Summary
Vibrio anguillarum serotype O1 is part of the natural flora in the aquatic habitat, but under certain circumstances it can cause terminal haemorrhagic septicemia in marine and fresh water fish due to the action of the anguibactin iron uptake system encoded by the virulence plasmid pJM1. This plasmid harbours the genes for the biosynthesis of the siderophore anguibactin and the ferric anguibactin transport proteins FatD, C, B and A encoded in the iron transport operon. The FatA protein is the outer membrane receptor for the ferric siderophore complex and the FatB lipoprotein provides the periplasmic domain for its internalization, whereas the FatC and D proteins are located in the cytoplasmic membrane and might play a role as part of the ABC transporter for internalization of the ferric siderophore. In this work we demonstrate the essential role of these two inner membrane proteins in ferric anguibactin transport and that the lipo-protein nature of FatB is not necessary for ferric anguibactin transport.
Introduction
Vibrio anguillarum is part of the natural flora of the aquatic habitat and can also be a pathogen causing terminal haemorrhagic septicemia in marine and fresh water fish (Actis et al., 1999). Some serotype O1 strains of this bacterium carry the pJM1 virulence plasmid harbouring genes encoding the siderophore anguibactin biosynthesis and transport proteins. This system is essential for V. anguillarum virulence as well for its survival in environments in which the iron concentration is limited (Crosa, 1980; Di Lorenzo et al., 2003).
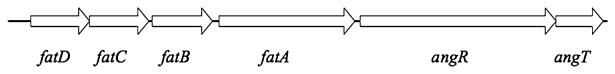
The iron transport and biosynthesis operon (ITBO) in the pJM1 plasmid of V. anguillarum 775(pJM1) encodes most of the genes necessary for anguibactin transport and biosynthesis (Tolmasky et al., 1988). There are six genes, fatA, B, C, D, angR and angT in this operon (Fig. 1). We have previously shown that FatB, a periplasmic binding lipoprotein, and FatA, the outer membrane anguibactin-specific receptor, are essential for ferric anguibactin transport (Actis et al., 1985; 1988; 1995; Lopez and Crosa, 2007; Lopez et al., 2007). AngR and AngT are involved in anguibactin biosynthesis while the former also regulates the expression of the ITBO operon (Salinas et al., 1989; Farrell et al., 1990; Tolmasky et al., 1993; Salinas and Crosa, 1995; Chen et al., 1996; Wertheimer et al., 1999). Our in silico analysis showed that FatC and FatD are polytopic integral cytoplasmic membrane proteins that could be involved in the translocation of ferric anguibactin across the cytoplasmic membrane (Koster et al., 1991). However, it has not been demonstrated as yet whether FatC and FatD are indeed involved on anguibactin transport due to the polar effect on fatB expression of the previously constructed mutants (Actis et al., 1995).
Fig. 1.
The iron transport and biosynthesis operon (ITBO) in the pJM1 plasmid of V. anguillarum 775(pJM1). FatC and FatD are polytopic integral cytoplasmic membrane proteins. FatB is a periplasmic binding lipoprotein. FatA is a ferric-anguibactin outer membrane receptor. AngR is involved in anugibactin biosynthesis and regulates the expression of the ITBO. AngT is involved in anguibactin biosynthesis.
In this work we demonstrate the essential role of these two inner membrane proteins in ferric anguibactin transport and that FatB does not need to be a lipoprotein to operate in ferric anguibactin-mediated transport.
Results and discussion
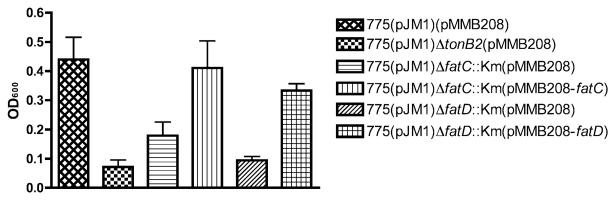
FatC and FatD are important for growth of V. anguillarum in iron-limiting condition
The bacterial strains and primers used in this work are listed in Table 1 and Table S1 respectively. To characterize the fatC and fatD genes we first mutated them in the wild-type strain 775(pJM1). For this task we combined flanking regions of the fatC or fatD genes by SOE PCR (Senanayake and Brian, 1995) with primers containing an Eco47III site in the middle of the deletion, and thereafter we cloned the product into pGEM-T easy vector (Promega). The kanamycin resistance cassette from pKD4 with SmaI site in both ends was ligated into the Eco47III site, and the insert DNA was subcloned into pDM4 (Milton et al., 1996). These plasmids were conjugated into V. anguillarum 775(pJM1) by triparental mating, and plasmid cointegrates were selected on tryptic soy agar with 1% NaCl (TSAS) supplemented with rifampacin (Rif) and chloramphenicol (Cm). Colonies thus obtained were cultured in tryptic soy broth with 1% NaCl (TSBS) without antibiotics. The mutants were then screened on TSAS with 15% sucrose and kanamycin (Km), and the mutations were confirmed by colony PCR and pJM1 DNA digestion. To complement the mutations the wild-type fatC or fatD genes with the upstream Shine-Dalgarno (SD) sequences were cloned into the pGEM-T easy vector and subcloned into pMMB208. The plasmids were conjugated into each of the V. anguillarum fatC and fatD mutants by triparental mating. The growth in minimal media of the wild-type and mutant strains under iron-limiting and -rich conditions was compared. Figure 2 clearly shows that under iron-limiting conditions (1 μM EDDA) both the fatC and fatD mutants grew much less than the wild-type strain whereas they behaved similarly under iron-rich conditions (10 μg ml−1 ferric ammonium citrate, FAC) (data not shown). Upon complementation with the wild-type genes, we observed the recovery of the mutants growth rate under iron-limiting conditions (Fig. 2). These results underscore the important role that the FatC and FatD proteins play in the ability of V. anguillarum to grow under iron starvation.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain or plasmid | Phenotype | Source or reference |
|---|---|---|
| Vibrio anguillarum strains | ||
| 775(pJM1) | Wild-type, Washington (serotype O1, pJM1) | Laboratory stock |
| 775(pJM1)-pMMB | 775(pJM1) harbouring pMMB208 | Naka et al. (2008) |
| HNVA-1 | 775(pJM1)ΔfatC::Km | This study |
| HNVA-2 | 775(pJM1)ΔfatD::Km | This study |
| CSL-71 | 775(pJM1) fatB::Km | This study |
| CSL-48 | 775(pJM1)ΔtonB2 | Lopez et al. (2009) |
| CC9-16 | Anguibactin deficient, anguibactin transport system deficient | Walter et al. (1983) |
| HNVA-3 | CC9-16ΔfatC::Km | This study |
| HNVA-4 | CC9-16ΔfatD::Km | This study |
| HNVA-5 | CC9-16 fatB::Km | This study |
| HNVA-6 | CC9-16ΔtonB2 | This study |
| Escherichia coli strains | ||
| S17-1 λpir | λ-pir lysogen; thi pro hsdR hsdM+ recA RP4 2-Tc:Mu-Km:Tn7(Tpr Smr) | Simon et al. (1983) |
| DH5α | F−, ϕ80lacZΔM15, endA1, recA1, hsdR17, (rK−mK+), supE44, thi-1, gyrA96, relA1, Δ(lacZYA-argF)U169, λ− | Laboratory stock |
| Plasmids | ||
| pGEM-T easy | Cloning vector Ampr | Promega |
| pCR2.1 | Cloning vector Ampr, Kmr | Invitrogen |
| pDM4 | Suicide plasmid sacB gene, R6K origin, Cmr | Milton et al. (1996) |
| pKD4 | Template plasmid, Kmr, Ampr | Datsenko and Wanner (2000) |
| pHN1 | pDM4-ΔfatC::Km | This study |
| pHN2 | pDM4-ΔfatD::Km | This study |
| pCL23 | pDM4-fatB::Km | This study |
| pMMB208 | A broad-host-range expression vector; Cmr IncQ lacIq Ptac; polylinker from M13mp19 | Morales et al. (1991) |
| pHN3 | pMMB208-fatC | This study |
| pHN4 | pMMB208-fatD | This study |
| pCL24 | pMMB208-fatB | This study |
| pCL25 | pMMB208-C23A fatB | This study |
Vibrio anguillarum strains were grown at 25°C in Tryptic soy broth (TSBS) or on agar (TSAS) supplemented with 1% NaCl, or in CM9 minimal medium (Crosa, 1980). Either ferric ammonium citrate (FAC) or ethylenediamine-di-(o-hydroxyphenyl acetic) acid (EDDA) was supplied in the CM9 medium to obtain iron-rich or -limiting conditions respectively. Escherichia coli were grown at 37°C in LB broth or agar. Antibiotics were used as following concentrations: 100 μg ml−1 rifampicin (Rif), 10 μg ml−1 chloramphenicol (Cm) and 200 μg ml−1 kanamycin (Km) for V. anguillarum, and 50 μg ml−1 ampicillin (Amp), 30 μg ml−1 chloramphenicol and 50 μg ml−1 kanamycin for E. coli.
Fig. 2.
Effect of mutations in either fatC or fatD on growth under iron-limiting conditions. Vibrio anguillarum strains were grown first in TSBS and then in CM9 minimal medium. A 40 μl aliquot of an overnight culture in CM9 (adjusted OD600 to 1) was inoculated into CM9 supplemented with 10 μg ml−1 Cm and 0.5 mM IPTG. EDDA (1μM) was added to achieve iron-limiting conditions. OD600 was measured after 24 h incubation at 25°C. Experiments were carried out three times, and the error bar shows the standard deviation.
FatC and FatD are essential for anguibactin-mediated iron transport
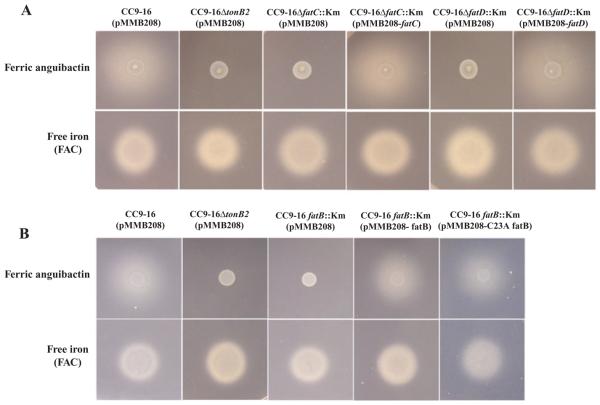
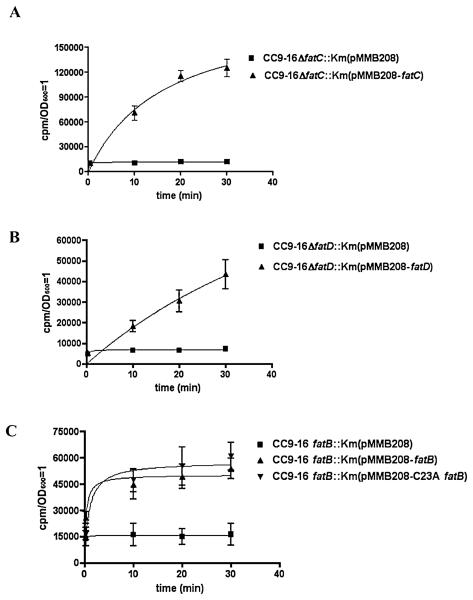
To assess whether the slower growth of the fatC and fatD mutants in iron-limiting conditions is due to a defect in the transport of ferric anguibactin we performed cross-feeding bioassays. By using the suicide vector mentioned above, we constructed the same fatC and fatD mutants in an anguibactin biosynthesis-deficient and transport-proficient strain, CC9-16. As shown in Fig. 3 we did not observe any growth halo with the fatC and fatD mutants in bioassays with spots of the anguibactin-producing strain 775(pJM1), whereas the wild-type positive control showed a distinct halo. A growth halo was indeed observed when the fatC or fatD mutants were complemented with the wild-type gene in trans. Negative and positive controls gave the expected results. These results demonstrate that fatC and fatD are indispensable for ferric-anguibactin iron transport, and even though FatC and FatD are very highly related (45% similarity at amino acid level) they cannot be replaced by each other, suggesting specific roles for these two proteins. Furthermore, we also performed iron uptake experiments as described previously (Crosa and Hodges, 1981; Welch and Crosa, 2005) to characterize their lack of functionality of the fatC and fatD mutants. The results in Fig. 4A (FatC) and B (FatD) clearly show that the FatC or FatD mutant does not transport ferric anguibactin while the complemented strains recover its transport. From these findings we conclude that in addition to fatA and fatB, fatD and fatC are also essential for ferric anguibactin transport since no chromosome gene can compensate for the deficiency when either of them is mutated.
Fig. 3. Bioassay for anguibactin uptake.
A. Bioassay to assess whether fatC or fatD is necessary for ferric anguibactin transport.
B. Bioassay anguibactin uptake of strains with the wild-type FatB and C23A FatB. Indicator strains were grown in TSBS, and then CM9 minimal medium. A 50 μl aliquot of an overnight culture was mixed with 20 μl of melted CM9 1.5% agar, 20 μM EDDA, 500 μM IPTG and 10 μg μl−1 Cm. After the agar became solid 5 μl of V. anguillarum 775(pJM1) culture (labelled as ferric anguibactin) and 1 μl of 1 mg ml−1 ferric ammonium citrate [labelled as free iron (FAC)] were spotted on each plate as sources of anguibactin and free iron respectively. Plates were incubated at 25°C for 24 h. The experiments were repeated three times, with consistent results. Strain CC9-16(pMMB208) was used as a positive control; strain CC9-16ΔtonB2(pMMB208), constructed as described (Lopez et al., 2009), was used as a negative control.
Fig. 4.
Kinetics of 55Fe-anguibactin uptake by V. anguillarum. Characterization of FatC (A), FatD (B) and FatB (C). Vibrio anguillarum strains grown to exponential phase (OD600 = 0.3–0.6) in CM9 were washed twice and resuspended in casamino acid-free CM9 containing the chelator sodium nitrilotriacetate at a concentration of 100 μM. The anguibactin siderophore was loaded with 55Fe by incubation with 55FeCl3(1 μCi ml−1) for 6 h and then mixing it with an equal volume of V. anguillarum in CM9 salts. At each time point 1 ml of mixture was withdrawn, filtered through a 0.45 μm filter (Millipore Corporation) and immediately washed twice with10 ml of 100 mM Sodium citrate. The filters were air-dried and the radioactivity was measured in a liquid scintillation counter. The values were normalized to an OD (OD600 = 1), and results were fitted using the GraphPad Prism4 program.
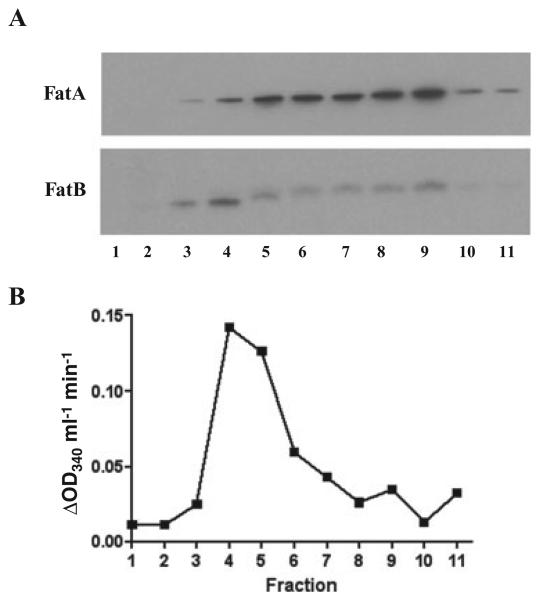
The FatB lipoprotein is anchored in the cytoplasmic membrane
The fatB gene encoding a periplasmic binding lipoprotein is essential for anguibactin transport and attaches to the membrane by its fatty acid tail (Actis et al., 1995). However, it is still unknown whether the FatB lipoprotein is attached to the outer or the cytoplasmic membrane. To answer this question total proteins from the wild-type strain were separated by sucrose gradient sedimentation. Each fraction thus obtained was used for Western blots using specific antibodies to FatB or FatA (outer membrane control). We also measured in each fraction the NADH oxidase activity, a control for cytoplasmic membrane proteins. Figure 5 shows the strongest intensity bands observed in fraction 4 for FatB and fraction 9 for FatA respectively. Furthermore, the strongest NADH oxidase activity was also observed in fraction 4 where the strongest FatB band was detected. Those results indicate that the lipoprotein FatB is anchored to the cytoplasmic membrane.
Fig. 5.
The FatB protein attaches to the cytoplasmic membrane. A. Western blots of proteins separated by sucrose density gradient centrifugation. Sucrose density gradient centerifugation was conducted as described by Nikaido (1994) with modification. One litre of V. anguillarum 775 overnight culture in CM9 broth was harvested and resuspended in 20 ml Hepes buffer (pH 7.4). After French pressure treatment and centrifugation, 1 ml of total protein was layered on top of a sucrose gradient (0.25 ml of saturated sucrose, 1.5 ml of 2.02 M sucrose, 5 ml of 1.44 M sucrose and 3 ml of 0.77 M sucrose) in the 14 × 89 mm polyallomer centrifuge tubes (Beckman). After 20 h ultracentrifugation at 4°C in the SW28 rotor (Beckman) at 100 000 g each 1 ml of fractions was collected from the top of the tube, and 3 μl (for FatA detection) and 15 μl (for FatB detection) of samples were used for Western bots. B. NADH oxidase activity of fractions obtained from sucrose gradient sedimentation. The NADH oxidase activity was measured as described by Osborn and colleagues (1972) with modification. Samples (30 μl) was mixed with 240 μl of assay buffer (50 mM Tris-HCl (pH 7.5), 0.2 mM dithiothreitol). After 5 min incubation at 25°C, 30 μl of 1.2 mM NADH was added, and the decrease in OD340 was measured at 25°C.
Is the lipoprotein nature of FatB essential for anguibactin transport?
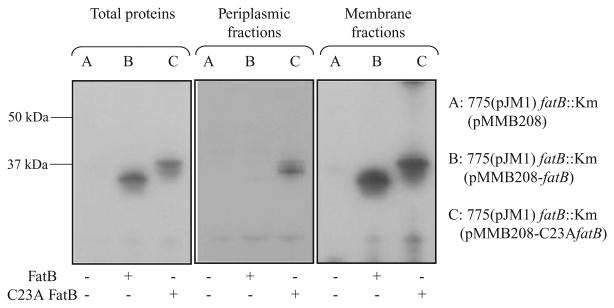
The lipoprotein nature of periplasmic binding proteins for siderophore transport is uncommon in Gram-negative bacteria. In addition to our finding with FatB, it was also shown in V. cholerae ViuP for vibriobactin as well as enterobactin transport (Wyckoff et al., 1999) and in Campylobacter jejuni and C. coli CeuE for enterobactin transport (Park and Richardson, 1995; Richardson and Park, 1995). In Gram-positives, such as the Bacillus subtilis FhuD protein required for the internalization of ferric-hydroxamates, is a lipoprotein that anchors to the outside of the cytoplasmic membrane, possibly due to the lack of an outer membrane (Schneider and Hantke, 1993). Bacterial lipoproteins are recognized by the type II signal peptidase that has a ‘lipobox’ recognition motif that is typically L3-[A/S/T]−2-[G/A]−1-C+1, where the signal peptidase cleaves in front of the +1 cysteine. The +1 cysteine is absolutely conserved in all bacterial lipoproteins (Braun and Wu, 1994; Hutchings et al., 2009). The thiol group of the cysteine is crucial to the lipoprotein biogenesis as the diacylglycerol lipid anchor is added in a thioether linkage (Braun and Wu, 1994). The sequence of the carboxy-terminal end of the FatB signal peptide is L-T-G-C, and the cysteine (position 23) residue should serve as the diacylglycerol lipid anchor (Actis et al., 1995). To test whether the lipoprotein nature of FatB is necessary for ferric anguibactin transport we replaced this cysteine for alanine. The wild-type fatB gene with its upstream region was cloned into the T-vector, and C23A fatB was created by site-directed mutagenesis using the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The wild-type fatB and C23A fatB were, respectively, subcloned into pMMB208, and subsequently conjugated into the fatB null mutant constructed by inserting the Km resistance cassette in the SalI site located in the middle of the fatB gene. To determine the localization of C23A FatB, we prepared total proteins, periplasmic proteins and membrane proteins from the fatB null mutant complemented with pMMB208 carrying either fatB, C23A fatB, or the empty vector and then identified with Western blots with anti-FatB serum. Figure 6 shows that C23A FatB fractionates now in the periplasm, whereas the wild-type FatB is not detected in this fraction. The wild-type FatB as well as C23A FatB can also be found in membrane fractions. Furthermore, in the total protein fraction C23A FatB has a higher molecular weight than the wild-type FatB possibly due to the lack of processing by the signal peptidase II. From these results we can speculate that the loss of the signal peptidase II recognition site due to the C23A mutation might have resulted in signal peptidase I cleavage of the C23A FatB N-terminal ends as reported in various systems (Cavard et al., 1987; Kempf et al., 1997; Yerushalmi et al., 2005). However, it is clear that the signal peptidase I cleavage is not perfect leading to the detection of C23A FatB in membrane factions. The prediction of the signal peptidase recognition site using the lipoP program (Juncker et al., 2003) showed that the change from C23 to A could lead to the creation of a signal peptidase I cleavage site between position 23 and 24 in C23A FatB.
Fig. 6.
Western blot detection of FatB in cell fractions of V. anguillarum carrying either the wild-type FatB or C23A FatB. Periplasmic proteins were extracted as described by Wunderlich and colleagues (1993) with some minor modifications. Briefly, an exponential phase bacterial culture in CM9 minimal medium with 10 μg ml−1 Cm and 0.5 mM IPTG was centrifuged. The pellets were washed with CM9 minimal medium and resuspended in 2 ml BBS/EDDA (200 mM boric acid/NaOH, pH 8.0, 160 mM NaCl, 1 mM EDTA) per gram cell. The suspension was incubated for 45 min at 4°C with gentle agitation, and centrifuged (27 000 g, 1 h, 4°C) to pellet the spheroplasts. The supernatant containing periplasmic proteins was carefully transferred into new tubes. To obtain total protein the bacterial cells prepared as described above were resuspended into 10 mM Tris buffer (pH 7.6), sonicated 5 × 5 s and centrifuged at 15 000 r.p.m. at 4°C for 5 min. Supernatants were transferred into new tubes and used as total proteins. Membrane proteins were obtained by centrifuging the total proteins for 1 h at 30 000 g, and after resuspension this step was repeated. The presence of FatB or C23A FatB in the periplasm fractions and in total proteins was determined by Western blotting using anti-FatB serum.
The next question was whether C23A FatB is still able to operate in ferric anguibactin transport. Mutation of the cysteine residue, indispensable for the lipoprotein nature, has been reported in other systems as well. In some cases the cysteine mutation affects the function of the protein (Cavard et al., 1987; Luirink et al., 1988; Fernandez et al., 1996), while in other cases the mutation does not affect or partially affect the function of proteins (Pugsley and Cole, 1987; Kornacker et al., 1991; Zhang et al., 1992; Yerushalmi et al., 2005). pMMB208 harbouring the wild-type fatB and C23A fatB were, respectively, conjugated into the null fatB mutant in a CC9-16 genetic background and bioassays were conducted. We observe in Fig. 3 that C23A fatB and the wild-type fatB complemented the null fatB mutant, showing growth halos around the spots with anguibactin. Negative and positive controls gave the expected results (see Fig. 3). From this experiment it is clear that C23A FatB is still able to function in ferric anguibactin transport. To further demonstrate that the FatB wild-type and C23A proteins behaved similarly at the transport level, we performed an iron uptake experiment as described previously (Crosa and Hodges, 1981; Welch and Crosa, 2005). The results in Fig. 4 clearly demonstrate that the wild-type FatB and C23A FatB transported the radioactive ferric anguibactin with similar kinetics. From these results we conclude that the lipoprotein nature of FatB is not essential for ferric anguibactin transport.
It is not clear what is the reason for the lipoprotein nature of FatB and its binding to the cytoplasmic membrane of the Gram-negative V. anguillarum. We recently demonstrated that the high affinity pJM1-encoded siderophore anguibactin system might have been acquired at some point during evolution, possibly horizontally (Alice et al., 2005; Naka et al., 2008), thus the FatB lipoprotein could have originated in bacteria in which the embedding of the periplasmic domain as a lipoprotein is essential for their life cycle.
Supplementary Material
Acknowledgements
This work was supported by Grant AI19018 from the National Institutes of Health to J.H.C.
Footnotes
Supporting information
Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Actis LA, Potter SA, Crosa JH. Iron-regulated outer membrane protein OM2 of Vibrio anguillarum is encoded by virulence plasmid pJM1. J Bacteriol. 1985;161:736–742. doi: 10.1128/jb.161.2.736-742.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Actis LA, Tolmasky ME, Farrell DH, Crosa JH. Genetic and molecular characterization of essential components of the Vibrio anguillarum plasmid-mediated iron-transport system. J Biol Chem. 1988;263:2853–2860. [PubMed] [Google Scholar]
- Actis LA, Tolmasky ME, Crosa LM, Crosa JH. Characterization and regulation of the expression of FatB, an iron transport protein encoded by the pJM1 virulence plasmid. Mol Microbiol. 1995;17:197–204. doi: 10.1111/j.1365-2958.1995.mmi_17010197.x. [DOI] [PubMed] [Google Scholar]
- Actis LA, Tolmasky ME, Crosa JH. Vibriosis. Vol. 3. Cab International Publishing; Wallingford, UK: 1999. [Google Scholar]
- Alice AF, Lopez CS, Crosa JH. Plasmid- and chromosome-encoded redundant and specific functions are involved in biosynthesis of the siderophore anguibactin in Vibrio anguillarum 775: a case of chance and necessity? J Bacteriol. 2005;187:2209–2214. doi: 10.1128/JB.187.6.2209-2214.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V, Wu HC. Lipoproteins: structure, function, biosynthesis and model for protein export. New Compr Biochem. 1994;27:319–341. [Google Scholar]
- Cavard D, Baty D, Howard SP, Verheij HM, Lazdunski C. Lipoprotein nature of the colicin A lysis protein: effect of amino acid substitutions at the site of modification and processing. J Bacteriol. 1987;169:2187–2194. doi: 10.1128/jb.169.5.2187-2194.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Wertheimer AM, Tolmasky ME, Crosa JH. The AngR protein and the siderophore anguibactin positively regulate the expression of iron-transport genes in Vibrio anguillarum. Mol Microbiol. 1996;22:127–134. doi: 10.1111/j.1365-2958.1996.tb02662.x. [DOI] [PubMed] [Google Scholar]
- Crosa JH. A plasmid associated with virulence in the marine fish pathogen Vibrio anguillarum specifies an iron-sequestering system. Nature. 1980;284:566–568. doi: 10.1038/284566a0. [DOI] [PubMed] [Google Scholar]
- Crosa JH, Hodges LL. Outer membrane proteins induced under conditions of iron limitation in the marine fish pathogen Vibrio anguillarum 775. Infect Immun. 1981;31:223–227. doi: 10.1128/iai.31.1.223-227.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Lorenzo M, Stork M, Tolmasky ME, Actis LA, Farrell D, Welch TJ, et al. Complete sequence of virulence plasmid pJM1 from the marine fish pathogen Vibrio anguillarum strain 775. J Bacteriol. 2003;185:5822–5830. doi: 10.1128/JB.185.19.5822-5830.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell DH, Mikesell P, Actis LA, Crosa JH. A regulatory gene, angR, of the iron uptake system of Vibrio anguillarum: similarity with phage P22 cro and regulation by iron. Gene. 1990;86:45–51. doi: 10.1016/0378-1119(90)90112-5. [DOI] [PubMed] [Google Scholar]
- Fernandez D, Dang TA, Spudich GM, Zhou XR, Berger BR, Christie PJ. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J Bacteriol. 1996;178:3156–3167. doi: 10.1128/jb.178.11.3156-3167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings MI, Palmer T, Harrington DJ, Sutcliffe IC. Lipoprotein biogenesis in Gram-positive bacteria: knowing when to hold 'em, knowing when to fold 'em. Trends Microbiol. 2009;17:13–21. doi: 10.1016/j.tim.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12:1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf B, Gade J, Bremer E. Lipoprotein from the osmoregulated ABC transport system OpuA of Bacillus subtilis: purification of the glycine betaine binding protein and characterization of a functional lipidless mutant. J Bacteriol. 1997;179:6213–6220. doi: 10.1128/jb.179.20.6213-6220.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornacker MG, Faucher D, Pugsley AP. Outer membrane translocation of the extracellular enzyme pullulanase in Escherichia coli K12 does not require a fatty acylated N-terminal cysteine. J Biol Chem. 1991;266:13842–13848. [PubMed] [Google Scholar]
- Koster WL, Actis LA, Waldbeser LS, Tolmasky ME, Crosa JH. Molecular characterization of the iron transport system mediated by the pJM1 plasmid in Vibrio anguillarum 775. J Biol Chem. 1991;266:23829–23833. [PubMed] [Google Scholar]
- Lopez CS, Crosa JH. Characterization of ferric-anguibactin transport in Vibrio anguillarum. Biometals. 2007;20:393–403. doi: 10.1007/s10534-007-9084-9. [DOI] [PubMed] [Google Scholar]
- Lopez CS, Alice AF, Chakraborty R, Crosa JH. Identification of amino acid residues required for ferric-anguibactin transport in the outer-membrane receptor FatA of Vibrio anguillarum. Microbiology. 2007;153:570–584. doi: 10.1099/mic.0.2006/001735-0. [DOI] [PubMed] [Google Scholar]
- Lopez CS, Peacock RS, Crosa JH, Vogel HJ. Molecular characterization of the TonB2 protein from the fish pathogen Vibrio anguillarum. Biochem J. 2009;418:49–59. doi: 10.1042/BJ20081462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luirink J, Hayashi S, Wu HC, Kater MM, de Graaf FK, Oudega B. Effect of a mutation preventing lipid modification on localization of the pCloDF13-encoded bacteriocin release protein and on release of cloacin DF13. J Bacteriol. 1988;170:4153–4160. doi: 10.1128/jb.170.9.4153-4160.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton DL, O'Toole R, Horstedt P, Wolf-Watz H. Flagellin A is essential for the virulence of Vibrio anguillarum. J Bacteriol. 1996;178:1310–1319. doi: 10.1128/jb.178.5.1310-1319.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales VM, Backman A, Bagdasarian M. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene. 1991;97:39–47. doi: 10.1016/0378-1119(91)90007-x. [DOI] [PubMed] [Google Scholar]
- Naka H, Lopez CS, Crosa JH. Reactivation of the vanchrobactin siderophore system of Vibrio anguillarum by removal of a chromosomal insertion sequence originated in plasmid pJM1 encoding the anguibactin siderophore system. Environ Microbiol. 2008;10:265–277. doi: 10.1111/j.1462-2920.2007.01450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Isolation of outer membranes. Methods Enzymol. 1994;235:225–234. doi: 10.1016/0076-6879(94)35143-0. [DOI] [PubMed] [Google Scholar]
- Osborn MJ, Gander JE, Parisi E, Carson J. Mechinism of assembly of the outer membrane of Salmonella typhimurium. Isolation and characterization of cytoplasmic and outer membrane. J Biol Chem. 1972;247:3962–3972. [PubMed] [Google Scholar]
- Park SF, Richardson PT. Molecular characterization of a Campylobacter jejuni lipoprotein with homology to periplasmic siderophore-binding proteins. J Bacteriol. 1995;177:2259–2264. doi: 10.1128/jb.177.9.2259-2264.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley AP, Cole ST. An unmodified form of the ColE2 lysis protein, an envelope lipoprotein, retains reduced ability to promote colicin E2 release and lysis of producing cells. J Gen Microbiol. 1987;133:2411–2420. doi: 10.1099/00221287-133-9-2411. [DOI] [PubMed] [Google Scholar]
- Richardson PT, Park SF. Enterochelin acquisition in Campylobacter coli: characterization of components of a binding-protein-dependent transport system. Microbiology. 1995;141:3181–3191. doi: 10.1099/13500872-141-12-3181. [DOI] [PubMed] [Google Scholar]
- Salinas PC, Crosa JH. Regulation of angR, a gene with regulatory and biosynthetic functions in the pJM1 plasmid-mediated iron uptake system of Vibrio anguillarum. Gene. 1995;160:17–23. doi: 10.1016/0378-1119(95)00213-p. [DOI] [PubMed] [Google Scholar]
- Salinas PC, Tolmasky ME, Crosa JH. Regulation of the iron uptake system in Vibrio anguillarum: evidence for a cooperative effect between two transcriptional activators. Proc Natl Acad Sci USA. 1989;86:3529–3533. doi: 10.1073/pnas.86.10.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Hantke K. Iron-hydroxamate uptake systems in Bacillus subtilis: identification of a lipoprotein as part of a binding protein-dependent transport system. Mol Microbiol. 1993;8:111–121. doi: 10.1111/j.1365-2958.1993.tb01208.x. [DOI] [PubMed] [Google Scholar]
- Senanayake SD, Brian DA. Precise large deletions by the PCR-based overlap extension method. Mol Biotechnol. 1995;4:13–15. doi: 10.1007/BF02907467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- Tolmasky ME, Actis LA, Crosa JH. Genetic analysis of the iron uptake region of the Vibrio anguillarum plasmid pJM1: molecular cloning of genetic determinants encoding a novel trans activator of siderophore biosynthesis. J Bacteriol. 1988;170:1913–1919. doi: 10.1128/jb.170.4.1913-1919.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmasky ME, Actis LA, Crosa JH. A single amino acid change in AngR, a protein encoded by pJM1-like virulence plasmids, results in hyperproduction of anguibactin. Infect Immun. 1993;61:3228–3233. doi: 10.1128/iai.61.8.3228-3233.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MA, Potter SA, Crosa JH. Iron uptake system medicated by Vibrio anguillarum plasmid pJM1. J Bacteriol. 1983;156:880–887. doi: 10.1128/jb.156.2.880-887.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch TJ, Crosa JH. Novel role of the lipopolysaccharide O1 side chain in ferric siderophore transport and virulence of Vibrio anguillarum. Infect Immun. 2005;73:5864–5872. doi: 10.1128/IAI.73.9.5864-5872.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheimer AM, Verweij W, Chen Q, Crosa LM, Nagasawa M, Tolmasky ME, et al. Characterization of the angR gene of Vibrio anguillarum: essential role in virulence. Infect Immun. 1999;67:6496–6509. doi: 10.1128/iai.67.12.6496-6509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich M, Jaenicke R, Glockshuber R. The redox properties of protein disulfide isomerase (DsbA) of Escherichia coli result from a tense conformation of its oxidized form. J Mol Biol. 1993;233:559–566. doi: 10.1006/jmbi.1993.1535. [DOI] [PubMed] [Google Scholar]
- Wyckoff EE, Valle AM, Smith SL, Payne SM. A multifunctional ATP-binding cassette transporter system from Vibrio cholerae transports vibriobactin and enterobactin. J Bacteriol. 1999;181:7588–7596. doi: 10.1128/jb.181.24.7588-7596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yerushalmi G, Zusman T, Segal G. Additive effect on intracellular growth by Legionella pneumophila Icm/Dot proteins containing a lipobox motif. Infect Immun. 2005;73:7578–7587. doi: 10.1128/IAI.73.11.7578-7587.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WY, Inouye M, Wu HC. Neither lipid modification nor processing of prolipoprotein is essential for the formation of murein-bound lipoprotein in Escherichia coli. J Biol Chem. 1992;267:19631–19635. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.