Abstract
Adeno-associated virus (AAV) type 2 is a human parvovirus whose replication is dependent upon cellular proteins as well as functions supplied by helper viruses. The minimal herpes simplex virus type 1 (HSV-1) proteins that support AAV replication in cell culture are the helicase-primase complex of UL5, UL8, and UL52, together with the UL29 gene product ICP8. We show that AAV and HSV-1 replication proteins colocalize at discrete intranuclear sites. Transfections with mutant genes demonstrate that enzymatic functions of the helicase-primase are not essential. The ICP8 protein alone enhances AAV replication in an in vitro assay. We also show localization of the cellular replication protein A (RPA) at AAV centers under a variety of conditions that support replication. In vitro assays demonstrate that the AAV Rep68 and Rep78 proteins interact with the single-stranded DNA-binding proteins (ssDBPs) of Ad (Ad-DBP), HSV-1 (ICP8), and the cell (RPA) and that these proteins enhance binding and nicking of Rep proteins at the origin. These results highlight the importance of intranuclear localization and suggest that Rep interaction with multiple ssDBPs allows AAV to replicate under a diverse set of conditions.
Adeno-associated virus (AAV) is a nonpathogenic human parvovirus with a biphasic life cycle (reviewed in reference 45). The linear, single-stranded DNA genome of AAV possesses inverted terminal repeats (ITRs) at either end that fold into hairpin structures and serve as the origin of replication. There are two open reading frames that encode the replication (Rep) and structural (Cap) proteins. The two large Rep proteins (Rep68 and Rep78) are required for replication and possess multiple activities, including specific DNA binding and site-specific endonuclease nicking at the viral ITR (45). Productive AAV infection in cell culture requires helper functions that can be supplied by coinfection with a second virus. In the absence of helper virus, the AAV genome stably integrates into the host genome. Helper viruses that enable efficient AAV replication include adenovirus (Ad) and viruses of the herpesvirus group, such as herpes simplex virus types 1 and 2 (HSV-1 and -2) (12), human herpesvirus 6 (51), and human cytomegalovirus (HCMV) (43). Replication of AAV can also be achieved in several cell lines by the addition of a variety of genotoxic agents (67-69), and autonomous replication has been demonstrated in cultured differentiating keratinocytes (44). A basic question in AAV biology is the nature of the helper effect supplied by the different helper viruses as well as by other permissive conditions.
Genetic analysis using mutant helper viruses and expression of individual helper virus genes has defined gene products that are required for efficient AAV replication. The helper functions of Ad are well characterized and are supplied by E1a, E1b, E2a, E4, and the VA RNA (36, 50). Specific functions have been assigned for these proteins and their predominant role appears to be in regulation of gene expression for AAV proteins. The product of the E2a gene is a single-stranded DNA-binding protein (ssDBP), referred to as Ad-DBP, that has been shown to play a direct role in aiding processivity of DNA synthesis during AAV replication (56). During AAV and Ad coinfection, it is the replication machinery of the host cell that is utilized for AAV replication. In vitro studies have identified cellular factors involved in AAV replication (46, 56). These include replication protein A (RPA), a heterotrimeric cellular complex that is an ssDBP involved in both replication and repair of cellular DNA (reviewed in reference 34).
The specific role of HSV-1 helper proteins in the AAV life cycle has not been extensively studied. Seven of the HSV-1 open reading frames encode factors essential for HSV-1 DNA replication. These include UL30/42 (DNA polymerase and accessory protein), UL9 (origin-binding protein), UL5/8/52 (helicase-primase), and UL29 (the ssDBP known as ICP8) (reviewed in reference 39). Using either HSV-1 with mutations in individual replication genes or transfections of different combinations of replication genes, Weindler and Heilbronn defined the minimal helper activities for AAV replication (60). The helicase-primase complex of UL5, UL8, and UL52 and the major DNA-binding protein ICP8 were all that was required to enable AAV replication in cell culture. In this context, the virus is utilizing the cellular replication machinery. In vitro studies have shown that the HSV-1 polymerase UL30 can also be used to replicate AAV in a reconstituted system of purified HSV-1 replication proteins (57). The helicase-primase complex was not required for UL30-dependent DNA synthesis on the AAV template in this in vitro system (57). AAV replication takes place through a rolling hairpin mechanism that utilizes a DNA primer to initiate synthesis and is exclusively mediated by leading-strand synthesis, suggesting that other functions of the HSV-1 helicase-primase complex may explain its requirement for AAV replication in cultured cells.
Many viral infections demonstrate remarkable spatial regulation, with the formation of structures within the nucleus, called replication centers, in which viral transcription and replication occur. Both Ad and HSV-1 helper viruses replicate at discrete intranuclear sites that can be observed by immunofluorescence using antibodies specific to their replication proteins (21, 37, 48). Cellular proteins involved in DNA replication colocalize to these discrete subnuclear sites of viral DNA synthesis (8, 13, 23, 64). Early during HSV-1 infection, small punctate nuclear structures, termed prereplicative sites or foci, are formed (49). The viral core proteins UL5, UL8, UL52, UL9, and ICP8 are essential for the formation of prereplicative sites in infected cells (13, 40). In cells transfected with expression vectors for UL5, UL8, UL52, and ICP8, discrete foci are formed but these do not require the viral UL9 protein (41, 52). Omission of any member of the helicase-primase complex results in ICP8 being detected in either a diffuse nuclear pattern or at a limited number of foci (41). In the presence of the viral polymerase UL30, the accessory protein UL42, and the origin-binding protein UL9, replication compartments can form that expand throughout the nucleoplasm (41, 52).
Our goal is to understand how a diverse set of helper activities can each achieve an intracellular environment that enables productive AAV replication. Ad proteins have been implicated in individual steps in the AAV life cycle, such as enhancement of second-strand synthesis on the AAV genome (24, 25) and activation of the AAV p5 promoter (15). We found no evidence of a role for the HSV-1 helper proteins in these steps of AAV infection (T. H. Stracker and M. D. Weitzman, unpublished data). Among the helper proteins from Ad and HSV-1, the two that appear most likely to be similar in function are the Ad-DBP protein and the HSV-1 ICP8 protein. These are both proteins that possess nonspecific single-stranded DNA-binding activity and are involved directly in viral DNA replication. A common feature among many DNA viruses is the interaction of an ssDBP with the viral origin-binding protein. For example, the ICP8 protein of HSV-1 interacts directly with the origin-binding protein UL9 (6, 7) and stimulates its helicase and ATPase activities (1, 5, 38). RPA is the ssDBP present in human cells, and it can be recruited for viral replication through direct interactions with origin-binding proteins such as EBNA1 of Epstein-Barr virus (71), E1 of bovine papillomavirus (29), and the large T antigen of simian virus 40 (SV40) (22, 61). AAV does not encode its own ssDBP, but Ad-DBP, ICP8, and RPA have all been implicated in AAV replication (46, 56, 57). Our laboratory has previously shown that the AAV Rep protein colocalizes with Ad-DBP at intranuclear viral replication centers during coinfection with Ad helper virus (62). We therefore investigated the localization of the different ssDBPs under a variety of conditions that support AAV replication and examined the physical and functional interactions of the ssDBPs with the AAV origin-binding Rep protein. By use of transfections, we show that the HSV-1 genes for UL5, UL8, UL52, and ICP8 are sufficient to establish AAV replication centers at which Rep and ICP8 colocalize. Mutants of the helicase-primase complex that fail to complement HSV-1 replication are able to support AAV replication in transfections of cultured cells. These mutants are still capable of forming discrete foci for ICP8 and Rep, suggesting that the role of the HSV-1 helicase-primase complex in AAV helper activity is to modulate the function or subnuclear localization of ICP8. In vitro replication assays confirm that the helicase-primase is not required and suggest that ICP8 plays an analogous role to Ad-DBP by increasing processivity of replication by the cellular polymerase. We further show that the AAV origin-binding proteins Rep68 and Rep78 are able to interact physically with the ssDBPs of Ad, HSV-1, and the host cell (RPA) and that these proteins enhance the DNA-binding and enzymatic activities of Rep. Together, our results suggest that Rep interactions with numerous ssDBPs allow AAV to utilize a diverse set of conditions for replication.
MATERIALS AND METHODS
Cell lines.
HeLa, Vero, and 293 cells were purchased from ATCC and maintained as monolayers in Dulbecco modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). All cultures were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Plasmids and transfections.
The AAV type 2 (AAV-2) genome was supplied by the plasmid pNTC244 (17). The adenovirus helper genes Ad-DBP, E1b55K, and E4orf6 were expressed from the HCMV promoter in expression vector pRK5 or pcDNA3.1 (Clontech). The HSV-1 helper proteins were supplied by expression vectors for the individual genes, as previously described (60). Mutants of UL5 and UL52 were generated by site-directed mutagenesis (4, 14). Subconfluent monolayers of cells were transfected by calcium phosphate precipitation according to standard protocols.
Viruses.
Wild-type Ad type 5 was propagated in 293 cells and purified by sequential rounds of ultracentrifugation in CsCl gradients. Titers of Ad were determined by plaque assays on 293 cells. Wild-type AAV-2 was generated by transfection of 293 cells with an AAV plasmid, pNTC244, together with the pXX6 plasmid, which supplies Ad helper functions (66). Virus was purified through iodixanol gradients as described elsewhere (72). The titer of AAV genome-containing particles per milliliter was determined by real-time PCR using SYBR Green I double-stranded DNA-binding dye and an ABI Prism 7700 sequence detection system (PE Biosystems). The KOS strain of HSV-1 was used as the wild-type virus and was propagated and titrated by plaque assay in Vero cells.
Hirt extractions and Southern blotting.
Low-molecular-weight episomal DNA was extracted from cell pellets by a modified version of the Hirt procedure, as previously described (24, 25). DNA was ethanol precipitated at −20°C overnight, resuspended in TE (10 mM Tris-1 mM EDTA), and digested with DpnI and XbaI at 37°C overnight. The DNA was electrophoresed in 0.8% agarose gels in 1× Tris-acetate-EDTA buffer at 75 V for 4.5 h. The gels were stained with ethidium bromide and denatured in 0.2 N HCl for 5 min. Gels were denatured and neutralized, and the DNA was transferred onto a nylon membrane in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) overnight. The membranes were cross-linked at 1,200 J and prehybridized at 65°C with Rapid-Hyb buffer (Amersham) for 30 min. The probe was added and hybridization continued at 65°C for another 3 to 4 h. The probe was a 4-kb plasmid fragment containing a partial AAV genome. Blots were washed twice with 1× SSC-0.1% sodium dodecyl sulfate (SDS) and 0.1× SSC-0.1% SDS at 65°C for 20 min and then with 0.1× SSC-0.5% SDS at 70°C for 15 min. Blots were exposed to X-ray film for autoradiography.
Immunofluorescence.
For immunofluorescence, cells were grown on coverslips in 24-well plates and infected with virus or transfected with plasmids. Infections were performed in 250 μl of DMEM-2%FBS for 2 h, with regular agitation. The medium was replaced with DMEM-10%FBS and incubation was allowed to proceed for the indicated time. HeLa cells were plated on glass coverslips for 24 h prior to transfection by calcium phosphate precipitation. For labeling of sites of active replication, infected cells were pulsed with bromodeoxyuridine (BrdU) (200 mM) for 15 min prior to fixation. After 40 h, cells were washed three times in phosphate-buffered saline (PBS) and fixed in 3.7% paraformaldehyde for 15 min. Cells were washed in PBS and permeabilized for 10 min in 0.1% Triton X-100 in PBS. Cells were incubated for 60 min at room temperature with primary antibodies diluted in PBS-B (5% bovine serum albumin in PBS). The primary antibodies and dilutions used were as follows: for Rep, rabbit polyclonal (1:250; gift of J. Trempe) and mouse monoclonal (1:200; gift of J. Samulski) antibodies; for RPA32, mouse monoclonal (1:300) and rabbit polyclonal (1:500 to 1:1,000) antibodies; for ICP8, mouse monoclonal 39S (1:300; ATCC) antibody; for Ad-DBP, mouse monoclonal (1:5,000; gift from A. Levine) and rabbit polyclonal (1:10,000) antibodies; and for BrdU, mouse monoclonal (1:32; Roche) antibody. Secondary antibodies were conjugated to fluorescein isothiocyanate and Texas red (both 1:200; Jackson). Nuclear DNA was stained with 4′,6′-diamidino-2-phenylindole (DAPI). Coverslips were mounted with Fluoromount-G (Southern Biotechnology Associates) and immunoreactivity was visualized by epifluorescence by use of a Nikon microscope in conjunction with a charge-coupled device camera (Cooke Sensicam). Deconvolution images were collected in series (12 to 15 images; 0.5-μm increments along the z axis) through the focal plane of the cell and deconvolved by use of the constrained iterative function of Slidebook software, with a maximum of 10 iterations. A single image slice is presented in the figures, representing a 0.5-μm cross section of the cell. Images were obtained in double or triple excitation mode and processed by use of SlideBook and Adobe Photoshop.
Protein purification.
The Rep68 and Rep78 proteins were expressed in Escherichia coli and purified as recombinant proteins with either a polyhistidine tag (Rep68H) or a maltose-binding protein (MBP) tag (MBPRep78), essentially as previously described (18, 70). The plasmid containing Rep68 protein fused to a C-terminal polyhistidine tag (Rep68H) was a gift from R. J. Samulski (70). The double mutation (Y121H/K340H) was introduced into the Rep68 gene in the His-tagged pQE70 vector (Qiagen) by use of the QuickChange site-directed mutagenesis kit (Stratagene).
The HSV-1 proteins were expressed by recombinant baculoviruses in Sf9 insect cells and purified as previously described (28). The Ad-DBP protein was purified from baculovirus-infected cells as previously described (53). The RPA heterotrimeric complex was overproduced in E. coli with the T7 expression system and purified to near homogeneity as described previously (29). The Oct-1 protein was purified as previously described (21a). The E. coli single-stranded DNA-binding protein (SSB) was purchased from Amersham Pharmacia. Purified proteins were visualized by denaturing SDS-polyacrylamide gel electrophoresis and staining with Coomassie brilliant blue, using standard protocols.
In vitro replication assay.
In vitro replication assays for AAV were performed as previously described (57), with HeLa cell extracts, recombinant Rep protein, and full-length open-ended AAV duplex DNA substrate in the presence of 32P-labeled dCTP. Aliquots of the radiolabeled products were separated by electrophoresis in a 0.8% agarose gel in Tris-borate-EDTA buffer and exposed to film for autoradiography.
ELISAs.
Enzyme-linked immunosorbent assays (ELISAs) were carried out in 96-well plates with purified proteins, as previously described (29). Micrococcal nuclease was included in the ELISAs to prevent DNA bridging. For each interaction, the assay was performed in duplicate and repeated at least twice and a representative experiment is presented in each figure.
Assays for Rep activities.
Electrophoretic mobility shift assays (EMSAs) were performed essentially as previously described (35, 47). The amount of purified Rep fusion protein (either MBPRep78 or Rep68H) is indicated for each experiment. In brief, reactions contained purified Rep proteins, the indicated amount of ssDBP, and approximately 1 to 2 ng of radiolabeled DNA substrate (1,000 cpm), which was either the AAV ITR in the hairpin configuration or a linear fragment containing the Rep recognition sequence (RRS). Reactions were performed in the standard binding buffer (47) at room temperature for 20 to 30 min and resolved by 5% polyacrylamide gel electrophoresis in 0.5× Tris-borate-EDTA buffer at 100 V for 3.5 h. The gels were dried and data were analyzed by PhosphorImager and ImageQuant software (Molecular Dynamics).
Endonuclease assays were performed essentially as previously described (18, 35, 47). The reaction mixtures contained 1,000 cpm of 32P-labeled AAV hairpin DNA in terminal resolution buffer (47). Wild-type and mutant Rep68H proteins were purified by virtue of the His tag and added into the reaction in the presence or absence of the indicated ssDBPs. Reactions were carried out for 1 h at room temperature and samples were boiled in SDS buffer prior to electrophoresis through a 5% polyacrylamide gel. The gels were dried and data were analyzed by PhosphorImager and ImageQuant software (Molecular Dynamics).
RESULTS
AAV colocalizes with HSV-1 sites in coinfected cells.
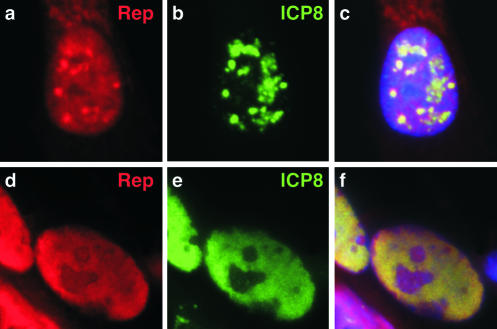
Members of our laboratory previously showed by immunofluorescence and in situ hybridization that AAV colocalizes with Ad replication centers (62). We have now used immunofluorescence to visualize viral proteins during coinfection of Vero cells with AAV-2 and its helper virus, HSV-1. A number of different patterns were observed for the spatial intracellular distribution of Rep and Cap proteins and these were reminiscent of those previously described by our laboratory (62) and others (33, 65) for infections with Ad as a helper. These patterns have been suggested to reflect stages during the progression of AAV infection (62, 65). We observed colocalization of signals for the AAV-2 Rep proteins and the HSV-1 ICP8 protein (Fig. 1). In some cells, sites of Rep accumulation in the nucleus partially overlapped with discrete sites of ICP8 (Fig. 1a to c). In cells infected with HSV-1 alone, only large globular replication centers were detected at this time point. The presence of small discrete replication centers during coinfection with AAV may reflect inhibition of HSV-1 replication by the AAV Rep protein, as previously reported (31). In other cells, the Rep and ICP8 proteins were both found diffusely spread throughout the nucleoplasm (Fig. 1d to f). Based on previous studies with Ad as a helper and analysis of viral replication by DNA hybridization (data not shown), we believe that the discrete centers represent early sites of AAV replication that colocalize with HSV-1 replication proteins and that the diffuse pattern represents late stages of AAV replication.
FIG. 1.
AAV colocalizes with HSV-1 sites in coinfected cells. Vero cells were coinfected with AAV-2 (1,000 genomes/cell) and HSV-1 (multiplicity of infection of 0.1 PFU/cell). Subcellular localization of viral proteins was visualized by indirect immunofluorescence at 12 h postinfection (a to c) and 16 h postinfection (d to f). The AAV-2 Rep proteins were detected with a rabbit polyclonal antibody and ICP8 of HSV-1 was visualized with a mouse monoclonal antibody. Nuclei were located by costaining DNA with DAPI, as shown in the merged images in the right columns.
AAV colocalizes with HSV-1 helper proteins at foci formed in transfected cells.
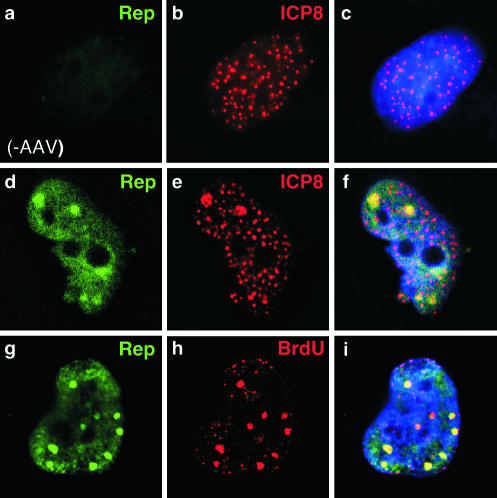
We next examined whether Rep and ICP8 proteins colocalize at sites formed upon transfection of viral genes. Infectious AAV can be generated by transfection of the AAV genome together with genes for the UL5, UL8, UL52, and ICP8 helper proteins (60). We transfected HeLa cells with this minimal set of HSV-1 helper proteins in the presence and absence of a plasmid containing the AAV-2 genome (16). We analyzed the localization of the AAV Rep and HSV-1 ICP8 proteins by immunofluorescence (Fig. 2). Transfection of expression plasmids for UL5, UL8, UL52, and ICP8 leads to formation of discrete ICP8 foci (Fig. 2b) which require the presence of all four HSV-1 proteins (14, 40, 41, 52). When the AAV plasmid was included in the transfection, we observed diffuse Rep localization in the nucleus, with some areas of concentration resembling the replication foci observed with helper virus coinfection (Fig. 2d). These sites of concentration overlapped with ICP8 staining (Fig. 2f), and we propose that they represent sites of AAV replication within the transfected cells. To determine if these sites of Rep concentration corresponded to sites of DNA synthesis, we pulse-labeled the cells with BrdU. In cells transfected with UL5, UL8, UL52, and ICP8, we observed BrdU accumulation in small discrete foci (data not shown). With the addition of the AAV plasmid, larger sites of BrdU localization appeared, which colocalized with accumulation of Rep protein (Fig. 2h). Therefore, Rep colocalized with ICP8 and BrdU at discrete sites that are presumed to be active centers for viral DNA synthesis.
FIG. 2.
AAV colocalizes with ICP8 at discrete sites formed by transfection of HSV-1 helper genes. Transfection of HeLa cells with plasmids expressing UL5, UL8, UL52, and ICP8 (a to c) resulted in formation of discrete sites of ICP8 accumulation, as detected with a mouse monoclonal antibody. Cotransfection of the AAV genome with ICP8 and the helicase-primase (d to i) results in the formation of larger foci, to which Rep (d) and ICP8 (e) localize in the nucleus (f). Pulse-labeling with BrdU followed by immunofluorescence using a mouse monoclonal antibody demonstrated that sites of Rep accumulation (g) are active sites of DNA synthesis (h). Nuclei were located by costaining DNA with DAPI, as shown in the merged images in the right columns.
The helicase-primase activity of the UL5-UL8-UL52 complex is not required for AAV replication.
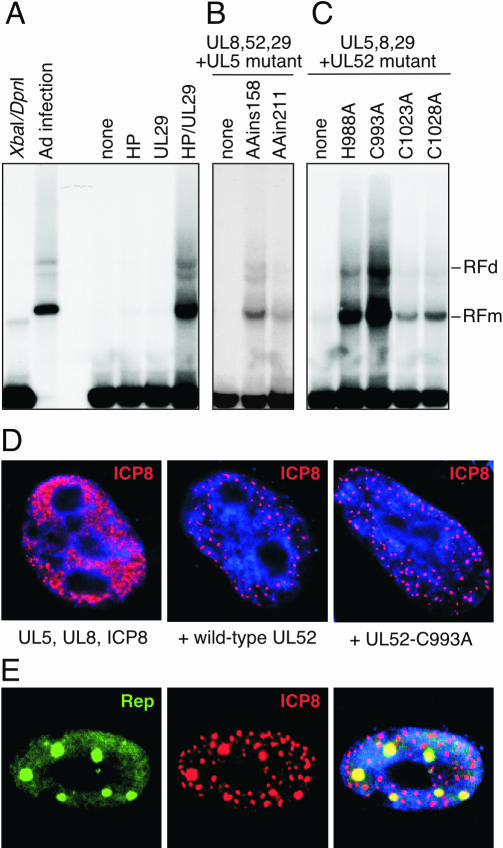
We asked whether the enzymatic functions of the helicase-primase are important for their helper activity by using mutants of UL5 and UL52. Two UL5 mutants were used which contained insertions of two alanine residues at positions 158 and 211 (AAins158 and AAins211, respectively). These mutants retain interactions with the other members of the primase-helicase complex, as determined by their ability to form foci upon transfection with UL52, UL8, and ICP8, but they fail to complement an HSV-1 UL5 mutant virus for replication (S. K. Weller, unpublished data). The UL52 protein contains a zinc finger motif that is highly conserved among primases (3). A double mutation in the conserved cysteine residues (C1023 and C1028) of the zinc finger results in proteins that can still form complexes with UL5 and UL8 but are severely defective in biochemical activities (3). A series of alanine substitution mutants were generated in UL52 (H988A, C993A, C1023A, and C1028A), and all of these produced proteins that were unable to complement replication of a UL52 mutant virus (14). The four HSV-1 genes, either wild-type or mutant, were transfected into cells with the cloned AAV genome and the degree of AAV replication was determined by Southern blot analysis of low-molecular-weight DNA extracted after 40 h (Fig. 3). No AAV replication was observed with just the UL5, UL8, and UL52 genes of the helicase-primase complex or with the UL29 gene alone (Fig. 3A), but in combination these four HSV-1 helper genes facilitated AAV replication and two characteristic bands that represent replicative intermediates of AAV in the monomer and dimer form (RFm and RFd) were observed with the AAV probe. Omission of UL5 or UL52 abolished AAV replication entirely (Fig. 3B and C). However, mutants of UL5 (Fig. 3B) and UL52 (Fig. 3C) that were unable to complement replication of mutant HSV-1 viruses were still able to provide some helper activity for AAV replication. The efficiency of replication varied between the different mutants but in every case the degree of replication was higher than that observed in the absence of either UL5 or UL52.
FIG. 3.
The helicase-primase activities of the UL5-UL8-UL52 complex are not required for AAV helper activity. Human HeLa cells were transfected with a clone of AAV-2 (pNTC244) alone or together with various combinations of vectors expressing wild-type or mutant versions of the four HSV-1 helper genes for UL5, UL8, UL52, and ICP8. Cells were harvested at 40 h posttransfection, and low-molecular-weight DNA was extracted and digested with XbaI and DpnI to remove the input plasmid. A Southern blot of the samples run on an agarose gel was hybridized with an AAV probe. The replicative intermediates in monomer or dimer form (RFm and RFd) are indicated on the right. (A) AAV replication requires both the UL5, UL8, and UL52 genes that encode the helicase-primase complex (HP) and the UL29 gene that encodes the ICP8 protein. Controls included the input AAV plasmid digested with XbaI and DpnI, the AAV plasmid transfected into Ad-infected cells as a positive control for AAV replication, and a transfection of the AAV plasmid without any HSV-1 helper genes (none). (B) AAV replication in transfections of HSV-1 helper genes without UL5 (none) or with mutants of UL5 (AAins158 and AAins211). (C) AAV replication in transfections of HSV-1 helper genes without UL52 (none) or with mutants of UL52 (H988A, C993A, C1023A, and C1028A). (D) The mutants of UL5 and UL52 are still able to assemble complexes at HSV prereplicative sites. Cells were transfected with the indicated viral genes and processed for immunofluorescence after 40 h. The UL29 gene product (ICP8) was detected by indirect immunofluorescence. In the absence of UL52, the ICP8 protein is detected in a diffuse pattern, but with either wild-type or mutant UL52, it is localized to discrete foci. All mutants tested showedformation of these discrete ICP8 centers. Nuclei were stained for DNA with DAPI, as shown in blue. (E) AAV Rep proteins colocalize with ICP8 at foci formed with mutant primase-helicase complexes. Cells were transfected with the AAV genome together with constructs expressing wild-type or mutant versions of the four HSV-1 genes. At 40 h, cells were processed for immunofluorescence with a rabbit polyclonal antibody against Rep and a monoclonal antibody against ICP8. The merged image is shown to the right, with costaining of nuclear DNA using DAPI. The example shown is for the UL52-C993A mutant, but all mutants tested showed similar discrete sites where Rep and ICP8 proteins colocalized.
Transfection and immunofluorescence were also used to analyze the mutant helicase-primase complex (Fig. 3D). In the absence of any member of the helicase-primase complex, the ICP8 protein was detected either in a diffuse nuclear pattern or in very few ICP8 foci, as previously described (41, 52). In the presence of the helicase-primase complex, the ICP8 protein was detected at numerous discrete foci. These foci were formed with all the mutants tested in the replication assay: one example is shown in Fig. 3D and the remainder were similar (14; data not shown). The localization of AAV replication, as determined by immunofluorescence of the Rep protein, was examined with the different HSV-1 mutants. In all cases, Rep colocalized with ICP8 in a similar pattern to that observed with the wild-type proteins (Fig. 3E and data not shown). Taken together, these results demonstrate that the enzymatic activities of the helicase-primase complex are not essential for AAV helper activity but suggest that the correct subnuclear localization of the UL5-UL8-UL52-ICP8 complex or recruitment of host factors may be important for AAV replication in a cellular context.
ICP8 protein enhances in vitro replication of AAV.
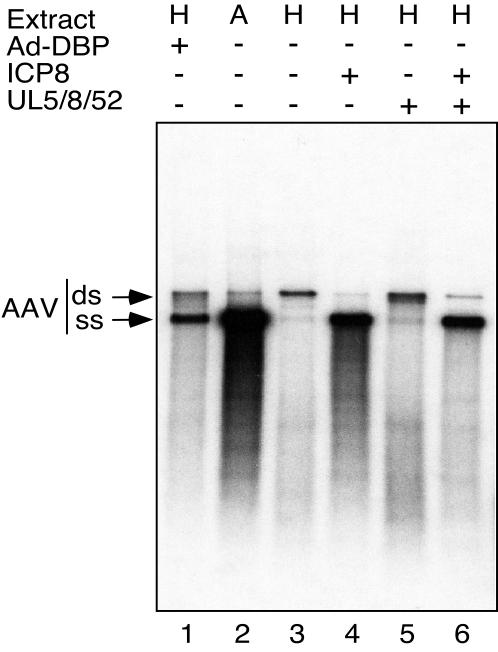
The study of AAV replication has been facilitated by the use of in vitro assays (46, 59). AAV DNA synthesis can be observed in an ori-dependent fashion in cell extracts supplemented with purified recombinant Rep protein (59). Extracts from Ad-infected cells are able to replicate AAV DNA with much greater processivity than uninfected cell extracts, resulting in a substantial enhancement in the production of full-length AAV replication products (55). This has been shown to be due to Ad-DBP and can be achieved by addition of the protein to an extract from uninfected cells (56). We employed an in vitro assay to assess a direct role for HSV-1 replication proteins in AAV replication (Fig. 4). For this assay, an extract from uninfected cells is supplemented with purified recombinant Rep78 protein, and replication of a linear duplex form of AAV DNA is assessed by incorporation of radiolabeled nucleotides (59). It has previously been reported that two full-length products are observed upon gel electrophoresis of the products of the in vitro AAV reaction (58). The upper band represents the full-length duplex DNA genome and a faster migrating band is thought to be made up of single-stranded genome molecules (58).
FIG. 4.
The product of the HSV-1 UL29 gene (ICP8) and the Ad-DBP protein fulfill analogous functions and provide a direct helper effect to enhance AAV DNA replication in an in vitro assay. In vitro replication assays were performed as described in Materials and Methods. Extracts used were made from Ad-infected HeLa cells (A) or uninfected HeLa cells (H). Extracts were supplemented with recombinant Rep78 protein, and replication of a linear AAV DNA molecule was monitored by the incorporation of radiolabeled nucleotides. Replication products were separated by gel electrophoresis and two full-length products were visible: the upper band represents the full-length duplex genome (ds) and the lower band represents single-stranded DNA progeny genomes (ss), as previously described (55). The effect of proteins from the helper viruses was assessed by the addition of purified Ad-DBP or the HSV-1 proteins UL5, UL8, UL52, and ICP8, as indicated.
Using an extract from uninfected HeLa cells for this assay resulted in minimal incorporation of radiolabeled nucleotides into full-length replication products and this was accompanied by a faint smear of shorter products (lane 3) which were shown to be newly synthesized DNA that is less than the full length (55). There was very little production of the full-length single-stranded progeny DNA in the uninfected HeLa cell extract. The presence of Ad-DBP, provided either from an Ad-infected extract (lane 2) or as a purified protein added to an uninfected extract (lane 1), increased the processivity of AAV replication. In the presence of Ad-DBP, the labeled DNA appeared predominantly in the faster migrating band, which agrees with previous studies and is consistent with the strand displacement model for AAV replication (56, 58). We assessed the effect of HSV-1 proteins by using the UL5, UL8, UL52, and ICP8 proteins purified from baculovirus extracts (28). The addition of ICP8 alone was sufficient to increase AAV replication in a similar fashion to Ad-DBP and generated the full-length single-stranded DNA progeny genomes (lane 4). In contrast, the addition of a mixture of UL5, UL8, and UL52 proteins to uninfected HeLa cell extracts had a minimal effect on AAV replication (lane 5). Combining ICP8 with the other three HSV proteins gave no additional increase in AAV replication (lane 6). Therefore, accumulation of single-stranded DNA is observed in the in vitro assay with the addition of single-stranded DNA-binding proteins ICP8 and Ad-DBP from helper viruses (Fig. 4) and also with the cellular ssDBP complex RPA (58), but not with a nonspecific E. coli SSB (58). These results suggest that the crucial HSV-1 helper protein is ICP8 and that it can play an analogous role to the Ad-DBP during AAV replication in vitro. The in vitro assay bypasses the need for subnuclear localization and hence the UL5, UL8, and UL52 proteins are not required for ICP8 to enhance replication. These results are consistent with the conclusion from the transfection experiments above, which show that the enzymatic functions of the complex are not required for AAV replication.
Cellular RPA colocalizes with Rep at AAV replication centers.
Our experiments in cell culture and in vitro demonstrated a key role for ssDBPs from Ad and HSV-1 helper viruses in AAV replication. The cellular RPA complex has been reported to be recruited to replication centers of HSV-1 and HCMV (26, 64). We therefore examined the localization of RPA in cells during AAV replication (Fig. 5).
FIG. 5.
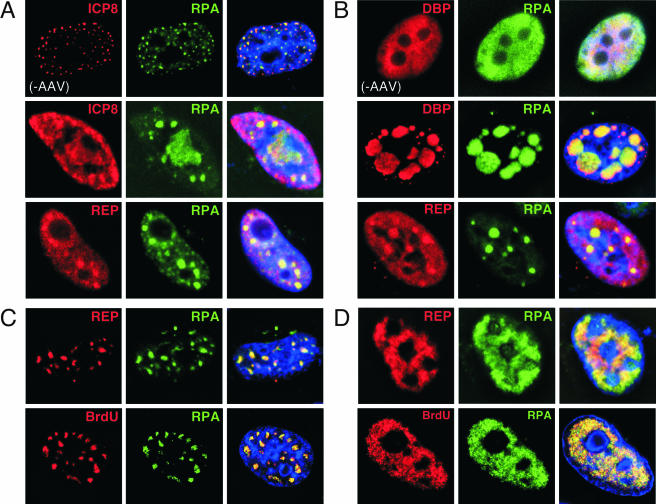
Accumulation of RPA at sites of AAV replication. Various combinations of Ad and HSV-1 proteins that allow replication of AAV were analyzed by indirect immunofluorescence after transfection of HeLa cells. (A) RPA colocalizes with sites formed by transfection of the helicase-primase and ICP8 of HSV-1. The ICP8 protein colocalized with RPA in cells transfected with the HSV-1 genes for UL5, UL8, UL52, and ICP8 (top row). The addition of an AAV plasmid resulted in some enlarged foci in which Rep and RPA colocalize. (B) RPA colocalizes at replication centers formed by Ad-DBP and AAV. Transfection of Ad-DBP alone results in diffuse nuclear expression and RPA is unaltered (top row). Cotransfection with an AAV genome results in the formation of discrete centers of replication in which RPA colocalizes with both DBP and Rep. (C and D) Transfection of expression vectors for the Ad helper proteins E1b55K and E4orf6 enables AAV replication in the absence of a viral ssDBP. Two major patterns of replication were observed in an equal number of cells, as shown. In both cases, Rep colocalized with RPA (top rows) and the replication centers were stained by Rep and BrdU (bottom rows). These images represent a single focal plane from a 0.5-μm cross section of the cell after deconvolution.
In actively dividing HeLa cells, the RPA complex can be visualized by immunofluorescence with an antibody against the RPA32 subunit and is detected diffusely throughout the nucleus, with accumulation at replication sites in S-phase cells (9). When cells were transfected with UL5, UL8, UL52, and ICP8, the RPA subunit was localized to the discrete sites that stain for ICP8 (Fig. 5A, top row). We then investigated whether this was also the case when AAV replication was supported by the minimal HSV-1 helper proteins. Addition of the AAV plasmid led to the formation of enlarged foci (Fig. 5A, middle row) in which Rep and RPA colocalized (Fig. 5A, bottom row).
We also examined RPA localization under conditions in which Ad genes supplied helper activity for AAV replication. We observed colocalization of RPA with the Rep protein during coinfection of AAV with the Ad helper virus (T. H. Stracker and M. D. Weitzman, unpublished data). We also used two different transfection approaches to analyze AAV replication in the presence of Ad helper proteins. For the first experiment, the gene for Ad-DBP was transfected, together with an AAV plasmid, and the localization of Rep, Ad-DBP, and RPA was assessed by immunofluorescence (Fig. 5B). Transfection of Ad-DBP alone in the absence of other adenoviral proteins can support replication of AAV to a similar level as that with the minimal HSV-1 helper proteins (Stracker and Weitzman, unpublished data). In the absence of AAV, Ad-DBP localization was diffusely nuclear and the distribution of RPA was unaltered (Fig. 5B, top row). The addition of the AAV plasmid led to the formation of discrete foci of Ad-DBP that also contained both Rep and RPA. In a second set of experiments, cells were transfected with expression vectors for the adenoviral helper proteins E4orf6 and E1b55K, which can also support AAV replication. For these experiments, no viral ssDBP was included. Two different patterns were observed in approximately equal numbers, and representative images are shown in Fig. 5C and D. Either the proteins appeared at discrete intranuclear sites (Fig. 5C) or the proteins were observed in a pattern that covered the nucleoplasm (Fig. 5D). We believe that these probably represent early and late stages of AAV replication, respectively, based on patterns seen during Ad coinfection. Rep and RPA again colocalized in transfected cells (Fig. 5C, top row) and sites of RPA concentration colocalized with BrdU upon pulse-labeling (Fig. 5C, bottom row).
Together, these data demonstrate that RPA is a component of AAV replication centers formed by transfection of minimal helper proteins from either Ad or HSV-1, regardless of whether a viral ssDBP is present. Other cellular proteins involved in the AAV life cycle may be similarly located at these sites.
Interaction of Rep with ssDBPs.
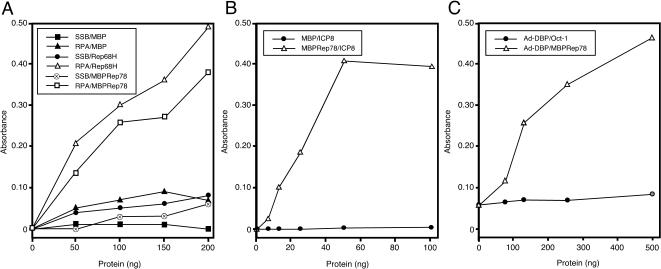
The data we have presented demonstrate colocalization of AAV Rep protein with three ssDBPs (RPA, ICP8, and Ad-DBP) at sites of AAV replication. We therefore investigated whether there was a physical interaction between Rep and these proteins. We used an ELISA to detect protein-protein interactions in vitro (Fig. 6). This type of assay has previously been employed to demonstrate interactions of RPA with the SV40 T antigen (22) and the papillomavirus E1 proteins (29). In the first set of experiments (Fig. 6A), ssDBPs were immobilized in a 96-well ELISA plate and challenged with increasing amounts of purified recombinant proteins. The immobilized proteins used were E. coli SSB and recombinant human RPA. These were challenged with MBP, His-tagged Rep68 (Rep68H) (70), or a fusion protein of Rep78 and MBP (MBPRep78) (18). There was no significant interaction observed for the control pairings of RPA and SSB with MBP. In contrast, both Rep68 and Rep78 interacted with human RPA but not with E. coli SSB. The reciprocal assay, adding increasing amounts of RPA to immobilized MBP or MBPRep78, also demonstrated an interaction of RPA with Rep (data not shown). Similar assays were performed with the HSV-1 ICP8 and Ad-DBP proteins (Fig. 6B and C). In all cases, specific interactions were detected with the AAV Rep protein in a dose-dependent manner. We were also able to demonstrate an interaction between Ad-DBP and Rep78 by immunoprecipitation assays (data not shown). Together, these data demonstrate that the three ssDBPs that enhance AAV replication (Ad-DBP, ICP8, and RPA) can each interact with the AAV Rep protein, whereas E. coli SSB, which does not facilitate AAV replication in the in vitro assay (56), does not interact with Rep.
FIG. 6.
The AAV Rep protein interacts with RPA, ICP8, and Ad-DBP. ELISAs were performed in which purified protein was immobilized in 96-well plates and then challenged with increasing amounts of a second purified protein. In each case, the first protein listed is the immobilized one and the second is the challenging protein. (A) Rep proteins specifically bind to immobilized RPA. Recombinant human RPA (250 ng) or E. coli SSB (250 ng) was immobilized and challenged with increasing amounts of bacterially purified MBP, MBPRep78, or Rep68H. Interactions were detected with rabbit polyclonal antibodies against MBP or Rep and horseradish peroxidase anti-rabbit antibody. (B) Recombinant ICP8 binding to immobilized MBPRep78. Purified MBPRep78 (250 ng) or MBP (250 ng) was bound to ELISA wells and challenged with ICP8. Binding was detected with an anti-ICP8 monoclonal antibody. (C) Rep binding to immobilized Ad-DBP. Purified Ad-DBP (500 ng) was bound to ELISA wells and challenged with increasing amounts of MBPRep78 or the POU homeodomain of Oct-1 as a control protein. Binding was detected with an anti-MBP monoclonal antibody or an Oct-1 antibody (21a).
ssDBPs enhance Rep functions in vitro.
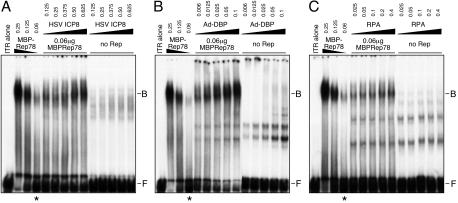
The Rep proteins perform a number of essential functions during AAV DNA replication (45). We assessed whether these were affected by the addition of interacting ssDBPs. The effect on DNA binding was assessed by including the proteins in an EMSA. In the first set of experiments, we examined MBPRep78 binding to the AAV ITR in its hairpin configuration (Fig. 7). We observed an overall enhancement of binding of the Rep fusion protein in the presence of HSV ICP8 (Fig. 7A), Ad-DBP (Fig. 7B), and RPA (Fig. 7C). We also examined the effect of ssDBPs on binding of Rep proteins to a linear double-stranded DNA substrate that contained the consensus sequence for Rep binding known as the RRS. Binding of MBPRep78 to the linear fragment was also enhanced by ICP8 and RPA, but not by E. coli SSB, suggesting that there is some specificity to the effect (Fig. 8A). Similar observations were made when His-tagged Rep68 (Rep68H) was examined for binding to the linear DNA fragment in the presence of ssDBPs (Fig. 8B and data not shown).
FIG. 7.
ssDBPs enhance Rep binding to the AAV hairpin ITR. EMSAs were performed with the 32P-labeled AAV terminal repeat hairpin DNA (1,000 cpm) incubated with the indicated amounts of MBPRep78 protein in the presence or absence of the ssDBPs. The numbers at the top represent the amounts of added protein, in micrograms. A constant amount of Rep (indicated by asterisks) was assessed for the effect of the ssDBPs. Rep binding was enhanced with HSV-1 ICP8 (A), Ad-DBP (B), and RPA (C). The lane marked “ITR alone” contains no added proteins. The positions of free DNA probe (F) and DNA probe bound in a Rep complex (B) are shown to the right.
FIG. 8.
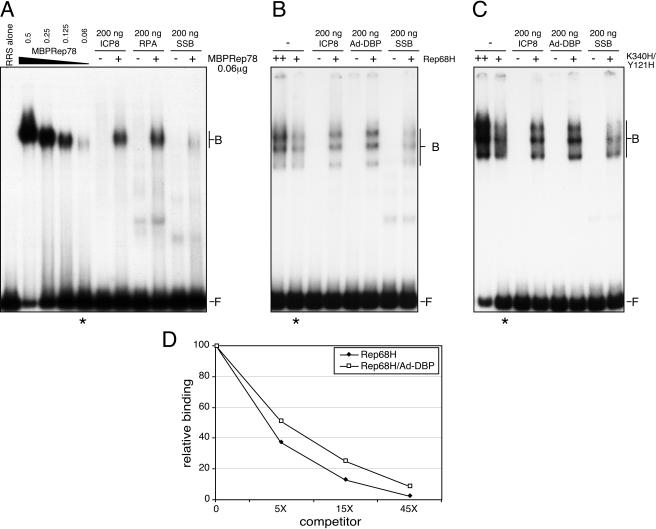
Enhanced binding by Rep proteins is specific and does not require helicase activity. (A) Effect of ssDBPs on binding of MBPRep78 to a linear DNA substrate containing the RRS. The radiolabeled RRS fragment (1,000 cpm) was incubated with MBPRep78 (the numbers at the top indicate the amounts of protein, in micrograms). The ssDBPs were incubated with the probe in the absence (−) or presence (+) of MBPRep78. Binding was enhanced by the HSV-1 ICP8 and RPA proteins but not by the E. coli SSB protein. A constant amount of Rep (indicated by the asterisk below) was assessed for the effect of the ssDBPs. The lane marked “RRS alone” does not contain any added protein. The positions of free DNA probe (F) and DNA probe bound in a Rep complex (B) are shown to the right. (B) Enhanced binding of Rep68 to the RRS fragment. Either 1 ng (+) or 2 ng (++) of His-tagged recombinant Rep68 protein (Rep68H) was incubated with a radiolabeled RRS fragment together with the indicated ssDBP. (C) ssDBPs increase binding of a mutant Rep protein that has lost helicase activity. A mutant Rep protein (Y121H/K340H) was incubated in an EMSA with a 32P-labeled linear DNA fragment containing the RRS, in the presence or absence of the indicated ssDBPs. (D) Complex dissociation was assessed with Rep68H alone or in the presence of Ad-DBP. The Rep68H protein was bound to the RRS fragment and then dissociation was assessed by the addition of increasing amounts of unlabeled competitor DNA to assembled complexes. Binding assays were analyzed by gel electrophoresis and relative binding was quantitated by PhosphorImager analysis of gels.
One possibility was that the helicase activity of the Rep protein unwound the double-stranded DNA substrate in the region of the RRS, allowing ssDBPs to bind to the exposed single-stranded DNA. To address this issue, we used purified Rep proteins that possess a mutation in the nucleotide triphosphate binding domain (K340H) or a double mutant that combines this with a mutation in the helicase active site (Y121H/K340H). These proteins have lost their helicase function (54). The ssDBPs (Ad-DBP, RPA, and ICP8) all enhanced binding to the RRS by both of these mutants, whereas E. coli SSB still had no effect (Fig. 8C and data not shown). This suggests that the increased shifts observed with the ssDBPs reflect an increase in Rep protein complex formation.
We used competition EMSAs to address the question of whether the Rep-RRS complexes were more stable in the presence of ssDBPs or whether there were simply more complexes forming. The specificity of the enhancement of binding by Rep was confirmed by competition experiments using unlabeled RRS or a mutant version of the RRS (mRRS) that is not bound by Rep (63). Even in the presence of the ssDBPs, no binding was observed for the mRRS and competition was only observed for the wild-type RRS sequence (data not shown). We then allowed Rep-RRS complexes to form, and these were chased with increasing amounts of RRS competitor DNA (5×, 15×, and 45×). We found that in both the presence and absence of Ad-DBP, the Rep-RRS complexes had similar dissociation rates (Fig. 8D). This indicated that the stabilities were similar, regardless of whether the ssDBP was present. We found that the addition of Ad-DBP at any time point in the reaction could enhance Rep binding (data not shown). This suggests that the specific enhancement by ssDBPs is due to an increase in formation of the Rep-DNA complex.
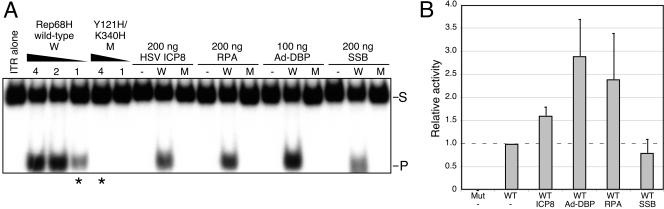
We also examined the effect of the ssDBPs on the endonuclease activity of Rep, using the ITR as a substrate for site-specific cleavage at the trs site (35). Rep nicking of the 5′-labeled ITR releases a short labeled product in this assay (Fig. 9A). The Ad-DBP, RPA, and ICP8 proteins all enhanced the Rep68 endonuclease activity, whereas E. coli SSB had no effect. The Y121H/K340H mutant did not nick this double-stranded DNA substrate, as it lacks the helicase activity required to unwind the DNA to facilitate transesterification (11). Addition of the ssDBPs did not enable this mutant to nick, suggesting that their effect is not through destabilization of the DNA double helix of the ITR. Quantitation of these data demonstrated a two- to threefold enhancement of Rep activity at the concentrations shown (Fig. 9B), which correlates to the observed enhancement of binding. Similar results were obtained with RPA and in vitro-translated Rep78 protein in a rabbit reticulocyte lysate (data not shown). These results suggest that enhanced binding of Rep in the presence of the ssDBPs results in an increase in nicking activity.
FIG. 9.
Enhancement of Rep endonuclease activity by ssDBPs. The trs nicking assay included 32P-labeled AAV terminal repeat hairpin DNA (1,000 cpm) and either wild-type or mutant purified Rep68H in the presence or absence of the ssDBPs. (A) A representative experiment demonstrating increased endonuclease activity of Rep68H in the presence of ICP8, RPA, and Ad-DBP, but not E. coli SSB. Purified recombinant proteins of wild-type Rep68H (W) or the Y121H/K340H mutant (M) were incubated with the ITR in the hairpin configuration. A titration of wild-type Rep68H showed nicking of the hairpin substrate (S) and release of the cleavage product (P). The mutant failed to nick the double-stranded hairpin ITR. The amount of Rep protein (in nanograms) is indicated above the lanes, and the asterisks below indicate the amounts of wild-type and mutant Rep proteins incubated with the ssDBPs. The amount of each ssDBP included is indicated at the top, in nanograms. Samples of the hairpin (ITR alone) and the ssDBPs in the absence of Rep protein were included as negative controls. (B) Quantitation of endonuclease activities. The amount of nicked product in each reaction was quantitated by PhosphorImager analysis and plotted relative to a constant amount of Rep protein (1 ng). At least three independent reactions were quantitated for each experimental condition.
DISCUSSION
The results of this study, together with previous analysis of AAV replication in Ad-infected cells (33, 62, 65), implicate discrete intranuclear sites as important for the helper functions for AAV replication. Although in this study we have not analyzed the location of AAV nucleic acid, based on our previous extensive characterization of the relative locations of AAV proteins and nucleic acid in Ad-infected cells, we believe that the Rep pattern will reflect that of AAV DNA. The helper viruses each initiate important structural rearrangements in the nucleus as they subvert host cellular mechanisms for viral production (reviewed in references 10 and 37). Cellular proteins involved in DNA replication, such as DNA polymerase alpha, PCNA, topoisomerase II, and tumor suppressors p53 and Rb are often selectively found localized to these viral replication centers (23, 26, 64). It is possible that AAV benefits from sequestration of cellular proteins into the replication centers of helper viruses. The transfection experiments with the HSV-1 helper proteins suggest that the UL5, UL8, UL52, and ICP8 proteins are sufficient to form a scaffold for the accumulation of cellular replication proteins that enable AAV replication. For HSV, recruitment of the viral polymerase to the scaffold of UL5, UL8, UL52, UL9, and ICP8 requires an active primase unit (14). However, in the experiments that we have described for the present study, recruitment of the cellular polymerase to allow replication of the AAV genome did not appear to require functional primase. Results from infections (30), transfections (60), and the in vitro replication assay (57) suggest that the HSV-1 polymerase may also be used for AAV replication in some circumstances. AAV replication may therefore have adapted to use replication components from the cell and also from different helper viruses. It will be interesting to examine AAV infection in the presence of genotoxic agents to determine whether replication takes place at specific sites and what cellular factors are involved.
The addition of RPA stimulates AAV replication in cellular extracts by increasing polymerase processivity on the AAV templates (56). It remains puzzling that although abundant in infected cells, RPA is not sufficient to replicate AAV in the absence of a helper virus or genotoxic stress. RPA activity is regulated by both protein-protein interactions and modification by phosphorylation, both of which are likely altered by virus infection. We have observed hyperphosphorylation of RPA32 during AAV replication, and this may affect its interactions (Stracker and Weitzman, unpublished data). By analogy to other viral origin-binding proteins that interact with RPA, it is likely that Rep binds directly to the RPA70 subunit, similar to SV40 T antigen, EBNA1 of EBV, and E1 of human papillomaviruses (29, 71). This would support a recent structure analysis of the N terminus of Rep, which found an unexpected structural relationship between Rep and other viral origin-binding proteins such as SV40 T antigen and papillomavirus E1 (32).
It is interesting that an analogous situation seems to exist with other parvoviruses. A recent report used a similar approach to ours to show that the parvoviral NS1 protein of the minute virus of mouse also interacts with the cellular RPA complex in vitro (19). In this case, RPA also enhanced unwinding of the origin nicked by the NS1 protein, and it was suggested that RPA may be required to form a functional replication complex for parvovirus replication. E. coli SSB had no effect on unwinding and it did not interact with NS1 in the ELISA (19). Another study examined the spatial organization of minute virus of mouse replication and found NS1 at discrete nuclear structures, where RPA and other cellular replication factors also accumulated (2). These results are completely consistent with our observations for the Rep protein of AAV.
The enhancement of Rep's DNA-binding and endonuclease activities by the ssDBPs is similar to the effect noted for the high-mobility group chromosomal protein 1, which also interacts with Rep (20). It is possible that the ssDBPs enhance binding by bringing Rep into contact with its substrate or by promoting multimerization of the protein. The increase in DNA binding could result from either promotion of complex formation or enhanced complex stability. Our competition EMSA results suggest that the former scenario is most likely. The enhanced levels of nicking are probably due to an increase in binding induced by the ssDBPs, as similar fold increases were observed. Enhancing the ability of Rep protein to bind to the ITR and nick the trs could contribute to the increase in viral DNA replication observed when these ssDBPs are added to cellular extracts (56). The cellular RPA protein could also contribute to complex formation between the viral genome and the AAVS1 integration locus during Rep-mediated targeted integration in the absence of a helper virus (63).
Studies of helper functions from different viruses will provide us with a better understanding of the cellular environment that promotes productive AAV replication. One area that will benefit from studies of the requirements for AAV propagation is the production of recombinant AAV (rAAV) vectors for gene therapy. AAV is a promising candidate that is being developed into a gene transfer vector for a wide range of applications. A thorough understanding of the basic biology of the virus is crucial to its successful development as a vector. Conventional production of rAAV is achieved by cotransfection of the AAV vector construct and the packaging plasmid into cells infected with a helper virus. One approach to improving rAAV production is to bypass the requirement for extensive purification of helper virus by using plasmid transfections instead of helper virus infections (27, 42, 66). Understanding the role of HSV-1 helper proteins in AAV replication will enable HSV-based transfection protocols. It is also conceivable that by combining features from different viruses it will be possible to improve upon current systems which use either Ad or HSV-1 alone.
Acknowledgments
We are grateful to J. Trempe, R. J. Samulski, and D. Knipe for antibodies and reagents. We thank W. Cordier and P. Polderman for technical assistance. We thank Toni Cathomen, Christian Carson, Caroline Lilley, Sarah Malpel, and Dianna Wilkenson for critical reading of the manuscript.
This work was supported by NIH grants AI43341 and AI51686 (M.D.W.) and by gifts from the Mary H. Rumsey and Irving A. Hansen foundations (M.D.W.). We acknowledge the James B. Pendleton Charitable Trust for providing the Pendleton Microscopy Facility. Work by Y.-M.L. and T.M. was supported by NIH grants GM56406 and AI01686 (T.M.). Work by S.C.-L. and S.K.W. was supported by NIH grant AI21747. T.H.S. was supported by an NIH Graduate Training Grant to UCSD and by fellowships from the H. A. and Mary K. Chapman Charitable Trust, the Legler Benbough Foundation, and the Salk Institute Association. G.D.C. was supported by an NIH National Research Service Award Institutional Research Training Grant.
REFERENCES
- 1.Arana, M. E., B. Haq, N. Tanguy Le Gac, and P. E. Boehmer. 2001. Modulation of the herpes simplex virus type-1 UL9 DNA helicase by its cognate single-strand DNA-binding protein, ICP8. J. Biol. Chem. 276:6840-6845. [DOI] [PubMed] [Google Scholar]
- 2.Bashir, T., J. Rommelaere, and C. Cziepluch. 2001. In vivo accumulation of cyclin A and cellular replication factors in autonomous parvovirus minute virus of mice-associated replication bodies. J. Virol. 75:4394-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas, N., and S. K. Weller. 1999. A mutation in the C-terminal putative Zn2+ finger motif of UL52 severely affects the biochemical activities of the HSV-1 helicase-primase subcomplex. J. Biol. Chem. 274:8068-8076. [DOI] [PubMed] [Google Scholar]
- 4.Biswas, N., and S. K. Weller. 2001. The UL5 and UL52 subunits of the herpes simplex virus type 1 helicase-primase subcomplex exhibit a complex interdependence for DNA binding. J. Biol. Chem. 276:17610-17619. [DOI] [PubMed] [Google Scholar]
- 5.Boehmer, P. E. 1998. The herpes simplex virus type-1 single-strand DNA-binding protein, ICP8, increases the processivity of the UL9 protein DNA helicase. J. Biol. Chem. 273:2676-2683. [DOI] [PubMed] [Google Scholar]
- 6.Boehmer, P. E., M. C. Craigie, N. D. Stow, and I. R. Lehman. 1994. Association of origin binding protein and single strand DNA-binding protein, ICP8, during herpes simplex virus type 1 DNA replication in vivo. J. Biol. Chem. 269:29329-29334. [PubMed] [Google Scholar]
- 7.Boehmer, P. E., and I. R. Lehman. 1993. Herpes simplex virus type 1 ICP8: helix-destabilizing properties. J. Virol. 67:711-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosher, J., A. Dawson, and R. T. Hay. 1992. Nuclear factor I is specifically targeted to discrete subnuclear sites in adenovirus type 2-infected cells. J. Virol. 66:3140-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brenot-Bosc, F., S. Gupta, R. L. Margolis, and R. Fotedar. 1995. Changes in the subcellular localization of replication initiation proteins and cell cycle proteins during G1- to S-phase transition in mammalian cells. Chromosoma 103:517-527. [DOI] [PubMed] [Google Scholar]
- 10.Bridge, E., and U. Pettersson. 1995. Nuclear organization of replication and gene expression in adenovirus-infected cells. Curr. Top. Microbiol. Immunol. 199:99-117. [DOI] [PubMed] [Google Scholar]
- 11.Brister, J. R., and N. Muzyczka. 1999. Rep-mediated nicking of the adeno-associated virus origin requires two biochemical activities, DNA helicase activity and transesterification. J. Virol. 73:9325-9336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buller, R. M., J. E. Janik, E. D. Sebring, and J. A. Rose. 1981. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J. Virol. 40:241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burkham, J., D. M. Coen, and S. K. Weller. 1998. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J. Virol. 72:10100-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrington-Lawrence, S. D., and S. K. Weller. 2003. Recruitment of polymerase to herpes simplex virus type 1 replication foci in cells expressing mutant primase (UL52) proteins. J. Virol. 77:4237-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang, L. S., Y. Shi, and T. Shenk. 1989. Adeno-associated virus P5 promoter contains an adenovirus E1A-inducible element and a binding site for the major late transcription factor. J. Virol. 63:3479-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chejanovsky, N., and B. J. Carter. 1989. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology 173:120-128. [DOI] [PubMed] [Google Scholar]
- 17.Chejanovsky, N., and B. J. Carter. 1990. Mutation of a consensus purine nucleotide binding site in the adeno-associated virus rep gene generates a dominant negative phenotype for DNA replication. J. Virol. 64:1764-1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiorini, J. A., M. D. Weitzman, R. A. Owens, E. Urcelay, B. Safer, and R. M. Kotin. 1994. Biologically active Rep proteins of adeno-associated virus type 2 produced as fusion proteins in Escherichia coli. J. Virol. 68:797-804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Christensen, J., and P. Tattersall. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 76:6518-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costello, E., P. Saudan, E. Winocour, L. Pizer, and P. Beard. 1997. High mobility group chromosomal protein 1 binds to the adeno-associated virus replication protein (Rep) and promotes Rep-mediated site-specific cleavage of DNA, ATPase activity and transcriptional repression. EMBO J. 16:5943-5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Bruyn Kops, A., and D. M. Knipe. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55:857-868. [DOI] [PubMed] [Google Scholar]
- 21a.de Jong, R. N., M. E. Mysiak, L. A. T. Meijer, M. van der Linden, and P. C. van der Vliet. 2002. Recruitment of the priming protein pTP and DNA binding occur by overlapping Oct-1 POU homeodomain surfaces. EMBO J. 21:725-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dornreiter, I., L. F. Erdile, I. U. Gilbert, D. von Winkler, T. J. Kelly, and E. Fanning. 1992. Interaction of DNA polymerase alpha-primase with cellular replication protein A and SV40 T antigen. EMBO J. 11:769-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ebert, S. N., D. Subramanian, S. S. Shtrom, I. K. Chung, D. S. Parris, and M. T. Muller. 1994. Association between the p170 form of human topoisomerase II and progeny viral DNA in cells infected with herpes simplex virus type 1. J. Virol. 68:1010-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrari, F. K., T. Samulski, T. Shenk, and R. J. Samulski. 1996. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 70:3227-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fisher, K. J., G. P. Gao, M. D. Weitzman, R. DeMatteo, J. F. Burda, and J. M. Wilson. 1996. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J. Virol. 70:520-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fortunato, E. A., and D. H. Spector. 1998. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J. Virol. 72:2033-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grimm, D., A. Kern, K. Rittner, and J. A. Kleinschmidt. 1998. Novel tools for production and purification of recombinant adeno-associated virus vectors. Hum. Gene Ther. 9:2745-2760. [DOI] [PubMed] [Google Scholar]
- 28.Hamatake, R. K., M. Bifano, W. W. Hurlburt, and D. J. Tenney. 1997. A functional interaction of ICP8, the herpes simplex virus single-stranded DNA-binding protein, and the helicase-primase complex that is dependent on the presence of the UL8 subunit. J. Gen. Virol. 78:857-865. [DOI] [PubMed] [Google Scholar]
- 29.Han, Y., Y. M. Loo, K. T. Militello, and T. Melendy. 1999. Interactions of the papovavirus DNA replication initiator proteins, bovine papillomavirus type 1 E1 and simian virus 40 large T antigen, with human replication protein A. J. Virol. 73:4899-4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Handa, H., and B. J. Carter. 1979. Adeno-associated virus DNA replication complexes in herpes simplex virus or adenovirus-infected cells. J. Biol. Chem. 254:6603-6610. [PubMed] [Google Scholar]
- 31.Heilbronn, R., A. Burkle, S. Stephan, and H. zur Hausen. 1990. The adeno-associated virus rep gene suppresses herpes simplex virus-induced DNA amplification. J. Virol. 64:3012-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hickman, A. B., D. R. Ronning, R. M. Kotin, and F. Dyda. 2002. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol. Cell 10:327-337. [DOI] [PubMed] [Google Scholar]
- 33.Hunter, L. A., and R. J. Samulski. 1992. Colocalization of adeno-associated virus Rep and capsid proteins in the nuclei of infected cells. J. Virol. 66:317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iftode, C., Y. Daniely, and J. A. Borowiec. 1999. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34:141-180. [DOI] [PubMed] [Google Scholar]
- 35.Im, D. S., and N. Muzyczka. 1990. The AAV origin binding protein Rep68 is an ATP-dependent site-specific endonuclease with DNA helicase activity. Cell 61:447-457. [DOI] [PubMed] [Google Scholar]
- 36.Janik, J. E., M. M. Huston, and J. A. Rose. 1981. Locations of adenovirus genes required for the replication of adenovirus-associated virus. Proc. Natl. Acad. Sci. USA 78:1925-1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Knipe, D. M. 1989. The role of viral and cellular nuclear proteins in herpes simplex virus replication. Adv. Virus Res. 37:85-123. [DOI] [PubMed] [Google Scholar]
- 38.Lee, S. S., and I. R. Lehman. 1997. Unwinding of the box I element of a herpes simplex virus type 1 origin by a complex of the viral origin binding protein, single-strand DNA binding protein, and single-stranded DNA. Proc. Natl. Acad. Sci. USA 94:2838-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lehman, I. R., and P. E. Boehmer. 1999. Replication of herpes simplex virus DNA. J. Biol. Chem. 274:28059-28062. [DOI] [PubMed] [Google Scholar]
- 40.Liptak, L. M., S. L. Uprichard, and D. M. Knipe. 1996. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol. 70:1759-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lukonis, C. J., and S. K. Weller. 1997. Formation of herpes simplex virus type 1 replication compartments by transfection: requirements and localization to nuclear domain 10. J. Virol. 71:2390-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsushita, T., S. Elliger, C. Elliger, G. Podsakoff, L. Villarreal, G. J. Kurtzman, Y. Iwaki, and P. Colosi. 1998. Adeno-associated virus vectors can be efficiently produced without helper virus. Gene Ther. 5:938-945. [DOI] [PubMed] [Google Scholar]
- 43.McPherson, R. A., L. J. Rosenthal, and J. A. Rose. 1985. Human cytomegalovirus completely helps adeno-associated virus replication. Virology 147:217-222. [DOI] [PubMed] [Google Scholar]
- 44.Meyers, C., M. Mane, N. Kokorina, S. Alam, and P. L. Hermonat. 2000. Ubiquitous human adeno-associated virus type 2 autonomously replicates in differentiating keratinocytes of a normal skin model. Virology 272:338-346. [DOI] [PubMed] [Google Scholar]
- 45.Muzyczka, N., and K. I. Berns. 2001. Parvoviridae: the viruses and their replication, p. 2327-2359. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 46.Ni, T. H., W. F. McDonald, I. Zolotukhin, T. Melendy, S. Waga, B. Stillman, and N. Muzyczka. 1998. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J. Virol. 72:2777-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Owens, R. A., J. P. Trempe, N. Chejanovsky, and B. J. Carter. 1991. Adeno-associated virus rep proteins produced in insect and mammalian expression systems: wild-type and dominant-negative mutant proteins bind to the viral replication origin. Virology 184:14-22. [DOI] [PubMed] [Google Scholar]
- 48.Pombo, A., J. Ferreira, E. Bridge, and M. Carmo-Fonseca. 1994. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 13:5075-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 50.Richardson, W. D., and H. Westphal. 1981. A cascade of adenovirus early functions is required for expression of adeno-associated virus. Cell 27:133-141. [DOI] [PubMed] [Google Scholar]
- 51.Thomson, B. J., F. W. Weindler, D. Gray, V. Schwaab, and R. Heilbronn. 1994. Human herpesvirus 6 (HHV-6) is a helper virus for adeno-associated virus type 2 (AAV-2) and the AAV-2 rep gene homologue in HHV-6 can mediate AAV-2 DNA replication and regulate gene expression. Virology 204:304-311. [DOI] [PubMed] [Google Scholar]
- 52.Uprichard, S. L., and D. M. Knipe. 1997. Assembly of herpes simplex virus replication proteins at two distinct intranuclear sites. Virology 229:113-125. [DOI] [PubMed] [Google Scholar]
- 53.van Breukelen, B., P. N. Kanellopoulos, P. A. Tucker, and P. C. van der Vliet. 2000. The formation of a flexible DNA-binding protein chain is required for efficient DNA unwinding and adenovirus DNA chain elongation. J. Biol. Chem. 275:40897-40903. [DOI] [PubMed] [Google Scholar]
- 54.Walker, S. L., R. S. Wonderling, and R. A. Owens. 1997. Mutational analysis of the adeno-associated virus Rep68 protein: identification of critical residues necessary for site-specific endonuclease activity. J. Virol. 71:2722-2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward, P., and K. I. Berns. 1996. In vitro replication of adeno-associated virus DNA: enhancement by extracts from adenovirus-infected HeLa cells. J. Virol. 70:4495-4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ward, P., F. B. Dean, M. E. O'Donnell, and K. I. Berns. 1998. Role of the adenovirus DNA-binding protein in in vitro adeno-associated virus DNA replication. J. Virol. 72:420-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ward, P., M. Falkenberg, P. Elias, M. Weitzman, and R. M. Linden. 2001. Rep-dependent initiation of adeno-associated virus type 2 DNA replication by a herpes simplex virus type 1 replication complex in a reconstituted system. J. Virol. 75:10250-10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ward, P., and R. M. Linden. 2000. A role for single-stranded templates in cell-free adeno-associated virus DNA replication. J. Virol. 74:744-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ward, P., E. Urcelay, R. Kotin, B. Safer, and K. I. Berns. 1994. Adeno-associated virus DNA replication in vitro: activation by a maltose binding protein/Rep 68 fusion protein. J. Virol. 68:6029-6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weindler, F. W., and R. Heilbronn. 1991. A subset of herpes simplex virus replication genes provides helper functions for productive adeno-associated virus replication. J. Virol. 65:2476-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Weisshart, K., P. Taneja, and E. Fanning. 1998. The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication. J. Virol. 72:9771-9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weitzman, M. D., K. J. Fisher, and J. M. Wilson. 1996. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J. Virol. 70:1845-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weitzman, M. D., S. R. Kyostio, R. M. Kotin, and R. A. Owens. 1994. Adeno-associated virus (AAV) Rep proteins mediate complex formation between AAV DNA and its integration site in human DNA. Proc. Natl. Acad. Sci. USA 91:5808-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilcock, D., and D. P. Lane. 1991. Localization of p53, retinoblastoma and host replication proteins at sites of viral replication in herpes-infected cells. Nature 349:429-431. [DOI] [PubMed] [Google Scholar]
- 65.Wistuba, A., A. Kern, S. Weger, D. Grimm, and J. A. Kleinschmidt. 1997. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol. 71:1341-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiao, X., J. Li, and R. J. Samulski. 1998. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 72:2224-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yakinoglu, A. O., R. Heilbronn, A. Burkle, J. R. Schlehofer, and H. zur Hausen. 1988. DNA amplification of adeno-associated virus as a response to cellular genotoxic stress. Cancer Res. 48:3123-3129. [PubMed] [Google Scholar]
- 68.Yakobson, B., T. A. Hrynko, M. J. Peak, and E. Winocour. 1989. Replication of adeno-associated virus in cells irradiated with UV light at 254 nm. J. Virol. 63:1023-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yakobson, B., T. Koch, and E. Winocour. 1987. Replication of adeno-associated virus in synchronized cells without the addition of a helper virus. J. Virol. 61:972-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Young, S. M., Jr., D. M. McCarty, N. Degtyareva, and R. J. Samulski. 2000. Roles of adeno-associated virus Rep protein and human chromosome 19 in site-specific recombination. J. Virol. 74:3953-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, D., L. Frappier, E. Gibbs, J. Hurwitz, and M. O'Donnell. 1998. Human RPA (hSSB) interacts with EBNA1, the latent origin binding protein of Epstein-Barr virus. Nucleic Acids Res. 26:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zolotukhin, S., B. J. Byrne, E. Mason, I. Zolotukhin, M. Potter, K. Chesnut, C. Summerford, R. J. Samulski, and N. Muzyczka. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6:973-985. [DOI] [PubMed] [Google Scholar]