Abstract
Introduction
Approximately 10–15% of all fractures of long bones heal with delay, prolonged immobilization and repetitive operative interventions. Despite intense investigations, the pathomechanism of impaired healing of skeletal tissue remains unclear. An important role in the pathomechanism of mal-union of close fractures plays subclinically proceeding infections.
Aim
The question arises whether colonization and proliferation of bacteria in the fracture gap could be related to the mutation of genes for factors regulating local antimicrobial response, such as pathogen recognizing receptors (PRR), cytokines and chemokines.
Methods
We carried out studies in patients with delayed long bone fractures estimating the frequency of mutation of genes crucial for pathogen recognition (TLR2, TLR4 and CD14), and elimination (CRP, IL-6, IL-1ra), as well as wound healing (TGF-β). The molecular milieu regulating healing process (IGF-1, COLL1a, TGF-β, BMP-2, and PDGF) was validated by Western blot analysis of the gap tissue.
Results
Microbiological investigations showed the presence of viable bacterial strains in 34 out of 108 gaps in patients with non-healing fractures (31.5%) and in 20 out of 122 patients with uneventful healing (16.4%) (P < 0.05). The occurrence of mutated TLR4 1/W but not 2/W gene was significantly higher (P < 0.05) in the non-healing infected than sterile group. In the non-healing infected group 1/W mutated gene frequency was also higher than in healing infected. In the TGF-β codon 10 a significantly higher frequency of mutated homozygote T and heterozygote C/T in the non-healing infected versus non-healing sterile subgroup was observed (P < 0.05). Similar difference was observed in the non-healing infected versus healing infected subgroup (P < 0.05). The CRP (G1059C), IL1ra (genotype 2/2), IL-6 (G176C), CD14 (G-159T), TLR2 (G2259A) and TLR4/2 (Thr399Ile) polymorphisms did not play evident role in the delay of fracture healing.
Conclusions
Individuals bearing the mutant TLR 4 gene 1/W (Asp299Gly) and TGF-β gene codon 10 mutant T and T/C allele may predispose to impaired pathogen recognition and elimination, leading to prolonged pathogen existence in the fracture gaps and healing delays.
Keywords: Bone fracture, Malunion, Infection, 16S rRNA, TLR4, CRP, IL1ra, IL-6, CCR2, CD14, TGF-β, TLR2, TLR4, Polymorphism
Introduction
Approximately 10–15% of all fractures heal in a delayed fashion with prolonged immobilization and repetitive operative interventions [1]. Despite intense investigations, the pathomechanism of impaired healing of skeletal tissue remains unclear. Clinical and experimental studies brought the knowledge of the molecular regulators as growth factors, cytokines and chemokines regulating cellular events proceeding in the fracture gap [2, 3]. They also showed markedly lowered or lack of expression of molecules crucial for fracture healing [4–6].
A special, until recently not fully recognized, role in the pathomechanism of non-union plays subclinically proceeding infection at the site of fractured bone. We reported previously about the presence of pathogens in closed fracture gaps without clinically evident inflammation. Using microbiological cultures, bacterial cells were isolated from 35% of closed fractures with delayed healing and also in 14% of uneventfully healing fractures [7].
The question arose what were the phenotypes of callus bacterial isolates and whether colonization and proliferation of bacteria in the fracture gap could be related to individual predilection depending on mutation of genes for molecular factors regulating local antimicrobial and inflammatory response, such as pathogen recognizing receptors (PRR), cytokines and chemokines.
We carried out bacteriological and genetic studies in patients with delayed and uneventful healing of long bone closed fractures. Callus was harvested from fracture gaps for microbial culture and detection of bacterial 16S RNA. The frequency of allele of genes crucial for pathogen recognition (toll-like receptors TLR2 and TLR4 and CD14) and elimination (CRP, interleukins IL-6 and IL-1ra), as well as wound healing (transforming growth factor, TGF-β) was estimated. The molecular protein milieu regulating healing including factors as insulin-like growth factor (IGF-1), collagen type 1 (COL1A), TGF-β, bone morphogenetic protein 2 (BMP-2), and platelet derived growth factor (PDGF) were validated in callus by western blot analysis.
Patients
Investigations were performed in randomly selected group of 295 patients, in an order as they were registered in our out-patients’ clinic. They were undergoing routine post-injury check-up 6 months after sustaining closed fractures of tibia, fibula or femur. There were 151 patients with delayed (group A) and 144 with uneventful bone healing (group B). The bone reparative process was evaluated clinically and with the use of imaging techniques. Lack of bone union was diagnosed 6 months after fracture, if there were (1) pathologic movements in fracture gap with pain and (2) non-union on standard X-ray pictures in two perpendicular planes. In uncertain cases the CT and NMR scans were used. Uneventful healing was recognized in cases without pathological movements in fracture gap, callus visible on at least 3 of 4 cortex layers on X-ray pictures taken in perpendicular position. Excluded were patients with: (1) open fractures, (2) massive contusion of soft tissues covering the fracture gap, (3) trophic lesions of soft tissues, (4) chronic inflammatory foci and (5) diseases requiring medication with immunosuppressive drugs. The control group for genetic studies consisted of 125 healthy blood donors from the ethnically same Polish population. Studies were approved by the Ethics Committee of the Medical University, Warsaw, according to the international regulations.
Harvesting of the tissue specimens for microbiological and protein studies
Tissue specimens were harvested during operative interventions: in group A correcting delayed bone union, and in group B during operative treatment of bone non-alignment. Skin was sterilized three-times using SkinSept (B. Braun, Meisungen, Germany) and strict antimicrobial regime was maintained during operation and tissue harvesting. Routinely performed bacteriological cultures from disposable scalpel blade, surgeon’s gloves and air in the operating theater validated the sterility of the operation field. Granulation tissue from the fracture gap and adjacent soft tissues were taken and transported in sterile, disposable containers (Blue Max, 352070, Becton–Dickinson, USA). They were cultured on culture media: BHI (Difco, USA), liquid Hemoline (bioMerieux, Paris, France), blood agar, Columbia agar enriched with 5% sterile gout serum, MacConkey’s agar, Chapman’s agar, and Sabouraud agar (Oxoid,Oxford, UK) for at least 14 days. Isolates were analyzed biochemically using ApiStaph, ATB G(−), ApiNG and Api30E (bioMerieux, Paris, France).
Venous blood for genetic investigations
DNA was isolated from leukocytes isolated from venous blood obtained from the cubital vein.
Isolation of DNA
Genomic DNA was isolated from whole blood using NucleoSpin Kit, according to the manufacturer’s instructions (Marcherey-Nagel, Dueren, Germany). Quality of isolated DNA was checked electrophoretically on 1% agarose gels with ethidium bromide. Quantification of DNA was performed spectrophotometrically on ND-1000 spectrophotometer (Nanodrop, Wilmington, USA).
RFLP-polymerase chain reaction analysis of gene polymorphism
Polymerase chain reaction (PCR) was performed employing FastStart Taq DNA polymerase (Roche, Mannheim, Germany) in a 25 μl reaction mixture containing 5 μl genomic DNA solution, 2.5 μl 10× buffer with MgCl2, 0.5 μl deoxyribonucleoside trisphosphate mix (Sigma-Aldrich, St. Louis, USA) and 30 pmol each forward and reverse primers (Oligo, Warsaw, Poland) (Table 1). Amplification was carried out in a Thermal Cycler (MJ Research, Watertown, USA).
Table 1.
Primers and parameters for PCR sequencing
| Gene | Primer sequence (from 5′ to 3′) | Parameters of PCR | Restriction enzymes |
|---|---|---|---|
| CRP |
F: GATCTGTGTGATCTGAGAAACCTCCT R: GAGGTACCAGAGACAGAGACGTG |
95°5′; 30 cycles (94°30″, 62°1′, 72°30″), 72°10′ | MaeIII |
| CCR2 |
F: GAAAGTGGATTGAACAAGGA R: CAGGTTGAGCAGGTAAATGT |
95°5′; 30 cycles (94°30″, 58°30″, 72°30″), 72°10′ | FokI |
| CD14 |
F: GTGCCAACAGATGAGGTTCA R: CGCAGCGGAAATCTTCATC |
95°5′; 30 cycles (94°30″, 58°30″, 72°30″), 72°10′ | HaeIII |
| TGF-β |
F: TCGCGGGTGCTGTTGTACA R: TTCAAGACCACCCACCTTCT |
94°5′; 35 cycles (94°30″, 58°30″, 72°30″), 72°10′ | FseI |
| TLR2 |
F: GCCTACTGGGTGGAGAACCT R: GGCCACTCCAGGTAGGTCTT |
95°10′; 35 cycles (94°30″, 60°30″, 72°30″), 72°10′ | AciI |
| TLR4 (1) |
F: GATTAGCATACTTAGACTACTACCTCCATG R: GATCAACTTCTGAAAAAGCATTCCCAC |
95°4′; 30 cycles (95°30″, 55°30″, 72°30″), 72°10′ | NcoI |
| TLR4 (2) |
F: GGTTGCTGTTCTGAAAGTGATTTTGGGAGAA R: ACCTGAAGACTGGAGAGTGAGTTAAATGCT |
95°4′; 30 cycles (95°30″, 55°30″, 72°30″), 72°10′ | HinfI |
| IL-6 |
F: CTGATTGGAAACCTTATTAAG R: TGACTTCAGCTTTACTCTTGT |
95°5′; 30 cycles (94°30″, 48°30″, 72°30″), 72°10′ | Hsp92II |
| IL-1ra |
F: CTCAGCAACACTCCTAT R: TCCTGGTCTGCAAGGTAA |
95°5′; 30 cycles (94°30″, 60°30″, 72°30″), 72°10′ | – |
| 16S RNA |
F: AGTTTGATCCTGGCTCAG R: GGACTACCAGGGTATCTAAT |
95°15′; 35 cycles (60°45″, 72°10″, 95°45″, 60°45″, 72°60″), 72°10′ | – |
F forward, R reverse
PCR analysis of bacterial 16S rRNA
16S rRNA genomic DNA was prepared following manufacturer’s protocol (High Pure PCR Template Preparation Kit, Roche, Mannheim, Germany). PCR was performed employing RedAllegroTaq DNA polymerase (Novazym, Poznan, Poland) in a 25 μl reaction mixture containing 5 μl genomic DNA solution, 2.5 μl 10× buffer with MgCl2, 0.5 μl deoxyribonucleoside trisphosphate mix (Sigma-Aldrich, St. Louis, USA), 0.5 μl anti-inhibitor (DNA-Gdansk, Poland), 5 μl GC-rich (Roche Mannheim, Germany) and 25 pmol each forward and reverse primers (Oligo, Warsaw, Poland). Amplification was carried out in a thermal cycler (MJ Research, Watertown, USA).
Western blotting
Callus protein sediment was prepared using Total Protein Extraction Kit (Millipore, Billerica, USA) according to manufacturer protocol. Sequenced proteins were moved from polyacrylamide gel to PVDF membrane (Porablot PVDF–PVDF membrane, Macherey–Nagel, Duren, Germany) using TRANS-BLOT SD, SEMI DRY TRANSFER CELL (Biorad, Hercules, USA), then incubated with I-grade antibody (BMP-2, Col1A1, PDGF-A, TGF-β1/2/3, and IGF-I, Santa Cruz Biotechnology, USA). Membrane was incubated with II-grade antibody conjugated with alkaline phosphatase (goat anti-rabbit AP, rabbit anti-goat AP, Santa Cruz Biotechnology, USA). For protein detection, membrane was incubated with Alkaline Phosphatase Conjugate Substrate Kit (Biorad, Hercules, USA). Protein expression was validated comparing obtained results with marker (Novex Sharp Protein Standard, Invitrogen, California, USA).
The results are presented as the ratio of target protein over GAPDH protein expression.
Statistical methods
The obtained results were analyzed statistically using standard χ2 and Fisher’s χ2 tests.
Results
Microbiological evaluation
Microbiological investigations showed the presence of viable bacterial strains in 34 out of 108 gaps (31.5%) in patients with non-healing fractures (group A), and in 20 of 122 gaps (16.4%) in patients with uneventful healing fractures (group B) (P < 0.05). Viable pathogens were isolated in 12 out of 55 biopsies of adjacent soft tissues (subcutaneous fat, striated muscle, and periosteum) in patients from group A (21.8%) and in 9 out of 43 patients from group B (20.9%; NS). In 80% of cases from group A and 59% from group B isolated pathogens belonged to staphylococci (P < 0.05). The most common strains were S. epidermidis and S. aureus. Occasionally Micrococcus spp., Enterococcus spp., and Pseudomonas spp. were isolated in both groups.
Amplification of bacterial 16S rRNA from fracture gaps was found positive in 59 out of 95 patients from group A (56.8%) and in 75 out of 115 from group B (65.4%, NS). Negative bacterial cultures with positive 16S rRNA amplifications were observed in 31.5% cases from group A and 50% in group B.
Evaluation of mutations
The genotype frequencies of the whole investigated and reference ethnic group did not differ by more than 5% meeting the conditions for statistical comparison of clinical groups. Also, there were no differences in the demographic data between patients from investigated groups A and B.
Analysis of TLR4 gene polymorphism
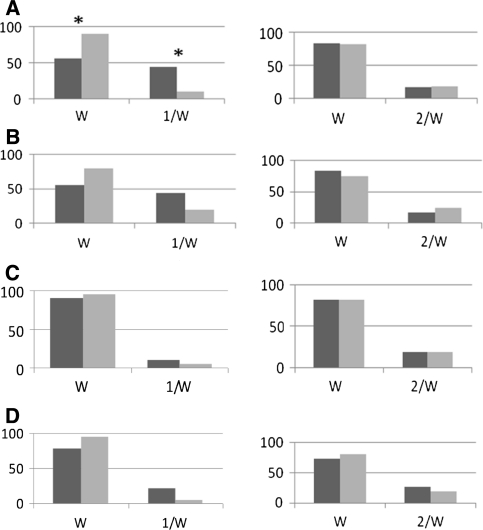
The frequency rate of 1/W (Asp299Gly) and 2/W (Thr399Ile) was estimated in the subgroups (a) non-healing infected versus non-healing sterile, (b) non-healing infected versus healing infected, (c) non-healing sterile versus healing sterile and (d) healing infected versus healing sterile, subgroups. The occurrence of mutated 1/W but not 2/W was significantly higher (P < 0.05) in subgroup (a) in the non-healing infected than sterile (P < 0.05) (Fig. 1a). In subgroup (b) 1/W mutated gene frequency was also higher in the non-healing infected than healing infected; however, the difference was not significant (Fig. 1b). There were no statistically significant differences in 1/W and 2/W frequency in group (c) of sterile non-healing and healing fractures (Fig. 1c) and healing infected and sterile (Fig. 1d). Taken together, mutation of TLR4 1/W gene was found highest in the infected non-healing subgroup.
Fig. 1.
Comparison of the frequency rate of TLR4 gene mutation in positions Asp299Gly (left) and Thr399Ile (right) marked as 1/W and 2/W, respectively, a in patients with delayed fracture healing and viable bacteria isolated from fracture gaps (darkly shaded bar) and patients with delayed healing and sterile fracture gaps (lightly shaded bar). W wild allele; 1/W, 2/W mutations (%), Chi-square test, *P < 0.05, b with delayed healing and bacteria isolated from fracture (darkly shaded bar) and patients with uneventful healing fractures also with bacteria (lightly shaded bar) (NS), c with delayed healing and sterile gaps (darkly shaded bar) and patients with uneventful healing fractures also with sterile gaps (lightly shaded bar) (NS), d with uneventful healing and bacteria isolated from fracture (darkly shaded bar) and uneventful healing fractures also without bacteria (lightly shaded bar) (NS)
Analysis of TGF-β gene polymorphism
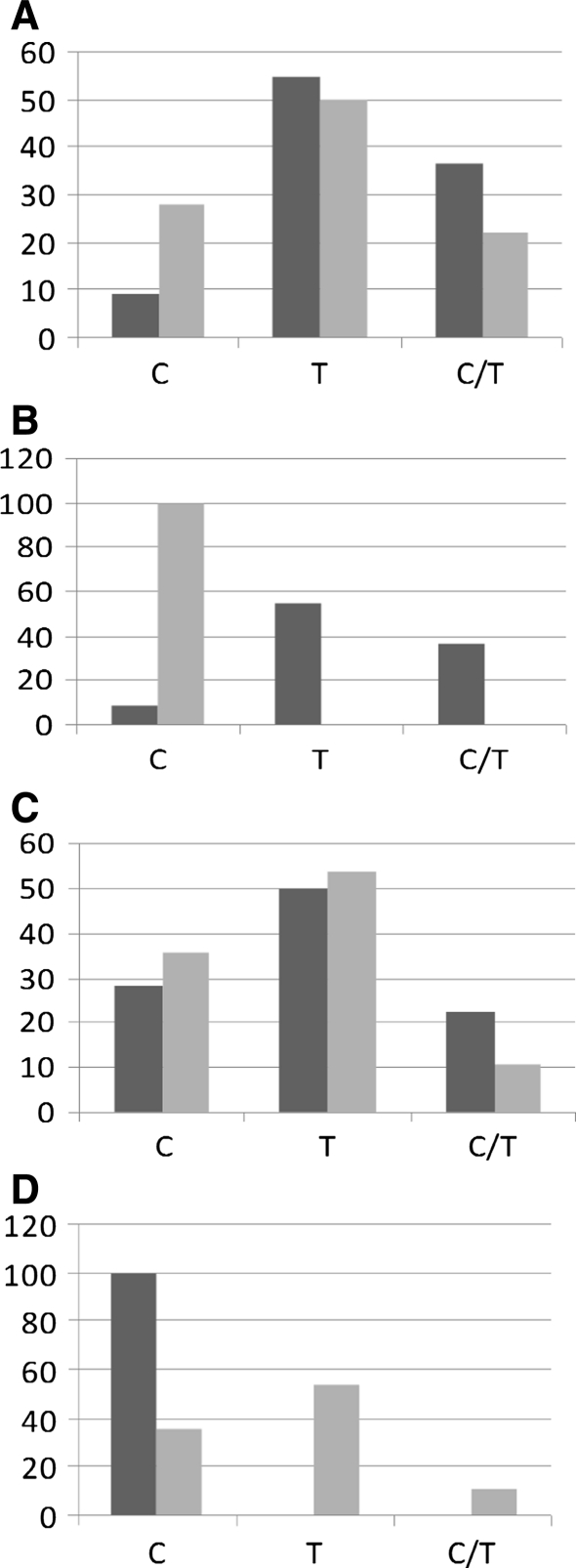
In the TGF-β codon 10 a significantly higher frequency of mutated homozygote T and heterozygote C/T in the non-healing infected versus non-healing sterile was observed (P < 0.05) (Fig. 2a). Similar difference was observed in the non-healing infected versus healing infected subgroup (P < 0.05) (Fig. 2b). In the subgroups without infection no differences between wild and mutated gene frequencies were seen (Fig. 2c), whereas in the healing subgroup dominated the wild homozygote C (Fig. 2d).
Fig. 2.
Comparison of the frequency rate of TGF-β codon 10 gene mutation a in patients with delayed fracture healing and viable bacteria isolated from fracture gaps (darkly shaded bar) and patients with delayed healing and sterile fracture gaps (lightly shaded bar).W wild allele; 1/W, 2/W mutations (%), Chi-square test, *P < 0.05, b with delayed healing and bacteria isolated from fracture (darkly shaded bar) and patients with uneventful healing fractures also with bacteria (lightly shaded bar) (NS), c with delayed healing and sterile gaps (darkly shaded bar) and patients with uneventful healing fractures also with sterile gaps (lightly shaded bar) (NS), d with uneventful healing and bacteria isolated from fracture (darkly shaded bar) and uneventful healing fractures also without bacteria (lightly shaded bar) (NS)
Analysis of other genes polymorphism
The genetic analysis of mutation frequency of genes for CRP (G1059C), IL1ra (genotype 2/2), IL-6 (G176C), CD14 (G-159T), TLR2 (G2259A) and CCR2 (G190A) did not reveal differences between (a) non-healing infected and non-healing sterile, (b) non-healing infected and healing infected, (c) non-healing sterile and healing sterile and (d) healing infected and healing sterile subgroups.
Proteins in fracture gaps
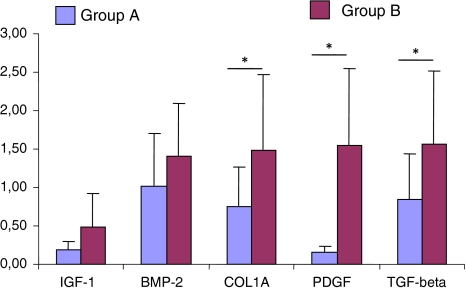
Western blot analysis of the fracture callus key factors playing role in the bone union process and skeletal tissue regeneration showed reduced expression of all investigated factors in the delayed healing fractures compared with healing uneventfully. Statistically significant changes were observed for TGF-β, PDGF and collagen type I [0.85 ± 0.59 vs. 1.56 ± 0.96; 0.16 ± 0.07 vs. 1.55 ± 1.00 and 0.75 ± 0.52 vs. 1.9 ± 0.49 (optical density ratio), respectively] (all P < 0.05) (Fig. 3). Insulin-like growth factor (IGF-1) and bone morphogenetic protein 2 (BMP-2) showed an insignificant decrease.
Fig. 3.
Western blot results of expression of IGF-1, BMP-2, Coll-1A, PDGF and TGF-β in fracture gap tissue of non-healing (group A) versus healing (group B) fractures (optical density units, n = 26, mean values ± SD). *P < 0.05, Chi-square test
Discussion
Delayed fracture repair with lack of contact between bone fragments including bone fragment loss was observed in above 10% of our clinical material. Approximately 35% of these cases could be attributed to unsuccessful surgery or immobilization. The remaining 65% suffered from delayed fracture repair without known causative factors. Since our previous data showed that fracture gaps of non-healing fractures are not sterile [7], we concentrated upon genetic background factors crucial for pathogen recognition and elimination, namely CRP (G1059C), IL1ra (genotype 2/2), IL-6 (G176C), CCR2 (G190A), CD14 (G-159T), TLR2 (G2259A) and TLR4 (Asp299Gly and Thr399Ile) and wound healing TGF-β (G25C). Mutation at the listed points could predispose for bacterial colonization of tissues.
In our studies, bacterial isolates were detected in 31% of non-healing gaps compared with 16% in the uneventfully healing fractures. Beside live bacterial cells, the presence of bacterial DNA (16S rRNA) was documented in around 60% in both groups. This material originated from both live and dead bacteria. Since frequency of microbial genetic material validated by 16S rRNA was similar in both A and B groups, but frequency of live bacteria differed significantly, we concluded that there was impairment of the mechanisms controlling live bacterial cell recognition and elimination. The question, whether pathogens regularly reside in deep tissues or colonize them after trauma, as well as how they can affect and are affected by host immune cells enabling them to survive without induction of inflammation, remains unanswered.
In all subgroups with viable pathogens isolated from fracture gaps the frequency of TLR4 mutated allel 1/W was higher, compared with subgroups with sterile fracture gaps. Each TLR enables recognition of a specific molecule. TLR4s are activated by LPS, HSP-60 and HSP-70. Also, staphylococcal exotoxin and leukocidin possess the capability to activate TLR4 [8]. These are mainly the live proliferating bacterial cells which are easily recognized by the host macrophage TLR4s [9]. Single nucleotide mutations of genes coding TLRs and their receptors are able to hinder the process of pathogen recognition and elimination. TLR4 mutations were reported to correspond with prolonged pathogen elimination [10] and increased number of infectious diseases, such as oral infections (periodontitis), recurrent lung infections and others [11, 12]. Montes et al. [13] showed an increased risk of musculoskeletal infections in Asp299Gly mutants of gene coding TLR4. In other studies single-nucleotide polymorphism of genes encoding CD14 (C-159T) and TLR4 (Asp299Gly) was reported to increase susceptibility of bacterial [14] and fungal infections [15].
The fracture gaps also contained bacterial genetic material (16S rRNA). Bacterial DNA of disintegrated microbes can be stored in tissue matrix or be present in local macrophages in a “dormant” state, not activating the host immune cells because of lack of direct physical contact with TLR4. However, activation may take place during tissue mechanical destruction and abundant recruitment of host immune cells. Subsequently bacterial genetic material will be eliminated. Mutation of TLR4 genes can make them ineffective in recognition of 16S RNA. Our cases with no TLR4 mutation and satisfactory bone union despite the fact that the presence of viable pathogens in the fracture gap proves abnormal function of the mutated TLR4 1/W gene.
TGF-β plays a key role in healing and regeneration of soft tissue wounds and bone fractures. Atilla et al. [16] and Babel et al. [17] reported C-allele bearer of the TGF-β gene for increased risk of periodontitis. TGF-β codon 25 gene polymorphisms (GG allele) were also reported to predispose to bacterial and viral infections [18]. Although the correlation of TGF-β codon 25 polymorphism with impaired healing was documented, such a correlation could not be found for codon 10 [19]. In our studies, the TGF-β codon 10 showed significantly higher frequency of mutated homozygote T and heterozygote C/T in the non-healing infected versus non-healing sterile (P < 0.05). Similar difference was observed in the non-healing infected versus healing infected subgroups. In the subgroups without infection no differences between wild and mutated gene frequencies were observed. Observed by us TGF-β genetic profile correlated with the expression of TGF-β proteins in gap tissue validated by western blot analysis. There was significantly less TGF-β protein in the non-healing infected compared with healing non-infected gap tissue. The same was observed for all the remaining proteins as IGF 1, BMP, Coll 1A and PDGF. This low protein expression might be the effect of enzymatic activity of bacterial cells as well as genetic defects.
We did not observe significant differences in frequency of mutated genes for CRP (G1059C), IL1ra (genotype 2/2), IL-6 (G176C), CD14 (G-159T), TLR2 (G2259A) and CCR2 (G190A) between the non-healing and healing fractures, although its has been reported that these mutant genes may play role in wound healing and inflammation.
IL-1ra polymorphism was reported to predispose to loosening of the orthopedic implants [20], most probably due to the impact on expression of molecular markers of systemic inflammation.
Kolundzić et al. [21] showed that IL-6 polymorphism G-174A increases the risk of implant loosing, but Malik et al. [22] found no correlation with IL-6 G-174C polymorphism and an increased risk of septic loosening of the hip endoprosthesis.
Two polymorphisms in the exon part of TLR2, which attenuate receptor signaling, enhance the risk of acute severe infections. Lorenz et al. [23] reported on the clinical importance of mutations in TLR2 and susceptibility to staphylococcal infection.
CCR2, chemokine MCP-1/CCL2 ligand, regulates infiltration of mononuclear cells to the sites of inflammation, playing the role in control and elimination of bacterial infections [24].
Taken together, we showed that individuals bearing T and C/T allele of TGF-β gene codon 10, and mutated TLR 4 gene W/1 (Asp299Gly) may be predisposed to impaired recognition and elimination of bacteria, leading to prolonged pathogen existence in fracture gaps and pathogen, depending on fracture healing delays. In our studies, CRP (G1059C), IL1ra (genotype 2/2), IL-6 (G176C), CD14 (G-159T), TLR2 (G2259A) and TLR4 (Thr399Ile) polymorphisms did not coincide with delay of bone fracture healing.
Acknowledgments
This work was financially supported by Ministry of Science (Grant No. 2 P05C 082 29).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Grzegorz Szczęsny, Phone: +48-22-6086410, FAX: +48-22-6685334, Email: g.szczesny@cmdik.pan.pl.
Waldemar L. Olszewski, Email: wlo@cmdik.pan.pl
References
- 1.Bostrom MP, Camacho NP. Potential role of bone morphogenetic proteins in fracture healing. Clin Orthop Relat Res. 1998;355(Suppl):S274–S282. doi: 10.1097/00003086-199810001-00028. [DOI] [PubMed] [Google Scholar]
- 2.Einhorn TA. The cell and molecular biology of fracture healing. Clin Orthop Relat Res. 1998;355(Suppl):S7–S21. doi: 10.1097/00003086-199810001-00003. [DOI] [PubMed] [Google Scholar]
- 3.Schindeler A, McDonald MM, Bokko P, et al. Bone remodeling during fracture repair: the cellular picture. Semin Cell Dev Biol. 2008;19:459–466. doi: 10.1016/j.semcdb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Schmidmaier G, Lucke M, Schwabe P, et al. Collective review: bioactive implants coated with poly(d, l-lactide) and growth factors IGF-I, TGF-beta1, or BMP-2 for stimulation of fracture healing. J Long Term Eff Med Implants. 2006;16:61–69. doi: 10.1615/jlongtermeffmedimplants.v16.i1.70. [DOI] [PubMed] [Google Scholar]
- 5.Linkhart TA, Mohan S, Baylink DJ. Growth factors for bone growth and repair: IGF, TGF-beta and BMP. Bone. 1996;19(1 Suppl):1S–12S. doi: 10.1016/S8756-3282(96)00138-X. [DOI] [PubMed] [Google Scholar]
- 6.Szczęsny G. Molecular aspects of bone healing and remodeling. Pol J Pathol. 2002;53:145–153. [PubMed] [Google Scholar]
- 7.Szczęsny G, Interewicz B, Swoboda-Kopeć E, et al. Bacteriology of callus of closed fractures of tibia and femur. J Trauma. 2008;65:837–842. doi: 10.1097/TA.0b013e3181469d44. [DOI] [PubMed] [Google Scholar]
- 8.Inden K, Kaneko J, Miyazato A, et al. Toll-like receptor 4-dependent activation of myeloid dendritic cells by leukocidin of Staphylococcus aureus. Microbes Infect. 2009;11:245–253. doi: 10.1016/j.micinf.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 9.McAleer JP, Vella AT. Understanding how lipopolysaccharide impacts CD4 T-cell immunity. Crit Rev Immunol. 2008;28:281–299. doi: 10.1615/critrevimmunol.v28.i4.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferwerda B, McCall MB, Verheijen K, et al. Functional consequences of toll-like receptor 4 polymorphisms. Mol Med. 2008;14:346–352. doi: 10.2119/2007-00135.Ferwerda. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rohde G, Klein W, Arinir U, et al. Association of the ASP299GLY TLR4 polymorphism with COPD. Respir Med. 2006;100:892–896. doi: 10.1016/j.rmed.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 12.Noack B, Görgens H, Lorenz K, et al. TLR4 and IL-18 gene variants in chronic periodontitis: impact on disease susceptibility and severity. Immunol Invest. 2009;38:297–310. doi: 10.1080/08820130902846290. [DOI] [PubMed] [Google Scholar]
- 13.Montes AH, Asensi V, Alvarez V, et al. The Toll-like receptor 4 (Asp299Gly) polymorphism is a risk factor for Gram-negative and haematogenous osteomyelitis. Clin Exp Immunol. 2006;143:404–413. doi: 10.1111/j.1365-2249.2005.03002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biebl A, Muendlein A, Kazakbaeva Z, et al. CD14 C-159T and toll-like receptor 4 Asp299Gly polymorphisms in surviving meningococcal disease patients. PLoS One. 2009;7(4):e7374. doi: 10.1371/journal.pone.0007374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van der Graaf CA, Netea MG, Morré SA, et al. Toll-like receptor 4 Asp299Gly/Thr399Ile polymorphisms are a risk factor for Candida bloodstream infection. Eur Cytokine Netw. 2006;17(1):29–34. [PubMed] [Google Scholar]
- 16.Atilla G, Emingil G, Köse T, et al. TGF-beta1 gene polymorphisms in periodontal diseases. Clin Biochem. 2006;39:929–934. doi: 10.1016/j.clinbiochem.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Babel N, Cherepnev G, Babel D, et al. Analysis of tumor necrosis factor-alpha, transforming growth factor-beta, interleukin-10, IL-6, and interferon-gamma gene polymorphisms in patients with chronic periodontitis. J Periodontol. 2006;77:1978–1983. doi: 10.1902/jop.2006.050315. [DOI] [PubMed] [Google Scholar]
- 18.Pereira FA, Pinheiro da Silva NN, Rodart IF, et al. Association of TGF-beta1 codon 25 (G915C) polymorphism with hepatitis C virus infection. J Med Virol. 2008;80:58–64. doi: 10.1002/jmv.21011. [DOI] [PubMed] [Google Scholar]
- 19.Bayat A, Bock O, Mrowietz U, et al. Genetic susceptibility to keloid disease and hypertrophic scarring: transforming growth factor beta1 common polymorphisms and plasma levels. Plast Reconstr Surg. 2003;111:535–543. doi: 10.1097/01.PRS.0000041536.02524.A3. [DOI] [PubMed] [Google Scholar]
- 20.Rollins BJ. Monocyte chemoattractant protein-1: a potential regulator of monocyte recruitment in inflammatory disease. Mol Med Today. 1996;2:198–204. doi: 10.1016/1357-4310(96)88772-7. [DOI] [PubMed] [Google Scholar]
- 21.Kolundzić R, Orlić D, Trkulja V, et al. Single nucleotide polymorphisms in the interleukin-6 gene promoter, tumor necrosis factor-alpha gene promoter, and transforming growth factor-beta1 gene signal sequence as predictors of time to onset of aseptic loosening after total hip arthroplasty: preliminary study. J Orthop Sci. 2006;11:592–600. doi: 10.1007/s00776-006-1069-y. [DOI] [PubMed] [Google Scholar]
- 22.Malik MH, Jury F, Bayat A, et al. Genetic susceptibility to total hip arthroplasty failure: a preliminary study on the influence of matrix metalloproteinase 1, interleukin 6 polymorphisms and vitamin D receptor. Ann Rheum Dis. 2007;66:1116–1120. doi: 10.1136/ard.2006.062018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenz E, Mira JP, Cornish KL, et al. A novel polymorphism in the toll-like receptor 2 gene and its potential association with staphylococcal infection. Infect Immun. 2000;68:6398–6401. doi: 10.1128/IAI.68.11.6398-6401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murdoch C, Finn A. Chemokine receptors and their role in inflammation and infectious diseases. Blood. 2000;95:3032–3034. [PubMed] [Google Scholar]