Summary
Vasa, a DEAD box helicase, is a germline marker that may also function in multipotent cells. In the embryo of the sea urchin Strongylocentrotus purpuratus, Vasa protein is posttranscriptionally enriched in the small micromere lineage, which results from two asymmetric cleavage divisions early in development. The cells of this lineage are subsequently set aside during embryogenesis for use in constructing the adult rudiment. Although this mode of indirect development is prevalent among echinoderms, early asymmetric cleavage divisions are a derived feature in this phylum. The goal of this study is to explore how vasa is regulated in key members of the phylum with respect to the evolution of the micromere and small micromere lineages. We find that although striking similarities exist between the vasa mRNA expression patterns of several sea urchins and sea stars, the time frame of enriched protein expression differs significantly. These results suggest that a conserved mechanism of vasa regulation was shifted earlier in sea urchin embryogenesis with the derivation of micromeres. These data also shed light on the phenotype of a sea urchin embryo upon removal of the Vasa-positive micromeres, which appears to revert to a basal mechanism used by extant sea stars and pencil urchins to regulate Vasa protein accumulation. Furthermore, in all echinoderms tested here, Vasa protein and/or message is enriched in the larval coelomic pouches, the site of adult rudiment formation, thus suggesting a conserved role for vasa in undifferentiated multipotent cells set aside during embryogenesis for use in juvenile development.
Introduction
Vasa, an ATP-dependent DEAD box helicase, is a faithful marker of the germline in numerous animals across phylogeny (Extavour and Akam, 2003; Wylie, 1999). Vasa was initially identified in Drosophila melanogaster as a component of the germ plasm, a specialized region of the cytoplasm that accumulates a select population of proteins and mRNA during oogenesis. Cells that inherit this cytoplasm during embryogenesis develop into the germline (Lasko and Ashburner 1988). In the germ plasm, vasa functions as a translational activator of the maternally inherited RNAs gurken and oskar (Markussen et al. 1995; Styhler et al. 1998; Tomancak et al. 1998). In D. melanogaster, the germ plasm dictates germline fate in a cell autonomous manner and is necessary for germline formation (Illmensee and Mahowald 1974); loss of vasa in this animal results in female sterility (Lasko and Ashburner 1988). Analogous structures to the germ plasm of D. melanogaster have been identified in several organisms, including Danio rerio, Xenopus laevis, and Caenorhabditis elegans, and in all cases vasa is an essential component (Komiya et al. 1994; Gruidl et al. 1996; Yoon et al. 1997). Vasa is also expressed in the germ cells of animals that do not use a germ plasm to specify their germline. These animals use inductive signals from surrounding cells to conditionally specify their germline (for a review, see Extavour and Akam 2003). In the mouse, for example, germ cells are specified at the proximal epiblast on embryonic day 6.5 by inductive signals from the extraembryonic tissue (Tam and Zhou 1996). Vasa expression begins in mouse germ cells during their migration to the gonadal ridge on embryonic day 10.5 (Toyooka et al. 2000) and loss of this gene function results in male sterility (Tanaka et al. 2000). Importantly, in some phyla vasa is not a strict germline marker, but is also expressed in somatic stem cells, which are capable of differentiating into both somatic and germline cells. For example, in mature Hydra polyps, vasa is expressed in the multipotent I cells, which give rise to both germ cells and three somatic cell types (Bosch and David 1987; Mochizuki et al. 2001; Rebscher et al. 2008). In the adult flatworm Macrostomum lignano, vasa is expressed in the totipotent neoblasts, which can give rise to all somatic cell types as well as germ cells (Reddien and Sanchez Alvarado 2004; Pfister et al. 2008). Thus, vasa may have a basal role in repressing differentiation, both in the germline and in select somatic stem cells.
During embryogenesis of the sea urchin, Strongylocentrotus purpuratus, vasa mRNA is uniformly distributed through blastula formation, followed by specific expression in the small micromere lineage during gastrulation (Juliano et al. 2006). By contrast, Vasa protein remains uniformly distributed through only the first three cleavage divisions (Voronina et al. 2008). The fourth cell division is unequal, thus producing a 16-cell embryo with 8 mesomeres, 4 macromeres, and 4 micromeres (Fig. 1). The newly formed micromeres are a signaling center that influences the fates of adjacent cells (Angerer and Angerer 2000). Vasa protein accumulation exhibits a major transition during early cleavage: at the eight-cell stage it is uniformly distributed, but during the fourth cleavage division it is rapidly enriched in the four micromeres (Voronina et al. 2008). The micromeres undergo a second unequal cleavage division resulting in further Vasa protein enrichment in the small micromeres. The Vasa-depleted large micromere descendants will ingress into the blastocoel to give rise to the larval skeleton (Fig. 1, shown in blue). The Vasa-enriched small micromere descendants, the only cells in the embryo to retain high levels of Vasa protein, remain at the vegetal plate of the blastula, are carried passively at the tip of the archenteron into the blastocoel during gastrulation, and are then incorporated into the left and right coelomic pouches of the larva, the site of adult rudiment formation (Fig. 1, shown in red).
Fig. 1.
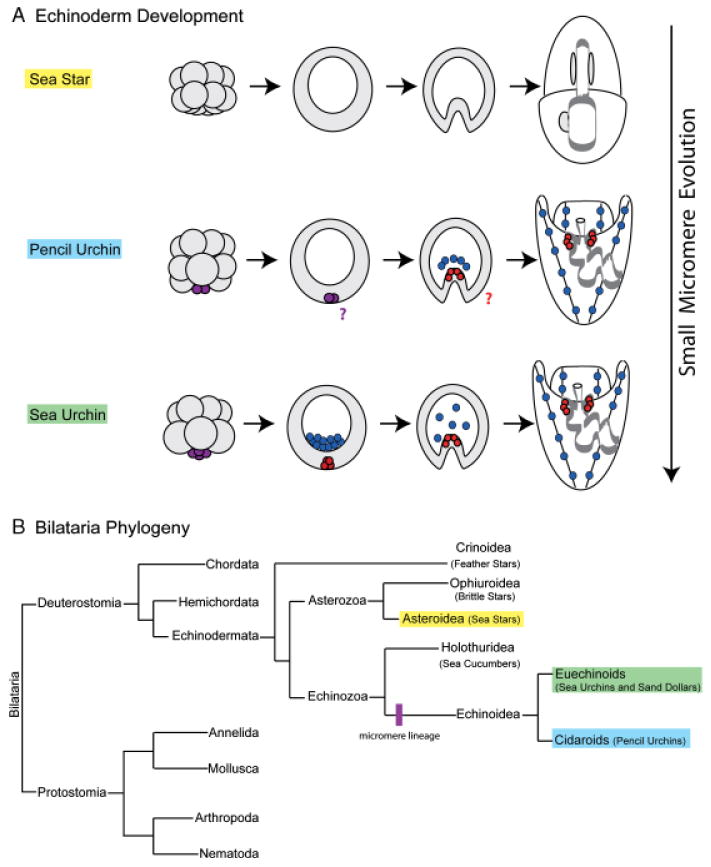
Echinoderm phylogeny and small micromere evolution. (A) Micromeres are a derived feature among echinoderms. The sea star embryo exhibits equal early cleavage divisions and therefore does not have a micromere lineage. The pencil urchin embryo has a variable number of micromeres (purple) that will give rise to the larval skeleton (blue); however, it is unclear whether the micromeres go through a second asymmetric division to give rise to small micromeres. In the sea urchin embryo, four micromeres (purple) asymmetrically divide to give rise to four large micromeres (blue) and four small micromeres (red). Large micromeres give rise to the larval skeleton. Small micromeres reside at the vegetal plate of the blastula, travel at the tip of archenteron in the gastrula, and divide only once before being incorporated into the left and right coelomic pouches of the larva. (B) Members of the phylum Echinodermata are a sister-group to the chordates. There are five extant classes of echinoderms: Crinoidea (feather stars), Ophiuroidea (brittle stars), Asteroidea (sea stars), Holothuridea (sea cucumbers), and Echinoidea. Members of the class Echinoidea, which have a micromere lineage, are split into the subclasses Euechinoids (sea urchins and sand dollars) and Cidaroids (pencil urchins) (Ettensohn et al. 2004).
S. purpuratus is a maximal indirect developer and embryogenesis culminates with the creation of a free-swimming and feeding microscopic larval form, which supports the developing adult rudiment. At metamorphosis the juvenile emerges and the majority of larval tissues are destroyed (Peterson et al. 1997). The cells that contribute to the adult rudiment in the S. purpuratus larva are termed “set-aside cells” because they are segregated away from differentiating cell populations during embryogenesis (Peterson et al. 1997). As the larva forms, the set-aside cells are deposited into the larval coelomic pouches where the adult rudiment will develop. Two sources of embryonic cells contribute to the larval coelomic pouches: a subset of the macromere descendents contributes approximately 60% of the initial coelomic pouch constituents and the Vasa-positive small micromere lineage contributes the remaining 40% (Cameron et al. 1991). During embryogenesis, the small micromeres exhibit a slow cell cycle as compared with surrounding differentiating cells due to an extended S-phase, likely reflecting their undifferentiated, quiescent state (Tanaka and Dan 1990). They divide only once before they are incorporated into the larval coelomic pouches where they resume proliferation (Tanaka and Dan 1990). Although it is clear that the small micromeres contribute to the adult rudiment, the exact fate of the small micromere lineage in the juvenile is unknown. However, due to both their slow cycle and the fact that they contribute entirely to adult tissues it has been hypothesized that the small micromeres are germ cell precursors (Pehrson and Cohen 1986). We find that the number of Vasa-positive cells in the coelomic pouches expand dramatically as the larva develops, and essentially all cells of the developing rudiment are Vasa-positive (C. Juliano and M. Yajima, unpublished data). Given that the small micromeres are not the sole contributors to the adult rudiment, we hypothesize that new vasa expression must account for this finding. Therefore, we favor the hypothesis that in the S. purpuratus embryo vasa is a broad marker of an undifferentiated state rather than a strict germline marker. Further, we hypothesize that the small micromere lineage is multipotent and will contribute to several cell types in the adult, perhaps including, but not necessarily limited to the germline. However, lineage-tracing experiments are required to ultimately resolve small micromere fate.
Vasa protein accumulation is regulated posttranscriptionally in the sea urchin embryo; vasa mRNA is uniformly distributed whereas the protein accumulates selectively at the fourth cleavage division. This requires unique control of the translation of the ubiquitous mRNA and/or stability of the Vasa protein in specific cell types. A remarkable form of vasa regulation is observed in embryos whose four Vasa-positive micromeres are removed at the 16-cell stage. These embryos recover all the cell types essential for adult formation, including the germline (Ransick et al. 1996). Importantly, this recovery is accompanied by the ubiquitous accumulation of Vasa protein from existing vasa mRNA (Voronina et al. 2008). If such embryos are allowed to develop, Vasa protein accumulation is once again restricted in the larvae; the larvae are able to metamorphose and grow into normal, fertile adults (Ransick et al. 1996; Voronina et al. 2008).
The sea urchin is a member of Echinodermata, a phylum that holds an informative place on the phylogenetic tree as a sister-group to chordates (Cameron et al. 2000) (Fig. 1B). To address the potential association of vasa with multipotent cell lineages further, we have examined vasa expression in key members of the phylum with respect to micromere evolution. The phylum consists of five extant and diverse classes: Crinoidea (feather stars), Asteroidea (sea stars), Ophiuroidea (brittle stars), Echinoidea (sea urchins and sand dollars), and Holothuroidea (sea cucumbers) (Fig. 1B). The micromeres are a recently acquired trait in this phylum and are found only in the embryos of class Echinoidea, which includes the euechinoids (sea urchins and sand dollars) and the cidaroids (pencil urchins) (Fig. 1B). The euechinoids and cidaroids likely diverged around 225 Ma during the Triassic period (Kier 1974). Evidence from the fossil record indicates that the cidaroids, or pencil urchins, have retained a primitive adult morphology and so it is thought that their embryonic development also reflects a basal characteristic of this class (Schroeder 1981). At the 16-cell stage the pencil urchin does develop micromeres, but their number varies between 1 and 4. Furthermore, these cells ingress as primary mesenchyme cells and form the larval skeleton, but this process is delayed until gastrulation (Schroeder 1981). Lineage tracing experiments in the pencil urchin embryo revealed that, like in the euechinoids, the micromeres are the precursors of the larval skeleton (Wray and McClay 1988). However, it is unclear whether the pencil urchin has a lineage analogous to the sea urchin small micromere lineage.
The other major class of echinoderms commonly used in developmental studies is the asteroids, or sea stars. The last common ancestor of the sea star and the sea urchin existed approximately 500 Ma in the Late Cambrian Period (Smith 1988; Wada and Satoh 1994). The sea star embryo undergoes equal early cleavage divisions, thus representing a basal form of development with respect to the micromere lineage. The embryo does not have a micromere lineage and the larva does not have a skeleton (Fig. 1). Recent work by Hinman and colleagues demonstrates that despite 500 million years of evolution several striking similarities remain in embryonic gene regulation between these two organisms (Hinman et al. 2003; Hinman and Davidson 2007). The goal of this study is to determine how key members of the phylum Echinodermata regulate vasa, especially with respect to the evolution of micromere formation. We look for both common and divergent features that will direct our understanding of multipotent cell origins in these maximally indirect developers.
Materials and Methods
Animals
Animal suppliers
Lytechinus variegatus, Eucidaris tribuloides—Ken Nedimeyer, Florida Keys; Heliocidaris tuberculata—Rudy Raff, Australia; Strongylocentrotus droebachiensis—Gary Wessel, Maine; Arbacia punctulata, Asterias forbesii—Marine Biological Laboratories, Woods Hole, MA; Echinarachnius parma—Jim Coffman, Mt. Desert, ME; Mellita quinquiesperforata—Beaufort, NC; Asterina miniata—Pat Leahy, Kerchoff Marine Laboratories, California Institute of Technology.
Animals were housed in aquaria with artificial seawater (ASW) at either 16°C or 22°C, to most closely match the animal's natural environment (ASW; Coral Life Scientific Grade Marine Salt; Energy Savers Unlimited Inc., Carson, CA, USA). Gametes were acquired either by 0.5 M KCl injection (sea urchins and pencil urchins) or dissection (sand dollars and sea stars). Eggs were collected in ASW and sperms were collected dry. To obtain embryos, fertilized eggs were cultured in ASW at temperatures between 16°C and 22°C. When early stage embryos were required for labeling, fertilization was performed in the presence of either 1 mm 3-amino-triazol (Sigma, St. Louis, MO, USA) or 10 mm para-aminobenzoic acid (Sigma) to inhibit cross-linking of the fertilization envelopes. Before fixing, envelopes were removed by passing the embryos through Nytex mesh of appropriate size for each species. Careful monitoring was required to ensure the integrity of the embryo.
Identification and cloning of vasa homologs
Two rounds of nested, degenerate PCR were used to clone partial sequences of the Et-vasa and Af-vasa homologs from cDNA derived from either oocytes (A. forbesii) or early embryos (E. tribuloides). Degenerate primers were designed to target the highly conserved DEAD box region. Primer sets were as follows: first round, 5′-ATHCCIGTIGARGTIWSIGG-3′ (forward), 5′-CKRTGIACRTAYTCRTCDAT-3′ (reverse); and second round, 5′-CCIACICCIGTICARAARTAY-3′ (forward), 5′-GGDATRTCIARICCICKIGC-3′ (reverse). A partial Am-vasa sequence was identified by a low-stringency hybridization to macroarrays. Vasa gene sequences were extended to full length using RACE (Ambion, Austin, TX, USA) and cloned into pGEMT-EZ (Promega, Madison, WI, USA) for sequencing. Identification of each homolog was confirmed by BLAST analysis (Altschul et al. 1997). Using PAUP, unrooted neighbor-joining phylograms were made from both full-length vasa sequences (shown) and the DEAD box region alone; bootstrap replicate values are from 1000 iterations (Swofford 2002).
Whole-mount RNA in situ hybridization (WMISH)
Lv-vasa, Et-vasa, Am-vasa, and Af-vasa fragments between 600 and 1000 nucleotides long were amplified by PCR or degenerate PCR from either ovary or early embryo cDNA and cloned into pGEMT-EZ (Promega). These plasmids were linearized using either SalI (T7 transcription) or NcoI (SP6 transcription) (Promega). Anti-sense DIG-labeled RNA probes were constructed using a DIG RNA-labeling kit (Roche, Indianapolis, IN, USA). WMISH experiments were performed as described previously (Minokawa et al. 2004), except alkaline phosphatase staining was carried out for either 6 or 22 h. A nonspecific DIG-labeled RNA probe complimentary to pSPT 18 (Roche) was used as a negative control. Samples were imaged on a Zeiss Axiovert 200M microscope equipped with a Zeiss color AxioCam MRc5 camera (Carl Zeiss Inc., Thornwood, NY, USA).
SDS-PAGE and immunoblot analysis
A. miniata embryos were cultured as described above and used to prepare embryo extracts. At the indicated developmental stages, 30 embryos of each stage were collected with a mouth pipette into microfuge tubes, pelleted, resuspended in heated SDS loading buffer, boiled for 5 min, and stored at − 80°C. For SDS-PAGE, samples were thawed at 37°C and DTT (Roche) was added at a final concentration of 5 mm. Samples were incubated at 100°C for 5 min, spun at 18,000 × g for 2 min, and then loaded onto Trisglycine, 4–20% gradient gels (NuSep, Frenchs Forest, NSW, Australia). After transfer to nitrocellulose, blots were probed with a polyclonal, affinity-purified antibody directed against Sp-vasa (see Voronina et al. 2008) at a concentration of 1 μg/ml. For visualization, blots were probed with an anti-rabbit-HRP secondary antibody diluted 1:5000 (Jackson ImmunoResearch Laboratories, West Grove, PA, USA), and visualized by standard ECL detection.
Whole-mount immunolabeling
Embryos were cultured as described above and samples were taken at indicated stages of development for whole-mount antibody labeling. For E. tribuloides, A. miniata, and A. forbesii, embryos were fixed in 4% paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA, USA)/ASW for 10 min at room temperature, extracted in 100% MeOH (− 20°C) for 1 min, washed three times with PBS-Tween, and stored at 4°C. Antibody labeling was preceded by a blocking step of at least 30 min in 4% sheep serum (Sigma)/PBS-Tween. For labeling, embryos were incubated overnight at 4°C with affinity-purified Sp-vasa antibody (Voronina et al. 2008) diluted in blocking buffer to a concentration of 10 μg/ml. The embryos were washed three times with PBS-Tween and then were incubated with anti-rabbit Cy3-conjugated antibody (Jackson ImmunoResearch Laboratories) diluted 1:300 in blocking buffer at room temperature for 1 h. The embryos were then washed three times with PBS-Tween, the second containing a 1:1000 dilution of a 10 mg/ml Hoechst stock solution (Molecular Probes, Carlsbad, CA, USA) for DNA labeling. For L. variegatus, H. tuberculata, S. droebachiensis, A. punctulata, E. parma, and M. quinquiesperforata, embryos were fixed as described for RNA WMISH (Minokawa et al. 2004). Vasa antibody labeling was performed as described above except that PBS-Tween was replaced with MOPS-Tween buffer (100 mm MOPS, pH 7.0, 500 mm NaCl, and 0.01% Tween). Labeled E. tribuloides, A. miniata, and A. forbesii embryos were visualized on an LSM 510 laser scanning confocal microscope (Carl Zeiss Inc.). Labeled L. variegatus, S. droebachiensis, A. punctulata, E. parma, and M. quinquiesperforata embryos were visualized on a TCS SP2 AOBS confocal scanning microscope (Leica Microsystems, Bannockburn, IL, USA). In all cases, secondary antibody-alone controls were conducted using the same experimental procedures described above, except the primary antibody incubation was replaced with an overnight incubation in block at 4°C. In all cases, the same confocal settings were used to examine protein expression within the developmental series of a species.
Results
Identification of echinoderm vasa homologs
Vasa was cloned from the pencil urchin, E. tribuloides, and two sea stars, A. miniata, and A. forbesii, allowing for a sequence comparison with both S. purpuratus and L. variegatus (Voronina et al. 2008). A phylogenetic analysis demonstrates that the identified sequences are true vasa homologs rather than the closely related DEAD box helicase PL10 (Fig. 2B). All five of the echinoderm vasa homologs identified contain the highly conserved DEAD box and helicase domains at their C-terminus. These two domains comprise the helicase core, which consists of 10 conserved motifs common to the DEAD box helicase family; these motifs are required for RNA binding, ATPase activity, and RNA unwinding (see supporting information Fig. S1; reviewed in Cordin et al. 2006). As is common in all vasa homologs across phylogeny, significant variability occurs at the N-terminus of the echinoderm vasa homologs identified here, including a variable number of CCHC zinc fingers (Fig. 2A and supporting information Fig. S1).
Fig. 2.
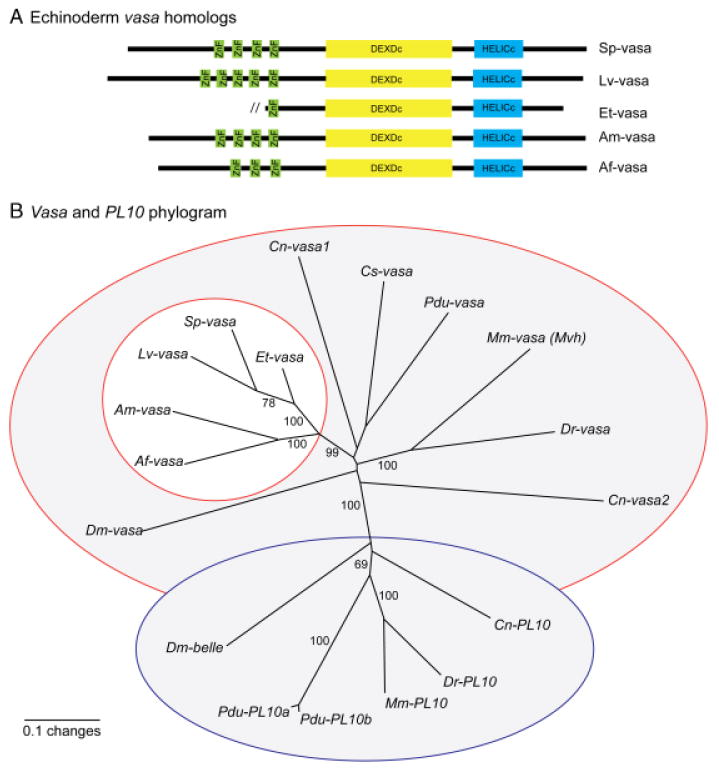
Phylogentic analysis of echinoderm vasa homologs. (A) Echinoderm vasa homologs have a variable number of zinc fingers (green) at the N-terminus. The helicase core at the C-terminus consists of the highly conserved DEXDc (yellow) and HELICc (blue) domains. Domains were defined by SMART (Schultz et al. 1998; Letunic et al. 2006). (B) An unrooted neighbor-joining phylogram demonstrates that the echinoderm vasa homologs (white circle) cluster with several other vasa homologs (red outline) rather than the DEAD box helicase PL10 (blue outline), a close relative of vasa. Numbers indicate bootstrap replicate values from 1000 iterations. Analysis was performed with both the full-length sequences (shown here) and the DEAD box regions alone, and the resulting phylograms were the same. Vasa and PL10 homologs are named as follows: Mus musculus (Mm-vasa, NP_034159), Danio rerio (Dr-vasa, NP_571132), Ciona savignyi (Cs-vasa, BAB12217), Drosophila melanogaster (Dm-vasa, CAA31405), Platy-nereis dumerilii (Pdu-vasa, AM048812), Hydra magnipapillata (Cn-vasa1, BAB13307), Hydra magnipapillata (Cn-vasa2, BAB13308), Mus musculus (Mm-PL10, NP_149068), D. rerio (Dr-PL10, NP_571016), D. melanogaster (Dm-belle, NP_536783), Platynereis dumerilii (Pdu-PL10a, AM048813), Platynereis dumerilii (Pdu-PL10b, AM048814), Hydra magnipapillata (Cn-PL10, BAB13306), Asterias forbesii (Af-vasa, FJ605736), Asterina miniata (Am-vasa, FJ605737), Eucidaris tribuloides (Et-vasa, FJ605738), Lytechinus variegatus (Lv-vasa, FJ605739), Strongylocentrotus purpuratus (Sp-vasa, FJ605740). Echinoderm vasa sequences have been submitted to NCBI.
The small micromere lineage of euechinoids specifically accumulates Vasa protein
Both sea urchins and sand dollars, collectively called euechinoids, have micromeres that form at the fourth cleavage division and subsequently generate a small micromere lineage. In three sea urchin species examined, L. variegatus, S. droebachiensis, and H. tuberculata, Vasa protein expression is enriched in the small micromere lineage early in embryogenesis, although the timing of enrichment varies (Fig. 3, A–C, supporting information Figs. S2 and S3). For example, whereas selective expression of Vasa protein is clearly seen in the micromeres at the 16-cell stage of S. purpuratus, it is only slightly enriched in the micromeres at the 16-cell stage during the embryogenesis of both L. variegatus and H. tuberculata (Fig. 3B and supporting information Fig. S3; Voronina et al. 2008). One sea urchin included in the study, A. punctulata, expresses Vasa protein uniformly in the embryo, followed by enrichment of Vasa protein in both coelomic pouches of the larva (Fig. 3D and supporting information Fig. S4). Although it has not been experimentally determined, the location of the Vasa-positive cells in the coelomic pouches of A. punctulata is consistent with their identity as small micromere descendents similar to the other sea urchins examined in this study.
Fig. 3.
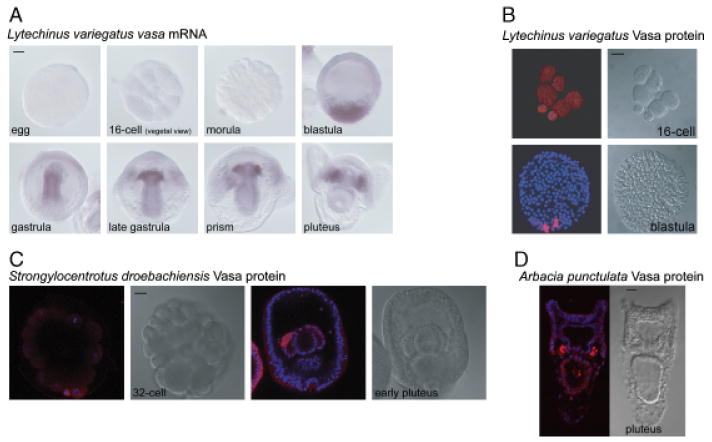
Vasa mRNA and protein is restricted to the small micromere lineage and larval coelomic pouches in sea urchins. (A) In Lytechinus variegatus, vasa mRNA is enriched at the vegetal plate of the blastula, in the gut of the gastrula, and finally in the small micromere lineage of the prism and pluteus larva. (B) In L. variegatus, Vasa protein is enriched in the micromeres of the 16-cell embryo and in the small micromeres of the blastula. (C) In Strongylocentrotus droebachiensis, Vasa protein accumulates in the small micromeres upon their formation, remains enriched in this lineage throughout embryogenesis, and is then enriched in the coelomic pouches of the pluteus larva. (D) In Arbacia punctulata, Vasa protein is enriched in the coelomic pouches of the pluteus larva. (B–D) Vasa protein (red), Hoechst (blue); Vasa immunofluorescence was performed with an antibody raised against Sp-Vasa (Voronina et al. 2008). Scale bars are 20 μm.
The euechinoid subclass also includes the sand dollars, two of which were examined in this study. Similar to several of the sea urchins examined, Vasa protein is enriched in the small micromere lineage during E. parma embryogenesis and then selectively accumulates in the coelomic pouches of the larva (Fig. 4A). However, in the sand dollar M. quinquiesperforata, Vasa protein is uniformly expressed through embryonic development and then becomes selectively enriched in the coelomic pouches of the larva, similar to what is seen in the sea urchin A. punctulata (Fig. 4B and supporting information Fig. S5). Overall, euechinoids appear to selectively accumulate Vasa protein in their small micromere lineage and/or their larval coelomic pouches, although the timing varies among different species (Table 1).
Fig. 4.
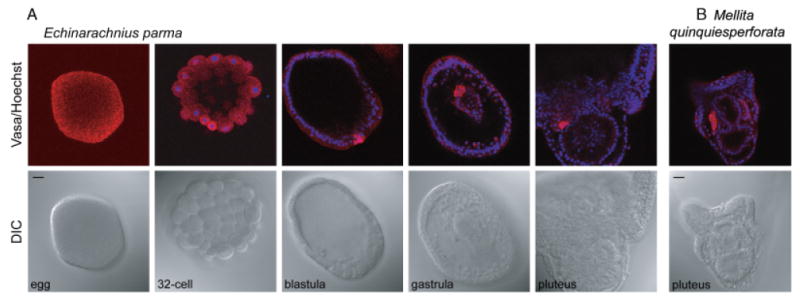
Vasa protein is enriched in the small micromere lineage and the larval coelomic pouches of sand dollars. (A) In Echinarachnius parma, Vasa protein is uniformly distributed in the egg and early cleavage stages, accumulates selectively in the small micromeres upon their formation, remains enriched in this lineage throughout embryogenesis, and is then enriched in the coelomic pouches of the pluteus larva. (B) In Mellita quinquiesperforata, Vasa protein is enriched in the coelomic pouches of the pluteus larva. (A–B) Vasa protein (red), Hoechst (blue); Vasa immunofluorescence was performed with an antibody raised against Sp-vasa (Voronina et al. 2008). Scale bars are 20 μm.
Table 1. Summary of vasa mRNA and protein expression characteristics during echinoderm embryogenesis.
| species | class | cleavage pattern | vasa mRNA enrichment | Vasa protein enrichment |
|---|---|---|---|---|
| Strongylocentrotus purpuratus | cleavage: uniform blastula: uniform/vegetal plate gastrula: small micromeres larva: coelomic pouches (Juliano et al., 2006) | micromeres/small micromeres, larval coelomic pouches (Voronina et al., 2008) | ||
| Strongylocentrotus droebachiensis | not determined | micromeres/small micromeres, larval coelomic pouches (Fig. 3 and supporting information Fig. S2) | ||
| Lytechinus variegatus | Echinoidea (Euechinoids: sea urchins and sand dollars) | Unequal divisions: micromeres and small micromeres | cleavage: undetectable blastula: vegetal plate gastrula: gut larva: coelomic pouches (Fig. 3) | micromeres/small micromeres (Fig. 3) larval coelomic pouches (data not shown) |
| Heliocidaris tuberculata | not determined | micromeres/small micromeres, larval coelomic pouches (supporting information Fig. S3) | ||
| Arbacia punctulata | not determined | uniform through embryogenesis, then enriched in larval coelomic pouches (Fig. 3 and supporting information Fig. S4) | ||
| Echinarachnius parma | not determined | micromeres/small micromeres, larval coelomic pouches (Fig. 4) | ||
| Mellita quinquiesperforata | not determined | uniform through embryogenesis, then enriched in larval coelomic pouches (Fig. 3 and supporting information Fig. S5) | ||
| Eucidaris tribuloides | Echinoidea (Cidaroids: pencil urchins) | Unequal divisions: micromeres | cleavage: unknown blastula: vegetal plate gastrula: gut larva: coelomic pouches (Fig. 5) | uniform through embryogenesis, then enriched in larval coelomic pouches (Fig. 5) |
| Asterina miniata | Asteroidea (sea stars) | Equal divisions | cleavage: uniform blastula: uniform/vegetal plate gastrula: gut outpocket larva: posterior enterocoel (Fig. 6) | uniform through embryogenesis, then enriched in larval posterior enterocoel (Fig. 7, supporting information Fig. S6) |
| Asterias forbesii | cleavage: unknown blastula: vegetal plate gastrula: gut/archenteron tip larva: pouches and PE (Fig. 6) | not determined | ||
Vasa mRNA accumulates more broadly than Vasa protein in euechinoid embryos
An analysis of vasa mRNA accumulation in L. variegatus reveals that the message is detectable by qPCR in eggs and early embryos (M. Lopez and D. McClay, unpublished data), although the message is not detectable by WMISH at these stages (Fig. 3A). By contrast, in S. purpuratus vasa, mRNA is highly expressed and uniformly accumulates in eggs and early embryogenesis as determined by WMISH (Juliano et al., 2006; Fig. 3A). The vasa mRNA in L. variegatus is first detected at the vegetal plate of the blastula by WMISH, is found throughout the gut in the gastrula, and then becomes restricted to the small micromere lineage of the late gastrula and the larval coelomic pouches (Fig. 3A). In S. purpuratus, vasa mRNA is also enriched at the blastula vegetal plate, but is then selectively enriched in the small micromere lineage early during gastrulation (Juliano et al. 2006). Despite these differences, like S. purpuratus, L. variegatus must also differentially regulate the expression of vasa message and/or protein by cell type. For example, in the blastula, mRNA can be found in a much wider domain at the vegetal plate than the protein, which is enriched specifically in the four small micromeres (Fig. 3B).
Vasa mRNA and protein accumulation in cidaroid development
In the E. tribuloides embryo, vasa mRNA accumulates at the vegetal plate of the blastula, throughout the developing gut of the gastrula, and then is enriched in both coelomic pouches after larval formation (Fig. 5A). This mRNA expression pattern is almost identical to that of the euechinoid L. variegatus; the only significant difference is the timing of specific mRNA accumulation, which occurs just before pouch formation in L. variegatus.
Fig. 5.
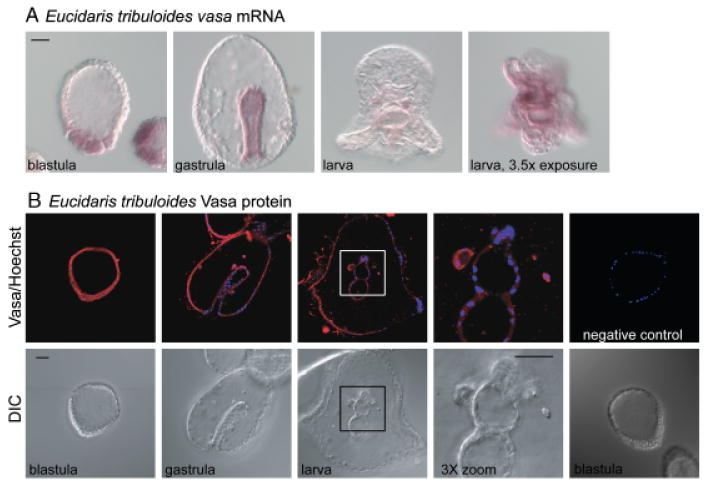
Vasa mRNA and protein are enriched in the coelomic pouches of the pencil urchin, Eucidaris tribuloides. (A) Vasa mRNA is enriched at the vegetal plate of the blastula, in the gut of the gastrula, and finally in the coelomic pouches of the larva. To visualize vasa mRNA in the coelomic pouches the time of exposure to the alkaline phosphatase reaction conditions was increased from approximately 6 to 22 h. (B) Vasa protein accumulates uniformly through embryogenesis and is then enriched in the coelomic pouches of the larva. Negative control excludes the primary antibody. Vasa protein (red), Hoechst (blue); Vasa immunofluorescence was performed with an antibody raised against Sp-vasa (Voronina et al. 2008). Scale bars are 20 μm.
Unlike several of the euechinoids examined, Vasa protein is not localized to any specific lineage during E. tribuloides embryogenesis, but rather accumulates uniformly (Fig. 5B). Enriched Vasa protein accumulation can first be detected in the coelomic pouches of a 1-week-old larva (Fig. 5B). Overall, members of the class Echinoidea have the following shared characteristics: (1) Vasa protein enrichment in the small micromere lineage and/or the coelomic pouches, (2) vasa mRNA enrichment at the vegetal plate followed by restriction to the small micromere lineage and/or the coelomic pouches, and (3) vasa mRNA and protein are not strictly coincident (Table 1).
Vasa mRNA accumulation in asteroid embryos shares similarities with echinoids
During early embryogenesis in A. miniata, vasa mRNA is uniformly distributed through the embryo until blastula formation (Fig. 6A). In the late blastula/early gastrula, vasa mRNA becomes highly enriched at the vegetal plate and remains localized to the blastopore as gastrulation proceeds (Fig. 6A). In the late gastrula vasa mRNA is localized to a small group of cells on the left side of the gut that then forms the posterior enterocoel (PE) of the larva (Fig. 6A; Byrne and Barker 1991). Based on both morphology and extirpation results, the cells of the PE were hypothesized to be the germ cells of the developing sea star (Inoue et al. 1992). The enrichment of vasa mRNA in the PE is therefore consistent with its potential role in germline determination. A similar vasa expression pattern is seen in the sea star A. forbesii with a few minor differences, the most significant being that vasa mRNA is not detected in the PE until after larval formation (Fig. 6B). The sea star vasa expression pattern is similar to the echinoids in that vasa mRNA is enriched at the vegetal plate of the blastula, followed by enrichment in a subset of cells in the gastrula and larva (Table 1).
Fig. 6.
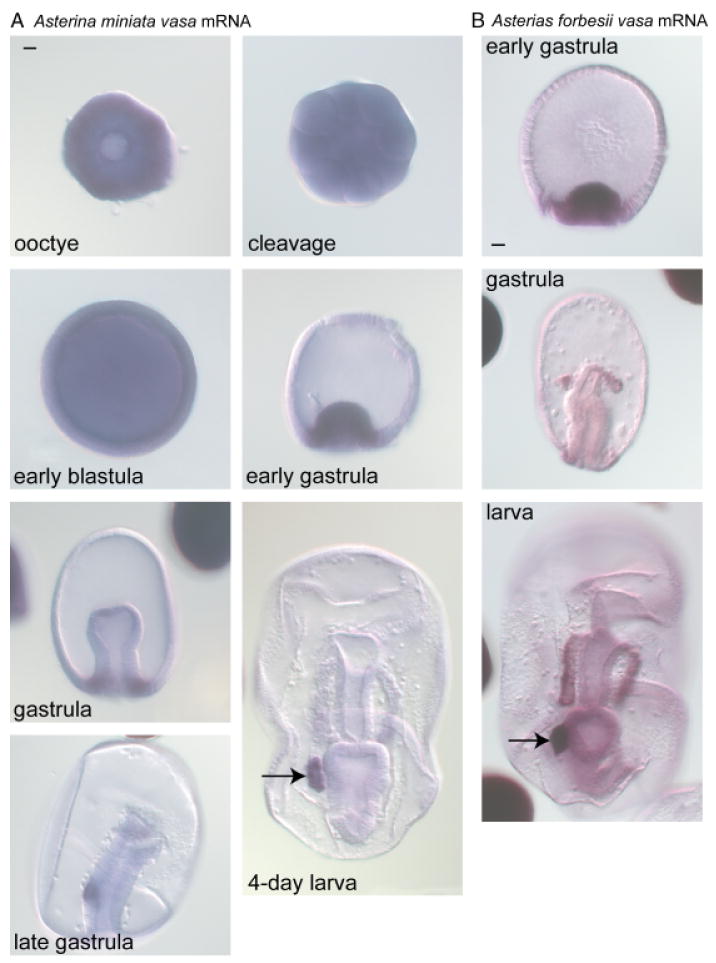
Vasa mRNA is enriched in the posterior enterocoel (PE) of the sea stars Asterina miniata and Asterias forbesii. (A) In A. miniata, vasa mRNA accumulates uniformly through blastula formation, is enriched in the vegetal region of the gastrula, and then accumulates selectively in the PE (arrow) of the larva. (B) In A. forbesii, vasa mRNA is enriched at the vegetal region of the early gastrula, at the tip of the archenteron in the gastrula, and in both the coelomic pouches and PE (arrow) of the larva. Scale bars are 20 μm.
Specific Vasa protein accumulation occurs in later stages of asteroid development
The antibody generated against Sp-vasa specifically recognizes the A. miniata vasa homolog, which has a predicted MW of 77 kDa, very similar in size to the S. purpuratus vasa homolog (MW of 79 kDa) (Voronina et al. 2008; Fig. 7A). Immunoblot analysis demonstrates that Vasa protein accumulates at a constant level throughout A. miniata embryogenesis and then decreases in the 1-week-old larva (Fig. 7A). Immunofluorescence analysis demonstrates that Vasa protein accumulates uniformly throughout embryogenesis and is then slightly enriched in the PE, the site of enriched vasa mRNA accumulation, of the 3-day-old larva (Fig. 7B and Supplemental Fig. S6). In a 6-day-old A. miniata larva, Vasa protein is highly enriched in the PE, which has flattened onto the side of the gut and joined the left coelomic pouch (Fig. 7, B and C).
Fig. 7.
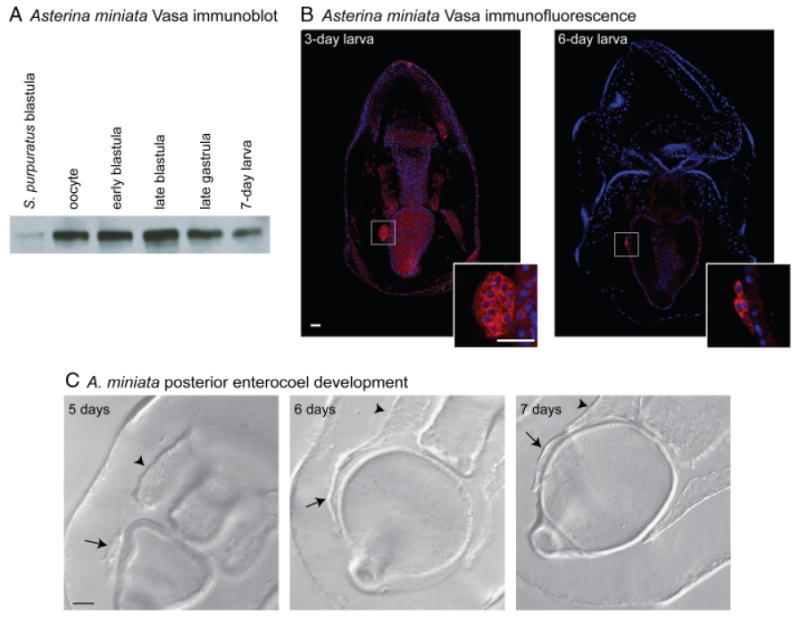
Vasa protein is restricted to the posterior enterocoel (PE) of the larva in the sea star Asterina miniata. (A) The Sp-vasa antibody recognizes an approximately 80 kDa protein, similar to the size of Sp-vasa (Voronina et al. 2008), in A. miniata embryonic extracts. Vasa protein is present in all stages of A. miniata embryogenesis; levels decrease in the late gastrula and larva (30 embryos loaded/lane). (B) Vasa protein is enriched in the PE of the 3-day-old and 6-day-old larva. PEs are shown at 3× zoom in insets. (A–B) Vasa protein (red), Hoechst (blue); Vasa immunofluorescence was performed with an antibody raised against Sp-Vasa (Voronina et al. 2008). Scale bars are 20 μm. (C) In A. miniata, the PE joins the left coelomic pouch after approximately 6 days of development. Arrows indicate the PE and arrowheads indicate the left coelomic pouch.
Discussion
Vasa protein expression in the small micromere lineage and/or coelomic pouches is conserved in euechinoids
Our previous work in S. purpuratus demonstrated that Vasa protein is enriched in the small micromeres upon their formation and then accumulates selectively in the larval coelomic pouches (Voronina et al. 2008). Here, we show that this is a conserved feature among euechinoids; however, the timing of enriched Vasa protein accumulation in the small micromere lineage and/or coelomic pouches occurs in a variable time frame ranging from shortly before small micromere formation to after larval formation. We hypothesize that the common euechinoid ancestor exhibited enriched Vasa protein early in development and that this feature was independently lost in the euechinoids, A. punctulata and M. quinquiesperforata, which exhibit larval Vasa protein restriction. Alternatively, early Vasa protein restriction may have been independently acquired in the five other euechinoids tested in this study (Table 1). In either case, this argues that the timing of Vasa protein restriction in this phylum is labile over short periods of evolutionary time. Future work will focus on understanding the molecular function(s) of Vasa in echinoderms, which will perhaps shed light on this observation. Furthermore, in S. purpuratus, enriched Vasa expression in the small micromere lineage is followed by Sp-nanos and Sp-seawi expression, which are genes known to function in germline determination and stem cell maintenance in other animals (Kobayashi et al. 1996; Cox et al. 1998; Forbes and Lehmann 1998; Tsuda et al. 2003; Wang and Lin 2004; Juliano et al. 2006; Suzuki et al. 2007). Therefore, it will be informative to examine the expression patterns of these genes in a variety of euechinoids in order to understand their relationship to specific Vasa protein accumulation in the small micromere lineage.
Asteroid larvae express vasa in the PE, a potential site of primordial germ cell accumulation
In both of the sea stars examined, A. miniata and A. forbesii, vasa mRNA is highly expressed in a group of cells originating from the gut that form the PE. Inoue et al. (1992) hypothesized that these were primordial germ cells based on larval location and characteristically enlarged nuclei. In support of this premise, they found that removing the PE from 2-day-old Asterina pectinifera larvae resulted in approximately 50% less presumptive germ cells in the 6-week-old larvae (Inoue et al. 1992). In the sea stars A. pectinifera, P. regularis, and A. miniata, the PE joins the left coelomic pouch early in larval development, suggesting that the vasa-positive PE in asteroids is analogous to the vasa-positive small micromere lineage of the echinoids (Byrne and Barker 1991; Inoue et al. 1992; Fig. 8). The coelomic pouches of the sea star also contain low levels of vasa mRNA, suggesting the presence of multipotent cells (Fig. 6). Perhaps in the sea star the vasa-positive primordial germ cells and the multipotent cells originate at separate places in the larva and then meet in the left coelomic pouch, whereas in the sea urchin the small micromeres have acquired both functions as a multipotent cell lineage, which contributes to the germ cell population later in development. Lineage tracing experiments are needed to definitively test this hypothesis.
Fig. 8.
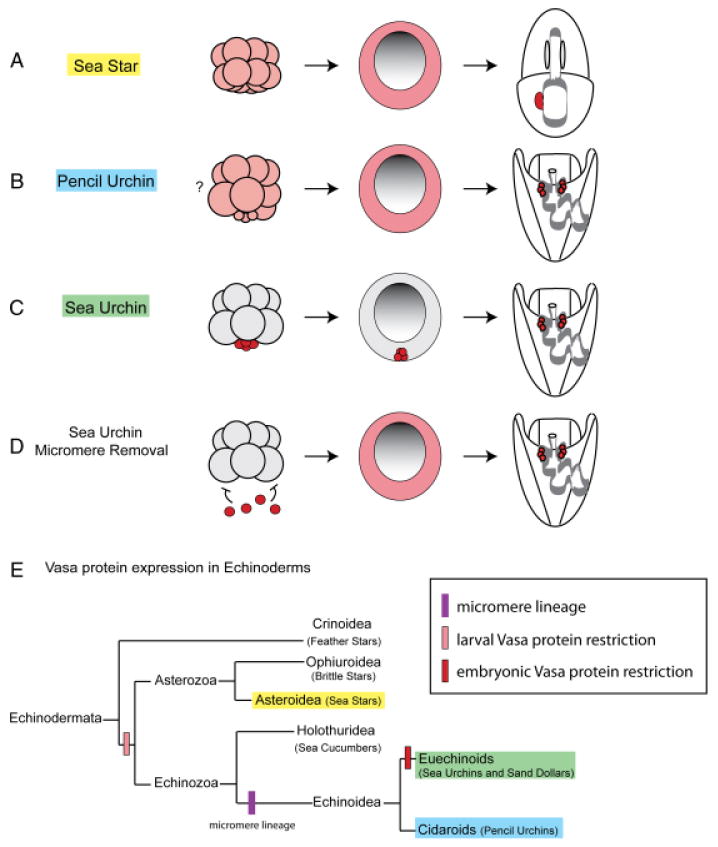
Vasa protein expression is restricted to the small micromeres in the sea urchin, but is inducible in other lineages. (A and B) In the pencil urchin and the sea star, Vasa protein is uniformly distributed during embryogenesis (pink) and then selectively enriched in the larva. (C) In the sea urchin, Vasa protein accumulates selectively in the micromeres and then the small micromeres (red). (D) Removing the micromeres from a 16-cell sea urchin embryo leads to uniform upregulation of Vasa protein (pink). This suggests that Vasa-positive micromeres normally send a repressive signal to the rest of the embryo that inhibits Vasa protein accumulation. Furthermore, the micromere-deleted sea urchin is capable of growing into a normal adult, suggesting that it reverts to an ancient mechanism of vasa regulation, used by the pencil urchin and the sea star, to restrict Vasa protein accumulation in the larva. (E) Vasa protein expression patterns presented here support the hypothesis that larval Vasa protein restriction is a basal trait among echinoderms, and that early restriction of Vasa protein to the micromere lineage is a derived feature of Euechinoids.
Micromere-deleted sea urchins may revert to a basal mechanism of vasa regulation
The Vasa-positive micromeres of the 16-cell sea urchin embryo are a signaling center that directs patterning of the early embryo (Angerer and Angerer 2000). Micromere-deleted embryos rapidly accumulate Vasa protein in all cells of the embryo similar to what is observed in normal pencil urchin and sea star embryos, and develop into normal, fertile adults (Ransick et al. 1996; Voronina et al. 2008). We hypothesize that loss of the micromeres removes a repressive signal, thus allowing upregulation of Vasa protein, which is likely accomplished by increasing the translation rate of existing ubiquitous vasa mRNA and/or a removal of Vasa protein degradation in nonmicromere cells (Fig. 8, A–D). In light of the present results, we suggest that when micromeres are removed, the sea urchin embryo reverts to a mechanism used by its echinoderm ancestors to first accumulate Vasa protein throughout the embryo and then restrict Vasa protein expression to re-create a new multipotent cell lineage.
The last common ancestor of the sea urchin and the sea star existed approximately 500 Ma. Despite this evolutionary distance, striking similarities remain in the expression of vasa mRNA, including enrichment at the vegetal plate, expression in the gut, and localized expression in the larva. However in the sea star, similar to the pencil urchin, Vasa protein accumulation is not specifically enriched until after larval formation. Therefore, given the late enriched expression of Vasa in both the pencil urchin and the sea star, we conclude that this represents the basal mechanism for regulating vasa among echinoderms (Fig. 8E). In both the pencil urchin and the sea star a mechanism must exist for clearing Vasa protein from the majority of cells in the larva to allow for enriched protein accumulation. It is possible that through the course of evolution this mechanism shifted temporally earlier in the sea urchin embryo and now restricts Vasa protein expression to the small micromere lineage. Furthermore, when this mechanism is disrupted by micromere removal, it re-establishes itself in the larva, allowing for enriched Vasa expression and multipotent cell fate specification.
Vasa protein degradation appears to be a conserved mechanism for directing the selective accumulation of genes to the germline across phylogeny. For example, early in Drosophila embryogenesis vasa mRNA is uniformly distributed whereas Vasa protein accumulates selectively in the future germline (Hay et al. 1990; Lasko and Ashburner, 1990). Accumulation of Vasa protein in the Drosophila germline is in part dependent upon protection from degradation by a deubiquitinating enzyme, thus implying that active degradation of Vasa protein may exist in the soma (Liu et al. 2003). During zebrafish embryogenesis, vasa mRNA is localized to the germ cells by 6 h, whereas Vasa protein remains uniform and then is enriched in the germline after 24 h of development (Braat et al. 2000; Knaut et al. 2000). Furthermore, the restriction of Vasa protein to the zebrafish germline depends on the Vasa ORF, thus suggesting protein turnover is required (Wolke et al. 2002). Future work on the sea urchin will focus on understanding the mechanism used to clear Vasa protein from non-small-micromere cells, and then testing the conservation of this mechanism in the sea star. These results will be of general interest given the conservation of protein restriction to the germline by protein turnover mechanisms (DeRenzo and Seydoux 2004).
Vasa may have a conserved function in multipotent cells during embryogenesis
During the embryonic development of a direct-developing animal, such as a mouse, the majority of cells undergo differentiation, thus creating a juvenile with features similar to the adult. In this case, it is essential that a totipotent population of cells, the germline, is set aside to give rise to the next generation. This mode of development is sharply contrasted by the maximal indirect development displayed by the majority of species belonging to phylum Echinodermata, including all of the species examined in this study (Wray and Bely 1994). In this mode of development, embryogenesis culminates in the creation of a larval form, which serves to feed and support the developing adult rudiment. Furthermore, multipotent, undifferentiated cells are set aside early in embryogenesis and these cells will give rise to the juvenile (Peterson et al. 1997). Our work demonstrates that Vasa protein enrichment in the set-aside cells, found in the larval coelomic pouches, is a conserved feature of echinoderms. Interestingly, this may be a conserved feature of Vasa expression in animals that segregate their germline from multipotent cells, as a similar result was recently found in the polychaete annelid, Platynereis dumerilii (Rebscher et al. 2007). In this polychaete, vasa mRNA is uniformly distributed during early cleavage stages and is then progressively restricted to the mesodermal posterior growth zone (MPGZ) of the larvae, which gives rise to all of the mesodermal tissue of the developing juvenile segments. By contrast, Vasa protein is segregated into the micromeres at the 38-cell stage, and finally to the 4d mesoblast lineage, which gives rise to the MPGZ. Importantly, Vasa-positive cells in the MPGZ give rise to both germ cells and to other somatic mesodermal cell types in the juvenile (Rebscher et al. 2007). Thus, our work, in combination with the polychaete annelid results, suggests a broader association of Vasa with multipotent cells in embryonic development. Indeed, in many direct-developing embryos the germ cells are the only cells set aside during embryogenesis. Thus, the functional role of Vasa in these two cases may be analogous.
Supplementary Material
Echinoderm vasa homolog sequence alignments. Echinoderm vasa homologs have a variable number of zinc fingers (green) at the N-terminus. The highly conserved DEXDc (yellow) and HELICc (blue) domains comprise the helicase core. The helicase core consists of 10 motifs common to members of the DEAD box helicase family. These motifs are required for ATP binding (Q motif and I), ATPase activity (II, III, and VI), helicase activity (III), RNA binding (Ia, Ib, IV, V, and VI), and protein interactions (GG doublet) (reviewed in Cordin et al. 2006). Sp-Strongylocentrotus purpuratus, Lv—Lytechinus variegatus, Et—Eucidaris tribuloides, Am—Asterina miniata, Af—Asterias forbesii
Vasa protein is enriched in the small micromere lineage and larval coelomic pouches of the sea urchin Strongylocentrotus droebachiensis. Vasa protein accumulates selectively in the small micromeres upon their birth, and is enriched in this lineage at the vegetal plate of the blastula and at the tip of the archenteron in the gastrula, followed by enrichment in the coelomic pouches of the pluteus. Negative control excludes the primary antibody. Vasa protein (red), Hoechst (blue); Vasa immunofluorescence was performed with an antibody raised against Sp-Vasa (Voronina et al. 2008). Scale bar is 20 μm.
Vasa protein is enriched in the small micromere lineage and the larval coelomic pouches of the sea urchin Heliocidaris tuberculata. Vasa protein is enriched in the micromeres of the 16-cell embryo, and then in the small micromere lineage at the vegetal plate of the blastula and at the tip of the archenteron in the gastrula, followed by enrichment in the coelomic pouches of the pluteus. Negative control excludes the primary antibody. Vasa protein (red), Hoechst (blue); Vasa immunofluorescence was performed with an antibody raised against Sp-Vasa (Voronina et al. 2008). Scale bar is 20 μm.
In the sea urchin Arbacia punctulata, Vasa protein accumulates uniformly through embryogenesis and is then enriched in the coelomic pouches of the pluteus larva. Negative control excludes the primary antibody. Vasa protein (red), Hoechst (blue); Vasa immunofluorescence was performed with an antibody raised against Sp-Vasa (Voronina et al. 2008). Scale bar is 20 μm.
In the sand dollar Mellita quinquiesperforata, Vasa protein accumulates uniformly through embryogenesis and is then enriched in the coelomic pouches of the pluteus larva. Negative control excludes the primary antibody. Vasa protein (red), Hoechst (blue); Vasa immunofluorescence was performed with an antibody raised against Sp-Vasa (Voronina et al. 2008). Scale bar is 20 μm.
In the sea star, Asterina miniata, Vasa protein accumulates uniformly through embryogenesis. Negative control excludes the primary antibody. Vasa protein (red), Hoe-chst (blue); Vasa immunofluorescence was performed with an antibody raised against Sp-Vasa (Voronina et al. 2008). Scale bar is 50 μm.
Acknowledgments
We thank all current and past members of PRIMO for helpful discussions, especially Ekaterina Voronina for excellent experimental assistance and advice. We thank Hyla Sweet for providing us with E. tribuloides cDNA, Jim Coffman for sending Echinarachnius parma, and Rudy Raff for fixed Heliocidaris tuberculata embryos. We thank Eric Gustafson, Casey Dunn, and anonymous reviewers for helpful suggestions on the manuscript. This work was supported by grants from the NIH and the NSF.
Footnotes
Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angerer LM, Angerer RC. Animal-vegetal axis patterning mechanisms in the early sea urchin embryo. Dev Biol. 2000;218:1–12. doi: 10.1006/dbio.1999.9553. [DOI] [PubMed] [Google Scholar]
- Bosch T, David C. Stem Cells of Hydra magnipapillata can differentiate into somatic cells and germ line cells. Dev Biol. 1987;121:182–191. [Google Scholar]
- Braat AK, van de Water S, Goos H, Bogerd J, Zivkovic D. Vasa protein expression and localization in the zebrafish. Mech Dev. 2000;95:271–274. doi: 10.1016/s0925-4773(00)00344-0. [DOI] [PubMed] [Google Scholar]
- Byrne M, Barker MF. Embryogenesis and larval development of the Asteroid Patiriella regularis by light and scanning electron microscopy. Biol Bull. 1991;180:332–345. doi: 10.2307/1542335. [DOI] [PubMed] [Google Scholar]
- Cameron CB, Garey JR, Swalla BJ. Evolution of the chordate body plan: new insights from phylogenetic analyses of deuterostome phyla. Proc Natl Acad Sci USA. 2000;97:4469–4474. doi: 10.1073/pnas.97.9.4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RA, Fraser SE, Britten RJ, Davidson EH. Macromere cell fates during sea urchin development. Development. 1991;113:1085–1091. doi: 10.1242/dev.113.4.1085. [DOI] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 1998;12:3715–3727. doi: 10.1101/gad.12.23.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRenzo C, Seydoux G. A clean start: degradation of maternal proteins at the oocyte-to-embryo transition. Trends Cell Biol. 2004;14:420–426. doi: 10.1016/j.tcb.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Ettensohn CA, Wessel GM, Wray GA. The invertebrate deuterostomes: an introduction to their phylogeny, reproduction, development, and genomics. Methods Cell Biol. 2004;74:1–13. doi: 10.1016/s0091-679x(04)74001-7. [DOI] [PubMed] [Google Scholar]
- Extavour CG, Akam M. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 2003;130:5869–5884. doi: 10.1242/dev.00804. [DOI] [PubMed] [Google Scholar]
- Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- Gruidl ME, et al. Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc Natl Acad Sci USA. 1996;93:13837–13842. doi: 10.1073/pnas.93.24.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN. Localization of vasa, a component of Drosophila polar granules, in maternal-effect mutants that alter embryonic anteroposterior polarity. Development. 1990;109:425–433. doi: 10.1242/dev.109.2.425. [DOI] [PubMed] [Google Scholar]
- Hinman VF, Davidson EH. Evolutionary plasticity of developmental gene regulatory network architecture. Proc Natl Acad Sci USA. 2007;104:19404–19409. doi: 10.1073/pnas.0709994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinman VF, Nguyen AT, Cameron RA, Davidson EH. Developmental gene regulatory network architecture across 500 million years of echinoderm evolution. Proc Natl Acad Sci USA. 2003;100:13356–13361. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illmensee K, Mahowald AP. Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc Natl Acad Sci USA. 1974;71:1016–1020. doi: 10.1073/pnas.71.4.1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Kiyomoto M, Shirai H. Germ cell differentiation in starfish: the posterior enterocoel as the origin of germ cells in Asterina pectinifera. Develop Growth Differ. 1992;34:413–418. doi: 10.1111/j.1440-169X.1992.00413.x. [DOI] [PubMed] [Google Scholar]
- Juliano CE, Voronina E, Stack C, Aldrich M, Cameron AR, Wessel GM. Germ line determinants are not localized early in sea urchin development, but do accumulate in the small micromere lineage. Dev Biol. 2006;300:406–415. doi: 10.1016/j.ydbio.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Kier PM. Evolutionary trends and their functional significance in the post-Paleozoic echinoids. J Paleont Paleont Soc Mem. 1974;5 48(suppl):1–95. [Google Scholar]
- Knaut H, Pelegri F, Bohmann K, Schwarz H, Nusslein-Volhard C. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J Cell Biol. 2000;149:875–888. doi: 10.1083/jcb.149.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Yamada M, Asaoka M, Kitamura T. Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature. 1996;380:708–711. doi: 10.1038/380708a0. [DOI] [PubMed] [Google Scholar]
- Komiya T, Itoh K, Ikenishi K, Furusawa M. Isolation and characterization of a novel gene of the DEAD box protein family which is specifically expressed in germ cells of Xenopus laevis. Dev Biol. 1994;162:354–363. doi: 10.1006/dbio.1994.1093. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. The product of the Drosophila gene vasa is very similar to eukaryotic initiation factor-4A. Nature. 1988;335:611–617. doi: 10.1038/335611a0. [DOI] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M. Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev. 1990;4:905–921. doi: 10.1101/gad.4.6.905. [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dansereau DA, Lasko P. Fat facets interacts with vasa in the Drosophila pole plasm and protects it from degradation. Curr Biol. 2003;13:1905–1909. doi: 10.1016/j.cub.2003.10.026. [DOI] [PubMed] [Google Scholar]
- Markussen FH, Michon AM, Breitwieser W, Ephrussi A. Translational control of oskar generates short OSK, the isoform that induces pole plasma assembly. Development. 1995;121:3723–3732. doi: 10.1242/dev.121.11.3723. [DOI] [PubMed] [Google Scholar]
- Minokawa T, Rast JP, Arenas-Mena C, Franco CB, Davidson EH. Expression patterns of four different regulatory genes that function during sea urchin development. Gene Expr Patterns. 2004;4:449–456. doi: 10.1016/j.modgep.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Nishimiya-Fujisawa C, Fujisawa T. Universal occurrence of the vasa-related genes among metazoans and their germ-line expression in Hydra. Dev Genes Evol. 2001;211:299–308. doi: 10.1007/s004270100156. [DOI] [PubMed] [Google Scholar]
- Pehrson JR, Cohen LH. The fate of the small micromeres in sea urchin development. Dev Biol. 1986;113:522–526. doi: 10.1016/0012-1606(86)90188-0. [DOI] [PubMed] [Google Scholar]
- Peterson KJ, Cameron RA, Davidson EH. Set-aside cells in maximal indirect development: evolutionary and developmental significance. Bioessays. 1997;19:623–631. doi: 10.1002/bies.950190713. [DOI] [PubMed] [Google Scholar]
- Pfister D, et al. Flatworm stem cells and the germ line: developmental and evolutionary implications of macvasa expression in Macrostomum lignano. Dev Biol. 2008;319:146–159. doi: 10.1016/j.ydbio.2008.02.045. [DOI] [PubMed] [Google Scholar]
- Ransick A, Cameron RA, Davidson EH. Postembryonic segregation of the germ line in sea urchins in relation to indirect development. Proc Natl Acad Sci USA. 1996;93:6759–6763. doi: 10.1073/pnas.93.13.6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebscher N, Volk C, Teo R, Plickert G. The germ plasm component vasa allows tracing of the interstitial stem cells in the cnidarian Hydractinia echinata. Dev Dyn. 2008;237:1736–1745. doi: 10.1002/dvdy.21562. [DOI] [PubMed] [Google Scholar]
- Rebscher N, Zelada-Gonzalez F, Banisch TU, Raible F, Arendt D. Vasa unveils a common origin of germ cells and of somatic stem cells from the posterior growth zone in the polychaete Platynereis dumerilii. Dev Biol. 2007;306:599–611. doi: 10.1016/j.ydbio.2007.03.521. [DOI] [PubMed] [Google Scholar]
- Reddien PW, Sanchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- Schroeder TE. Development of a “primitive” sea urchin (Eucidaris tribuloides): irregularities in the hyaline layer, micromeres, and primary mesenchyme. Biol Bull. 1981;161:141–151. [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AB. Fossil evidence for the relationships of extant echinoderm classes and their times of divergence. In: Paul C, Smith A, editors. Echinoderm Phylogeny and Evolutionary Biology. Clarendon; Oxford: 1988. pp. 85–97. [Google Scholar]
- Styhler S, Nakamura A, Swan A, Suter B, Lasko P. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development. 1998;125:1569–1578. doi: 10.1242/dev.125.9.1569. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Tsuda M, Saga Y. Functional redundancy among Nanos proteins and a distinct role of Nanos2 during male germ cell development. Development. 2007;134:77–83. doi: 10.1242/dev.02697. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and other methods) Version 4. Sinauer Associates; Sunderland, MA: 2002. [Google Scholar]
- Tam PP, Zhou SX. The allocation of epiblast cells to ectodermal and germ-line lineages is influenced by the position of the cells in the gastrulating mouse embryo. Dev Biol. 1996;178:124–132. doi: 10.1006/dbio.1996.0203. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Dan K. Study of the lineage and cell cycle of small micromeres in embryos of the sea urchin, Hemicentrotus pulcherrimus. Dev Growth Differ. 1990;32:145–156. doi: 10.1111/j.1440-169X.1990.00145.x. [DOI] [PubMed] [Google Scholar]
- Tanaka SS, et al. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev. 2000;14:841–853. [PMC free article] [PubMed] [Google Scholar]
- Tomancak P, Guichet A, Zavorszky P, Ephrussi A. Oocyte polarity depends on regulation of gurken by Vasa. Development. 1998;125:1723–1732. doi: 10.1242/dev.125.9.1723. [DOI] [PubMed] [Google Scholar]
- Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T. Expression and intracellular localization of mouse Vasahomologue protein during germ cell development. Mech Dev. 2000;93:139–149. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- Tsuda M, et al. Conserved role of nanos proteins in germ cell development. Science. 2003;301:1239–1241. doi: 10.1126/science.1085222. [DOI] [PubMed] [Google Scholar]
- Voronina E, et al. Vasa protein expression is restricted to the small micromeres of the sea urchin, but is inducible in other lineages early in development. Dev Biol. 2008;314:276–286. doi: 10.1016/j.ydbio.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Satoh N. Phylogenetic relationships among extant classes of echinoderms, as inferred from sequences of 18S rDNA, coincide with relationships deduced from the fossil record. J Mol Evol. 1994;38:41–49. doi: 10.1007/BF00175494. [DOI] [PubMed] [Google Scholar]
- Wang Z, Lin H. Nanos maintains germline stem cell self-renewal by preventing differentiation. Science. 2004;303:2016–2019. doi: 10.1126/science.1093983. [DOI] [PubMed] [Google Scholar]
- Wolke U, Weidinger G, Koprunner M, Raz E. Multiple levels of posttranscriptional control lead to germ line-specific gene expression in the zebrafish. Curr Biol. 2002;12:289–294. doi: 10.1016/s0960-9822(02)00679-6. [DOI] [PubMed] [Google Scholar]
- Wray GA, Bely AE. The evolution of echinoderm development is driven by several distinct factors. Development. 1994;(suppl):97–106. [PubMed] [Google Scholar]
- Wray GA, McClay DR. The origin of spicule-forming cells in a ‘primitive’ sea urchin (Eucidaris tribuloides) which appears to lack primary mesenchyme cells. Development. 1988;103:305–315. doi: 10.1242/dev.103.2.305. [DOI] [PubMed] [Google Scholar]
- Wylie C. Germ cells. Cell. 1999;96:165–174. doi: 10.1016/s0092-8674(00)80557-7. [DOI] [PubMed] [Google Scholar]
- Yoon C, Kawakami K, Hopkins N. Zebrafish vasa homologue RNA is localized to the cleavage planes of 2- and 4-cell-stage embryos and is expressed in the primordial germ cells. Development. 1997;124:3157–3165. doi: 10.1242/dev.124.16.3157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Echinoderm vasa homolog sequence alignments. Echinoderm vasa homologs have a variable number of zinc fingers (green) at the N-terminus. The highly conserved DEXDc (yellow) and HELICc (blue) domains comprise the helicase core. The helicase core consists of 10 motifs common to members of the DEAD box helicase family. These motifs are required for ATP binding (Q motif and I), ATPase activity (II, III, and VI), helicase activity (III), RNA binding (Ia, Ib, IV, V, and VI), and protein interactions (GG doublet) (reviewed in Cordin et al. 2006). Sp-Strongylocentrotus purpuratus, Lv—Lytechinus variegatus, Et—Eucidaris tribuloides, Am—Asterina miniata, Af—Asterias forbesii
Vasa protein is enriched in the small micromere lineage and larval coelomic pouches of the sea urchin Strongylocentrotus droebachiensis. Vasa protein accumulates selectively in the small micromeres upon their birth, and is enriched in this lineage at the vegetal plate of the blastula and at the tip of the archenteron in the gastrula, followed by enrichment in the coelomic pouches of the pluteus. Negative control excludes the primary antibody. Vasa protein (red), Hoechst (blue); Vasa immunofluorescence was performed with an antibody raised against Sp-Vasa (Voronina et al. 2008). Scale bar is 20 μm.
Vasa protein is enriched in the small micromere lineage and the larval coelomic pouches of the sea urchin Heliocidaris tuberculata. Vasa protein is enriched in the micromeres of the 16-cell embryo, and then in the small micromere lineage at the vegetal plate of the blastula and at the tip of the archenteron in the gastrula, followed by enrichment in the coelomic pouches of the pluteus. Negative control excludes the primary antibody. Vasa protein (red), Hoechst (blue); Vasa immunofluorescence was performed with an antibody raised against Sp-Vasa (Voronina et al. 2008). Scale bar is 20 μm.
In the sea urchin Arbacia punctulata, Vasa protein accumulates uniformly through embryogenesis and is then enriched in the coelomic pouches of the pluteus larva. Negative control excludes the primary antibody. Vasa protein (red), Hoechst (blue); Vasa immunofluorescence was performed with an antibody raised against Sp-Vasa (Voronina et al. 2008). Scale bar is 20 μm.
In the sand dollar Mellita quinquiesperforata, Vasa protein accumulates uniformly through embryogenesis and is then enriched in the coelomic pouches of the pluteus larva. Negative control excludes the primary antibody. Vasa protein (red), Hoechst (blue); Vasa immunofluorescence was performed with an antibody raised against Sp-Vasa (Voronina et al. 2008). Scale bar is 20 μm.
In the sea star, Asterina miniata, Vasa protein accumulates uniformly through embryogenesis. Negative control excludes the primary antibody. Vasa protein (red), Hoe-chst (blue); Vasa immunofluorescence was performed with an antibody raised against Sp-Vasa (Voronina et al. 2008). Scale bar is 50 μm.


