Abstract
Baculovirus GP64 is a low-pH-dependent membrane fusion protein required for virus entry and cell-to-cell transmission. Recently, GP64 has generated interest for practical applications in mammalian systems. Here we examined the membrane fusion function of GP64 from Autographa californica multiple nucleopolyhedrovirus (AcMNPV) expressed in mammalian cells, as well as its capacity to functionally complement a mammalian virus, human respiratory syncytial virus (HRSV). Both authentic GP64 and GP64/F, a chimeric protein in which the GP64 cytoplasmic tail domain was replaced with the 12 C-terminal amino acids of the HRSV fusion (F) protein, induced low-pH-dependent cell-cell fusion when expressed transiently in HEp-2 (human) cells. Levels of surface expression and syncytium formation were substantially higher at 33°C than at 37°C. The open reading frames (ORFs) encoding GP64 or GP64/F, along with two marker ORFs encoding green fluorescent protein (GFP) and β-glucuronidase (GUS), were used to replace all three homologous transmembrane glycoprotein ORFs (small hydrophobic SH, attachment G, and F) in a cDNA of HRSV. Infectious viruses were recovered that lacked the HRSV SH, G, and F proteins and expressed instead the GP64 or GP64/F protein and the two marker proteins GFP and GUS. The properties of these viruses, designated RSΔsh,g,f/GP64 or RSΔsh,g,f/GP64/F, respectively, were compared to a previously described HRSV expressing GFP in place of SH but still containing the wild-type HRSV G and F proteins (RSΔsh [A. G. Oomens, A. G. Megaw, and G. W. Wertz, J. Virol., 77:3785-3798, 2003]). By immunoelectron microscopy, the GP64 and GP64/F proteins were shown to incorporate into HRSV-induced filaments at the cell surface. Antibody neutralization, ammonium chloride inhibition, and replication levels in cell culture showed that both GP64 proteins efficiently mediated infectivity of the respective viruses in a temperature-sensitive, low-pH-dependent manner. Furthermore, RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F replicated to higher levels and had significantly higher stability of infectivity than HRSVs containing the homologous HRSV G and F proteins. Thus, GP64 and a GP64/HRSV F chimeric protein were functional and efficiently complemented an unrelated human virus in mammalian cells, producing stable, infectious virus stocks. These results demonstrate the potential of GP64 for both practical applications requiring stable pseudotypes in mammalian systems and for studies of viral glycoprotein requirements in assembly and pathogenesis.
The budded virus (BV) form of baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) acquires its envelope by budding through the host cell plasma membrane and is responsible for systemic spread of the virus through the insect host (for review see reference 3). The major glycoprotein species in the envelope of the BV phenotype is GP64, a low-pH-mediated membrane fusion protein that exists as a trimer with a molecular mass of approximately 175 kDa (4, 44, 60). GP64 is an essential viral protein required for entry and cell-to-cell transmission of AcMNPV in vitro and in vivo (3, 7, 21, 35, 60). Besides GP64, another type of baculoviral low-pH-activated membrane fusion protein has been described, the F protein (3, 25, 45, 60). All baculoviruses contain an F homolog but only the so-called group I nucleopolyhedroviruses, to which the prototype AcMNPV belongs, contain a GP64 gene. Evolutionary studies suggest that GP64 was acquired relatively recently, after which it functionally displaced the baculovirus F protein (45). Surprisingly, GP64 has low but significant homology with the envelope proteins of orthomyxoviruses from the Thogotovirus genus (36), a group of tick-transmitted viruses that are able to replicate in both ticks and vertebrates. Unlike the thogotoviruses, baculoviruses do not replicate in vertebrate cells; however, AcMNPV is known to enter and efficiently deliver genes to a wide variety of cell types, including mammalian cells (2, 6, 8, 10, 13, 22, 53, 54, 59). These findings have generated interest in the use of AcMNPV as a safe viral vector for gene delivery in humans (24). Recently, specific targeting of AcMNPV to mammalian cell types was demonstrated by fusing ligands to the N terminus or transmembrane domain of GP64, and it was shown that GP64 could mediate entry of a lentivirus into mammalian cell types (30, 41). GP64 is substantially less cytotoxic than the vesicular stomatitis Indiana virus (VSIV) G protein that has been used in many pseudotyping approaches (22, 30). The GP64 protein has also become a model for fusion pore formation and is being exploited for eukaryotic surface display applications (5, 11, 14, 15, 18, 28, 37, 41, 46, 47). However, in spite of the attractive features of GP64 and a range of proposed practical applications, GP64 expression and function in mammalian cells have been only minimally explored.
In this study we investigated the expression and membrane fusion function of GP64 and a GP64 chimeric protein in mammalian cells, as well as the capacity of these proteins to functionally complement a mammalian virus lacking all of its transmembrane glycoproteins. For the latter purpose, we used human respiratory syncytial virus (HRSV) because GP64 complementation may provide a valuable tool to aid future studies of the roles of the HRSV glycoproteins in assembly and pathogenesis. HRSV, a nonsegmented negative-strand RNA virus that causes severe lower respiratory tract disease in infants and children, expresses 11 known proteins, three of which have been characterized as transmembrane glycoproteins: SH (a small hydrophobic protein of unknown function), attachment protein G, and fusion protein F (1, 12, 23, 63). Previous work has shown that the SH protein is dispensable for growth of HRSV in cell culture (9). In addition, infectious HRSVs lacking the SH and G genes, or in which the SH, G, and F genes were replaced by the VSIV G protein, were recovered (29, 43, 57, 58). In this work, we deleted the SH, G, and F open reading frames (ORFs) from a cDNA of the prototypic HRSV A2 strain and replaced them with that of AcMNPV GP64. In addition to authentic GP64, we also tested a chimeric GP64 protein, termed GP64/F, in which the cytoplasmic tail domain (CTD) was replaced with the 12 C-terminal amino acids of the HRSV F protein, which constitute approximately half of the predicted F CTD. This was done because, for several viruses, the CTD plays an important role in membrane fusion, glycoprotein incorporation, budding, or morphology (27, 38, 42, 48, 50, 51, 56, 65). Along with GP64 or GP64/F, the ORFs encoding enhanced green fluorescent protein (GFP; Clontech) and β-glucuronidase (GUS) were inserted in place of the other two genes to keep the number of genes constant within the engineered genomes, as gene position relative to a single 3′ promoter is important for control of gene expression in negative-strand RNA viruses (64). Infectious, engineered HRSVs expressing GP64 as their only viral transmembrane glycoprotein were recovered from the constructed cDNAs by using techniques previously described (43, 52). The ability of GP64 to induce membrane fusion and to mediate entry, propagation, and stability of an unrelated paramyxovirus in mammalian cell types was examined.
MATERIALS AND METHODS
Cells and antibodies.
HEp-2 and Vero 76 (Vero) cells were acquired from the American Type Culture Collection and were grown in standard growth medium containing 3% (Vero) or 5% (HEp-2) fetal bovine serum. Although we predominantly used Vero cells for this study, the transient-expression analysis (Fig. 1C and D) was done in HEp-2 cells due to significantly better transfection rates. Monoclonal antibodies (MAbs) 6 (MAb 6) and 19 were provided by Geraldine Taylor (Institute for Animal Health, Compton, United Kingdom), MAb L9 by Ed Walsh (University of Rochester School of Medicine, Rochester, N.Y.),and MAbs AcV1 and 5 by Gary Blissard (Boyce Thompson Institute at Cornell University, Ithaca, N.Y.).
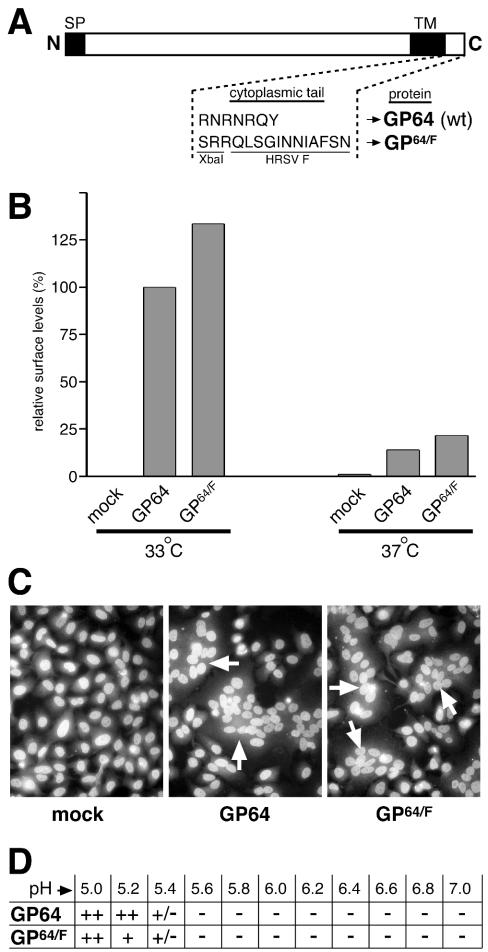
FIG. 1.
Transient-expression and membrane fusion function of GP64 and chimeric protein GP64/F. (A) Composition of the C-terminal region of GP64 and chimeric protein GP64/F. In GP64/F, the 7-amino-acid GP64 CTD was replaced by the 12 C-terminal amino acids of the HRSV F protein via an introduced XbaI site (SP, signal peptide; TM, transmembrane domain). (B) Relative levels of GP64 and GP64/F at the cell surface. HEp-2 cells were transfected with plasmids expressing GP64 and GP64/F or mock transfected and were incubated at 33 or 37°C. At 34 h posttransfection, cells were chilled and incubated with anti-GP64 antibody AcV1 on ice, and relative protein surface levels were determined in duplicate by using CELISA as described previously (34, 42). Expression levels were normalized to that of GP64 at 33°C. (C) HEp-2 cells transfected as above were exposed to PBS, pH 5.0, for 3.5 min, incubated in normal growth medium at 33°C for 4 h, and fixed with paraformaldehyde. Cells were stained with Hoechst reagent and were examined by fluorescence microscopy (magnification, ×400). Arrows indicate syncytia. (D) pH threshold for membrane fusion. Transfected HEp-2 cells were exposed to PBS with a pH ranging from 5 to 7 for 3.5 min and were processed as above. Syncytia containing at least five nuclei were scored with phase-contrast microscopy (++, >150 syncytia per well; +, 40 to 150 syncytia per well; +/−, between 2 and 5 syncytia, −, no syncytia observed).
Construction of a chimeric GP64/HRSV F protein.
The AcMNPV GP64 gene was kindly provided by Gary Blissard. A truncated GP64 ORF (amino acids 1 to 505) flanked by a 3′ XbaI restriction site was generated by using PCR. Nucleotides encoding the C-terminal 12 amino acids (residues 563 to 574) of the HRSV F protein preceded by three nucleotides encoding an additional arginine residue (to ensure proper membrane anchoring) were amplified by using PCR creating a 5′ XbaI site. The introduced restriction site was utilized to ligate the two fragments, resulting in chimeric ORF GP64/F (Fig. 1A).
Construction of viral cDNAs and recovery of infectious engineered viruses.
Engineered cDNAs were generated based on a cDNA of the A2 strain of HRSV constructed and described previously (43). Briefly, by use of standard cloning techniques, two shuttle vectors were constructed that contained three ORFs (GFP-GP64-GUS or GFP-GP64/F-GUS) separated by authentic HRSV intergenic junctions and flanked by unique restriction sites FseI and AscI. The GFP ORF included in the genomes was that of enhanced GFP (Clontech). These shuttle vectors were cloned into a cDNA backbone, from which SH/G/F had been deleted, also containing FseI and AscI restriction sites, in order to yield cDNAs that varied from wild-type (wt) HRSV only in the content of ORFs at gene positions 6, 7, and 8 (pRSΔsh,g,f/GP64 and pRSΔsh,g,f/GP64/F [Fig. 2A]). Modified areas of the engineered cDNAs were verified by nucleotide sequencing. Infectious viruses were recovered from cDNA as described previously (43). Viral RNAs were harvested from cells infected with the engineered virus stocks at pass 3, amplified by reverse transcriptase PCR, and verified by nucleotide sequence analysis across cloning junctions and in modified areas. Pass 3 stocks were used for the experiments described, except for previously published virus RSΔsh, for which pass 5 stocks were used.
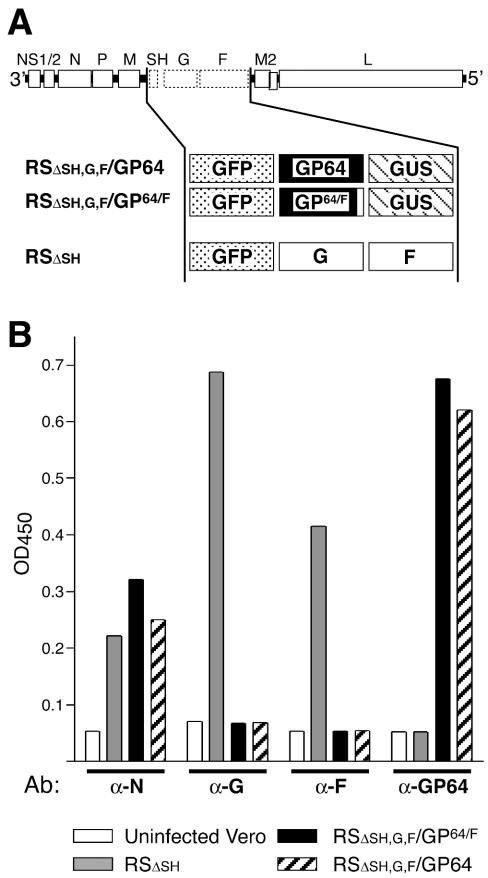
FIG. 2.
Composition of engineered virus genomes and glycoprotein expression. (A) Genome content of engineered cDNAs. In virus RSΔsh,g,f/GP64 or RSΔsh,g,f/GP64/F, the SH, G, and F ORFs were deleted from the cDNA of the HRSV A2 strain as described previously and were replaced with the ORF of either GP64 or GP64/F, respectively, along with ORFs encoding marker proteins GFP and GUS. Properties of these viruses were compared to virus RSΔsh, a similarly engineered HRSV derived from the same A2 backbone, also containing GFP but having the homologous G and F ORFs in their native genome location (43). (B) Transmembrane glycoprotein expression by the engineered viruses by CELISA. Duplicate samples of cells infected with the viruses indicated were fixed and permeabilized at 28 h postinfection, and protein expression was measured with antibodies against G (L9), F (MAb 19), GP64 (AcV5), or nucleocapsid protein (N) (MAb 6) as a control, by CELISA as previously described (34, 42). OD450, optical density at 450 nm.
Transient-expression and membrane fusion analysis.
HEp-2 cells were plated in six-well dishes and were transfected with plasmids encoding GP64 or GP64/F by using Lipofectin (Invitrogen) for 8 h at 37°C and were then incubated at 33 or 37°C. At 34 h posttransfection, infected cultures were examined for glycoprotein surface expression or were subjected to a syncytium formation assay. Relative surface levels of GP64 and GP64/F were measured with cell enzyme-linked immunosorbent assay (CELISA) as previously described, with minor modifications (34, 42). Briefly, plates were chilled and cells were incubated with MAb AcV1 for 1 h on ice. Cells were washed extensively with cold phosphate-buffered saline (PBS) to remove unbound antibody and were fixed with cold 4% paraformaldehyde for 10 min on ice and were then shifted to room temperature and incubated for another 15 min. Next, cells were incubated with a horseradish peroxidase conjugated secondary antibody, washed, and incubated in 1 ml of 3,3′,5,5′ tetramethylbenzidine substrate (Pierce). At various times after addition of substrate, 100-μl aliquots were taken and added to 2 M sulfuric acid in a 96-well plate to stop the reaction and the optical density at 450 nm was determined in an ELISA plate reader. For membrane fusion analysis, duplicate wells of cells at 34 h posttransfection were incubated for 3.5 min in PBS, pH 5.0. After 3.5 min, cells were incubated in normal growth medium at 33°C for 4 h and were fixed with 4% paraformaldehyde. Fixed cells were stained for 5 min with Hoechst reagent (Molecular Probes) and were photographed on an Olympus IX70 inverted microscope. To determine the pH threshold for fusion, HEp-2 cells were transfected as above and were exposed to PBS with a pH ranging from 5 to 7 for 3.5 min at room temperature. Cells were then incubated for 4 h in normal growth medium and fixed in 4% paraformaldehyde. Syncytia containing five or more nuclei were counted with phase-contrast microscopy.
Neutralization of infectivity by antibodies and inhibition of entry by ammonium chloride.
For antibody neutralization analysis, 1.5 × 105 PFU of each of the engineered viruses was preincubated with MAb AcV1 or MAb 19 at a range of concentrations for 70 min at room temperature and was then used to infect 5 × 105 Vero cells for 1.5 h at 37°C. Cells were washed, incubated for 22 h at 33°C, and examined for infectious virus by measurement of GFP expression from the viral genome in duplicate with flow cytometry (FACSCalibur; Becton Dickinson) as previously described, using 100,000 events per sample (43). To calculate relative entry, the number of cells expressing GFP in the presence of antibody was divided by the number of GFP-expressing cells in the absence of antibody and was multiplied by 100. For inhibition of entry analysis, Vero cells were infected with each of the engineered viruses at a multiplicity of infection of approximately 0.3 for 1.5 h in the presence of 0, 2, or 10 mM ammonium chloride. Ammonium chloride concentrations were maintained during postinfection incubation. At 22 h postinfection, cells were processed for flow cytometry as described above. Relative entry was calculated by dividing the number of cells expressing GFP in the presence of ammonium chloride by the number of GFP-expressing cells in the absence of ammonium chloride and multiplying by 100.
Kinetics of virion release from endosomes.
To determine the kinetics of virion release from endosomes, Vero cells (n = 0.4 × 106/well) were chilled to 4°C. Prechilled virus suspensions containing approximately 5 × 105 PFU were added and were incubated for 90 min at 4°C. Inoculum was then removed and replaced with normal growth medium warmed to 37°C (time zero). Ten millimolar ammonium chloride was added at the time intervals indicated in Fig. 4B. For time zero samples, ammonium chloride was added when the inoculum was replaced with medium. Cells were incubated for 20 h at 37°C, and the percentage of cells expressing GFP from the viral genome was determined with flow cytometry and relative entry was calculated as described above.
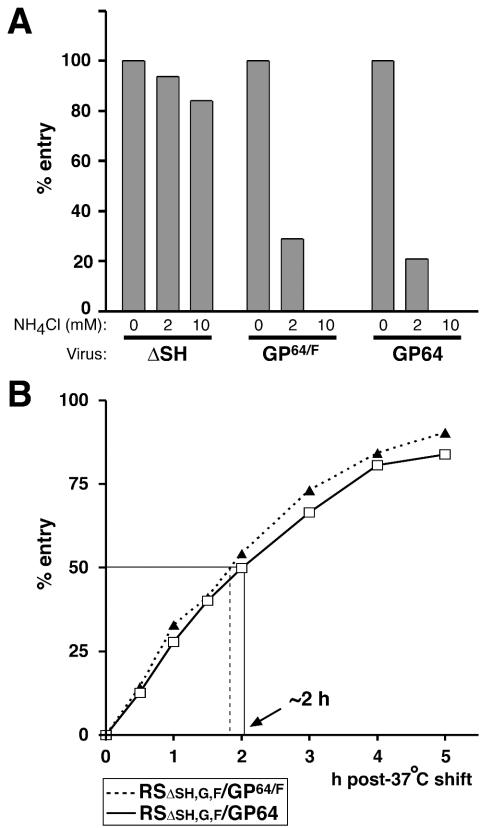
FIG. 4.
Effect of ammonium chloride (NH4Cl) on virus entry. (A) Vero cells were infected for 1.5 h with viruses RSΔsh, RSΔsh,g,f/GP64/F, and RSΔsh,g,f/GP64 in the presence or absence of NH4Cl and were further incubated at 33°C, maintaining the NH4Cl concentrations used during virus adsorption. At 22 h postinfection, cells were trypsinized, fixed, and examined for infectivity by measuring GFP expression by flow cytometry in duplicate as described previously (43). Bars represent the number of GFP-expressing cells relative to the number of GFP-expressing cells in the absence of NH4Cl. NH4Cl concentrations used are indicated in millimolars. (B) Timing of virion release from endosomes. Viruses RSΔsh,g,f/GP64/F and RSΔsh,g,f/GP64 were bound to Vero cells at 4°C for 90 min and were then replaced with medium prewarmed to 37°C (time zero). At time zero or at 30- to 60-min intervals thereafter, ammonium chloride was added to the infected cells. At 20 h post-37°C shift, cells were harvested and the percentage of GFP-expressing cells was determined by flow cytometry as above. To calculate relative entry, the percentage of GFP-expressing cells of duplicate samples was compared to the percentage of GFP-expressing cells in the absence of ammonium chloride and was multiplied by 100. The time at which 50% of the bound virus was released from endosomes is indicated with an arrow.
Immunoelectron microscopy (IEM).
Vero cells infected with each of the engineered viruses at a multiplicity of infection of approximately 3 were fixed for 30 min at room temperature with 3% paraformaldehyde, 0.5% glutaraldehyde, and 2 mM calcium chloride in PBS at 27 h postinfection. Cells were then incubated in 0.1% sodium dodecyl sulfate for 5 min at room temperature to enhance epitope recognition by MAb AcV5 and were blocked for 10 min in 50 mM glycine and subsequently 30 min in 2% bovine serum albumin, 0.1% cold-water fish skin gelatin, and 1% normal goat serum (Aurion). Cells were then incubated with antibody AcV5 in PBS and 0.1% bovine serum albumin, washed, and incubated with a secondary goat anti-mouse antibody conjugated to 6-nm gold (Aurion). Cells were washed four times in PBS and were then fixed for 30 min in 2% glutaraldehyde and washed again in PBS. Next, cells were fixed in 1% osmium tetroxide for 60 min, washed, dehydrated in a series of ethanol baths, and infiltrated in Polybed 812. Cut sections were lightly stained with uranyl acetate before examination and acquisition of images with a Hitachi H-7000 transmission electron microscope.
Growth curves.
Vero and HEp-2 cells in six-well plates were infected at a multiplicity of infection of 0.5 for 1.5 h at 37°C and washed two times and were then incubated in 2 ml of normal growth medium at 37 or 33°C. At 0 h postinfection and at 24-h intervals thereafter, one well for each condition was harvested by scraping the cells into the supernatant and viral titers were determined in duplicate, without freezing the samples, by 50% tissue culture infective dose (TCID50) based on GFP expression from the viral genome as described previously (43). To determine the percentage of infectivity associated with the cellular and supernatant fractions, the supernatant of infected cells at 24 and 48 h postinfection was drawn off; an equal volume of fresh growth medium was then added to the cells and cells were then scraped into the medium, followed by gentle pipetting. The titers of both the cellular and supernatant derived virus suspensions were determined by TCID50. The percentage of infectivity associated with the supernatant and the cell fraction was determined by dividing the respective titers by the combined titer of supernatant and cell-derived virus × 100.
Stability of infectivity analysis.
Virus stocks for each of the engineered viruses and virus wt A2 were generated simultaneously by infecting Vero cells at a multiplicity of infection of <0.2. Stocks were harvested by scraping cells into the supernatant, followed by gentle but extensive pipetting and removal of cell debris by low-speed centrifugation. Stocks were distributed in 100-μl aliquots and were stored at 4°C. In addition to undiluted stocks, diluted stocks were prepared by mixing undiluted stocks 1:6 with the supernatant of uninfected Vero cells grown in parallel and harvested identically to the virus stocks. Titers of all prepared stocks were determined in triplicate immediately after harvest and also at 3.5 and 7 days postharvest by TCID50. This was done based on GFP expression for the engineered viruses and by an immunoassay based on the F protein for virus A2, as previously described (43). As a control, at every time point, titers of virus RSΔsh were determined by both methods described above; the two methods yielded identical viral titers (not shown). Stocks with similar starting concentrations were selected for continuation of the stability experiment to minimize concentration effects. Viral titers of these stocks were subsequently determined at weeks 2, 3, 4, 6, and 8 after harvest as described above. Relative titers were calculated by dividing the TCID50 score at each time point by the titer determined at the time of harvest and multiplying by 100.
RESULTS
Transient-expression and membrane fusion function of GP64 in mammalian cells.
As a first step to determine the ability of GP64 to function in mammalian cells, we expressed it independently in HEp-2 cells and assayed its membrane fusion capacity. For that purpose, the AcMNPV GP64 ORF was placed behind a cytomegalovirus promoter in plasmid vector pRc-CMV (Invitrogen). Because of its intended use as a substitute glycoprotein to mediate HRSV infectivity, in addition a chimeric form of GP64 having its own CTD replaced with a portion of that of the HRSV F protein was tested. The HRSV F protein is responsible for fusion and entry at the plasma membrane in a non-pH-dependent manner; by analogy to other negative-strand viruses, its CTD may play a role in the virus life cycle (27, 50, 51, 56). However, the predicted F protein CTD is approximately 22 amino acids, significantly longer than that of GP64; therefore, only the nucleotides encoding the C-terminal 12 amino acids of HRSV F were used to replace the nucleotides encoding the CTD of GP64 (amino acids 506 to 512) (GP64/F [Fig. 1A]). GP64, when expressed alone in insect cells, was previously shown to induce syncytium formation after brief exposure to low pH (4); this function is thought to mimic the events that occur after acidification of the endosome upon entry of baculovirus BV virions. Plasmids expressing GP64 or GP64/F were transfected into HEp-2 cells to examine simultaneously the relative surface expression levels of GP64 or GP64/F and their ability to cause membrane fusion (Fig. 1B and C). To measure surface levels, transfected cells were incubated at both 37 and 33°C because of the difference in temperatures typically used to grow these viruses: 37°C for HRSV in mammalian cells and 28°C for AcMNPV in insect cells. At 34 h posttransfection cells were incubated on ice with anti-GP64 MAb AcV1, which recognizes a conformation-dependent epitope, and relative GP64 and GP64/F surface levels were measured by CELISA as described previously (34, 42) (Fig. 1B). Similar levels of GP64 and GP64/F were detected, indicating that both proteins were expressed and present at the cell surface in a native conformation. However, the amount of protein at the cell surface was significantly greater at 33°C than at 37°C. To assay membrane fusion capacity, duplicate wells whose contents were incubated at 33 or 37°C were exposed to PBS, pH 5.0, for 3.5 min at 34 h posttransfection and their contents were incubated again for 4 h in normal growth medium. Cells were then fixed in 4% paraformaldehyde, stained with Hoechst (Molecular Probes) to visualize nuclei, and examined for syncytium formation with a fluorescence microscope (Fig. 1C). At 33°C, cells transfected with either GP64 or GP64/F plasmid displayed extensive syncytium formation, showing that both proteins are fusion competent. Though present at the surface at similar levels (Fig. 1B), more syncytia per well were observed for authentic GP64 than for cells transfected with GP64/F (not shown). In contrast to the situation at 33°C, few syncytia formed in transfected cells incubated at 37°C, in agreement with the reduced levels of GP64 and GP64/F at the cell surface at that temperature.
In insect cells, the pH threshold of AcMNPV GP64 for membrane fusion was reported to be approximately 5.5 (66). Because CTDs have been implicated in the capacity of some viral membrane fusion proteins to cause syncytium formation, the pH threshold for GP64-induced membrane fusion in HEp-2 cells was examined. Both GP64- and GP64/F-transfected cells were incubated at 33°C and were exposed for 3.5 min to PBS with a pH ranging from 5.0 to 7.0 (Fig. 1D). Extensive syncytium formation was observed at pH 5.0; in contrast, at pH 5.4 syncytia were present but rare. At pH 5.6 or higher, syncytia were no longer observed, indicating that the pH threshold in HEp-2 cells was similar to that observed in insect cells and that alteration of the GP64 CTD did not affect this threshold (Fig. 1D).
Generation and characterization of engineered HRSVs expressing GP64.
To test whether GP64 could functionally complement an unrelated virus lacking all of its transmembrane glycoproteins, infectious HRSVs were generated in which all three HRSV glycoprotein ORFs were deleted and were replaced with ORFs encoding GP64 or GP64/F along with two placekeeper ORFs (GFP and GUS) as previously described (Fig. 2A) (43). In this manner, both the number and positions of genes were kept as in a wt HRSV, as the level of gene expression of nonsegmented negative-strand RNA viruses is controlled by the position of a gene relative to a single 3′ promoter (64). Briefly, two shuttle vectors were constructed containing three ORFs (GFP-GP64-GUS or GFP-GP64/F-GUS) separated by authentic HRSV intergenic junctions and flanked by unique restriction sites. These shuttle vectors were cloned into a cDNA, with a deletion of SH/G/F, containing matching restriction sites in order to yield cDNAs for the recovery of viruses that varied from wt HRSV only in the content of ORFs at gene positions 6, 7, and 8 (pRSΔsh,g,f/GP64 and pRSΔsh,g,f/GP64/F, respectively) (Fig. 2A). As a comparison, a previously engineered virus was used in which the SH ORF was replaced with GFP but which contained wt HRSV G and F ORFs and replicated in a manner similar to that of wt HRSV in cell culture (RSΔsh [Fig. 2A]) (43). The GP64- and GP64/F-containing cDNAs were transfected into HEp-2 cells previously infected with MVA-T7, along with plasmids encoding the HRSV N, P, L, and M2-1 proteins, and infectious viruses were recovered from the supernatant and were designated RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F, respectively (Fig. 2A). Virus stocks were generated in Vero cells at 33°C, yielding titers of approximately 107 PFU/ml. The genomes of virus stocks were verified by reverse transcriptase PCR across the modified areas followed by sequence analysis at pass 3 (data not shown).
Expression of GP64 and/or HRSV G and F proteins in cells infected by each of the engineered viruses was examined. Infected Vero cells were fixed with paraformaldehyde at 28 h postinfection and were permeabilized with 0.1% sodium dodecyl sulfate. Cells were then incubated with antibodies against GP64 (AcV5), HRSV G (L9), HRSV F (MAb 19), or HRSV nucleocapsid (N) protein (MAb 6) as a control, and protein expression was measured by using CELISA (Fig. 2B). In cells infected with viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F, the HRSV N protein, GP64, and GP64/F proteins were detected but not the HRSV G and F proteins, consistent with the gene content of these viruses. In contrast, virus RSΔsh-infected cells demonstrated abundant expression of the HRSV N, G, and F proteins but not GP64 or GP64/F. All viruses expressed GFP, as monitored by fluorescence microscopy (not shown), and GFP expression was previously shown to provide an indicator of infectivity that correlated with the number of PFU (43). These results confirmed that engineered viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F expressed GP64 and GP64/F, respectively, as their only transmembrane glycoprotein.
Infectivity of viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F is mediated by GP64.
To determine whether the infectivity of viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F was mediated by GP64 and GP64/F, respectively, the effect on virus entry of a characterized neutralizing anti-GP64 MAb (AcV1) (62) was examined. Given quantities (1.5 × 105 PFU) of each of the engineered viruses were preincubated with MAb AcV1 at various concentrations for 70 min at room temperature and were then used to infect 5 × 105 Vero cells. After infection cells were washed, incubated for 22 h at 33°C, trypsinized, and fixed, and the percentage of infected cells was determined by assaying for cells in which GFP was expressed from the engineered HRSV genomes by flow cytometry as previously described (43) (Fig. 3A). MAb AcV1 inhibited entry of the GP64-containing viruses in a concentration-dependent manner, while virus RSΔsh, which contains the HRSV G and F proteins and no GP64, was unaffected. In contrast, in the same assay MAb19, an antibody specific for the HRSV F protein, inhibited infectivity of virus RSΔsh but not of viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F (Fig. 3B).
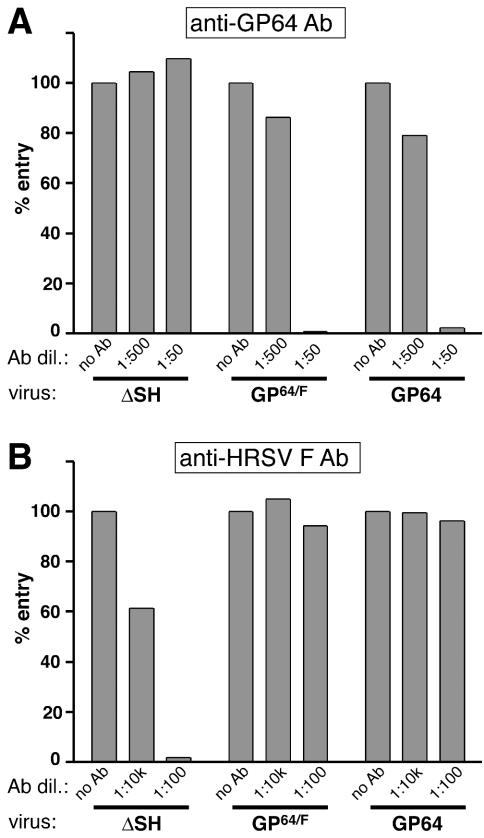
FIG. 3.
Neutralization of infectivity. Viruses RSΔsh, RSΔsh,g,f/GP64/F, and RSΔsh,g,f/GP64 were preincubated with anti-GP64 antibody (Ab) AcV1 (A) or anti-HRSV F antibody MAb 19 (B) and were used to infect Vero cells. After 22 h of incubation at 33°C, cells were trypsinized and examined for viral infectivity by quantitating GFP expression from the viral genome by flow cytometry as previously described (43). Bars represent relative entry, i.e., the number of GFP positive cells relative to the number of GFP-expressing cells in the absence of antibody × 100, of duplicate samples. Antibody dilutions used are indicated.
The HRSV G and F proteins mediate pH-independent entry at the plasma membrane, while entry mediated by the GP64 protein requires a low-pH step (4, 43, 55, 61). To determine whether replacement of the HRSV G and F proteins with GP64 or GP64/F rendered virus entry sensitive to compounds that buffer the endosomal pH, Vero cells were infected with the engineered viruses in the presence or absence of ammonium chloride (Fig. 4A). Ammonium chloride is known to buffer the endosomal pH within 1 min after addition to the cell medium and does not affect binding, endocytosis, or membrane fusion (33, 39). The presence of ammonium chloride blocked entry of viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F but not of virus RSΔsh. This indicated that, in contrast to virus RSΔSH, the entry pathway of the GP64- and GP64/F-containing viruses required a low-pH step. The data for Fig. 3 and 4A together indicate that infectivity of viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F was mediated by GP64 in a low-pH-dependent fashion.
Kinetics of virion release from endosomes.
Because of the fast-acting buffering capacity of ammonium chloride, kinetics of nucleocapsid release from the endosome after internalization of a virus can be accurately determined by addition of ammonium chloride to cells at time intervals after synchronous virus binding (19, 33, 39). The kinetics of endosomal release depend upon the pH at which a membrane fusion protein is triggered to induce fusion between the viral and endosomal membranes (33). The half time of AcMNPV nucleocapsid release from the endosome after internalization in insect cells was reported to be approximately 25 min (21). In a mammalian cell type, PK1-LLC (Pk1), half time of release of AcMNPV was approximately 50 min (59). We determined the timing of endosomal release for viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F in Vero cells (Fig. 4B). Virus was bound to cells at 4°C for 90 min and was then replaced with medium prewarmed to 37°C (time zero). Ammonium chloride was added to the infected cells at time zero or at 30- to 60-min intervals thereafter. At 20 h post-37°C shift, cells were harvested and the percentage of cells expressing GFP from the viral genome was determined by flow cytometry as previously described (43). The percentage of GFP-expressing cells of each sample was compared to the percentage of GFP-expressing cells in the absence of ammonium chloride to calculate relative entry and the time at which 50% of virions were released from endosomes. For both GP64-containing viruses, the half time of endosomal release was approximately 2 h (Fig. 4B). This was significantly slower than that reported for GP64-mediated endosomal release of AcMNPV in insect and PK1 cells (see above). The similar half time of release between viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F appears to be in agreement with the identical pH threshold for membrane fusion induced by GP64 and GP64/F, as determined in Fig. 1D.
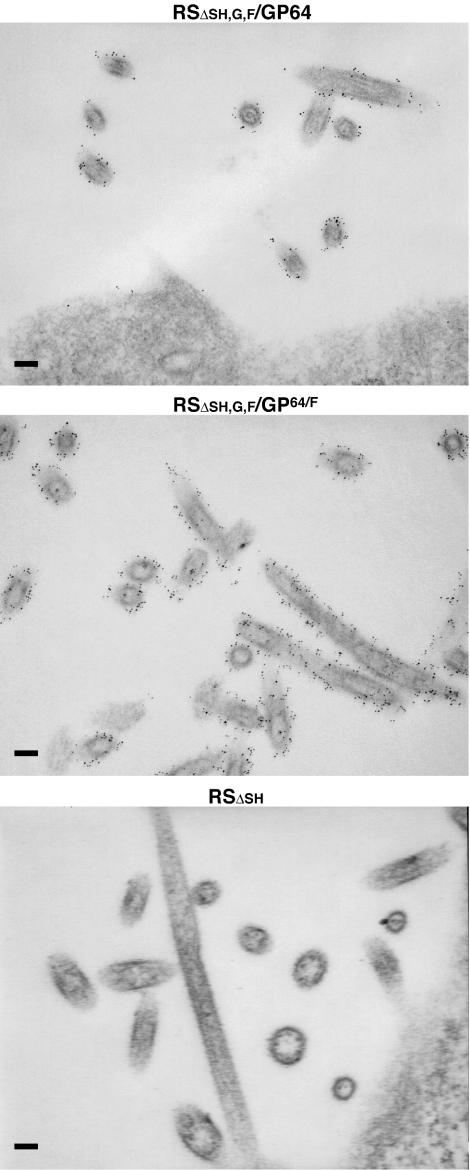
GP64 and GP64/F incorporate into HRSV-induced filaments at the cell surface.
The GP64 and GP64/F proteins efficiently mediated infectivity of their respective viruses (Fig. 3 and 4), indicating that these proteins incorporated into HRSV virions. The morphology of the infectious HRSV unit is not well defined. Both filaments of 4 to 10 μm in length and 80 to 150 nm in diameter and spherical structures of the same diameter, which may or may not represent filament cross-sections, are observed in great abundance at the surface of infected cells (17, 49). Because HRSV infectivity was shown to be predominantly cell associated and because the G and F proteins concentrate in the observed cell surface structures (43, 49), it is generally believed that these structures represent infectious virus particles. We have previously demonstrated that an HRSV in which all three transmembrane glycoprotein genes were replaced with that of VSIV G still induced the formation of cell surface filaments and that the VSIV G protein incorporated into these structures and colocalized with the HRSV F protein, when expressed simultaneously (43). However, in that case, the VSIV G protein contained the full predicted CTD of the HRSV F protein (approximately 22 amino acids). Here we examined (i) whether virally induced filaments were formed in the absence of any HRSV-specific glycoprotein components and (ii) whether GP64/F and authentic GP64 targeted to HRSV-induced filaments by IEM (Fig. 5). Vero cells at 27 h postinfection were fixed and incubated with MAb AcV5, followed by a secondary antibody conjugated to 6-nm gold. As a negative control, cells infected with virus RSΔsh were subjected to the same procedure. IEM analysis of RSΔsh-infected cells displayed the typical long-filament-containing and spherical structures (Fig. 5). On higher magnification, spikes extending from the membrane surrounding the structures were visible (data not shown); other studies have shown that these spikes consist of the HRSV G and F surface glycoproteins (17, 26). In cells infected with viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F, abundant filamentous and spherical particles of dimensions similar to those of wt and RSΔSH virus were observed; these viral structures were labeled across the entire surface after incubation with anti-GP64 antibodies and a gold-conjugated secondary antibody, whereas particles at the surface of virus RSΔsh-infected cells were not (compare top panels of Fig. 5 to bottom panel). Thus, both GP64 and GP64/F incorporated into HRSV-induced structures at the cell surface. GP64 gold labeling was slightly more intense for virus RSΔsh,g,f/GP64/F, suggesting that inclusion of the 12 C-terminal amino acids of HRSV may aid the incorporation process.
FIG. 5.
IEM of virus-infected cells. Vero cells infected with engineered virus RSΔsh, RSΔsh,g,f/GP64/F, or RSΔsh,g,fF/GP64 were fixed at 27 h postinfection, incubated with anti-GP64 antibodies followed by a 6-nm gold-conjugated secondary antibody, and prepared for IEM as described in Materials and Methods. Size bar, 100 nm.
The effect of temperature on replication of viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F.
The ability of the engineered viruses to replicate in mammalian cell cultures was examined (Fig. 6). Vero and HEp-2 cells were infected at a multiplicity of infection of 0.5, and the production of infectious progeny virus was measured at 1-day intervals. Because GP64 cell surface and membrane fusion levels were higher at reduced temperatures (Fig. 1), growth characteristics at 37 and 33°C were compared. wt HRSV is known to remain predominantly cell associated (31, 43, 49); to maximize viral yields virus was harvested by scraping cells into the supernatant followed by gentle pipetting. Viral titers were determined by TCID50 based on the expression of GFP, as described previously (43). Virus RSΔsh was previously shown to replicate in Vero and HEp-2 cells to levels similar to that of a wt HRSV (43). In Vero cells, virus RSΔsh replicated to similar levels at 33° and 37°C, albeit peak levels were reached sooner at 37°C, presumably due to the more rapid metabolism of cells at that temperature (Fig. 6A). For viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F, production of infectious virus was consistently higher at 33°C than at 37°C, in particular on days 4 to 6 postinfection (Fig. 6A). This difference was even more pronounced in HEp-2 cells, where the levels of infectious virus produced at 33°C were 100- to 1,000-fold higher than those at 37°C. Viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F amplified very poorly at 37°C in this cell type, and the onset of virus production at 33°C appeared somewhat delayed compared to virus RSΔsh. The observed temperature sensitivity correlated with the increased levels of GP64 at the cell surface at reduced temperature (Fig. 1). Minor differences in replication were observed between the viruses containing authentic GP64 or GP64/F; however, at 37°C virus RSΔsh,g,f/GP64/F replicated to moderately higher levels than did virus RSΔsh,g,f/GP64, indicating that the presence of the F protein CTD may confer an advantage.
FIG. 6.
Virus replication in Vero and HEp-2 cells at 37 and 33°C. (A) Vero and HEp-2 cells were infected with the engineered viruses for 1.5 h at 37°C at a multiplicity of 0.5, washed, and then incubated at 37 or 33°C. Immediately after virus adsorption and at 1-day intervals thereafter, cells were scraped into the supernatants and titers of virus suspensions were determined in duplicate by TCID50, as previously described (43). (B) Vero cells were infected as above, and supernatant and cells were harvested separately at 24 and 48 h postinfection. Virus titers were determined in duplicate as above, and the percentage of infectivity relative to the total (supernatant + cell associated) was calculated (cell, cell-associated infectivity; supt, supernatant-associated infectivity).
We also examined whether replacement of the HRSV G and F proteins with GP64 affected the predominantly cell-associated nature of HRSV infectivity. The supernatants and cell fractions of infected Vero cells were collected separately at a time when virus production was ongoing but when no significant cell lysis had yet occurred (24 and 48 h postinfection), and virus titers were compared (Fig. 6B). In agreement with previous results, the majority of RSΔsh infectivity (approximately 98 or 99%) was associated with the cellular fraction (43). Infectivity of the GP64-containing engineered HRSVs was also predominantly cell associated, even though the baculovirus BV phenotype, of which GP64 is the major envelope glycoprotein, is efficiently secreted into the supernatant and is stable in solution. Thus, GP64 could efficiently mediate infection of HRSV in two different mammalian cell types but did not alter the cell-associated nature of HRSV infectivity.
Stability of the GP64-containing engineered viruses.
GP64 is a major structural component of the relatively stable baculovirus BV phenotype. Stocks of baculovirus AcMNPV BV can be stored at 4°C for more than a year with infectivity virtually unchanged. In contrast, wt HRSV infectivity is notoriously unstable (16, 20). Infectivity declines even when viruses are stored frozen, and this is a significant problem for HRSV research and storage of potential vaccines. The cause of this instability is not known. Chemicals such as magnesium chloride and HEPES are generally added to increase stability (16, 20). We asked whether the presence of GP64 might influence the stability of HRSV. Stocks of viruses RSΔsh, RSΔsh,g,f/GP64, and RSΔsh,g,f/GP64/F were prepared simultaneously and identically from Vero cells infected at low multiplicity. Although deletion of the SH gene has no impact on replication of HRSV in cell culture (9, 29), a possible role in virion stability has never been examined. Therefore, the wt A2 strain of HRSV (from which the engineered viruses were derived) also was included in this experiment. Infected cells were scraped into the medium, and this was followed by gentle pipetting and removal of cell debris by low-speed centrifugation. Harvested stocks were divided into 100-μl aliquots and were transferred to 4°C for storage, and virus titers at day 0 were determined in triplicate. To avoid possible effects of concentration on virus stability, some stocks were diluted to reach similar starting concentrations (average in TCID50 per milliliter at time zero: RSΔsh, 3.4 × 107; RSΔsh,g,f/GP64/F, 3.2 × 107; RSΔsh,g,f/GP64, 3.2 × 107; and A2, 2.1 × 108). For these stocks, remaining infectivity was determined in triplicate by TCID50 at 1-week intervals after storage at 4°C and was compared to the infectivity measured at the day of harvest (Fig. 7). The infectivities of viruses RSΔsh and A2 declined rapidly; more than 50% of the infectivity measured at the time of harvest was lost within 3.5 days at 4°C, and after 8 weeks of storage less than 4% of infectivity remained. In contrast, titers of viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F did not decline upon storage. Even after 8 weeks at 4°C, infectivity was near 100% of that measured at the time of harvest. These data show that incorporation of GP64 in place of the homologous glycoproteins improved the stability of HRSV and suggest that the HRSV transmembrane glycoproteins may be a factor in the instability of HRSV infectivity.
FIG. 7.
Stability of infectivity. Vero cells were infected with viruses RSΔsh, RSΔsh,g,f/GP64/F, RSΔsh,g,f/GP64, and wt A2, and stocks were generated by scraping cells into the supernatant followed by gentle pipetting and removal of cell debris. Stocks were aliquoted and stored at 4°C. Titers were determined by TCID50 at the day of harvest, after 3.5 days, and at weekly intervals thereafter. Infectivities were plotted as a percentage of the TCID50 at the start of the experiment (week 0). Error bars represent standard deviations from the mean of triplicate samples.
DISCUSSION
The advantageous combination of having entry and exit functions within the same protein and low cell toxicity prompted us to examine the expression and function of GP64 in mammalian cells and to characterize its capacity to functionally complement HRSV, a mammalian virus. The results demonstrate that GP64 can efficiently replace the functions of heterologous viral glycoproteins and can provide stability to an unstable mammalian virus in cell culture and indicate that temperature of incubation is an important factor in its ability to do so.
GP64 membrane fusion in transfected mammalian cells.
Insect and mammalian cells differ in the capacity to glycosylate proteins, in particular in their ability to perform sialylation (32). Despite these and other differences such as pH and temperature, GP64, provided to a lentivirus from transfected mammalian cells, was shown to mediate entry, implying that GP64 was functional (30). Here direct evidence is provided that GP64 is expressed at the surface of mammalian cells and induces membrane fusion after exposure to low pH (Fig. 1B and C). While, for some viral envelope proteins, alterations in the CTD have been shown to affect the ability to induce membrane fusion (38, 40, 48, 65), a chimeric GP64 containing the HRSV F CTD in place of its own also efficiently induced membrane fusion. Moreover, the pH threshold of fusion for GP64 and GP64/F was unchanged from that of GP64 in insect cells (Fig. 1D). In contrast, it was necessary to lower the temperature of incubation from 37 to 33°C for optimal membrane fusion and surface expression. Although the underlying reasons for the temperature sensitivity have not been examined here, one possible explanation is misfolding and/or degradation of GP64 at 37°C; GP64 is commonly expressed under insect physiological conditions (28°C) and may have strong structure/function constraints (34). Synthesis and function were not examined at temperatures lower than 33°C; hence, it is possible that surface levels and membrane fusion capacity will further increase at lower temperatures.
HRSV infectivity mediated by GP64.
When the homologous HRSV glycoprotein ORFs were replaced with those of GP64 or GP64/F, infectious viruses were recovered that expressed GP64 or GP64/F as their only viral transmembrane glycoprotein. These viruses were effectively neutralized with GP64-specific antibodies. Similar results were obtained when the SH, G, and F ORFs were replaced with that of the VSIV G protein carrying the full HRSV F CTD (43). Thus, in cell culture, any potential assembly-related functions provided by the HRSV transmembrane glycoproteins appear to be replaceable by heterologous viral glycoproteins. Consistent with the above-mentioned low-pH-mediated membrane fusion capacity in transfected cells, GP64-mediated HRSV entry required a low-pH step (Fig. 4A). This is in contrast to wt HRSV, which is thought to enter at the plasma membrane and is not sensitive to compounds that buffer the endosomal pH (43, 55). Thus, GP64 and GP64/F are able to convert the entry pathway of HRSV from a pH-independent to a pH-dependent one. Although GP64-mediated infectivity of HRSV was efficient (Fig. 6), the rate of nucleocapsid release for viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F (half time of approximately 2 h) was markedly lower than that reported for AcMNPV in insect and mammalian cells (half times of approximately 25 and 50 min, respectively) (Fig. 4B) (21, 59). Among the possible explanations for the observed difference are cell-type-specific differences or suboptimal GP64 function at 37°C. A third possibility may relate to the morphology of the HRSV infectious unit. Infectivity is not well defined; however, it is lost after filtration though 0.45-μm-pore-size filters, suggesting that the long filaments observed may be a predominant infectious form of HRSV (49). If this is the case, the length of the filaments also may be a factor in limiting the rate of entry via the endocytic pathway.
The results of the growth curves in Fig. 6 appear to correlate with the temperature sensitivity of GP64 surface expression and membrane fusion (Fig. 1B and C). Amplification of viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F at 33°C was substantially higher than at 37°C for both cell types examined, while replication of virus RSΔsh was not. Although temperature is thus an important factor for GP64-mediated infectivity, the difference in replication efficiency at 37 and 33°C was much more pronounced in HEp-2 cells than in Vero cells, suggestive of cell-type-specific effects. Whether the observed differences are related to the expression, maturation, or incorporation of GP64 or other aspects of HRSV replication remains to be determined.
GP64 incorporation into HRSV-induced filaments.
Following wt HRSV entry and replication, the G and F proteins are seen by electron microscopy to concentrate in cell-associated, virus-induced filamentous structures at the cell surface that are believed to represent virus particles (17, 26, 43). In the present work, we inserted into an SH,G,F-depleted HRSV genome an ORF encoding a heterologous viral transmembrane glycoprotein carrying only a partial version of the HRSV F CTD (GP64/F) or no HRSV CTD at all (authentic GP64). Both resulting engineered viruses (RSΔsh,g,f/GP64/F and RSΔsh,g,f/GP64) induced abundant cell surface filaments as well as spheres that contained GP64/F and GP64, respectively (Fig. 5). It is likely that the observed GP64 or GP64/F-containing structures represent infectious virions, as the bulk of the infectivity of viruses RSΔsh,g,f/GP64/F and RSΔsh,g,f/GP64 remained cell associated (Fig. 6) and anti-GP64 antibodies neutralized infectivity (Fig. 3). Since both GP64 and GP64/F efficiently mediated HRSV infectivity (Fig. 6), it can be concluded that replacement of the GP64 CTD with a heterologous CTD is not required for incorporation of GP64 into HRSV filaments or infectivity in cell culture. However, as observed by IEM, the levels of GP64/F incorporated in the viral filaments were slightly higher than those of GP64. In addition, in the growth analysis (Fig. 6), amplification of virus RSΔsh,g,f/GP64/F at 37°C was modestly but consistently higher than that of virus RSΔsh,g,f/GP64, in particular during the first 24 h of incubation in Vero cells. Thus, although not required in this system, the presence of the 12 C-terminal amino acids of the HRSV F protein may confer a minor advantage. The absence of a significant advantage for the HRSV F CTD in cell culture does not exclude the possibility of a role for this domain in wt HRSV replication in vivo. The fact that infectivity of these viruses remains associated with the cells in the presence of GP64 and absence of SH, G, and F suggests that HRSV components other than its transmembrane glycoproteins are determinants of the cell-associated nature.
Interestingly, in the baculovirus BV phenotype, it appears that GP64 is localized exclusively at the ends of the rod-shaped virion. If the distribution of GP64 in baculovirus virions is indeed polarized, the relatively even distribution of GP64 across the HRSV filaments might suggest that, in the BV or insect cell membranes, viral or cellular factors are in place to concentrate GP64 at the ends of the rod-shaped BV particle and/or to exclude it elsewhere in the viral membrane.
GP64 and stability of HRSV.
Yields of viruses RSΔsh,g,f/GP64 and RSΔsh,g,f/GP64/F from Vero cells at late times postinfection were approximately 10-fold higher than that of virus RSΔsh (Fig. 6). It is not known whether this is predominantly a result of increased virus production, increased virus stability, or both. However, the results depicted in Fig. 7 showed a substantial increase in stability of HRSV infectivity when SH, G, and F are replaced with GP64 or GP64/F. The implications of these findings are twofold: (i) GP64 may provide a tool to improve the stability of heterologous viruses, similar to the use of VSIV G complementation of heterologous viruses for the generation of high-titer stocks, and (ii) the HRSV G and/or F proteins may be a source of, or contributing factor to, the unstable nature of HRSV infectivity, which is a significant problem in the HRSV field (16, 20).
In conclusion, GP64 and a GP64 chimeric protein carrying a partial HRSV F CTD were functional in two mammalian cell lines. Moreover, both proteins provided functions that allowed efficient, temperature-sensitive amplification of an unrelated human virus. In addition, infectivity of the GP64-complemented HRSVs was substantially more stable than that of wt HRSV. These results highlight the potential of GP64 for vaccine- or gene therapy-related applications in mammalian systems and as a substitute entry/exit protein for studies of the viral transmembrane glycoproteins in assembly of HRSV virions.
Acknowledgments
We thank the members of the G. W. Wertz and L. A. Ball laboratories for helpful discussions during preparation of the manuscript, Leigh Millican of the UAB imaging facility for technical support, and Gary Blissard for providing the GP64 construct and antibodies.
This work was supported by Public Health Service grant AI20181 from the NIH.
REFERENCES
- 1.Anderson, K., A. M. King, R. A. Lerch, and G. W. Wertz. 1992. Polylactosaminoglycan modification of the respiratory syncytial virus small hydrophobic (SH) protein: a conserved feature among human and bovine respiratory syncytial viruses. Virology 191:417-430. [DOI] [PubMed] [Google Scholar]
- 2.Bilello, J. P., E. E. Cable, R. L. Myers, and H. C. Isom. 2003. Role of paracellular junction complexes in baculovirus-mediated gene transfer to nondividing rat hepatocytes. Gene Ther. 10:733-749. [DOI] [PubMed] [Google Scholar]
- 3.Blissard, G. W. 1996. Baculovirus-insect cell interactions. Cytotechnology 20:73-93. [DOI] [PubMed] [Google Scholar]
- 4.Blissard, G. W., and J. R. Wenz. 1992. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 66:6829-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boublik, Y., P. Di Bonito, and I. M. Jones. 1995. Eukaryotic virus display: engineering the major surface glycoprotein of the Autographa californica nuclear polyhedrosis virus (AcNPV) for the presentation of foreign proteins on the virus surface. Bio/Technology 13:1079-1084. [DOI] [PubMed] [Google Scholar]
- 6.Boyce, F. M., and N. L. Bucher. 1996. Baculovirus-mediated gene transfer into mammalian cells. Proc. Natl. Acad. Sci. USA 93:2348-2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braunagel, S. C., and M. D. Summers. 1994. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology 202:315-328. [DOI] [PubMed] [Google Scholar]
- 8.Brusca, J., M. Summers, J. Couch, and L. Courtney. 1986. Autographa californica nuclear polyhedrosis virus efficiently enters but does not replicate in poikilothermic vertebrate cells. Intervirology 26:207-222. [DOI] [PubMed] [Google Scholar]
- 9.Bukreyev, A., S. S. Whitehead, B. R. Murphy, and P. L. Collins. 1997. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J. Virol. 71:8973-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carbonell, L. F., and L. K. Miller. 1987. Baculovirus interaction with nontarget organisms: a virus-borne reporter gene is not expressed in two mammalian cell lines. Appl. Environ. Microbiol. 53:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chernomordik, L., E. Leikina, M.-S. Cho, and J. Zimmerberg. 1995. Control of baculovirus gp64-induced syncytium formation by membrane lipid composition. J. Virol. 69:3049-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, P. L., Y. T. Huang, and G. W. Wertz. 1984. Nucleotide sequence of the gene encoding the fusion (F) glycoprotein of human respiratory syncytial virus. Proc. Natl. Acad. Sci. USA 81:7683-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Condreay, J. P., S. M. Witherspoon, W. C. Clay, and T. A. Kost. 1999. Transient and stable gene expression in mammalian cells transduced with a recombinant baculovirus vector. Proc. Natl. Acad. Sci. USA 96:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst, W., R. Grabherr, D. Wegner, N. Borth, A. Grassauer, and H. Katinger. 1998. Baculovirus surface display: construction and screening of a eukaryotic epitope library. Nucleic Acids Res. 26:1718-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst, W. J., A. Spenger, L. Toellner, H. Katinger, and R. M. Grabherr. 2000. Expanding baculovirus surface display. Modification of the native coat protein gp64 of Autographa californica NPV. Eur. J. Biochem. 267:4033-4039. [DOI] [PubMed] [Google Scholar]
- 16.Fernie, B. F., and J. L. Gerin. 1980. The stabilization and purification of respiratory syncytial virus using MgSO4. Virology 106:141-144. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs, H., and T. Bachi. 1975. Scanning electron microscopical demonstration of respiratory syncytial virus antigens by immunological markers. J. Ultrastruct. Res. 52:114-119. [DOI] [PubMed] [Google Scholar]
- 18.Grabherr, R., W. Ernst, C. Oker-Blom, and I. Jones. 2001. Developments in the use of baculoviruses for the surface display of complex eukaryotic proteins. Trends Biotechnol. 19:231-236. [DOI] [PubMed] [Google Scholar]
- 19.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 20.Gupta, C. K., J. Leszczynski, R. K. Gupta, and G. R. Siber. 1996. Stabilization of respiratory syncytial virus (RSV) against thermal inactivation and freeze-thaw cycles for development and control of RSV vaccines and immune globulin. Vaccine 14:1417-1420. [DOI] [PubMed] [Google Scholar]
- 21.Hefferon, K. L., A. G. Oomens, S. A. Monsma, C. M. Finnerty, and G. W. Blissard. 1999. Host cell receptor binding by baculovirus GP64 and kinetics of virion entry. Virology 258:455-468. [DOI] [PubMed] [Google Scholar]
- 22.Hofmann, C., V. Sandig, G. Jennings, M. Rudolph, P. Schlag, and M. Strauss. 1995. Efficient gene transfer into human hepatocytes by baculovirus vectors. Proc. Natl. Acad. Sci. USA 92:10099-10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, Y. T., P. L. Collins, and G. W. Wertz. 1985. Characterization of the 10 proteins of human respiratory syncytial virus: identification of a fourth envelope-associated protein. Virus Res. 2:157-173. [DOI] [PubMed] [Google Scholar]
- 24.Huser, A., and C. Hofmann. 2003. Baculovirus vectors: novel mammalian cell gene-delivery vehicles and their applications. Am. J. Pharmacogenomics 3:53-63. [PubMed] [Google Scholar]
- 25.IJkel, W. F. J., M. Westenberg, R. W. Goldbach, G. W. Blissard, J. M. Vlak, and D. Zuidema. 2000. A novel baculovirus envelope fusion protein with a proprotein convertase cleavage site. Virology 275:30-41. [DOI] [PubMed] [Google Scholar]
- 26.Jeffree, C. E., H. W. Rixon, G. Brown, J. Aitken, and R. J. Sugrue. 2003. Distribution of the attachment (G) glycoprotein and GM1 within the envelope of mature respiratory syncytial virus filaments revealed using field emission scanning electron microscopy. Virology 306:254-267. [DOI] [PubMed] [Google Scholar]
- 27.Jin, H., G. P. Leser, J. Zhang, and R. A. Lamb. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 16:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaba, S. A., J. C. Hemmes, J. W. van Lent, J. M. Vlak, V. Nene, A. J. Musoke, and M. M. van Oers. 2003. Baculovirus surface display of Theileria parva p67 antigen preserves the conformation of sporozoite-neutralizing epitopes. Protein Eng. 16:73-78. [DOI] [PubMed] [Google Scholar]
- 29.Karron, R. A., D. A. Buonagurio, A. F. Georgiu, S. S. Whitehead, J. E. Adamus, M. L. Clements-Mann, D. O. Harris, V. B. Randolph, S. A. Udem, B. R. Murphy, and M. S. Sidhu. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA 94:13961-13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar, M., B. P. Bradow, and J. Zimmerberg. 2003. Large-scale production of pseudotyped lentiviral vectors using baculovirus GP64. Hum. Gene Ther. 14:67-77. [DOI] [PubMed] [Google Scholar]
- 31.Levine, S., and R. Hamilton. 1969. Kinetics of the respiratory syncytial virus growth cycle in HeLa cells. Arch. Gesamte Virusforsch. 28:122-132. [DOI] [PubMed] [Google Scholar]
- 32.Marchal, I., D. L. Jarvis, R. Cacan, and A. Verbert. 2001. Glycoproteins from insect cells: sialylated or not? Biol. Chem. 382:151-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marsh, M., and A. Helenius. 1989. Virus entry into animal cells. Adv. Virus Res. 36:107-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Monsma, S. A., and G. W. Blissard. 1995. Identification of a membrane fusion domain and an oligomerization domain in the baculovirus GP64 envelope fusion protein. J. Virol. 69:2583-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Monsma, S. A., A. G. Oomens, and G. W. Blissard. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 70:4607-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morse, M. A., A. C. Marriott, and P. A. Nuttall. 1992. The glycoprotein of Thogoto virus (a tick-borne orthomyxo-like virus) is related to the baculovirus glycoprotein GP64. Virology 186:640-646. [DOI] [PubMed] [Google Scholar]
- 37.Mottershead, D. G., K. Alfthan, K. Ojala, K. Takkinen, and C. Oker-Blom. 2000. Baculoviral display of functional scFv and synthetic IgG-binding domains. Biochem. Biophys. Res. Commun. 275:84-90. [DOI] [PubMed] [Google Scholar]
- 38.Mulligan, M. J., G. V. Yamshchikov, G. D. Ritter, Jr., F. Gao, M. J. Jin, C. D. Nail, C. P. Spies, B. H. Hahn, and R. W. Compans. 1992. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J. Virol. 66:3971-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohkuma, S., and B. Poole. 1978. Fluorescence probe measurement of the intralysosomal pH in living cells and the perturbation of pH by various agents. Proc. Natl. Acad. Sci. USA 75:3327-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ohuchi, M., C. Fischer, R. Ohuchi, A. Herwig, and H. D. Klenk. 1998. Elongation of the cytoplasmic tail interferes with the fusion activity of influenza virus hemagglutinin. J. Virol. 72:3554-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ojala, K., D. G. Mottershead, A. Suokko, and C. Oker-Blom. 2001. Specific binding of baculoviruses displaying gp64 fusion proteins to mammalian cells. Biochem. Biophys. Res. Commun. 284:777-784. [DOI] [PubMed] [Google Scholar]
- 42.Oomens, A. G., and G. W. Blissard. 1999. Requirement for GP64 to drive efficient budding of Autographa californica multicapsid nucleopolyhedrovirus. Virology 254:297-314. [DOI] [PubMed] [Google Scholar]
- 43.Oomens, A. G., A. G. Megaw, and G. W. Wertz. 2003. Infectivity of a human respiratory syncytial virus lacking the SH, G, and F proteins is efficiently mediated by the vesicular stomatitis virus G protein. J. Virol. 77:3785-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oomens, A. G., S. A. Monsma, and G. W. Blissard. 1995. The baculovirus GP64 envelope fusion protein: synthesis, oligomerization, and processing. Virology 209:592-603. [DOI] [PubMed] [Google Scholar]
- 45.Pearson, M. N., and G. F. Rohrmann. 2002. Transfer, incorporation, and substitution of envelope fusion proteins among members of the Baculoviridae, Orthomyxoviridae, and Metaviridae (insect retrovirus) families. J. Virol. 76:5301-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plonsky, I., M. S. Cho, A. G. Oomens, G. Blissard, and J. Zimmerberg. 1999. An analysis of the role of the target membrane on the Gp64-induced fusion pore. Virology 253:65-76. [DOI] [PubMed] [Google Scholar]
- 47.Plonsky, I., and J. Zimmerberg. 1996. The initial fusion pore induced by baculovirus GP64 is large and forms quickly. J. Cell Biol. 135:1831-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ritter, G. D., Jr., M. J. Mulligan, S. L. Lydy, and R. W. Compans. 1993. Cell fusion activity of the simian immunodeficiency virus envelope protein is modulated by the intracytoplasmic domain. Virology 197:255-264. [DOI] [PubMed] [Google Scholar]
- 49.Roberts, S. R., R. W. Compans, and G. W. Wertz. 1995. Respiratory syncytial virus matures at the apical surfaces of polarized epithelial cells. J. Virol. 69:2667-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmitt, A. P., G. P. Leser, D. L. Waning, and R. A. Lamb. 2002. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J. Virol. 76:3952-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schnell, M. J., L. Buonocore, E. Boritz, H. P. Ghosh, R. Chernish, and J. K. Rose. 1998. Requirement for a non-specific glycoprotein cytoplasmic domain sequence to drive efficient budding of vesicular stomatitis virus. EMBO J. 17:1289-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schnell, M. J., T. Mebatsion, and K. K. Conzelmann. 1994. Infectious rabies viruses from cloned cDNA. EMBO J. 13:4195-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shoji, I., H. Aizaki, H. Tani, K. Ishii, T. Chiba, I. Saito, T. Miyamura, and Y. Matsuura. 1997. Efficient gene transfer into various mammalian cells, including non-hepatic cells, by baculovirus vectors. J. Gen. Virol. 78:2657-2664. [DOI] [PubMed] [Google Scholar]
- 54.Song, S. U., S. H. Shin, S. K. Kim, G. S. Choi, W. C. Kim, M. H. Lee, S. J. Kim, I. H. Kim, M. S. Choi, Y. J. Hong, and K. H. Lee. 2003. Effective transduction of osteogenic sarcoma cells by a baculovirus vector. J. Gen. Virol. 84:697-703. [DOI] [PubMed] [Google Scholar]
- 55.Srinivasakumar, N., P. L. Ogra, and T. D. Flanagan. 1991. Characteristics of fusion of respiratory syncytial virus with HEp-2 cells as measured by R18 fluorescence dequenching assay. J. Virol. 65:4063-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takimoto, T., T. Bousse, E. C. Coronel, R. A. Scroggs, and A. Portner. 1998. Cytoplasmic domain of Sendai virus HN protein contains a specific sequence required for its incorporation into virions. J. Virol. 72:9747-9754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Techaarpornkul, S., N. Barretto, and M. E. Peeples. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 75:6825-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Teng, M. N., S. S. Whitehead, and P. L. Collins. 2001. Contribution of the respiratory syncytial virus G glycoprotein and its secreted and membrane-bound forms to virus replication in vitro and in vivo. Virology 289:283-296. [DOI] [PubMed] [Google Scholar]
- 59.van Loo, N. D., E. Fortunati, E. Ehlert, M. Rabelink, F. Grosveld, and B. J. Scholte. 2001. Baculovirus infection of nondividing mammalian cells: mechanisms of entry and nuclear transport of capsids. J. Virol. 75:961-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Volkman, L. E. 1986. The 64K envelope protein of budded Autographa californica nuclear polyhedrosis virus. Curr. Top. Microbiol. Immunol. 131:103-118. [DOI] [PubMed] [Google Scholar]
- 61.Volkman, L. E., and P. A. Goldsmith. 1985. Mechanism of neutralization of budded Autographa californica Nuclear Polyhedrosis virus by a monoclonal antibody: inhibition of entry by adsorptive endocytosis. Virology 143:185-195. [DOI] [PubMed] [Google Scholar]
- 62.Volkman, L. E., P. A. Goldsmith, R. T. Hess, and P. Faulkner. 1984. Neutralization of budded Autographa californica NPV by a monoclonal antibody: identification of the target antigen. Virology 133:354-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wertz, G. W., P. L. Collins, Y. Huang, C. Gruber, S. Levine, and L. A. Ball. 1985. Nucleotide sequence of the G protein gene of human respiratory syncytial virus reveals an unusual type of viral membrane protein. Proc. Natl. Acad. Sci. USA 82:4075-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wertz, G. W., V. P. Perepelitsa, and L. A. Ball. 1998. Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc. Natl. Acad. Sci. USA 95:3501-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao, Q., and R. W. Compans. 1995. Differences in the role of the cytoplasmic domain of human parainfluenza virus fusion proteins. J. Virol. 69:7045-7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, S. X., Y. Han, and G. W. Blissard. 2003. Palmitoylation of the Autographa californica multicapsid nucleopolyhedrovirus envelope glycoprotein GP64: mapping, functional studies, and lipid rafts. J. Virol. 77:6265-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]