR-605 gene subcloning; and Xiaobin Luo and Xiaoyan Luo contributed to the western blot analysis and qRT–PCR measurements.
Footnotes
The authors declare that they have no conflict of interest.
References
- Barak Y, Juven T, Haffner R, Oren M (1993) mdm2 expression is induced by wild type p53 activity. EMBO J 12: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, Sedivy JM, Kinzler KW, Vogelstein B (1998) Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282: 1497–1501 [DOI] [PubMed] [Google Scholar]
- Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT (2007) Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 26: 745–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickens MP, Fitzgerald R, Fischer PM (2010) Small-molecule inhibitors of MDM2 as new anticancer therapeutics. Semin Cancer Biol 20: 10–18 [DOI] [PubMed] [Google Scholar]
- Eischen CM, Lozano G (2009) p53 and MDM2: antagonists or partners in crime? Cancer Cell 15: 161–162 [DOI] [PubMed] [Google Scholar]
- Ericson K, Gan C, Cheong I, Rago C, Samuels Y, Velculescu VE, Kinzler KW, Huso DL, Vogelstein B, Papadopoulos N (2010) Genetic inactivation of AKT1, AKT2, and PDPK1 in human colorectal cancer cells clarifies their roles in tumor growth regulation. Proc Natl Acad Sci USA 107: 2598–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Xiao J, Yang B, Sun Q, Lin H, Bai Y, Yang L, Wang H, Wang Z (2006) A single decoy oligodeoxynucleotides targeting multiple oncoproteins produces strong anti-cancer effects. Mol Pharmacol 70: 1621–1629 [DOI] [PubMed] [Google Scholar]
- Han H, Wang H, Long H, Nattel S, Wang Z (2001) Oxidative preconditioning and apoptosis in L-cells: roles of protein kinase B and mitogen-activated protein kinases. J Biol Chem 276: 26357–26364 [DOI] [PubMed] [Google Scholar]
- Harris SL, Levine AJ (2005) The p53 pathway: positive and negative feedback loops. Oncogene 24: 2899–2908 [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M (1997) Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299 [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ (2007) A microRNA component of the p53 tumour suppressor network. Nature 447: 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H (2010) The miR-34 family in cancer and apoptosis. Cell Death Differ 17: 193–199 [DOI] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 420: 25–27 [DOI] [PubMed] [Google Scholar]
- Keimling M, Wiesmüller L (2009) DNA double-strand break repair activities in mammary epithelial cells––influence of endogenous p53 variants. Carcinogenesis 30: 1260–1268 [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH (1997) Regulation of p53 stability by Mdm2. Nature 387: 299–303 [DOI] [PubMed] [Google Scholar]
- Lacroix M, Leclercq G (2004) Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat 83: 249–289 [DOI] [PubMed] [Google Scholar]
- Lee I, Ajay SS, Yook JI, Kim HS, Hong SH, Kim NH, Dhanasekaran SM, Chinnaiyan AM, Athey BD (2009) New class of microRNA targets containing simultaneous 5′-UTR and 3′-UTR interaction sites. Genome Res 19: 1175–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Xiao J, Luo X, Xu C, Gao H, Wang H, Yang B, Wang Z (2007) Overexpression HERG K+ channel gene mediates cell-growth signals on activation of oncoproteins Sp1 and NF-κB and inactivation of tumor suppressor Nkx3.1. J Cell Physiol 212: 137–147 [DOI] [PubMed] [Google Scholar]
- Lu Y, Xiao J, Lin H, Bai Y, Luo X, Wang Z, Yang B (2009) Complex antisense inhibitors offer a superior approach for microRNA research and therapy. Nucleic Acids Res 37: e24–e33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Lin H, Pan Z, Xiao J, Zhang Y, Lu Y, Yang B, Wang Z (2008) Overexpression of Sp1 and downregulation of miR-1/miR-133 activates re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. J Biol Chem 283: 20045–20052 [DOI] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ (1992) The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69: 1237–1245 [DOI] [PubMed] [Google Scholar]
- Morishita R, Sugimoto T, Aoki M, Kida I, Tomita N, Moriguchi A, Maeda K, Sawa Y, Kaneda Y, Higaki J, Ogihara T (1997) In vivo transfection of cis element ‘decoy' against nuclear factor-kappaB binding site prevents myocardial infarction. Nat Med 13: 894–899 [DOI] [PubMed] [Google Scholar]
- Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, Bentwich Z, Oren M (2007) Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell 26: 731–743 [DOI] [PubMed] [Google Scholar]
- Rokhlin OW, Scheinker VS, Taghiyev AF, Bumcrot D, Glover RA, Cohen MB (2008) MicroRNA-34 mediates AR-dependent p53-induced apoptosis in prostate cancer. Cancer Biol Ther 7: 1288–1296 [DOI] [PubMed] [Google Scholar]
- Shangary S, Wang S (2009) Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu Rev Pharmacol Toxicol 49: 223–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G, Hermeking H (2007) Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle 6: 1586–1593 [DOI] [PubMed] [Google Scholar]
- Vassilev LT (2007) MDM2 inhibitors for cancer therapy. Trends Mol Med 13: 23–31 [DOI] [PubMed] [Google Scholar]
- Wade M, Wang YV, Wahl GM (2010) The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol 20: 299–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang Y, Cao L, Han H, Wang J, Yang B, Nattel S, Wang Z (2002) HERG K+ channel: a regulator of tumor cell apoptosis and proliferation. Cancer Res 62: 4843–4848 [PubMed] [Google Scholar]
- Weber L (2010) Patented inhibitors of p53-Mdm2 interaction (2006–2008). Expert Opin Ther Pat 20: 179–191 [DOI] [PubMed] [Google Scholar]
- Welch C, Chen Y, Stallings RL (2007) MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene 26: 5017–5022 [DOI] [PubMed] [Google Scholar]
- Xiao J, Luo X, Lin H, Zhang Y, Lu Y, Wang N, Zhang Y, Yang B, Wang Z (2007) MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J Biol Chem 282: 12363–12367 [DOI] [PubMed] [Google Scholar]
- Xu C, Lu Y, Lin H, Xiao J, Wang H, Luo X, Li B, Wang Z, Yang B (2007) The muscle-specific microRNAs miR-1 and miR-133 produce opposing effects on apoptosis via targeting HSP60/HSP70 and caspase-9 in cardiomyocytes. J Cell Sci 120: 3045–3052 [DOI] [PubMed] [Google Scholar]
- Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, Zhang Y, Xu C, Bai Y, Wang H, Chen G, Wang Z (2007) The muscle-specific microRNA miR-1 causes cardiac arrhythmias by targeting GJA1 and KCNJ2 genes. Nat Med 13: 486–491 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.