Molecular mechanism of Ena/VASP-mediated actin-filament elongation
Ena/VASP proteins have important functions in actin-dependent processes. A model for the actin elongation activity of Ena/VASP based on the affinity and saturation state of WH2-domain-mediated actin monomer binding is presented.
Keywords: actin assembly, cell motility, formin, TIRF microscopy, WH2 motif
Abstract
Ena/VASP proteins are implicated in a variety of fundamental cellular processes including axon guidance and cell migration. In vitro, they enhance elongation of actin filaments, but at rates differing in nearly an order of magnitude according to species, raising questions about the molecular determinants of rate control. Chimeras from fast and slow elongating VASP proteins were generated and their ability to promote actin polymerization and to bind G-actin was assessed. By in vitro TIRF microscopy as well as thermodynamic and kinetic analyses, we show that the velocity of VASP-mediated filament elongation depends on G-actin recruitment by the WASP homology 2 motif. Comparison of the experimentally observed elongation rates with a quantitative mathematical model moreover revealed that Ena/VASP-mediated filament elongation displays a saturation dependence on the actin monomer concentration, implying that Ena/VASP proteins, independent of species, are fully saturated with actin in vivo and generally act as potent filament elongators. Moreover, our data showed that spontaneous addition of monomers does not occur during processive VASP-mediated filament elongation on surfaces, suggesting that most filament formation in cells is actively controlled.
Introduction
The precise control of actin-filament elongation in eukaryotic cells is fundamental to establish coordinated cell movement driven by the formation of protrusive structures like filopodia and lamellipodia, to assemble the contractile ring at the cleavage furrow during cell division and to coordinate endocytosis and phagocytosis (Chhabra and Higgs, 2007; Chesarone and Goode, 2009; Faix et al, 2009; Insall and Machesky, 2009). The only proteins known so far that directly enhance filament elongation by interaction with the growing barbed end and recruitment of monomeric actin for polymerization are formins and Ena/VASP proteins. Ena/VASP family members were previously shown to localize in a protrusion-dependent manner to lamellipodia tips (Rottner et al, 1999) and to influence the length of actin filaments as well as their apparent branching density within lamellipodia (Bear et al, 2002), and Listeria monocytogenes comet tails (Plastino et al, 2004). Ena/VASP proteins are required for the formation of filopodia in mammals and Dictyostelium (Schirenbeck et al, 2006; Applewhite et al, 2007; Dent et al, 2007) and were also shown to enhance the actin-driven propulsion of L. monocytogenes (Laurent et al, 1999; Loisel et al, 1999; Geese et al, 2002) as well as of beads coated with ActA (Samarin et al, 2003). Additionally, they are implicated in neuritogenesis and cortex development (Kwiatkowski et al, 2007, 2009) as well as in tumour development and progression (Hu et al, 2008; Philippar et al, 2008).
Ena/VASP proteins display a conserved tripartite architecture encompassing an N-terminal Ena/VASP homology 1 (EVH1) domain required for subcellular targeting followed by a central proline-rich domain implicated in recruitment of profilin–actin complexes (Jonckheere et al, 1999; Ferron et al, 2007), and a C-terminal EVH2 domain mediating tetramerization and interaction with monomeric and filamentous actin (Bachmann et al, 1999; Hüttelmeier et al, 1999; Breitsprecher et al, 2008). The G-actin-binding site (GAB) within the EVH2 domain displays close sequence homologies to WASP homology 2 (WH2) motifs, which are present in many actin regulators (Supplementary Figure S1; Paunola et al, 2002; Dominguez, 2007, 2009). In addition, the adjacent F-actin-binding site (FAB) was also proposed to posses WH2-like properties (Dominguez, 2007, 2009).
While both Ena/VASP proteins and formins accelerate actin filament barbed end elongation in vitro, the underlying molecular mechanisms are different (Faix and Grosse, 2006; Breitsprecher et al, 2008; Chesarone and Goode, 2009; Dominguez, 2010). Formin dimers remain processively associated with the growing filament barbed end by virtue of their dimeric FH2 domain while inserting thousands of actin monomers, thereby efficiently protecting the filament from heterodimeric capping proteins (CPs) (Zigmond et al, 2003; Harris et al, 2004; Schirenbeck et al, 2005; Kovar et al, 2006). Moreover, formin-mediated enhancement of filament elongation strictly depends on the recruitment of profilin–actin complexes by the adjacent proline-rich FH1 domain (Chang et al, 1997; Sagot et al, 2002; Kovar et al, 2006).
Surface-immobilized VASP captures growing barbed ends (Pasic et al, 2008) and, when clustered on the surface at sufficiently high density, can translocate processively with the growing filament end (Breitsprecher et al, 2008). Unlike formins, profilin appears dispensable for VASP-mediated filament elongation in vitro (Samarin et al, 2003; Schirenbeck et al, 2006; Breitsprecher et al, 2008). A notable property of VASP is the different influence of CPs when VASP is free in solution or clustered on the surface of a bead. Upon clustering, VASP triggers processive filament elongation at high concentrations of CP that block VASP-mediated filament elongation in solution (Breitsprecher et al, 2008). Collectively, this suggests that clustered VASP tetramers cooperate in tethering and elongating actin filaments at surfaces, such as on the cell membrane and the surface of pathogens such as L. monocytogenes (Laurent et al, 1999; Breitsprecher et al, 2008; Footer et al, 2008). Although the filament elongation activity of VASP could be attributed to its GAB and FAB motifs, the underlying general mechanisms of VASP-mediated actin assembly remained obscure, as human VASP (hVASP) showed a drastically reduced elongating activity in vitro when compared with the orthologue from the highly motile soil amoeba Dictyostelium discoideum (DdVASP) (Breitsprecher et al, 2008).
To dissect the molecular mechanism of Ena/VASP-mediated filament elongation, we employed a domain shuffling approach, replacing the GAB, FAB and their connecting linker region of hVASP by those of the fast-elongating DdVASP and by WH2 motifs from other actin-binding proteins. Our results allowed us to develop a quantitative mathematical model for an affinity based, WH2-domain-mediated actin assembly used by Ena/VASP proteins, whereby the filament elongation rate is correlated to the saturation of the GAB with actin monomers.
Results
VASP, Mena and EVL enhance filament elongation to similar extends
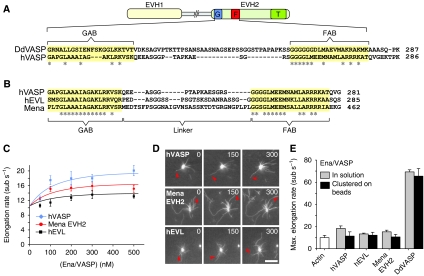
Previously, it was shown that hVASP only weakly accelerates actin elongation in vitro, whereas the Dictyostelium orthologue DdVASP enhanced the growth of single filaments about seven-fold (Breitsprecher et al, 2008). Mammalian cells express two additional Ena/VASP proteins, referred to as Ena (enabled) and EVL (Ena/VASP like), the latter of which is abundantly expressed in migrating neutrophils, suggesting that this particular paralogue might mediate faster filament elongation than hVASP. In search for the underlying cause of different filament elongation rates, we compared DdVASP and the three mammalian Ena/VASP proteins on the domain level and found that the lengths of the linkers separating the GAB and FAB motifs in hVASP, EVL, Mena and DdVASP differ considerably, encompassing 18, 25, 33 and 40 residues, respectively (Figure 1A and B). Recently, it was shown that the length of the linkers separating the three WH2 motifs in the protein Cobl was critical for its actin nucleation activity (Ahuja et al, 2007). Since models of VASP-mediated actin assembly propose that a GAB-bound actin monomer is handed over directly to the barbed end of the FAB-bound filament (Ferron et al, 2007; Breitsprecher et al, 2008; Dickinson, 2009), we assumed that the short 18 residues linker of hVASP might impair this transfer and hence cause the lower elongation activity compared with DdVASP. Whereas the GAB and FAB of DdVASP differ substantially from the mammalian Ena/VASP proteins, the sequences of GAB and FAB motifs of the latter are almost identical (Figure 1A and B; Supplementary Figure S1). This suggests that mammalian Ena/VASP proteins display comparable actin-binding properties, in turn making them well-suited candidates to investigate the effects of the linker lengths on the filament elongation rate.
Figure 1.
Effects of hVASP, Mena and hEVL on actin-filament elongation. (A) Domain organization of Ena/VASP proteins and sequence alignment of the corresponding GAB-linker-FAB region within the EVH2 domains of DdVASP and hVASP. The GAB and FAB are highlighted in yellow. Identical amino acids within these motifs are marked with an asterisk. G, GAB; F, FAB; T, tetramerization domain. (B) Sequence alignment of the GAB-linker-FAB region of hVASP, hEVL and Mena. The linker length differs in all three proteins. (C) Elongation rates of 1.3 μM actin (30% Oregon-Green (OG) labelled) in the presence of different concentrations of hVASP, Mena EVH2 and hEVL determined by single-filament TIRFM in TIRF buffer (see Materials and methods). (D) TIRFM micrographs of the assembly of 1.3 μM actin (30% OG labelled) on beads saturated with hVASP, Mena EVH2 and hEVL in the presence of 80 nM CP. Arrows indicate growing filaments. Time is indicated in seconds, scale=10 μm. (E) Comparison of the maximal elongation rates of 1.3 μM actin (30% OG labelled) for the three mammalian Ena/VASP paralogues and DdVASP determined by TIRFM in solution or immobilized on beads. Elongation rates are presented as mean values±s.e.m.
We employed TIRF microscopy to quantify the effects of purified Ena/VASP isoforms on single actin-filament elongation in vitro. Control actin filaments grew spontaneously from 1.3 μM G-actin (30% OG labelled), with an average rate of 10.5 subunits s−1 (sub s−1) in our assays. Similar to hVASP, the EVH2 domain from Mena and full-length EVL-bundled actin filaments and enhanced their elongation rate, both in solution and when clustered on beads in the presence of CP (Supplementary Movie 1). However, Mena and EVL again only slightly increased the elongation rate of actin filaments by ∼1.5-fold like hVASP (Figure 1C and D). Thus, despite different linkers, all three mammalian Ena/VASP paralogues possess low actin-filament elongation activities and mediate considerably slower elongation rates when compared with DdVASP in vitro (Figure 1E).
Domain shuffling of GAB and FAB motifs reveals the molecular requirement for fast filament elongation
Replacement of the GAB and FAB from hVASP (hGAB and hFAB) by the corresponding motifs from DdVASP (DdGAB and DdFAB) (Figure 2A) allowed hVASP to assemble actin filaments with significantly higher elongation rates when compared to the wild-type protein. Chimera hVASP DdGABFAB mediated virtually the same elongation rates as DdVASP, both in solution and when clustered on beads, enhancing filament elongation rates up to ∼70 sub s−1 (Figure 2B–D; Supplementary Movies 2 and 3). Almost identical results were obtained with hVASP DdGAB-L-FAB, additionally containing the entire linker region (L) of DdVASP, corroborating our previous finding that the linker region does not affect filament elongation (Figure 2C). Most notably, chimera hVASP DdGAB already enhanced the filament elongation rate up to 41 sub s−1 in solution and up to 49 sub s−1 on saturated beads, respectively, pointing towards a key role of actin monomer recruitment during VASP-mediated filament elongation (Supplementary Movies 2 and 3). In contrast, chimera hVASP DdFAB mediated only a moderate acceleration of the elongation rate up to 23.2 sub s−1 in solution and 19.2 sub s−1 on beads when compared with hVASP, suggesting that the contribution of the FAB motif to filament elongation is smaller than that of the GAB motif (Figure 2B–D; Supplementary Movie 4). The differential enhancement of the filament elongation rate by the VASP chimeras was confirmed by seeded pyrene actin polymerization assays (Supplementary Figure S2).
Figure 2.
Replacement of the GAB and FAB in hVASP with the corresponding DdVASP motifs markedly accelerates VASP-mediated filament elongation. (A) Scheme of hVASP chimeras bearing different domains of DdVASP. DdVASP is shown in colour and hVASP is shown in greyscale. G, GAB; F, FAB; T, tetramerization domain. (B) TIRFM micrographs of the assembly of 1.3 μM actin (30% OG labelled) in TIRF buffer containing 500 nM of the chimera are indicated. Time is indicated in seconds, scale=10 μm. (C) Elongation rates of the chimeras in solution in a concentration rage from 25 nM to 1 μM. (D) (Left) TIRFM micrographs of the assembly of 1.3 μM actin (30% OG labelled) in TIRF buffer in the presence of 200 nM CP on beads saturated with the hVASP chimeras are indicated. Time is shown in seconds, scale=10 μm. (Right) Plots of the lengths of individual filaments versus time yield filament elongation rates. Elongation rates are presented as mean values±s.e.m.
The GAB has a pivotal role in specifying Ena/VASP elongation properties
Since the transplantation of the DdGAB motif into the hVASP backbone was already sufficient to enhance actin-filament elongation 4–5-fold, we hypothesized that GAB-mediated actin monomer recruitment determines the filament elongation rates driven by Ena/VASP proteins. Therefore, we sought to analyse the affinities of DdGAB and hGAB peptides for G-actin under buffer conditions that were used in the TIRF experiments. Binding of GAB peptides to OG-actin monomers caused a 10–20% increase in OG-fluorescence, which was instrumental to detect the formation of GAB–actin complexes by fluorescence titrations. Determination of the equilibrium dissociation constants (KD) for DdGAB and hGAB peptides to latrunculin-A (LatA) sequestered Mg2+–ATP–actin in the presence of 50 mM KCl showed that their affinities towards G-actin differed by as much as ∼20-fold, yielding KD values of 0.63 and 10.3 μM, respectively (Figure 3A–C). Concomitant binding of LatA to actin had virtually no effect on the GAB–actin interaction (Figure 3C). Competition experiments with OG labelled and unlabelled actin in G-buffer revealed that both the DdGAB and hGAB peptides bound ∼1.7–2-fold stronger to OG-actin than to unlabelled actin (data not shown). The KD value determined for the DdEVH2–actin interaction was virtually identical to the KD value obtained for the DdGAB peptide, showing that the monomer affinity of the GAB motif is not changed within the VASP tetramer (Supplementary Figure S3). In addition to the equilibrium constants, we analysed the kinetics of actin monomer binding to the EVH2 domains of hVASP and DdVASP by stopped-flow analysis, yielding kon values of 53 μM−1 s−1 for the DdEVH2 domain and significantly lower rates of ∼12–18 μM−1 s−1 for the hEVH2 domain, respectively (Figure 3D).
Figure 3.
Actin-binding properties of the GAB motifs and EVH2 domains of hVASP and DdVASP. (A) Determination of the KD value of the DdGAB–actin interaction by fluorescence titration of 100 nM OG-actin (left) and 500 nM OG-actin (right) with the DdGAB peptide at the buffer conditions is indicated (for details see Materials and methods section). The solid lines represent calculated binding isotherms. (B) Determination of the KD value of the hGAB–actin interaction by fluorescence titration of 300 nM OG-actin (left) and 3 μM OG-actin (right) with the hGAB peptide at conditions as in (A). The solid lines represent calculated binding isotherms. (C) Summary of KD values determined from experiments similar as shown in (A, B) at the conditions indicated. KD values for the GAB–Ca2+–ATP–actin interaction are given in brackets. KD values obtained under conditions used in TIRF assays (50 mM KCl, Mg2+–ATP–actin) are shown in bold. (D) (Left) Kinetics of the DdEVH2–G-actin interaction. Time course of the binding of DdEVH2 to latA-sequestered, Mg2+–ATP–OG-actin was monitored by a stopped-flow apparatus at a final actin concentration of 2.5 μM actin and the DdEVH2 concentrations are indicated. Noisy curves represent experimental data, and solid lines are fits for a reversible, bimolecular reaction with KD=0.6 μM, yielding kon=53 μM−1 s−1. (Right) kon values of the interaction of the hEVH2 and DdEVH2 domain with OG-actin. Due to the low affinity of the hGAB, high actin concentrations had to be used (10 μM final), which resulted in more noisy signals (not shown) that could not be fitted as accurately as those for the higher-affinity DdEVH2.
As expected, the DdGAB peptide inhibited actin-filament nucleation in pyrene assays according to its relatively high affinity to monomers, but did not noticeably sequester actin monomers even at a high molar excess (Supplementary Figure S4), indicating that binding of the GAB to monomers does not interfere with filament elongation. This finding is in line with previous results showing that WH2 motifs display profilin-like properties during barbed end assembly, and that they rapidly dissociate from the monomer after its incorporation into the filament (Hertzog et al, 2002, 2004). Collectively, these results let us hypothesize that the faster filament elongation mediated by DdVASP compared with hVASP is mainly caused by a higher affinity for monomers, resulting in a significantly higher saturation of the VASP tetramer at low actin concentrations typically used in in vitro polymerization assays.
Replacement of the GAB of hVASP by other WH2 motifs reveals a saturation-based mechanism of VASP-mediated actin assembly
To further investigate the dependence of the filament elongation rates on the affinity of the G-actin-binding motif, we replaced the GAB in hVASP with foreign WH2 core motifs from WASP-interacting protein (WIP), which binds monomers with a high affinity in G-buffer (KD=0.16 μM), and the WH2 motif from thymosin β4 (Tβ4), which displays a much weaker G-actin-affinity (KD=3.1 μM) (Chereau et al, 2005). We found that the affinity of the WIP-WH2-peptide to Mg2+–ATP–actin in the presence of 50 mM KCl was rather high, yielding a KD value of 0.84 μM (Figure 4B). Unfortunately, Tβ4 binding to OG-actin did not result in a detectable change in fluorescence (data not shown) as previously reported for pyrene- or NBD-labelled actin (Hertzog et al, 2002). Notwithstanding, we noted that the KD value of the interaction of Mg2+–ATP–actin with full-length Tβ4 in the presence of 50 mM KCl was previously determined to be 1.4 μM (De La Cruz et al, 2000). Assuming that the affinity of the Tβ4–WH2 motif for actin under our polymerization conditions is about a factor 4 lower (as reported for G-buffer; Chereau et al, 2005), we estimated the KD for the Tβ4–WH2–actin interaction to be in the range of ∼5.6 μM.
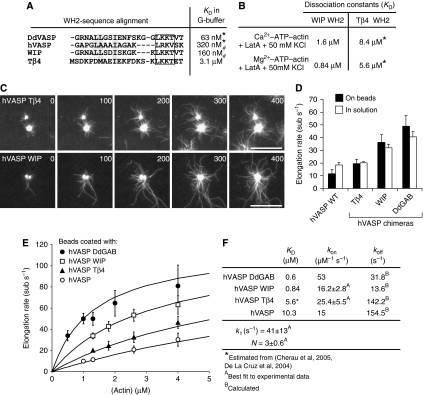
Figure 4.
Analysis of hVASP WH2 chimeras reveals VASP-mediated filament elongation is enhanced by saturable monomer binding to GAB sites. (A) Sequence alignment of the WH2 motifs is indicated. In the chimeric proteins, the GAB of hVASP was replaced by the WH2 motifs from Tβ4 and WIP. KD values of the actin–WH2 interaction in G-buffer where either determined in this study (*) or by Chereau et al (2005) (#). Conserved hydrophobic residues are highlighted in grey and the conserved LxxV/T motifs (x=basic amino acid) are boxed. (B) KD values determined as for Figure 3A and B under the conditions are indicated. (*) The KD values for the Tβ4–WH2 interaction were estimated on the basis of previous studies (De La Cruz et al, 2000; Hertzog et al, 2002; Chereau et al, 2005). (C) TIRFM micrographs of the assembly of 1.3 μM actin (30% OG labelled) on beads saturated with hVASP Tβ4 and hVASP WIP in TIRF buffer in the presence of 80 nM CP. Both chimeras processively elongate actin filaments. Time is indicated in seconds, scale=20 μm. (D) Elongation rates mediated by hVASP and the chimeras hVASP Tβ4, hVASP WIP and hVASP DdGAB at 1.3 μM G-actin either with 500 nM of the VASP constructs in solution or in the presence of 80 nM CP on saturated beads. Number of analysed filaments >20 for bead assays and >40 for assays in solution. Elongation rates are presented as mean values±s.e.m. (E) Elongation rates obtained from TIRF assays with beads coated with different hVASP constructs in the presence of 80 nM CP at the actin concentrations indicated. Solid lines represent best fits of the experimental data using the mathematical model for processive filament elongation by immobilized VASP as described in Figure 5 and the Materials and methods section. Elongation rates are presented as mean values±s.e.m. (F) Parameters derived from fitting of the data are shown in (E).
Based on these different affinities, we tested the filament elongation properties of chimeras hVASP WIP and hVASP Tβ4. Remarkably, both WH2 chimeras promoted actin assembly in an Ena/VASP-mediated manner, enhancing filament elongation in solution and processively elongating actin filaments in the presence of CP when clustered on polystyrene beads (Figure 4C; Supplementary Movie 5). As hypothesized, chimera hVASP WIP-mediated fast elongation rates both in solution and on beads (32.1 and 36.3 sub s−1, respectively), whereas chimera hVASP Tβ4 enhanced filament elongation only moderately to 20.1 and 19.5 sub s−1, respectively (Figure 4D). As shown in Figure 4E, the elongation rates mediated by VASP clustered on beads increased in order of their affinities for actin monomers. Notably, the rates appeared to approach saturation at higher actin concentrations, as can be most clearly seen for chimeras hVASP DdGAB and hVASP WIP. Thus, these results let us propose that the elongation rate of processive filament growth on VASP-saturated beads reaches a maximum rate when the available GAB regions are saturated (actin concentration ≫KD), suggesting that all Ena/VASP proteins are potent filament elongators at high actin monomer concentrations.
Model of VASP-mediated actin-filament elongation
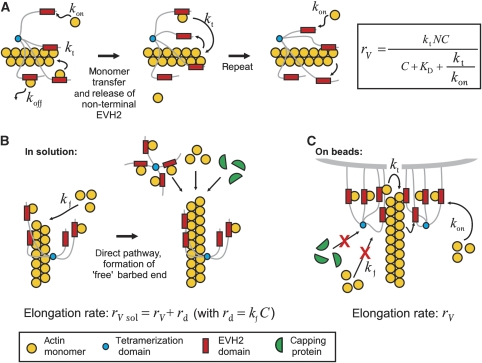
The dependence of processive VASP-mediated filament elongation on its saturation with actin monomers can be explained by a mathematical model based on the ‘actoclampin' model of actin-filament end-tracking proteins (Dickinson and Purich, 2002; Dickinson et al, 2004), in which processive elongation is achieved by multivalent affinity-modulated interactions between the VASP tetramer and the filament tip (Figure 5). At steady state, one VASP subunit of a tetramer binds to the filament terminal subunit by its GAB, leaving a number (N) of free GABs to capture monomers from solution (concentration C) with rate constant kon. Captured monomers either dissociate (at rate constant koff=kon × KD) or are transferred to the tip and irreversibly incorporated with rate constant kt. As shown in the Materials and methods section and Figure 5A, this kinetic model derives the VASP-mediated elongation rate rV to be
Figure 5.
Mathematical model of VASP-mediated actin-filament elongation. (A) General mechanism of VASP-mediated filament elongation. A VASP tetramer is attached to the filament barbed end by at least one EVH2 domain during filament elongation. The free EVH2 domains recruit actin monomers from solution with an on-rate constant kon, and subsequently either transfer the subunit onto the barbed end with a transfer rate kt or release the actin monomer back into solution with an off-rate koff. This model also assumes that upon binding of the GAB-associated monomer to the barbed end, the already bound GAB is quickly (or simultaneously) released from the now penultimate subunit so that it becomes immediately available to capture another monomer. By this cycle, the VASP tetramer is able to processively track the elongating filament tip while cyclically maintaining one GAB bound to the terminal subunit and three GABs free to capture monomers from solution. (B) VASP-mediated actin-filament elongation in solution. When VASP is free in solution, both, processive association of VASP with the barbed end and spontaneous addition of free actin monomers via the direct pathway with an independent transfer rate constant kf can occur. Free barbed ends produced by the direct pathway are accessible either for other VASP tetramers, actin monomers or, if present, capping proteins. The average elongation rate is therefore the sum of the VASP-mediated filament elongation rate and the rate of the direct pathway. (C) VASP-mediated filament elongation on surfaces. Upon clustering, the high density of VASP at the surface leads to processive association of VASP with the filament end, efficiently blocking binding of capping proteins and also preventing spontaneous addition of monomers via the direct pathway. Hence, filament elongation is exclusively fueled by actin monomers recruited and transferred by VASP.
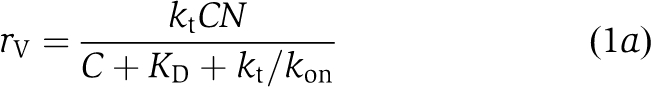 |
In Figure 4E, model predictions show excellent agreement with elongation rates observed on beads saturated with chimeras hVASP WIP, hVASP Tβ4, hVASP DdGAB and hVASP WT over the widest possible range of actin concentrations that is applicable to TIRF assays (0.5–4 μM actin). Importantly, the model anticipates a saturation of the rate with respect to monomer concentration, which will be achieved when all GAB sites are occupied with actin. All four curves were fitted simultaneously by weighted least-squares regression with only four fitted parameters: the number of GABs N, the transfer rate constant kt, which was shown to have nearly the same value for all hVASP constructs (Supplementary Figure S5A), and the values of kon only for the WIP and Tβ4 chimeras (for fitted values see Figure 4F). Interestingly, global fitting of the experimental data yielded N=3, suggesting that a single VASP tetramer operates at a filament tip during filament elongation.
Our model also implies that processive filament elongation by VASP may not only occur with VASP clustered on beads, but also with soluble VASP tetramers. Consistently, a monomeric DdVASP mutant was previously shown to be not functional in solution (Breitsprecher et al, 2008). Noting that half-maximal elongation rates are already achieved at relatively low tetramer concentrations (∼25–50 nM), we estimate by assuming an on-rate constant of ∼20 μM−1 s−1 for VASP binding to the barbed end that a single VASP tetramer might be processively associated with the filament end for ∼1–2 s. This would correspond to the incorporation of 70–140 subunits, which is in the range of the resolution limit of our TIRF setup (Supplementary Figure S6; Breitsprecher et al, 2008). A non-processive monomer transfer by single VASP tetramers transporting monomers from solution would require an at least one order of magnitude higher on-rate constant. For example, an on-rate constant of tetramer binding to a filament tip of 300 μM−1 s−1 would be required to add monomers at 60 s−1 from a 50-nM solution of VASP tetramers carrying four monomers at a time. Such a large rate rather supports the processive model over a shuttling mechanism. Notably, these calculations were very recently confirmed by experimental data showing short tracks of processive filament elongation by single VASP tetramers in solution using TIRF microscopy (Hansen and Mullins, 2010).
We have previously shown that filament elongation can be readily inhibited by CP, and that single VASP tetramers are only transiently associated with the barbed end (Breitsprecher et al, 2008). Therefore, we hypothesize that monomers are also added spontaneously to the filament end in a parallel pathway to VASP-mediated filament elongation. For monomer addition from solution, which we refer to as the direct pathway, the elongation rate would then be rd=kf × C, where rd is the elongation rate and kf is the rate constant of spontaneous actin monomer addition (neglecting the dissociation rate, which should be negligibly small <1 s−1 for ATP–actin (Pollard et al, 2000)). Thus, the total elongation rate observed with VASP in solution is the sum of the VASP-mediated filament elongation rate and the rate resulting from the direct pathway, and hence described by rV sol=rV+rd (Figure 5B). We speculate that the direct pathway creates free barbed ends, which in turn are accessible for actin monomers, other VASP tetramers or barbed end binding proteins such as CP (Figure 5B). However, when VASP is clustered on surfaces, CP is prevented from binding to barbed ends, suggesting that the direct pathway is largely inhibited under these conditions, probably due to sterical hindrance, allowing filament elongation exclusively by VASP-mediated monomer delivery (Figure 5C). Consistently, independent fitting of the elongation rates mediated by VASP proteins clustered on beads under consideration of the direct pathway yielded very low kf values between 0 and 0.25 μM−1 s−1 (Supplementary Figure S5A) and was almost equally well fitted with kf=0 (Figure 4E). Moreover, comparison of elongation rates mediated by the slow-elongating hVASP over a range of actin concentrations showed that the rates obtained with hVASP in solution depended much more strongly on the actin monomer concentration when compared with the rates on hVASP-coated beads, yielding a rate constant for spontaneous monomer addition of kf=8 μM−1 s−1 (Supplementary Figure S7). Consistently, global fitting of the data for VASP-coated beads employing this kf value was impossible (Supplementary Figure S5B), supporting that the direct pathway is inhibited during processive filament elongation by clustered VASP.
Role of the FAB motif in VASP-mediated filament elongation
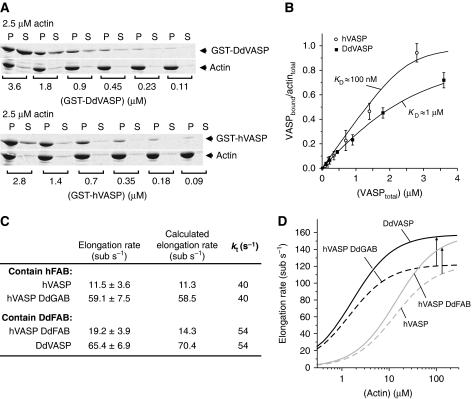
For lower-affinity Ena/VASP proteins, the elongation rate is determined entirely by the actin monomer affinity, the number of monomer binding sites (N), and the transfer rate kt. Since KD and N should be the same for hVASP and hVASP-DdFAB, the higher assembly rate by hVASP-DdFAB could be due to a faster transfer rate kt, perhaps arising from a lower affinity of the DdFAB motif for F-actin. Assuming that the FAB motif determines the affinity of VASP for F-actin and that VASP binds to F-actin in a 1:1 ratio (Bachmann et al, 1999; Laurent et al, 1999), we estimated the KD value for the hFAB to be in the range of ∼100 nM by high speed co-sedimentation assays (Figure 6A and B). DdVASP bound to F-actin with a significantly lower affinity, yielding a KD value of about 1 μM (Figure 6A and B). From these values, we estimated that individual DdFAB motifs remain bound to F-actin roughly 20% of the time during VASP-mediated elongation whereas hFAB motifs spent 40% of the time on the filament (see Supplementary data for calculations). Assuming that monomer incorporation by the adjacent GAB motif is only possible when the FAB motif is detached, this could account for the higher rates mediated by chimeras bearing the DdFAB relative to those containing the hFAB. The slightly higher elongation rates mediated by Ena/VASP proteins harbouring the DdFAB motif compared with equivalent constructs with the hFAB motif could indeed be fitted with a higher kt value (Figure 6C). This suggests that the FAB modulates filament elongation depending on its dwell time on the side of the filament, thus affecting the rate of monomer transfer to the barbed end. Consistently, deletion of the FAB motif from both Mena and VASP resulted in faster propulsion of L. monocytogenes in reconstituted MVD7 cells (Geese et al, 2002), and a DdVASPΔFAB mutant mediated slightly higher elongation rates in vitro when compared with the wild-type protein (Breitsprecher et al, 2008). Thus, a lower affinity of the FAB for F-actin might not only result in higher elongation rates under in vitro TIRF conditions, but could also enhance the maximal elongation rate of VASP-mediated filament elongation under saturating conditions (Figure 6D). What is clearly needed to understand the role of the FAB in this process in more detail are structural data of the FAB in complex with the filament.
Figure 6.
Function of the FAB motif in VASP-mediated filament elongation. (A) High-speed co-sedimentation analysis on the binding of GST-hVASP and GST-DdVASP to F-actin. In all, 2.5 μM actin was polymerized in polymerization buffer supplemented with VASP at the concentrations indicated. After centrifugation at 180 000 g for 30 min, supernatants and pellets were analysed by SDS–PAGE. (B) KD values of the VASP–F-actin interaction were estimated from densiometric analyses of the amounts of GST-VASP constructs in the pellets (bound VASP) and supernatants (free VASP) from experiments as shown in (A), and by assuming that VASP binds to F-actin in a 1:1 ratio (Samarin et al, 2003). Error bars represent s.e.m. from three experiments. (C) Potential role of the F-actin affinity of the FAB on VASP-mediated filament elongation. Taking into account that different FAB motifs might alter kt depending on their F-actin affinity, experimental data could be fitted with fixed KD and kon values, yielding a higher kt value for constructs bearing the DdFAB motif (kt values are highlighted in bold). Elongation rates were determined by TIRF microscopy and are presented as mean values±s.e.m. (D) Calculated effect of higher kt values on VASP-mediated filament elongation. Dashed lines represent calculated values for VASP constructs bearing the higher-affinity hFAB motif using kt=40 s−1. Solid lines represent calculated values for constructs with the lower-affinity DdFAB using kt=54 s−1. The arrows indicate that the faster kt increases the maximal elongation rate of VASP-mediated actin assembly by ∼30% under saturating conditions.
Discussion
Here, we unraveled an affinity-based mechanism by which Ena/VASP proteins differentially enhance actin-filament elongation in solution and on functionalized surfaces, and present a testable mathematical model for Ena/VASP-mediated actin-filament elongation. The comparison of hVASP chimeras encompassing WH2 motifs with different actin affinities revealed that filament elongation rates result from direct binding and incorporation of actin monomers by their WH2-like G-actin-binding motifs, and moreover, that their saturation with G-actin correlates directly with the enhancement of the filament elongation rate. Our results clearly explain the differences in the actin polymerizing activities of Ena/VASP proteins from different species in vitro and demonstrate that Ena/VASP proteins from evolutionary distant organisms employ a common mechanism to promote actin-filament assembly.
In the ‘actoclampin' model of tethered elongation (Dickinson et al, 2002; Dickinson, 2009), the hypothetical multimeric actoclampin protein mediates filament elongation by two adjacent actin-binding modules, one processively tracking and tethering the growing filament end and the other binding and delivering monomeric actin for elongation. Rate-limiting factors for filament elongation are therefore the translocation speed of the filament-binding modules at the barbed end, the on-rate of monomer binding, the kinetics of monomer transfer and GAB release from the barbed end or the number of monomers recruited by the monomer-binding modules. This model can in principle be applied to the action of both, formins and Ena/VASP proteins. However, due to the considerable structural and biochemical differences between their interactions with G- and F- actin, these two protein families employ different molecular mechanisms to enhance filament elongation (Kovar et al, 2006; Ferron et al, 2007; Breitsprecher et al, 2008; Chesarone and Goode, 2009): formins processively track the growing barbed end of a filament by virtue of their dimeric FH2 domains, which, however, have only negligible affinities towards G-actin (Pring et al, 2003; Kovar et al, 2006). Notably, FH2-assembled actin filaments grow slower than spontaneously assembled actin filaments, consistent with a rate-limiting effect of the translocating filament-binding module on the filament elongation rate in the actoclampin model (Kovar and Pollard, 2004; Dickinson, 2009; Paul and Pollard, 2009). This inhibitory effect of the FH2 domain is quantified by the ‘gating factor', which describes the fraction of time the formin spends in the open state, allowing actin monomer incorporation (Paul and Pollard, 2009). Impeded filament elongation by the FH2 domain is compensated by the FH1 domain, which recruits monomers as profilin–actin complexes that are subsequently transferred to the FH2-bound barbed end to enhance filament elongation (Kovar et al, 2006). The elongation rate of formin-mediated actin assembly was accordingly shown to correlate with the number of profilin-binding sites within the FH1 domain, and hence the number of recruited monomers (Paul and Pollard, 2008).
In contrast, Ena/VASP tetramers are composed of small G- and F-actin-binding modules, and are not expected to processively track the barbed end of the filament in the same way as the dimeric FH2 domain of formins (Breitsprecher et al, 2008; Dominguez, 2009). Additionally, profilin was shown to be dispensable for VASP-mediated actin assembly in vitro (Breitsprecher et al, 2008; Hansen and Mullins, 2010), suggesting that monomer recruitment is achieved directly by its WH2-like motifs. Since insertion of other WH2 motifs with different G-actin affinities into the hVASP backbone was sufficient to accelerate filament elongation to different extents according on their G-actin affinity, we propose that direct monomer recruitment is key for rapid Ena/VASP-mediated filament elongation. Our findings show that the function of the GAB in filament elongation can be mimicked by other WH2 motifs, and that a general modular arrangement of a G-actin-binding WH2 motif and the FAB from VASP is sufficient to generate actin-filament elongators that allow fast and processive filament elongation in the presence of CP when clustered on surfaces.
Importantly, the elongation rates mediated by Ena/VASP proteins were shown to correlate with the affinity of their GAB to actin monomers at low actin concentrations. Since the actin affinity of the hGAB motif is rather low and moreover strongly depends on the ionic strength of the reaction buffer, it is not surprising that previous studies reported incoherent results on the impact of vertebrate Ena/VASP proteins on filament barbed end elongation in in vitro studies using relatively low concentrations of actin and VASP in buffers with different salt concentrations (Skoble et al, 2001; Samarin et al, 2003; Barzik et al, 2005; Pasic et al, 2008).
The good agreement between the experimental data and the mathematical model presented here supports a processive mechanism whereby multiple free GABs of the tetrameric VASP capture actin monomers from solution and transfer them onto the filament tip (Figure 5), which is similar to the filament end-tracking mechanism (Dickinson et al, 2004). Notably, individual filaments elongated from VASP-coated beads at roughly the same rates as with VASP in solution (Figures 1, 2 and 4; Breitsprecher et al, 2008), suggesting that the number of involved GABs (N) is a fixed value, irrespective of whether VASP is immobilized on a surface or in solution. Interestingly, taken together, our data strongly suggest that filament elongation is mediated by a single VASP tetramer at any given time. These findings are consistent with a recent study by Hansen and Mullins (2010) reporting processive filament elongation by a single VASP tetramer in solution. On beads, filament elongation could be similarly accomplished by one VASP tetramer, but since clustering is required for processive elongation (Breitsprecher et al, 2008), our experiments cannot rule out the possibility that in a given geometric context four clustered VASP tetramers could operate together, with one EVH2 domain tethering the filament while the other three EVH2 domains each participate in monomer delivery around a central actin filament.
Together, our model does not only explain the different elongation rates mediated by Ena/VASP in vitro, but also anticipates that all Ena/VASP proteins are potent filament elongators in vivo, where the concentration of polymerization-competent actin is much larger than under in vitro conditions (Figure 7) (Pollard et al, 2000). It has been previously proposed that Ena/VASP enhances protrusion of lamellipodia and propulsion of ActA-coated beads and Listeria in reconstituted motility assays indirectly by preventing capping of barbed ends by CP, by lowering the number of Arp2/3-dependent filament branches or by mediating rapid attachment-detachment cycles of actin filaments to allow both the binding of F-actin and insertion of monomers by Brownian motion (Laurent et al, 1999; Bear et al, 2000, 2002; Samarin et al, 2003). We show here that these findings are more readily explained by enhanced actin-filament elongation rates mediated by processive interaction of Ena/VASP proteins with filament barbed ends, consistent with the actoclampin end-tracking motor hypothesis (Dickinson and Purich, 2002; Dickinson et al, 2002, 2004; Dickinson, 2009). Finally, since our model implies that spontaneous addition of actin monomers is likely not to occur during processive VASP-mediated filament elongation on surfaces, and since Ena/VASP proteins are abundantly found at sites of active actin assembly, it seems plausible that most if not all actin-filament elongation in the cell is directly controlled by end-tracking proteins.
Figure 7.
Calculation of the saturation dependence of filament elongation by Ena/VASP proteins. Comparison of the calculated elongation rates for processive filament elongation mediated by hVASP WT and hVASP DdGAB over a large range of actin concentrations. Grey boxes indicate G-actin concentrations typically used in TIRF assays or present in biomimetic motility assays (Samarin et al, 2003), as well as physiological G-actin concentrations, for instance in the lamellipodium tip (Koestler et al, 2009). Different G-actin affinities of the GAB motifs result in markedly different elongation rates mediated by chimeras hVASP and hVASP DdGAB in TIRF assays. However, both proteins are expected to be saturated with actin monomers and to enhance filament elongation to the same extent at very high monomer concentrations, for example under in vivo conditions.
Materials and methods
In vitro TIRF microscopy
Time lapse total internal reflection fluorescence microscopy (TIRFM) was essentially performed as described (Breitsprecher et al, 2008). Images from an Olympus IX-81 inverted microscope were captured every 5 or 10 s with exposures of 100 ms with a Hamamatsu Orca-R2 CCD camera (Hamamatsu Corp., Bridgewater, NJ). The pixel size corresponded to 0.11 μm.
If not stated otherwise, reactions in TIRF assays contained 1.3 μM actin (30% OG labelled). Prior to the experiments, Ca2+–ATP–actin was converted to Mg2+–ATP–actin by addition of 10 × Mg-exchange buffer (1 mM MgCl2, 10 mM EGTA, pH 7.4). Polymerization experiments were performed in TIRF buffer (10 mM imidazole, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 0.2 mM ATP, 10 mM DTT, 15 mM glucose, 20 μg ml−1 catalase, 100 μg ml−1 glucose oxidase and 0.25% methylcellulose (4000 cP), pH 7.4). Assays with functionalized beads additionally contained 0.05% (w/v) microspheres. Saturation of the beads with Ena/VASP proteins was confirmed by SDS–PAGE prior to the experiments. The recorded data were analysed with ImageJ software either by manually tracking filaments or by tracking barbed ends using the plugin MtrackJ. Every experiment was repeated at least three times. On beads, total lengths of growing filaments were measured for at least 10 time frames. Filament growth rates were diagrammed as plots of length versus time and the average elongation rate in subunits s−1 was calculated from linear regressions of the slopes. Carboxylated 2 μm-diameter polystyrene microspheres (Polyscience) were coated with 5 μM of the different VASP constructs according to Samarin et al (2003). The highest applicable actin concentration in TIRF assays was 4 μM since high background fluorescence prevented the analysis of single actin filaments at higher concentrations.
Mathematical model of VASP-mediated filament elongation
The saturating dependence on substrate concentration and dependence on affinity is reminiscent of the Michaelis–Menten model for enzyme kinetics, in which reversible binding of the substrate (in this case, actin monomers) is followed by irreversible conversion to product (F-actin subunits). We assume each filament tip remains associated with a number (N) of GAB regions that are available for capturing monomers from solution (at concentration, C) with forward rate constant kon (μM−1 s−1). Captured monomers are then either transferred to the filament tip at rate kt (s−1) or released back into solution with off-rate koff (s−1). Let n be the (average) number of the N total GAB regions occupied by bound monomers. The steady-state balance equation describing for n is then
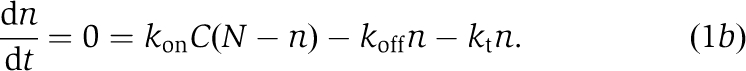 |
Solving for n and noting the rate of elongation is r=ktn yields
 |
where we have introduced the equilibrium dissociation constant, KD≡koff/kon. Saturation of binding sites occurs for large C relative to KD+kt/kon, such that r≈ktN.
Fitting the model to the rate data and estimation of unknown parameters
The model was simultaneously fit to all data to estimate the parameter set β=(kt, N, kon, WIP, kon, TB4) by weight least-squares regression, that is by minimizing the function
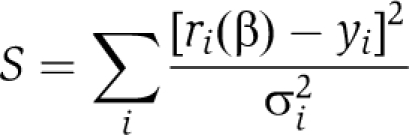 |
where ri (β) is rate calculated from Equation (2), yi is the measured rate (i=1 to M data points), and σi is the uncertainty in the mean for measurement i. The variance–covariance matrix of the least-squares estimate 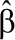 , was calculated from
, was calculated from
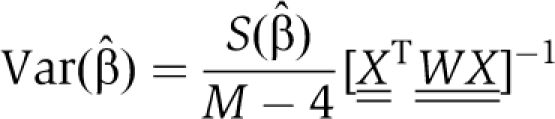 |
where the matrices are defined by  and Wi,i=σi−2, Wi,j≠i=0.
and Wi,i=σi−2, Wi,j≠i=0.
Actin monomer binding assays
Synthetic peptides DdGAB (GRNALLGSIENFSKGGLKKTVT) and hGAB (GLAAAIAGAKLRKVSK) were obtained from the Helmholtz Centre for Infection Research (Braunschweig), and the WIP (GRNALLSDISKGKKLKKTVT) and Tβ4 (MSDKPDMAEIEKFDKSKLKKTET) peptides were purchased from Peptide Specialty Laboratories. Except for Tβ4, the binding of WH2 peptides to OG-actin resulted in a 10–20% increase in OG-fluorescence, which was used to detect the formation of WH2–actin complexes. Fluorescence titrations were carried out in a Jasco FP-6500 fluorimeter. Excitation wavelength was 490 nm and the OG-fluorescence was detected at an emission wavelength of 520 nm. All titrations were carried out at 22°C in either G-buffer or G-buffer supplemented with the KCl concentrations indicated and a five-fold molar excess of LatA (Biomol) to actin. After each addition, the solution was allowed to reequilibrate for 100 s. If stated, Ca2+–ATP–actin was converted to Mg2+–ATP–actin by addition of 10 × Mg-exchange buffer prior to the experiment. Reactions contained 0.02% Tween 20 to avoid unspecific protein adsorption. Dissociation equilibrium constants (KD) were obtained by non-linear least-square fitting utilizing the program package BPCfit (Witte et al, 2008). The equilibrium concentration of the complex Cequ was calculated using the following equation:
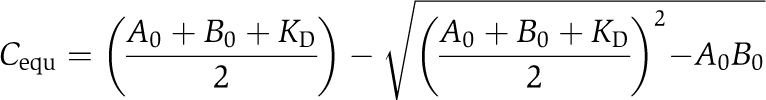 |
where A0 and B0 denote the total concentrations of OG-actin and WH2 peptide, respectively.
Stopped-flow experiments
The kinetics of the interaction of EVH2 domains and OG-actin were monitored by detecting the increase in OG-fluorescence using a Hi-tech Scientific SF-61 DX single mixing stopped-flow system, equipped with a 515-nM cutoff filter. All reactions were carried out with Mg2+–ATP OG-actin in G-buffer supplemented with 50 mM KCl, 0.02% Tween 20 and a five-fold molar excess of LatA over actin monomers. For experiments with the DdEVH2 domain, a final concentration of 2.5 μM OG-actin was used. For hEVH2, 10 μM OG-actin was used. Experiments were repeated 10–15 times for each EVH2 concentration, and the experimental data were accumulated for fitting. Association rate constants (kon) were obtained by non-linear least-square fitting of a reversible bimolecular reaction model (A+B↔C) to the data utilizing the program package BPCfit (Witte et al, 2008). The time course of the complex concentration C(t) was calculated using the following equation:
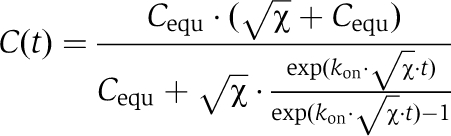 |
with:
where A0 and B0 denote the total concentrations of OG-actin and EVH2, respectively and Cequ denotes the equilibrium concentration of the complex, which was calculated using the KD values determined for the interaction of OG-actin and the WH2 peptides (v.i.).
Supplementary Material
Acknowledgments
We thank Dr W Tegge (Helmholtz Centre for Infection Research, Braunschweig) for providing VASP synthetic peptides, Sarah Heissler (Hannover Medical School) for advice on stopped-flow measurements, Annette Breskott for excellent technical assistance and Dr D Ushakov for critical reading of the manuscript. This work was supported by grants to TEBS (666/3-1) and JF (330/4-2) from the Deutsche Forschungsgemeinschaft, and a grant to JVS (P-21292-B09) from the Austrian Science Fund FWF.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, Qualmann B (2007) Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell 131: 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applewhite DA, Barzik M, Kojima S, Svitkina TM, Gertler FB, Borisy GG (2007) Ena/VASP proteins have an anti-capping independent function in filopodia formation. Mol Biol Cell 18: 2579–2591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann C, Fischer L, Walter U, Reinhard M (1999) The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem 274: 23549–23557 [DOI] [PubMed] [Google Scholar]
- Barzik M, Kotova TI, Higgs HN, Hazelwood L, Hanein D, Gertler FB, Schafer DA (2005) Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem 280: 28653–28662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Loureiro JJ, Libova I, Fässler R, Wehland J, Gertler FB (2000) Negative regulation of fibroblast motility by Ena/VASP proteins. Cell 101: 717–728 [DOI] [PubMed] [Google Scholar]
- Bear JE, Svitkina TM, Krause M, Schafer DA, Loureiro JJ, Strasser GA, Maly IV, Chaga OY, Cooper JA, Borisy GG, Gertler FB (2002) Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109: 509–521 [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Kiesewetter AK, Linkner J, Urbanke C, Resch GP, Small JV, Faix J (2008) Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J 27: 2943–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P (1997) Cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol 137: 169–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R (2005) Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci USA 102: 16644–16649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, Goode BL (2009) Actin nucleation and elongation factors: mechanisms and interplay. Curr Opin Cell Biol 21: 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra ES, Higgs HN (2007) The many faces of actin: matching assembly factors with cellular structures. Nat Cell Biol 9: 1110–1121 [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Ostap EM, Brundage RA, Reddy KS, Sweeney HL, Safer D (2000) Thymosin-β4 changes the conformation and dynamics of actin monomers. Biophys J 78: 2516–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent EW, Kwiatkowski AV, Mebane LM, Philippar U, Barzik M, Rubinson DA, Gupton S, Van Veen JE, Furman C, Zhang J, Alberts AS, Mori S, Gertler FB (2007) Filopodia are required for cortical neurite initiation. Nat Cell Biol 9: 1347–1359 [DOI] [PubMed] [Google Scholar]
- Dickinson RB (2009) Models for actin polymerization motors. J Math Biol 58: 81–103 [DOI] [PubMed] [Google Scholar]
- Dickinson RB, Caro L, Purich DL (2004) Force generation by cytoskeletal filament end-tracking proteins. Biophys J 87: 2838–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RB, Purich DL (2002) Clamped-filament elongation model for actin-based motors. Biophys J 82: 605–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RB, Southwick FS, Purich DL (2002) A direct-transfer polymerization model explains how the multiple profilin-binding sites in the actoclampin motor promote rapid actin-based motility. Arch Biochem Biophys 406: 296–301 [DOI] [PubMed] [Google Scholar]
- Dominguez R (2007) The beta-thymosin/WH2 fold: multifunctionality and structure. Ann N Y Acad Sci 1112: 86–94 [DOI] [PubMed] [Google Scholar]
- Dominguez R (2009) Actin filament nucleation and elongation factors--structure-function relationships. Crit Rev Biochem Mol Biol 44: 351–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R (2010) Structural insights into de novo actin polymerization. Curr Opin Struct Biol 20: 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J, Breitsprecher D, Stradal TE, Rottner K (2009) Filopodia: complex models for simple rods. Int J Biochem Cell Biol 41: 1656–1664 [DOI] [PubMed] [Google Scholar]
- Faix J, Grosse R (2006) Staying in shape with formins. Dev Cell 10: 693–706 [DOI] [PubMed] [Google Scholar]
- Ferron F, Rebowski G, Lee SH, Dominguez R (2007) Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J 26: 4597–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footer MJ, Lyo JK, Theriot JA (2008) Close packing of Listeria monocytogenes ActA, a natively unfolded protein, enhances F-actin assembly without dimerization. J Biol Chem 283: 23852–23862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geese M, Loureiro JJ, Bear JE, Wehland J, Gertler FB, Sechi AS (2002) Contribution of Ena/VASP proteins to intracellular motility of listeria requires phosphorylation and proline-rich core but not F-actin binding or multimerization. Mol Biol Cell 13: 2383–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SD, Mullins RD (2010) VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol 191: 571–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ES, Li F, Higgs HN (2004) The mouse formin, FRLα, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J Biol Chem 279: 20076–20087 [DOI] [PubMed] [Google Scholar]
- Hertzog M, van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, Didelot G, Preat T, Knossow M, Guittet E, Carlier MF (2004) The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell 117: 611–623 [DOI] [PubMed] [Google Scholar]
- Hertzog M, Yarmola EG, Didry D, Bubb MR, Carlier MF (2002) Control of actin dynamics by proteins made of beta-thymosin repeats: the actobindin family. J Biol Chem 277: 14786–14792 [DOI] [PubMed] [Google Scholar]
- Hu LD, Zou HF, Zhan SX, Cao KM (2008) EVL (Ena/VASP-like) expression is up-regulated in human breast cancer and its relative expression level is correlated with clinical stages. Oncol Rep 19: 1015–1020 [PubMed] [Google Scholar]
- Hüttelmeier S, Harbeck B, Steffens O, Messerschmidt T, Illenberger S, Jockusch BM (1999) Characterization of the actin binding properties of the vasodilator-stimulated phosphoprotein VASP. FEBS Lett 451: 68–74 [DOI] [PubMed] [Google Scholar]
- Insall RH, Machesky LM (2009) Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell 17: 310–322 [DOI] [PubMed] [Google Scholar]
- Jonckheere V, Lambrechts A, Vandekerckhove J, Ampe C (1999) Dimerization of profilin II upon binding the (GP5)3 peptide from VASP overcomes the inhibition of actin nucleation by profilin II and thymosin β4. FEBS Lett 447: 257–263 [DOI] [PubMed] [Google Scholar]
- Koestler SA, Rottner K, Lai F, Block J, Vinzenz M, Small JV (2009) F- and G-actin concentrations in lamellipodia of moving cells. PLoS One 4: e4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD (2006) Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124: 423–435 [DOI] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD (2004) Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA 101: 14725–14730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski AV, Garner CC, Nelson WJ, Gertler FB (2009) Cell autonomous defects in cortical development revealed by two-color chimera analysis. Mol Cell Neurosci 41: 44–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski AV, Rubinson DA, Dent EW, Edward van Veen J, Leslie JD, Zhang J, Mebane LM, Philippar U, Pinheiro EM, Burds AA, Bronson RT, Mori S, Fässler R, Gertler FB (2007) Ena/VASP is required for neuritogenesis in the developing cortex. Neuron 56: 441–455 [DOI] [PubMed] [Google Scholar]
- Laurent V, Loisel TP, Harbeck B, Wehman A, Grobe L, Jockusch BM, Wehland J, Gertler FB, Carlier MF (1999) Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes. J Cell Biol 144: 1245–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF (1999) Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 401: 613–616 [DOI] [PubMed] [Google Scholar]
- Pasic L, Kotova T, Schafer DA (2008) Ena/VASP proteins capture actin filament barbed ends. J Biol Chem 283: 9814–9819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD (2008) The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol 18: 9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD (2009) Energetic requirements for processive elongation of actin filaments by FH1FH2-formins. J Biol Chem 284: 12533–12540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunola E, Mattila PK, Lappalainen P (2002) WH2 domain: a small, versatile adapter for actin monomers. FEBS Lett 513: 92–97 [DOI] [PubMed] [Google Scholar]
- Philippar U, Roussos ET, Oser M, Yamaguchi H, Kim HD, Giampieri S, Wang Y, Goswami S, Wyckoff JB, Lauffenburger DA, Sahai E, Condeelis JS, Gertler FB (2008) A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev Cell 15: 813–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plastino J, Olivier S, Sykes C (2004) Actin filaments align into hollow comets for rapid VASP-mediated propulsion. Curr Biol 14: 1766–1771 [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD (2000) Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct 29: 545–576 [DOI] [PubMed] [Google Scholar]
- Pring M, Evangelista M, Boone C, Yang C, Zigmond SH (2003) Mechanism of formin-induced nucleation of actin filaments. Biochemistry 42: 486–496 [DOI] [PubMed] [Google Scholar]
- Rottner K, Behrendt B, Small JV, Wehland J (1999) VASP dynamics during lamellipodia protrusion. Nat Cell Biol 1: 321–322 [DOI] [PubMed] [Google Scholar]
- Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D (2002) An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol 4: 626–631 [DOI] [PubMed] [Google Scholar]
- Samarin S, Romero S, Kocks C, Didry D, Pantaloni D, Carlier MF (2003) How VASP enhances actin-based motility. J Cell Biol 163: 131–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirenbeck A, Arasada R, Bretschneider T, Stradal TE, Schleicher M, Faix J (2006) The bundling activity of vasodilator-stimulated phosphoprotein is required for filopodium formation. Proc Natl Acad Sci USA 103: 7694–7699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirenbeck A, Bretschneider T, Arasada R, Schleicher M, Faix J (2005) The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat Cell Biol 7: 619–625 [DOI] [PubMed] [Google Scholar]
- Skoble J, Auerbuch V, Goley ED, Welch MD, Portnoy DA (2001) Pivotal role of VASP in Arp2/3 complex-mediated actin nucleation, actin branch-formation, and Listeria monocytogenes motility. J Cell Biol 155: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte G, Fedorov R, Curth U (2008) Biophysical analysis of Thermus aquaticus single-stranded DNA binding protein. Biophys J 94: 2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond SH, Evangelista M, Boone C, Yang C, Dar AC, Sicheri F, Forkey J, Pring M (2003) Formin leaky cap allows elongation in the presence of tight capping proteins. Curr Biol 13: 1820–1823 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.