Abstract
The 5′ portion of the Sindbis virus (SIN) genome RNA is multifunctional. Besides initiating translation of the nonstructural polyprotein, RNA elements in the 5′ 200 bases of the SIN genome RNA, or its complement at the 3′ end of the negative-strand intermediate, play key roles in the synthesis of both negative- and positive-strand RNAs. We used here a combination of genetic and biochemical approaches to further dissect the functions of this sequence. Replacement of the SIN 5′ end in defective-interfering (DI) and genome RNAs with sequences from a distantly related alphavirus, Semliki Forest virus (SFV), resulted in nonviable chimeras. The addition of five nucleotides from the 5′ terminus of SIN restored negative-strand RNA synthesis in DI genomes but not their replication in vivo. Pseudorevertants of various SFV-SIN chimeras were isolated, and suppressor mutations were mapped to AU-rich sequences added to the 5′ end of the original SFV 5′ sequence or its “deleted” versions. Early pseudorevertants had heterogeneous 5′ termini that were inefficient for replication relative to the parental SIN 5′ sequence. In contrast, passaging of these pseudorevertant viral populations in BHK cells under competitive conditions yielded evolved, more homogeneous 5′-terminal sequences that were highly efficient for negative-strand synthesis and replication. These 5′-terminal sequences always began with 5′-AU, followed by one or more AU repeats or short stretches of oligo(A). Further analysis demonstrated a positive correlation between the number of repeat units and replication efficiency. Interestingly, some 5′ modifications restored high-level viral replication in BHK-21 cells, but these viruses were impaired for replication in the cells of mosquito origin. These studies provide new information on sequence determinants required for SIN RNA replication and suggest new strategies for restricting cell tropism and optimizing the packaging of alphavirus vectors.
The alphavirus genus of the family Togaviridae is comprised of about 30 known members (30). Sindbis virus (SIN), a prototype member of the genus, has been extensively studied to obtain information about fundamental issues of alphavirus replication, and the results from studies of this virus are largely applicable to other members of the genus (36).
The SIN genome is a single-stranded RNA of positive polarity of about 12 kb in length. Like most cellular mRNAs, it is capped at the 5′ end and contains a 3′-terminal poly(A) tail (36). Upon delivery into the cells, this RNA serves as the template for synthesis of nonstructural proteins (nsPs) that form the replicase-transcriptase complex required for genome RNA replication and subgenomic 26S RNA transcription. The latter RNA is coterminal with the 3′ one-third of the genome and encodes the viral structural proteins (35). Replication of the viral genome and transcription of the subgenomic RNA are highly regulated processes. Early in SIN infection, viral nsPs are synthesized as two polyproteins, P123 and P1234. The latter is cleaved in cis by a papain-like protease in nsP2 to generate P123 and nsP4, the RNA-dependent RNA polymerase (RdRp). This partially processed protein complex, P123/nsP4, efficiently synthesizes full-length negative-strand intermediates but is inefficient for positive-strand RNA synthesis. Further processing of P123 to nsP1, nsP2, and nsP3, performed in trans by nsP2 and nsP2-containing polyproteins, leads to the formation of a replicase consisting of nsP1, nsP2, nsP3, and nsP4. This complex is efficient for the synthesis of positive-sense genome and subgenomic RNAs (13-15, 32, 33). Both of these RNAs are required during the later stages of infection, when high levels of structural proteins and genome RNAs are needed for efficient formation of progeny virus.
Changes in replicase composition that correlate with regulated synthesis of the three RNA species suggest the use of different promoter elements. Initially, putative promoters were postulated on the basis of sequence conservation among alphaviruses, and their importance was later confirmed by extensive mutagenesis studies. One 24-nucleotide (nt) conserved sequence element (CSE) was identified upstream of and including the start of the subgenomic RNA (16, 23). The complement of this sequence in the negative strand is required for initiation of subgenomic mRNA transcription. A second CSE, a sequence of 19 nt immediately preceding the 3′ poly(A) tail, was proposed as a core promoter for synthesis of the genome-length, negative-strand RNA (11, 24). However, the same CSE is present at the 3′ end of the alphavirus subgenomic RNAs, and the absence of subgenomic intermediate implied that the 19-nt CSE was not sufficient for promoter activity. Other RNA elements essential for replication were found at the 5′ end of SIN genome RNA and include the 5′ UTR and a 51-nt CSE (6, 21, 22) located in the nsP1 coding sequence. Replacement of the SIN 5′ untranslated region (UTR) by heterologous 5′ UTRs derived from Semliki Forest virus (SFV) abolished RNA replication in vivo and negative-strand RNA synthesis in vitro (6). The 5′ end of the SIN genome was also shown to bind a limiting component(s) essential for negative-strand initiation (6). Thus, SIN genome replication requires an interaction between the 3′ and the 5′ ends of the genome to initiate negative-strand synthesis. The factors responsible for this interaction remain to be defined. Panhandle structures have been visualized by electronic microscopy and characterized biophysically (4), but perfectly complementary sequences near the genomic 5′ and 3′ termini are not present. Protein factors, in particular those of the host translation apparatus, have also been proposed as candidates to bring the 5′ and 3′ ends in close proximity, as seen in translation of cellular and viral mRNAs (6, 10). The 51-nt CSE (replication enhancer) increases RNA replication, and its integrity is more important for SIN replication in mosquito cells than in mammalian cells (22).
In addition to the roles of the 5′ end of the SIN genome RNA in translation and negative-strand initiation, the complement of this signal-intense sequence functions in positive-strand synthesis. Mutations have been identified that block RNA replication in vivo but do not affect (or even enhance) RNA synthesis in the in vitro system (where only negative-strand synthesis occurs) (6).
To further probe the function of the SIN 5′ UTR in RNA replication, we designed SIN genomes with 5′ UTRs from heterologous alphavirus, SFV. Some of these chimeras were lethal for virus replication, but viable variants containing compensating mutations could be selected. To characterize the effects of these mutations and pseudoreversions on different steps in RNA replication, we used a combination of in vitro and in vivo approaches, including an assessment of fitness in different host cell environments. Our data further define the elements in the 5′ terminus of SIN genome RNA needed for efficient replication in mammalian cells.
MATERIALS AND METHODS
Cell cultures.
BHK-21 cells were obtained from the American Type Culture Collection, Rockville, Md. These cells were propagated in alpha minimal essential medium (α-MEM) supplemented with 10% fetal bovine serum (FBS) and vitamins. Mosquito C710 cells were obtained from Henry Huang (Washington University, St. Louis, Mo.). They were propagated in Dulbecco modified Eagle medium supplemented with 10% heat-inactivated FBS and 10% tryptose phosphate broth.
Plasmid constructs.
All plasmid constructions were prepared by standard recombinant DNA techniques. The parental p5′SIN/DI and pToto1101 plasmids coding the defective-interfering (DI) RNA and infectious SIN viral genome were described elsewhere (6, 29). Briefly, the p5′SIN/DI contained the promoter for SP6 RNA polymerase, followed by 425 nt of the SIN 5′ end, 7335 to 7646 nt of the SIN genome encoding the promoter of the subgenomic RNA, a TCTAGACC sequence, an 1,654-nt sequence encoding the entire luciferase gene, and a 421-nt sequence containing the SIN 3′ UTR, poly(A), followed by an EcoRI restriction site. p5′SFV/DI and p5′SFVSL1/DI, described elsewhere (6), had essentially the same sequence as p5′SIN/DI, except the 5′ fragment of SIN (nt 1 to 40) was replaced by nt 1 to 65 and nt 1 to 27 of SFV, respectively. The same modifications in the 5′ UTR were made to design p5′SFV/SIN and p5′SFVSL1/SIN that contained cDNAs of the entire genome of SIN with the SFV-derived 5′ end. For all other constructs, fragments containing modified 5′ UTRs were generated by PCR amplification and cloned into p5′SIN/DI or pToto1101 by using the SacI restriction site, located upstream of the SP6 promoter, and the MfeI site located in the SIN 5′ UTR. All of the 5′ UTR-coding fragments were sequenced before we performed the SP6 transcription reactions. The sequences of the designed heterologous 5′ UTRs are shown in the figures and are described in Results.
The TSG/Pac replicon of SIN used as donor of the replicative complex has been described previously (5).
RNA transcriptions.
Plasmids were purified by centrifugation in CsCl gradients. Prior to transcription, the DI RNA genome-coding and viral genome-coding plasmids were linearized by treatment with EcoRI and XhoI, respectively. RNAs were synthesized by SP6 RNA polymerase in the presence of cap analog (29). The yield and integrity of transcripts were monitored by gel electrophoresis under nondenaturing conditions. For electroporation, aliquots of transcription reactions were used without additional purification, and for the in vitro replication system the RNAs were purified by phenol-chloroform extraction and ethanol precipitation.
In vivo replication assay.
Testing of DI RNAs replication was performed by using a previously described in vivo replication system. Briefly, BHK-21 cells were coelectroporated with 4 μg of TSG/Pac replicon, supplying the RdRp, and 6 μg of DI RNAs (17, 18). Transfected cells were divided into four dishes and incubated at 37°C. After 4 h, cells in one dish were lysed in luciferase lysis buffer (Promega), and luciferase activity was assessed by using the luciferase assay system according to recommendations of the manufacturer (Promega). To rule out the possibility that lower Luc activity was a result of spontaneous mutations in the Luc gene introduced during the cloning procedure, the intracellular RNAs in the second dish were metabolically labeled with [3H]uridine (20 μCi/ml) in the presence of 1 μg of ActD/ml at between 1 and 4 h posttransfection. RNAs were isolated and analyzed by gel electrophoresis in the previously described conditions (1).
In vitro negative-strand RNA synthesis assay.
Polymerase extracts from the cells infected with vaccinia viruses expressing SIN P123 and nsP4 were prepared and used for in vitro negative-strand RNA synthesis as previously described (14). RNAs were isolated by phenol-chloroform extraction and ethanol precipitated. Then RNAs were denatured with glyoxal and then separated by electrophoresis and visualized by autoradiography. The quantitation of radioactivity was performed by using a Storm 860 PhosphorImager (Molecular Dynamics).
Infectious center assay.
In standard experiments, 1 μg of in vitro-synthesized, full-length RNA transcripts (or 4 μg of 5′SFV/SIN and 5′SFVSL1/SIN RNA) was used per electroporation. Tenfold dilutions of electroporated BHK-21 cells were seeded in six-well tissue culture plates containing 5 × 105 naive cells per well. After 1 h of incubation at 37°C in an 5% CO2 incubator, cells were overlaid with 2 ml of 0.5% Ultra-Pure agarose (Invitrogen) containing MEM supplemented with 3% FBS. Plaques were stained with crystal violet after 2 days of incubation at 37°C. The remaining cells were seeded into 100-mm dishes and incubated until the cytopathic effect (CPE) was observed.
Sequencing of the 5′ ends of viral genomes.
The 5′ ends of the selected SIN variants were sequenced on plaque-purified viruses. Plaques were isolated from the agarose overlay during the infectious center assay before being stained with crystal violet. Viruses were eluted in 1.5 ml of α-MEM supplemented with 10% FBS for several hours, and 0.5 ml was used to infect 5 × 105 BHK-21 cells in 35-mm dishes. After 1 h of virus adsorption at 37°C, an additional 1 ml of α-MEM-10% FBS was added, and the incubation was continued until the CPE was observed. Then, media were harvested, and RNAs were isolated from the cells by TRIzol by using the procedure recommended by the manufacturer (Invitrogen). DNA fragments representing the 5′-terminal sequences of virus isolates were synthesized by using a commercially available FirstChoice RLM-RACE (rapid amplification of cDNA ends) kit according to the procedure recommended by the manufacturer (Ambion). The primer used for cDNA synthesis was complementary to nt 417 to 435 of the SIN genome; the primers used for PCR amplification were (i) complementary to nt 241 to 260 of SIN genome and (ii) the 5′ RACE inner primer supplied with the kit. Fragments were purified by agarose gel electrophoresis and cloned into the plasmid pRS2. Multiple, independent clones were sequenced to determine variations in the 5′ end of the genomes.
Viral growth analysis.
BHK-21 and C710 cells were seeded at a concentration of 5 × 105 and 106 cells/35-mm dish, respectively. After 4 h of incubation at appropriate temperature, monolayers were infected at the multiplicities of infection (MOIs) indicated in the figure legends for 1 h, washed three times with phosphate-buffered saline, and overlaid with 1 ml of complete medium. At the indicated times postinfection, media were replaced by fresh media, and virus titers in the harvested samples were determined by plaque assay on BHK-21 cells (12).
RESULTS
Importance of 5′-terminal nucleotides of SIN for positive-strand RNA synthesis.
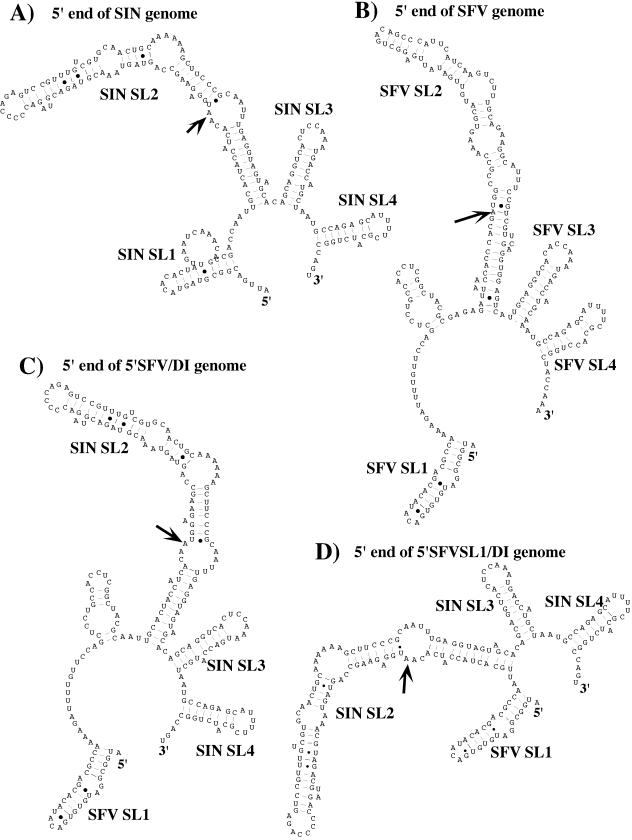
In a previous study, we made a number of modifications in the 5′ UTR of SIN-specific DI RNAs and demonstrated that replacement of the SIN 5′ UTR (Fig. 1A) by a heterologous 5′ UTR derived from the distantly related alphavirus, SFV (Fig. 1B and C), or by the SFV 5′-terminal stem-loop structure SL1 (Fig. 1D), produced RNAs incapable of replication (6). The most striking observation was that RNAs with these heterologous 5′ UTRs were highly inefficient templates for negative-strand RNA synthesis. To continue the investigation of the 5′ sequence elements involved in both positive- and negative-strand RNA synthesis, we designed several chimeric, luciferase-coding DI RNAs that contained the first five nucleotides of the SIN genome upstream of the 5′-terminal stem-loop structure SL1 of SFV (Fig. 2). In the 5ntSIN-SFV/DI RNA, 5′-terminal SIN nucleotides were followed by SL1 and an interstem fragment of SFV fused with SL2, SL3, and SL4 of SIN. The 5ntSIN-SFVSL1/DI genome was similar, but the interstem fragment between SFVSL1 and SINSL2 was deleted. As in the SIN genome, the 5′-terminal nucleotides in both RNAs were predicted by M-fold not to be involved in SL1 stem formation (data not shown).
FIG. 1.
Computer-predicted secondary structures of the 5′ end of SIN genome present in 5′SIN/DI RNA (A), 5′ end of SFV genome (B), 5′ end of 5′SFV/DI RNA (C), and 5′ end of 5′SFVSL1/DI RNA (D). Arrows indicate the initiating AUG codons. The computer-predicted stem-loop structures are indicated as SL1 to SL4. The SL3 and SL4 represent the 51-nt CSE.
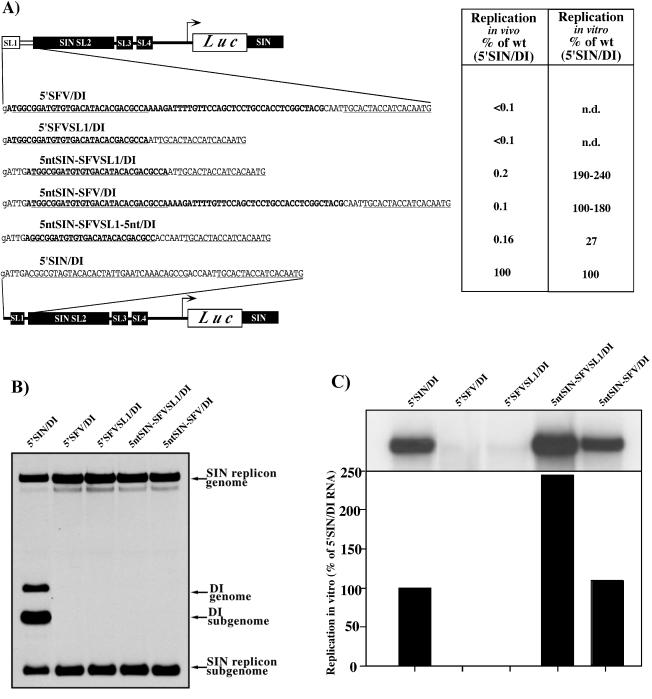
FIG. 2.
Replication of the DI RNAs with the 5′ ends containing SFV sequences. (A) Schematic representation and nucleotide sequences of the designed 5′ ends in the DI RNAs and their replication activity in vivo and in vitro. SFV and SIN sequences are shown in boldface and plain type, respectively. Underlined type represents the sequences predicted to form the stem-loop structures. Lowercase type indicates an additional 5′ nucleotide resulting from the SP6 in vitro transcription. (B) Autoradiograph of the dried gel, with the RNAs metabolically labeled with [3H]uridine after coelectroporation of the substrate DI RNAs with SIN replicon. The detailed conditions of in vivo experiments are described in Materials and Methods. (C) Autoradiograph of the dried gel with the 32P-labeled RNAs isolated from in vitro negative-strand RNA synthesis reactions. The lower panel shows the amount of radioactivity contained in the displayed RNA bands as measured by using a phosphorimager.
The addition of these five SIN nucleotides dramatically increased in vitro replication of the chimeric DI RNAs (Fig. 2A and C), yielding synthesis of negative-strand RNAs similar to that observed on 5′SIN/DI RNAs with homologous, SIN-derived, 5′ and 3′ ends. However, the addition of the SIN-specific sequence did not markedly improve replication of these DI RNAs in vivo (Fig. 2A and B). These chimeras replicated 1,000-fold less well than the DI RNAs with the authentic SIN 5′ UTR, although they were 10- to 20-fold more efficient than their counterparts without the five SIN 5′-terminal nucleotides (Fig. 2A and B). This finding suggested that, by adding these five SIN 5′-terminal nucleotides, we restored the activity of the promoter for negative-strand but not for positive-strand RNA synthesis.
In an attempt to increase positive-strand RNA synthesis in vivo, we modified the interstem region between SFV SL1 and SIN SL2 in the context of 5ntSIN-SFVSL1/DI (Fig. 2A). Insertion of three additional nucleotides made the 3′-terminal sequence of the negative-strand of 5ntSIN-SFVSL1-5nt/DI RNA capable of forming a stem structure that was similar, as determined by computer prediction, to the structure at the 3′ terminus of the SIN negative-strand template. This modification also did not increase the level of DI RNA replication in vivo, compared to 5ntSIN-SFVSL1/DI (Fig. 2A), and significantly reduced negative-strand RNA synthesis in vitro. This suggests that the determinants for positive-strand RNA synthesis are more complex than simple addition of a few 3′-terminal SIN-specific nucleotides to the negative-strand intermediate.
Selection of viable SIN pseudorevertants.
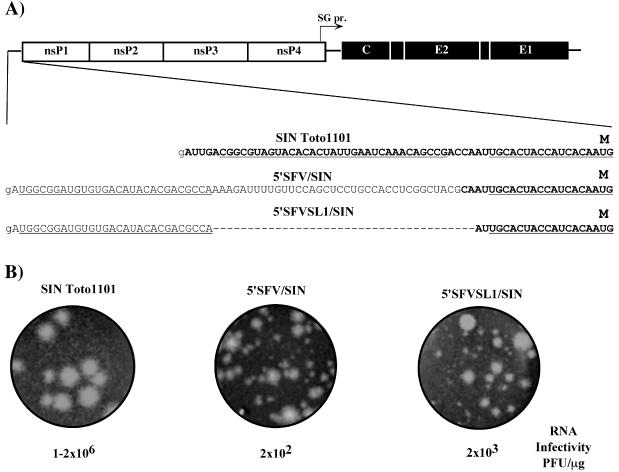
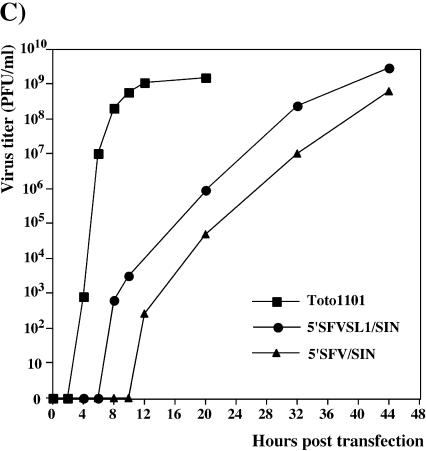
Selection of revertants and pseudorevertants of impaired mutants, has been a powerful tool in alphavirus genetics (3, 9). Given the advantage of using infectious virus for selection of rare variants, we designed a panel of chimeric viral genomes (Fig. 3A). 5′SFV/SIN and 5′SFVSL1/SIN had 5′ sequences that were the same as those previously described for 5′SFV/DI and 5′SFVSL1/DI RNAs, respectively (see Materials and Methods for further details). In vitro-synthesized RNAs were transfected into BHK-21 cells to assess their infectivity and to generate virus stocks. In an infectious center assay, 5′SFV/SIN and 5′SFVSL1/SIN RNAs were 4 and 3 orders of magnitude less infectious, respectively, than the SIN Toto1101 parent and produced heterogeneous plaques (Fig. 3B). In addition, both 5′SFV/SIN and 5′SFVSL1/SIN exhibited significant delays in growth after electroporation of transcript RNA (Fig. 3C). These observations suggested that heterologous 5′ ends impaired RNA replication to levels insufficient for productive infection. Delayed CPE and the low levels of plaque formation in infectious center assay were most likely due to the emergence of variants with increased replicative ability, because the 5′SFV/SIN and 5′SFVSL1/SIN RNAs demonstrated the same translational efficiency in vitro (in the rabbit reticulocyte lysate translation system), where the subsaturating amounts of RNAs were used (data not shown).
FIG. 3.
Selection of SIN variants with heterologous 5′ ends capable of replication in BHK-21 cells. (A) Nucleotide sequences of the 5′ ends of the RNAs used for electroporation. SFV and SIN sequences are shown in plain and in boldface type, respectively. Underlined type represents the sequences predicted to form the stem-loop structures. Lowercase type indicates an additional 5′ nucleotide resulting from the SP6 in vitro transcription. (B) Infectivity of in vitro-synthesized RNAs and morphology of plaques in an infectious center assay (see Materials and Methods for details). (C) Growth curves of viruses after transfection of 4 μg of in vitro-synthesized, full-length RNA transcripts of SIN Toto1101, 5′SFV/SIN, and 5′SFVSL1/SIN RNAs into BHK-21 cells. Aliquots were taken at the indicated time points postelectroporation, and virus titers were determined by using a standard plaque assay on BHK-21 cells.
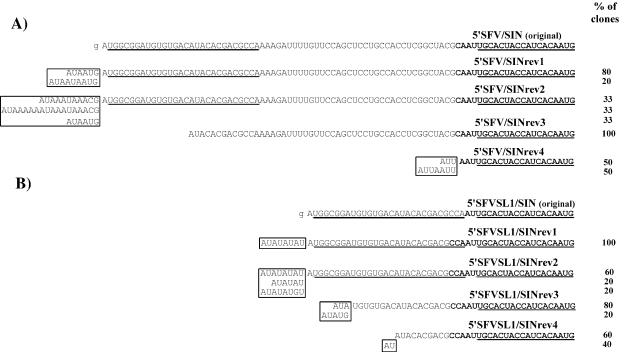
We randomly selected four plaques each for 5′SFV/SIN and 5′SFVSL1/SIN from the infectious center assay. The 5′ ends of these isolates were subjected to 5′ RACE, cloned into a plasmid, and sequenced. Multiple modifications were identified in the 5′ UTRs (Fig. 4). In some isolates, SFV SL1 was partially or completely deleted; others retained the stem-loop structure. The majority had additional 5′ sequences that were not present in the original RNA transcripts. A notable common feature was the presence of short sequences enriched with adenosine and uridine. All sequenced plaques shared the 5′-terminal AU, followed by repeating combinations of A's and U's: (AAU)1-2, (AU)1-4, [(A)3-6U]2-3, and (UAAU)1-2. The genome of 5′SFV/SINrev3 lacked additional bases but began with the sequence 5′-AUACA that was generated by deletion of 16 bases from the 5′ terminus.
FIG. 4.
Nucleotide sequences of the heterologous 5′ ends in the genomes of randomly selected plaque isolates of SIN pseudorevertants. Four micrograms of in vitro-synthesized, full-length RNA transcripts of 5′SFV/SIN (A) and 5′SFVSL1/SIN (B) RNAs were transfected into BHK-21 cells, and plaques were randomly selected directly during the infectious center assay. The 5′ ends of viral genomes were sequenced as described in Materials and Methods. SFV and SIN sequences are shown in plain and in boldface type, respectively. Underlined type represents the sequences predicted to form the stem-loop structures. Lowercase type indicates an additional 5′ nucleotide resulting from the SP6 in vitro transcription (present in the original in vitro-synthesized RNA). Boxes indicate heterologous sequences found in the 5′ termini. Multiple plasmids were sequenced for each plaque isolate, and the percentage of clones containing each acquired nucleotide sequence is indicated.
Apparent heterogeneity in the number of AU-rich repeats was found between different plasmid clones derived from each plaque isolate (Fig. 4, sequences in the boxes). To rule out the possibility that this heterogeneity was an artifact, the same method of the 5′ RACE was applied to determine 5′ sequences for plaque isolates of the SIN parent, Toto1101. Four plaques were selected from the infectious center assay, the RNA was subjected to 5′ RACE, and multiple plasmids were sequenced. All clones contained only the 5′ nucleotide sequence of SIN Toto1101 and lacked the additional guanosine that is present on in vitro transcripts synthesized by SP6 RNA polymerase (and eliminated during replication).
The results suggest that SIN chimeras with heterologous 5′ ends were able to acquire multiple modifications in the 5′ UTRs that presumably increased replication. Plaque-purified viruses also contained heterogeneous 5′-terminal sequences, indicating ongoing evolution of the 5′ promoter elements within individual plaques.
Replication of DI RNAs with modified 5′ UTRs.
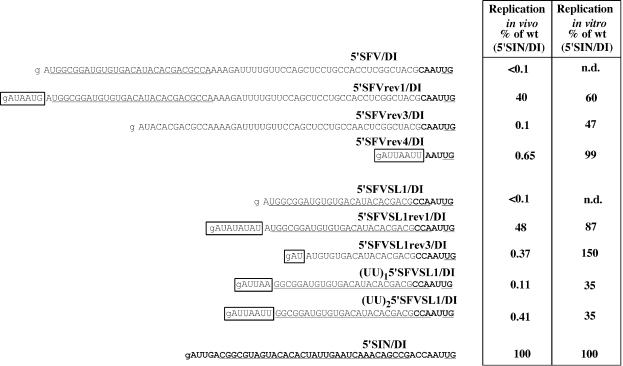
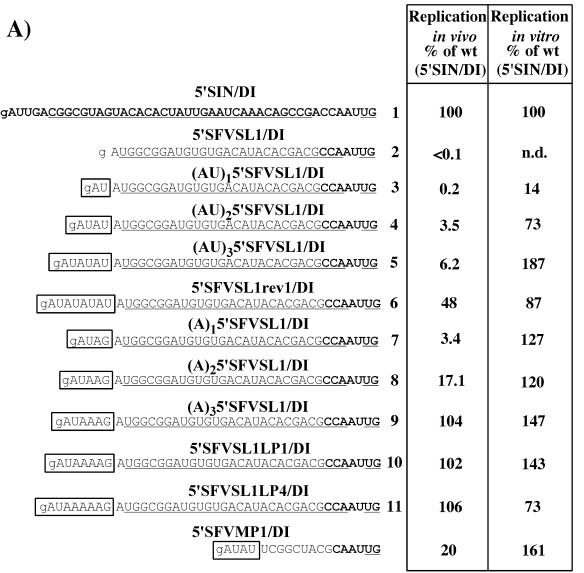
To directly test the ability of the 5′ UTRs identified in the pseudorevertants screen to drive replication, these fragments were first cloned into 5′SIN/DI RNA to replace the original SIN 5′ UTR (Fig. 5). The (UU)15′SFVSL1/DI and (UU)25′SFVSL1/DI RNAs were designed to test the effect of AUUAA and AUUAAUU sequences, which were found in 5′SFV/SINrev4 and were most similar to the authentic SIN 5′ terminus. The resulting DI RNAs were evaluated for their ability to replicate in vivo and in vitro by using previously described assays (6; see also Materials and Methods). All of these modified 5′ UTRs restored negative-strand RNA synthesis to the level that was 35 to 150% of that observed for 5′SIN/DI RNA (Fig. 5). However, the same RNAs demonstrated a wide spectrum of replication efficiencies in vivo, ranging from 0.1 to 48% of the 5′SIN/DI RNA parent (Fig. 5). This indicates that, for many of these sequences, the promoter for positive-strand RNA synthesis is still significantly impaired. Both 5′-AUUAA and 5′-AUUAAUU were inefficient for in vivo RNA replication, supporting the idea that it is not the presence of A's and U's alone but also their particular combination that determines the positive-strand promoter activity.
FIG. 5.
Replication of the DI RNAs with the 5′ ends derived from the 5′SFV/SIN and the 5′SFVSL1/SIN pseudorevertants. Nucleotide sequences of the designed 5′ ends in the DI RNAs, their replication activity in vivo, based on luciferase assay, and the in vitro negative-strand synthesis activity of the templates are shown. SFV and SIN sequences are in plain and boldface type, respectively. Underlining represents the sequences predicted to form the stem-loop structures. Boxes indicate the heterologous sequences acquired by viral pseudorevertants. Lowercase type indicates an additional 5′ nucleotide resulting from the SP6 in vitro transcription. These data represent one of a series of repeated experiments that generated very reproducible data. n.d., an undetectable level of negative-strand RNA synthesis.
Selection of SIN variants with more efficient 5′ promoter elements.
To identify the sequences that could support the efficient SIN RNA replication, the original viral stocks, generated after transfection of 5′SFV/SIN and 5′SFV SL1/SIN, were serially passaged under conditions where variants were competing with each other. Three passages were performed on BHK-21 cells at a high MOI (20 PFU/cell), followed by one passage at an MOI of 0.1 and an additional passage at an MOI of 0.01 PFU/cell. After passaging, the virus stocks formed more homogeneous plaques on BHK-21 cells than did stocks harvested directly after the RNA transfection. The 5′SFVSL1/SIN-derived virus contained only large plaque-forming variants (plaque size was indistinguishable from SIN Toto1101 [data not shown]); the 5′SFV/SIN virus still contained variants generating medium-sized plaques. Four plaques were randomly selected in each virus preparation (two large and two medium-sized plaques in 5′SFV/SIN-derived stock and four large plaques in 5′SFVSL1/SIN-derived stock), and the 5′-terminal sequences were analyzed (Fig. 6). Sequencing results confirmed that more-homogeneous viral populations had been selected. Variations in the 5′ ends of viruses within individual plaque-purified stocks were also minor. Furthermore, we did not detect a single variant with the same 5′-terminal sequence as seen in the primary isolates (Fig. 4).
FIG. 6.
Nucleotide sequences in the genomes of 5′SFV/SIN and 5′SFVSL1/SIN pseudorevertants after serial passaging. Viral libraries generated after electroporation of in vitro-synthesized RNAs were passaged three times on BHK-21 cells at a high MOI (20 PFU/cell), followed by one passage at an MOI of 0.1 and one passage at an MOI of 0.01 PFU/cell. Four plaques were randomly selected in both 5′SFV/SIN (A)- and 5′SFVSL1/SIN (B)-derived virus stocks. The 5′ ends of virus genomes were sequenced as described in Materials and Methods. SFV and SIN sequences are shown in plain and boldface type, respectively. Underlined type represents the sequences predicted to form the stem-loop structures. Lowercase type indicates an additional 5′ nucleotide resulting from the SP6 in vitro transcription (that is present in the original in vitro-synthesized RNA). Boxes indicate heterologous sequences found in the 5′ termini. Multiple plasmids were sequenced for each plaque isolate, and the percentage of clones containing each acquired nucleotide sequence is indicated.
The 5′ UTRs of the passaged isolates exhibited a number of similar features. The genomes of large-plaque-forming variants of both 5′SFV SL1/SIN- and 5′SFV/SIN-derived viruses started with 5′-AU, followed by oligo(A) (with minor variations in length between three and eight residues) and then by U (in 5′SFV/SIN-derived large-plaque-forming isolates). Similar to some of the pseudorevertants shown in Fig. 4A, these sequences were followed by additional guanosine, which could have arisen from the original SP6 transcripts (all of the plasmids contained the nonnative G in the +1 position of the promoter to ensure efficient transcription with SP6 polymerase), and by the unmodified SL1 sequence of the SFV5′ end. Only the 5′SFV/SIN MP1 and MP2 pseudorevertants (which were essentially the same) had different 5′ UTRs. These isolates formed considerably smaller plaques but were present in significant concentrations even after six passages. Their genomes also contained 5′-AU, followed by one or three AU repeats, but the entire SFV SL1 and major part of the interstem region between SL1 and SL2 was deleted. In the original stocks, as well as in the stocks after passaging, we did not detect pseudorevertants with the wild-type SIN 5′ sequence (5′-AUUGA or 5′-AUUGG).
Replication of DI RNAs with different repeating elements in the 5′ UTRs.
To evaluate the contribution of these sequences to RNA replication and, in particular, to test the effect of the repeating elements, we synthesized a set of 5′SFVSL1/DI RNAs containing 5′-AU, followed by different numbers of repeats: (AU)n or (A)n (Fig. 7A). These recombinant DI RNA genomes were analyzed for their ability to replicate in vivo and in vitro (see Materials and Methods). The efficiency of both positive- and negative-strand RNA synthesis was dependent on the number of repeats. Five AUs, followed by SFVSL1, in 5′SFVSL1rev1/DI yielded an RNA with 48% efficiency of the SIN 5′ UTR. In the in vitro assay, three AU repeats were even more efficient for negative-strand RNA synthesis than the authentic SIN 5′ terminus. Interestingly, two AU repeats in the context of 5′SFVMP1/DI RNA functioned 100-fold more efficiently than when they were fused with SFV SL1 in (AU)15′SFVSL1/DI construct, indicating that both the very 5′ terminus and downstream sequence of the 5′UTR strongly affect the activity of the promoter for positive-strand RNA synthesis.
FIG. 7.
Replication of the DI RNAs with different repeating elements in the 5′ ends. (A) Nucleotide sequences of the designed 5′ ends in the DI RNAs, replication activity of the RNAs in vivo, based on luciferase assay, and the in vitro negative-strand synthesis. SFV and SIN sequences are shown in plain and boldface type, respectively. Underlining represents the sequences predicted to form the stem-loop structures. Lowercase type indicates an additional 5′ nucleotide resulting from the SP6 in vitro transcription. Boxes indicate heterologous sequences that were not present in 5′SFV/SIN and 5′SFVSL1/SIN genomes. (B) Autoradiograph of the dried gel with the RNAs metabolically labeled with [3H]uridine after coelectroporation of the substrate DI RNAs with SIN replicon into BHK-21 cells. The detailed conditions of in vivo experiments are described in Materials and Methods. Numbers designated for DI RNAs in panel A correspond to the numbers on lanes on the gel as follows: 1, 5′SIN/DI; 2, 5′SFVSL1/DI; 3, (AU)15′SFVSL1/DI; 4, (AU)25′SFVSL1/DI; 5, (AU)35′SFVSL1/DI; 6, 5′SFVSL1rev1/DI; 7, (A)15′SFVSL1/DI; 8, (A)25′SFVSL1/DI; 9, (A)35′SFVSL1/DI; 10, 5′SFVSL1LP1/DI; and 11, 5′SFVSL1LP4/DI. These data represent one of two experiments that generated very similar data.
Similarly, increasing the number of adenosine residues between the 5′-AU and SFVSL1 augmented replication of the DI RNAs to the level of 5′SIN/DI both in vitro and in vivo. However, analysis of RNAs synthesized in vivo by gel electrophoresis revealed that (A)35′SFVSL1/DI, 5′SFVSL1LP1/DI, and 5′SFVSL1LP4/DI RNAs, containing three, four, and five adenosines, respectively, (and 5′SFVSL1rev1/DI, containing the highest number, i.e., five AU repeats at the 5′ terminus) replicated not only as efficiently as 5′SIN/DI but also significantly interfered with replication of the helper genome, TSG/Pac (Fig. 7B, lanes 6, 9, 10, and 11). This suggests that these RNAs had a higher affinity for the trans-acting replicase than the 5′SIN/DI parent. The difference was not detected in the luciferase assay perhaps due to strong interference of these DIs with TSG/Pac replication, leading to limiting levels of active replicase and RNA accumulation.
Thus, sequences of the repeating elements in the 5′ terminus and the number of repeats determined the efficiency of the RNA replication. Their activity was also strongly dependent on the downstream RNA sequence and/or higher-order RNA structure.
Replication of SIN with modified 5′ ends.
The results described above revealed an apparent discrepancy between the ability of primary viral pseudorevertants to grow in BHK-21 cells versus the low level of replication observed for DI RNAs with identical 5′ ends. One potential explanation for these differences could be in the presence of the additional mutations in the genomes of pseudorevertants conferring a positive impact on virus replication.
To rule out this possibility and to test the replication of recombinant viruses in different cell types, we replaced the 5′ UTR of SIN with (i) selected sequences found in the primary pseudorevertants and (ii) sequences identified after additional passaging (with various number of repeating elements) (Fig. 8A). The in vitro-synthesized RNAs were transfected into BHK-21 cells, and an infectious center assay revealed that all of the reconstructed RNAs were essentially as infectious as the RNA of SIN Toto1101 (Fig. 8A). Only 5′SFVrev3/SINr RNA demonstrated a noticeably lower infectivity, but the difference was minor.
FIG. 8.
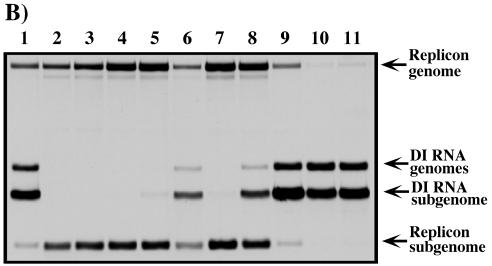
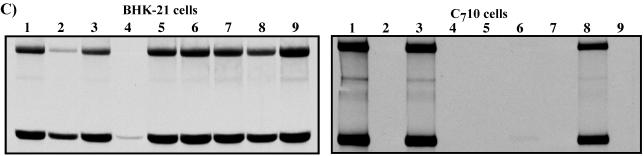
Single-step growth curves and replication of viral RNAs in different cell types. (A) 5′-Terminal sequences of the constructed SIN variants and infectivity of the in vitro-synthesized RNAs in the infectious center assay. The suffix “r” indicates that the genome was constructed in a cDNA form and used for generating virus stock. SFV and SIN sequences are shown in plain and boldface type, respectively. Underlining represents the sequences predicted to form the stem-loop structures. Lowercase type indicates an additional 5′ nucleotide resulting from the SP6 in vitro transcription. Boxes indicate heterologous sequences that were not present in the original 5′SFV/SIN and 5′SFVSL1/SIN genomes. (B) BHK-21 and C710 cells were seeded 4 h prior to infection on 35-mm dishes (5 × 105 cells/dish and 106 cells/dish, respectively). Infections were performed at an MOI of 1 PFU/cell for 1 h at 37 or 30°C (for BHK-21 or C710 cells, respectively) with shaking. The cells were then washed with the corresponding growth media, 1 ml of complete BHK-21 or C710 medium was added, and incubation was continued at 37 or 30°C, respectively. The media were harvested and replaced at the indicated time points. Released virus titers were measured by plaque assay on BHK-21 cells. (C) Autoradiograph of the dried gels with the RNAs metabolically labeled with [3H]uridine. Equal numbers of BHK-21 and C710 cells (5 × 105 and 1 × 106 cells, respectively) were infected with different SIN variants at MOIs of 5 PFU/cell (for BHK-21 cells) and 20 PFU/cell (for C710 cells). The RNAs were metabolically labeled with [3H]uridine (30 μCi/ml for BHK-21 and 40 μCi/ml for C710 cells) in the presence of 1 μg of ActD/ml at between 2 and 6 h postinfection of BHK-21 cells and between 12 and 21 h postinfection of C710 cells. Numbers designated for viruses in panels A and B correspond to the numbers on lanes on the gels shown on panel C as follows: 1, SIN Toto1101; 2, 5′SFVrev4/SINr; 3, 5′SFV/SIN/MP1r; 4, 5′SFVrev3/SINr; 5, 5′SFVrev1/SINr; 6, (AU)15′SFVSL1/SINr; 7, 5′SFVSL1rev1/SINr; 8, (A)15′SFVSL1/SINr; and 9, 5′SFVSL1/SIN/LP4r.
Virus stocks prepared after RNA electroporation were used to test virus growth on both BHK-21 and mosquito C710 cells (Fig. 8B). Some viruses with the 5′ ends found in the primary isolates (5′SFVrev1/SINr and 5′SFVSL1rev1/SINr) or in the isolates after competitive passaging (5′SFV/SIN/MP1r and 5′SFVSL1/SIN/LP4r) grew on BHK-21 cells as well as SIN Toto1101. 5′SFVrev4/SINr and 5′SFVrev3/SINr viruses displayed significantly slower growth kinetics that correlated with the dramatically lower ability of corresponding DI RNAs to replicate in vivo (Fig. 5). Synthesis of viral genomic RNAs in BHK-21 cells (Fig. 8C) paralleled the virus growth rates. The 5′SFVrev4/SINr and 5′SFVrev3/SINr RNAs replicated 4-and 20-fold less efficiently than SIN Toto1101 (lanes 2 and 4, respectively). All of the other RNAs replicated at levels similar to Toto1101.
Evaluation of growth and RNA replication of the viruses in C710 cells revealed significant differences between the variants (Fig. 8B and C). Surprisingly, only 5′SFV/SIN/MP1r and (A)15′SFVSL1/SINr were able to replicate at rates similar to that of SIN Toto1101. All of the other viruses propagated much less efficiently. In particular, the 5′SFVSL1/SINLP4r, which had the highest growth rate and the highest level of RNA replication in BHK-21 cells, was incapable of replication in mosquito cells (Fig. 8C, lanes 9). Similarly, the lower number of AU repeats in the genome of (AU)15′SFVSL1/SINr, compared to 5′SFVSL1rev1/SINr, allowed this virus to replicate far more efficiently in mosquito cells.
Taken together, these data indicated that the modified 5′ ends of the pseudorevertants were sufficient to rescue replication of SIN with an SFV-specific 5′ end in BHK-21 cells. However, these adaptations were host cell dependent and often not beneficial for replication in mosquito cells.
DISCUSSION
One of the important features of the 5′ cis-acting elements of SIN is the presence of sequence(s) serving as part of the promoters for the synthesis of both negative- and positive-strand genome-length RNAs. Sequences at or near the 5′ terminus make genomic, but not subgenomic, RNAs competent for replication (6, 21, 22, 38). At the same time, these overlapping functions in a relatively short sequence at the 5′ end of genome RNA (or its complement at the 3′ end of negative-strand RNA), as well as its role in translation initiation, make analysis of this element challenging. In order to exclude the effect of translational efficiency of the RNA templates from our analysis of the promoters for both negative- and positive-strand synthesis, we applied recently designed complementary in vivo and in vitro approaches, in which replication of the tested templates do not depend on their translational efficiency (6). We also used in our experiments BHK-21 cells that are believed to be deficient in alpha/beta interferon signaling and have not been reported as developing antiviral reaction.
The replacement of the SIN 5′ UTR in DI RNAs by the 5′ UTR derived from SFV made these RNAs incompetent for replication (6; see also Fig. 2). The SIN replicative enzymes did not synthesize detectable levels of negative-strand on this template in vitro and, as expected, replication in vivo was very inefficient. Most importantly, the SFV 5′ end was incapable of binding host and/or SIN-specific protein factors required for initiation of the negative-strand synthesis (6).
Previously, it was shown for both SIN and rubella virus that 5′-terminal nucleotides are critical for RNA replication (21, 27). Based on these data, we positioned five 5′-terminal nucleotides of SIN genome upstream of the SFV 5′ stem-loop-forming sequence to increase the level of DI RNA replication. This modification increased negative-strand RNA synthesis in vitro, elevating it to the level found for 5′SIN/DI RNA. However, the 5ntSIN-SFV/DI and 5ntSIN-SFVSL1/DI RNAs replicated in vivo 3 orders of magnitude less efficiently than RNA containing SIN 5′ UTR. Further modification of the 5′ end by inserting an additional sequence to mimic the RNA structure at the 3′ end of the negative-strand SIN template did not increase DI RNA replication in vivo. The 5ntSIN-SFVSL1-5nt/DI RNA still replicated 1,000-fold less efficiently than 5′SIN/DI RNA. Thus, although the addition of the SIN 5′-terminal nucleotides restored negative-strand RNA synthesis, their complement at the 3′ end of the negative strand did not function for efficient initiation of positive-strand RNA synthesis.
Since we were unable to predict which modifications were required for positive-strand promoter activity in the negative-strand template, we utilized an alternative strategy to define these elements. Specifically, we predicted that selection and analysis of pseudorevertants with enhanced replication and promoter activity would generate useful information. Fortunately, the 5′SFV/SIN and 5′SFVSL1/SIN RNAs were 4 and 3 orders of magnitude less infectious than SIN Toto1101 RNA (based on the infectious center plaque assay), indicating that these genomes were defective but still capable of generating pseudorevertants. Analysis of randomly selected variants revealed a variety of modifications in the 5′-terminal sequences, suggesting that a library of replication-competent pseudorevertants evolved upon transfection of the RNAs into the cells. Moreover, each plaque isolate of virus was not homogeneous and had a variety of similar 5′-terminal sequences differing in numbers of repeating elements, which indicated continued evolution. The presence of deletions in the SFV-specific sequences was not surprising; this provides a simple means of eliminating SFV sequences that have a negative effect on SIN RNA replication. However, the appearance of new repeating elements was unexpected. All variants retained the 5′-terminal AU that is conserved among alphaviruses. This dinucleotide was followed by repeats of (AU), (AAUU) or (A), and one or two extra (U)'s were found inside the oligo(A) sequences in some sequenced variants. In a previous study, we determined the consequences of substituting the 5′ UTR of bovine viral diarrhea virus (BVDV) with that of hepatitis C virus (7). In this case, replication-competent variants with additional 5′ sequences were also selected but appeared to have arisen from template switching by the RdRp during RNA replication (7). This template switching allowed the apparent capture of RNA sequences from cellular mRNAs that could function in BVDV RNA replication. Based on the results of the infectious center assay tests, in the present study, the efficiency of acquiring adaptive 5′ sequences was 2 to 3 orders of magnitude higher than in the case of the hepatitis C virus-BVDV chimeras. Although acquisition of these sequences from cellular mRNAs cannot be excluded, it looks more likely that the SIN RdRp can generate random AU-rich sequences at the 3′ end of the negative-strand RNA. Such sequences would then be positively selected based on their ability to promote positive-strand genome RNA synthesis. In support of this idea, previous studies have shown that SIN has the ability to generate U-rich sequences at the 3′ ends of defective viral genomes and to regenerate a 3′-terminal poly(A) tail (8, 28). In addition, a recent study has shown that 3′-UA extensions to the negative strand (5′-AU in the positive-strand genome RNA) emerge and suppress certain nonaromatic substitutions for the N-terminal Tyr residue of nsP4 (31). This additional enzymatic activity can continuously, but with low efficiency, generate new sequences on the SIN 5′ UTR (most probably by adding them to the 3′ end of the negative strand). However, immediately upon synthesis the genomes with these modifications have to compete with the efficient promoters of SIN genomes for the replicative enzymes. By using SIN genomes with SFV-specific 5′ UTRs or with mutations in nsP4 (31), it became possible to downregulate viral replication and to remove the continuous selective pressure of efficiently replicating natural SIN templates. Both systems provided an opportunity to detect the formation of different 5′-terminal sequences that had ability to drive RNA replication.
After replacement of the SFV sequences in 5′SFVSL1/DI and 5′SFV/DI RNAs by the variants' 5′ ends, these new chimeras became efficient templates for negative-strand RNA synthesis in vitro. However, the same DI genomes were still impaired for replication in vivo in a system where DI RNAs and the helper genome compete for replicative enzymes. Under these conditions, replication of DI RNAs ranged from 0.1 and 48% of the 5′SIN/DI RNA level. Different abilities of the modified 5′ ends to support DI RNA replication suggested the possibility that, upon transfection of 5′SFV/SIN and 5′SFVSL1/SIN viral RNAs into the cells, a library of SIN variants with heterogeneous 5′ termini was generated. Some of these appeared to be capable of replication at levels sufficient for virus spread and CPE development, and we detected them as plaque-forming viruses. However, ineffective replication of the DI RNAs with the 5′ ends identified in the genomes of randomly selected viruses (except 5′SFVrev1/DI and 5′SFVSL1rev1/DI) indicated that a major fraction of the identified 5′ UTRs encoded less-efficient promoters than the natural SIN 5′ end. Thus, poorly replicating 5′SFV/SIN and 5′SFVSL1/SIN RNAs generated variants with a variety of 5′UTRs that were sufficient, but not necessarily optimal, for virus replication.
To identify sequences that allowed higher levels of SIN replication in BHK-21 cells, we applied an additional two-step selection. First, virus populations, harvested after transfection of in vitro-synthesized 5′SFV/SIN and 5′SFVSL1/SIN RNAs, were serially passaged at a high MOI, creating an environment where viral genomes competed with each other for replicative enzymes and/or host factors, as occurs during viral genome-DI RNA replication. In the second step, viruses were passaged at a low MOI to select variants with the highest growth rates. The resulting stocks showed less heterogeneity at the 5′ ends of their genome RNAs. The majority of randomly picked plaques contained viruses with the entire SFV SL1 containing a short acquired upstream sequence 5′-AU(A)3U or 5′-AU(A)4. We detected more than four adenosine residues in only one plaque variant (5′SFVSL1/SIN LP4). Apparently, the addition of 5′-AU(A)n (where n > 2) to the SFV 5′ end transformed an inefficient positive-strand promoter into a highly competitive sequence, effectively functioning in RNA replication. One variant resulted from a replacement of the 5′-terminal 56 nucleotides of SFV by a short 5′-(AU)2 repeat. The presence of AU repeats in SIN genomes is not new. The 5′-AUAU sequence was first detected at the 5′ end of a naturally occurring SIN DI RNA containing a portion of tRNAAsp instead of the natural 5′ terminus (20, 37, 38). This DI RNA replicates efficiently and interferes with SIN replication, indicating the efficient recruitment of the SIN replicase. As mentioned above, 5′-AU and 5′-AUA were also found in SIN pseudorevertants harboring deleterious substitutions for the N-terminal tyrosine residue of nsP4 (31). The complement of this short sequence in the negative-strand must be a key determinant for initiation of positive-strand synthesis. Similar to the results of Shirako et al. (31), the heterologous AU-rich sequences that we identified were predicted to be unpaired in the 3′ end of the negative-strand template (data not shown), and this might be advantageous for initiation of the positive-strand RNA synthesis.
The 5′-terminal sequences found in passaged virus were tested in the context of DI RNAs. These 5′ termini led to efficient production of both positive- and negative-strand DI RNAs and effective competition with the helper construct in vivo. The efficiency of negative- and positive-strand RNA synthesis generally correlated with the number of the 5′ repeats. Three to five 5′-AU repeats, followed by SFV SL1, led to negative-strand RNA synthesis similar to that of the natural SIN 5′ UTR. In vivo, five 5′-AU repeats were needed to drive high levels of DI RNA replication and efficient interference with the helper replicon RNA. Similarly, one A residue in the 5′-terminal AU(A)nGAU sequence [(A)15′SFVSL1/DI construct] was sufficient to make the DI RNA efficient template for negative-strand RNA synthesis in vitro, but more than three adenosines were required for high-level DI RNA replication in vivo.
Similar results were obtained with viruses reconstructed with the 5′-terminal sequences found in pseudorevertants. Compared to parental 5′SFV/SIN and 5′SFVSL1/SIN RNAs, the modified 5′-UTR sequences increased the specific infectivity of in vitro-synthesized RNAs in BHK cells essentially to the level of SIN Toto1101. These results suggest that the 5′ modifications, and not mutations elsewhere, were responsible for enhanced replication and CPE. Growth rates of generated viruses in BHK cells were also similar to those of SIN Toto1101. Interestingly, replication of the reconstructed viral RNAs was found to be cell type specific. An increase in the numbers of 5′-AU repeats or adenosines upstream of 5′SFVSL1 structure was beneficial for virus growth in BHK-21 cells but was deleterious for growth in mosquito cells. Only one pseudorevertant, 5′SFV/SIN MP1, was capable of growing in both BHK-21 and C710 cells as efficiently as SIN Toto1101. Thus, 5′ UTRs apparently have different requirements for optimal replication in mammalian versus insect cells, and this effect is likely achieved though binding of host factors with different sequence specificity in the cells of mosquito and vertebrate origin.
The structure of SIN replicative complex and the mechanism of recognition of the promoter elements in SIN genome and its replicative intermediate are not completely understood yet. Previously, it was demonstrated that the 3′ ends of the negative strands of SIN and SFV genomes bind the same host proteins, and one of these proteins was later identified as a mosquito homolog of La autoantigen (25, 26). Its chicken homolog was also suggested as SIN RNA-binding protein (26). Nevertheless, the complement of SFV 5′ UTR in the negative-strand RNA was incapable of functioning as a promoter for SIN RdRp. Apparently, the very 3′ terminus of the negative-strand template was not properly presented to SIN replicase, even if it contained five SIN-specific nucleotides (Fig. 2A). Binding of host cell proteins to viral RNA might play an important role(s) in stabilizing SIN RdRp and/or directing it to the 3′ end of the negative strand. However, melting of the 3′-terminal secondary structure and recognition of the sequences on the 3′ end of the negative-strand genome for initiation of the positive-strand RNA synthesis appear to be a function of SIN nsP4 protein (31). In agreement with this hypothesis, the results of our study suggest that in chimeric viruses, SIN nsP4 is probably incapable of melting the GC-rich stem-loop derived from SFV, and new variants, containing the deletions and/or short unpaired RNA fragments enriched with adenosines and uridines, are generated and further selected. The new RNA elements strongly increased both positive- and negative-strand RNA synthesis, but their enhancing effect on replication depended on the cells used in the experiments. The upregulation of replication also depended on the number of generated repeats and their nucleotide sequencses. Based on analysis of functioning of the heterologous 5′ ends, we propose that structure of SIN RdRp or its positioning may not be the same in mammalian and insect cells, most likely due to significant variations in host factors involved in replication. This different positioning, in turn, requires a distinct 5′ terminus for initiation of the positive-strand RNA synthesis. However, this hypothesis needs further experimental support.
The authentic SIN 5′ UTR appears to be a successful compromise that allows efficient replication of viral genome by SIN RdRp and virus growth in both vertebrate and invertebrate cells. It has undoubtedly been selected to function efficiently in nature, because a successful arbovirus must be able to replicate to high levels in amplifying vertebrate hosts and develop persistent infection in mosquitoes. This complex cycle, which has been optimized over countless transmission cycles, requires replication in multiple cell types within both hosts. In addition, it was recently demonstrated that sequence of the 5′ end is critical for resistance of another alphavirus, Venezuelan equine encephalitis virus, to innate immune response (34, 40), and this factor, certainly, can cause additional selective pressure.
In conclusion, we have shown that SIN is capable of adapting to deleterious heterologous 5′ UTR sequences through deletion and generation of new 5′-terminal sequences containing short repeats enriched in adenosine and uridine. These sequences have a strong impact on both negative- and positive-strand RNA synthesis and their replication-enhancing ability depends on particular downstream sequences in the 5′ UTR and the nature or host cells (host protein factors) used in the experiments. These modifications can have differential effects on synthesis of negative- and positive-strand RNAs. Initiation of the negative-strand RNA synthesis relies more on the length of the 5′ unpaired terminus than on its particular sequence. However, for positive-strand synthesis, the SIN RdRp can utilize a more limited number of repeats with particular sequences. Besides enhancing understanding of this multifunctional RNA element, our results have possible practical applications. Alphavirus-based vectors are being widely used for protein production and as possible vaccine delivery vehicles (2, 19, 39). Using the approaches outlined here, it should be possible to select 5′ variants with restricted host range and perhaps cell tropism. In addition, by varying the 5′ sequence elements in defective helper and replicon RNAs, we can modulate RNA replication and the synthesis of trans-acting replicase and structural proteins with the goal of enhancing the production of packaged replicon RNAs.
Acknowledgments
We thank Peter Mason and Scott Weaver for critical reading of the manuscript.
This study was supported by Public Health Service grants AI50537 (I.F.) and AI24134 (C.M.R.).
REFERENCES
- 1.Bredenbeek, P. J., I. Frolov, C. M. Rice, and S. Schlesinger. 1993. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J. Virol. 67:6439-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, M., K. F. Hu, B. Rozell, C. Orvell, B. Morein, and P. Liljestrom. 2002. Vaccination with recombinant alphavirus or immune-stimulating complex antigen against respiratory syncytial virus. J. Immunol. 169:3208-3216. [DOI] [PubMed] [Google Scholar]
- 3.Davis, N. L., K. W. Brown, G. F. Greenwald, A. J. Zajac, V. L. Zacny, J. F. Smith, and R. E. Johnston. 1995. Attenuated mutants of Venezuelan equine encephalitis virus containing lethal mutations in the PE2 cleavage signal combined with a second-site suppressor mutation in E1. Virology 212:102-110. [DOI] [PubMed] [Google Scholar]
- 4.Frey, R. K., D. L. Gard, and J. H. Strauss. 1979. Biophysical studies on circle formation by Sindbis virus 49S RNA. J. Mol. Biol. 132:1-18. [DOI] [PubMed] [Google Scholar]
- 5.Frolov, I., E. Agapov, T. A. Hoffman, Jr., B. M. Prágai, M. Lippa, S. Schlesinger, and C. M. Rice. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73:3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frolov, I., R. Hardy, and C. M. Rice. 2001. cis-Acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frolov, I., M. S. McBride, and C. M. Rice. 1998. cis-acting RNA elements required for replication of bovine viral diarrhea virus-hepatitis C virus 5′ nontranslated region chimeras. RNA 4:1418-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.George, J., and R. Raju. 2000. Alphavirus RNA genome repair and evolution: molecular characterization of infectious sindbis virus isolates lacking a known conserved motif at the 3′ end of the genome. J. Virol. 74:9776-9785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grakoui, A., R. Levis, R. Raju, H. V. Huang, and C. M. Rice. 1989. A cis-acting mutation in the Sindbis virus junction region which affects subgenomic RNA synthesis. J. Virol. 63:5216-5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imataka, H., A. Gradi, and N. Sonenberg. 1998. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 17:7480-7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuhn, R. J., Z. Hong, and J. H. Strauss. 1990. Mutagenesis of 3′ nontranslated region of Sindbis virus RNA. J. Virol. 64:1465-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemm, J. A., R. K. Durbin, V. Stollar, and C. M. Rice. 1990. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J. Virol. 64:3001-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lemm, J. A., and C. M. Rice. 1993. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J. Virol. 67:1905-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lemm, J. A., and C. M. Rice. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol. 67:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemm, J. A., T. Rümenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levis, R., S. Schlesinger, and H. V. Huang. 1990. Promoter for Sindbis virus RNA-dependent subgenomic RNA transcription. J. Virol. 64:1726-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liljeström, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 18.Liljeström, P., S. Lusa, D. Huylebroeck, and H. Garoff. 1991. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the small 6,000-molecular-weight membrane protein modulates virus release. J. Virol. 65:4107-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lundstrom, K. 2002. Alphavirus-based vaccines. Curr. Opin. Mol. Ther. 4:28-34. [PubMed] [Google Scholar]
- 20.Monroe, S. S., and S. Schlesinger. 1983. RNAs from two independently isolated defective interfering particles of Sindbis virus contain a cellular tRNA sequence at their 5′ ends. Proc. Natl. Acad. Sci. USA 80:3279-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niesters, H. G. M., and J. H. Strauss. 1990. Defined mutations in the 5′ nontranslated sequence of Sindbis virus RNA. J. Virol. 64:4162-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niesters, H. G. M., and J. H. Strauss. 1990. Mutagenesis of the conserved 51 nucleotide region of Sindbis virus. J. Virol. 64:1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ou, J. H., C. M. Rice, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1982. Sequence studies of several alphavirus genomic RNAs in the region containing the start of the subgenomic RNA. Proc. Natl. Acad. Sci. USA 79:5235-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ou, J. H., E. G. Strauss, and J. H. Strauss. 1981. Comparative studies of the 3′ terminal sequences of several alphaviruses. Virology 109:281-289. [DOI] [PubMed] [Google Scholar]
- 25.Pardigon, N., E. Lenches, and J. H. Strauss. 1993. Multiple binding sites for cellular proteins in the 3′ end of Sindbis alphavirus minus-sense RNA. J. Virol. 67:5003-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pardigon, N., and J. H. Strauss. 1996. Mosquito homolog of the La autoantigen binds to Sindbis virus RNA. J. Virol. 70:1173-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pugachev, K. V., and T. K. Frey. 1998. Effects of defined muations in the 5′ nontranslated region of rubella virus genome RNA on virus viability and macromolecule synthesis. J. Virol. 72:641-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raju, R., M. Hajjou, K. R. Hill, V. Botta, and S. Botta. 1999. In vivo addition of poly(A) tail and AU-rich sequences to the 3′ terminus of the Sindbis virus RNA genome: a novel 3′-end repair pathway. J. Virol. 73:2410-2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlesinger, S., and M. J. Schlesinger. 2001. Togaviridae: the viruses and their replication, p. 895-916. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 31.Shirako, Y., E. G. Strauss, and J. H. Strauss. 2003. Modification of the 5′ terminus of Sindbis virus genomic RNA allows nsP4 RNA polymerases with nonaromatic amino acids at the N terminus to function in RNA replication. J. Virol. 77:2301-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shirako, Y., and J. H. Strauss. 1990. Cleavage between nsP1 and nsP2 initiates the processing pathway of Sindbis virus nonstructural polyprotein P123. Virology 177:54-64. [DOI] [PubMed] [Google Scholar]
- 33.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus strand RNA synthesis. J. Virol. 185:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spotts, D. R., R. M. Reich, M. A. Kalkhan, R. M. Kinney, and J. T. Roehrig. 1998. Resistance to alpha/beta interferons correlates with the epizootic and virulence potential of Venezuelan equine encephalitis viruses and is determined by the 5′ noncoding region and glycoproteins. J. Virol. 72:10286-10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1984. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133:92-110. [DOI] [PubMed] [Google Scholar]
- 36.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsiang, M., S. S. Monroe, and S. Schlesinger. 1985. Studies of defective interfering RNAs of Sindbis virus with or without tRNAAsp sequences at their 5′ termini. J. Virol. 54:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsiang, M., B. G. Weiss, and S. Schlesinger. 1988. Effects of 5′-terminal modifications on the biological activity of defective interfering RNAs of Sindbis virus. J. Virol. 62:47-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuji, M., C. C. Bergmann, Y. Takita-Sonoda, K. Murata, E. G. Rodrigues, R. S. Nussenzweig, and F. Zavala. 1998. Recombinant Sindbis viruses expressing a cytotoxic T-lymphocyte epitope of a malaria parasite or of influenza virus elicit protection against the corresponding pathogen in mice. J. Virol. 72:6907-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.White, L. J., J. G. Wang, N. L. Davis, and R. E. Johnston. 2001. Role of alpha/beta interferon in Venezuelan equine encephalitis virus pathogenesis: effect of an attenuating mutation in the 5′ untranslated region. J. Virol. 75:3706-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]