Abstract
In medical research, it is common to have doubly censored survival data: origin time and event time are both subject to censoring. In this paper, we review simple and probability-based methods that are used to impute interval censored origin time and compare the performance of these methods through extensive simulations in the one-sample problem, two-sample problem and Cox regression model problem. The use of a bootstrap procedure for inference is demonstrated.
Keywords: bootstrap, Cox regression model, interval censoring, Kaplan–Meier curve, logrank test
1. Introduction
Most statistical methods developed for the analysis of event time data assume that the origin time is known, but allow the event to be censored. Data that are censored both at the origin and at the event time are referred to as doubly censored data. HIV studies have provided many examples for doubly censored data. In this paper, we are interested in the distribution of time from HIV infection to death. The exact time of HIV infection is usually interval censored and death is subject to right censoring. This is the doubly censored situation considered here. However, that the term ‘doubly censored data’ is also used for situations where both the origin and the event time are interval-censored, for example, in De Gruttola and Lagakos [1] and Sun [2].
Doubly censored data can, in principle, be analysed using a maximum likelihood approach, but this approach can be challenging, both numerically and theoretically, particularly when covariates are involved. Maximum likelihood has been applied to the regression analysis of doubly censored data in Kim et al. [3], with both origin and event time being interval censored, using the discrete proportional hazards model. For the continuous proportional hazards model, Sun et al. [4] propose an estimating equation procedure to estimate the regression parameters and show that the estimator is asymptotically unbiased and normally distributed. The procedure is difficult to implement and can be intractable when the sample size is large. In addition, the method is challenging to implement when the covariates are interval censored, as is the case in our motivating example of Xiang et al. [5]. In contrast, if the origin time (HIV infection time) can be imputed reasonably, the missing value of the covariate for this study (age at the time of HIV infection) will be imputed simultaneously, then the analysis of doubly censored data with imputation is a straightforward analysis of the right-censored data.
Imputation is a general method for missing-data problems. One simple approach is to impute infection time using the right limit of the interval in which the infection time is censored. This typically corresponds to the date of diagnosis or to the date of study entry, and is expedient when no negative diagnostic test precedes the first positive test, as in Xiang et al. [5] and Tillmann et al. [6]. Another common approach is to impute infection time using the midpoint of the interval [7–9]. Law and Brookmeyer [10], however, have shown that, under certain distributional assumptions consistent with the data from the studies of HIV disease, Kaplan–Meier estimates of survival based on this method are considerably biased when censoring intervals are longer than two years. Yet another method of imputation is by the left limit of the interval. In many HIV studies, however, including our motivating example, the left limit may correspond to either date of birth or a date before the HIV epidemic emerged, and in this case the left-point imputation is likely to be unreasonable. For this reason, the left-limit imputation is not included in the following sections.
Other imputation methods impute the infection time of a subject based on Ĝ, an estimate of the marginal distribution G of HIV infection time. For example, Gauvreau et al. [11] adopt the self-consistency algorithm of Turnbull [12] to estimate G, and then impute the expected infection time based on Ĝ conditional on the subject’s interval. Goggins et al. [13] suggest a Monte Carlo EM algorithm to estimate G, and then repeatedly impute infection times based on random draws from Ĝ conditional on the subjects’ intervals. The estimated distribution of HIV infection time, Ĝ, is treated as if known when imputing infection times.
Pan [14] uses the approximate Bayesian bootstrap scheme [15,16] to take B bootstrap samples Db from the original data D, b = 1, …, B, then obtain Ĝb using the self-consistency algorithm, and then repeatedly impute B infection times based on random draws from Ĝb, b = 1, …, B. Finally, the results are combined using Rubin’s multiple imputation (MI) formula [17]. Geskus [18] compares the midpoint imputation, the conditional mean imputation, and MI methods for the bias and mean-squared error (MSE) of the estimator of Kaplan–Meier curves. In his simulation study, under some distributional assumptions for one-sample data, the conditional mean imputation stands out as the preferred method.
In Section 2, both simple imputations and probability-based imputations are outlined, and the MI inference procedure and bootstrap inference procedure are introduced. In Sections 3 and 4, simulations are described and the numerical performance of the different imputation methods is compared. Section 5 presents conclusions and further discussion.
2. Imputation methods
For simplicity, let HIV infection be the origin event and death the endpoint event. Let Xi and Yi denote HIV infection time and death time for subject i, i = 1, …, n. Assume Xi is interval censored Xi ∈ [Li, Ri]. We assume Yi is possibly right censored as in Sun et al. [4], Goggins et al. [13], and Pan [14]. Imputation methods can be classified into simple imputation methods and probability-based imputation methods.
2.1. Simple imputation methods
Right-point imputation refers to imputing the infection time by the right limit Ri of the interval and is denoted by RIGHT; midpoint imputation refers to imputing the infection time by the midpoint of the interval [Li, Ri] as (Li + Ri)/2 and is denoted by MID. When HIV infection and death both occur between two successive screening tests, that is Li < Xi < Yi < Ri, the MID uses midpoint of the interval [Li, Yi] as the infection time.
2.2. Probability-based imputation methods
Probability-based imputation requires estimating the distribution G for HIV infection time Xi based on observed intervals. The non-parametric maximum likelihood estimator (NPMLE) of G with interval censored data is fully developed in the statistical literatures. Groeneboom and Wellner [19] characterize the NPMLE and propose an iterative convex minorant algorithm for computing the estimate. Turnbull [12] proposes a self-consistency algorithm which can be realized as an application of the EM algorithm introduced by Dempster et al. [20]. The details of these algorithms can be found in Sun [21]. Turnbull’s self-consistency algorithm is used throughout this paper due to its simplicity of implementation.
Assume that the infection time X is a discrete random variable with a set of possible values x = {x1, x2, …, xm} associated with a set of probabilities g = (g1, g2, …, gm), respectively, where x1 < x2 < … < xm. Suppose for subject i, there are ri possible values of infection time yi = (yi1, …, y) ∈ [Li, Ri] associated with probabilities pi = (pi1, …, piri), i = 1, …, n. Note that both yi and pi are subsets of x and g, respectively. Let hi = (hi1, …, hiri), the conditional probability for subject i taking the value is yik is , k = 1, …, ri, conditioning on the interval [Li, Ri] and g.
Conditional mean imputation has been previously used [11,18] but conditional median and conditional mode appear to be new methods for imputation.
2.2.1. Conditional mean imputation (MEAN)
For subject i, the expected time of infection is . Therefore, infection time Xi can be imputed by .
2.2.2. Conditional median imputation (MEDIAN)
Infection time Xi is imputed by the median of yi weighted by the probability vector hi. In case the median is not unique, X̂i is taken as the average of medians.
2.2.3. Conditional mode imputation (MODE)
Infection time Xi is imputed by the mode of yi: the value corresponding to the maximum probability among hi. That is X̂i = yik, where k =max1≤k≤ri {ĥik}. In case the mode is not unique, X̂i is taken as the average of modes.
2.2.4. Multiple imputation
MI is a commonly used method. For m = 1, …, M, randomly sample from yi with replacement using the conditional probability vector hi as weight. Let D denote the original data set with interval-censored origin event, D̂m denote the data set that replaces the interval censored origin event by the mth imputation. Then D̂m is analysed using the regular right censored data method. Let θ̂m be the estimate of the parameter of interest obtained from the mth imputed data set D̂m, m = 1, …, M. The MI estimate of θ is .
2.2.5. Random imputation (RAND)
Randomly sample one value of yik from the vector yi using the conditional probability vector hi as weight. Then the infection time Xi is imputed by yik. This is a special case of MI, where M = 1.
2.3. Bootstrap inference procedure
The imputation methods in the previous section provide ways to estimate population parameters of interest for doubly censored data. To derive standard errors, Rubin’s variance estimation formula [17] has been used in MI inference [14,22,23]. The formula adds an expression for between-imputation variance to an expression for average within-imputation variance, to incorporate imputation uncertainty. An alternative bootstrap inference procedure for doubly censored data is introduced here.
Suppose B bootstrap samples Db, b = 1, …, B, are generated from the doubly censored data D. An estimate θ̂ of parameter θ is computed based on the imputed data D̂ by imputing infection times using a method described in Sections 2.1 and 2.2. A 100(1 − α)% empirical bootstrap confidence internal (EBCI) of θ for the chosen imputation procedure is given by [θ̂l, θ̂u], where θ̂l and θ̂u are the empirical 100(α/2) and 100(1 − α/2) percentiles of the bootstrap distribution of θ̂ [24].
Since estimation with right-censored data consumes very little in computing time, the bootstrap procedure for doubly censored data described above will not be computationally intensive, making it potentially attractive in practice.
2.4. Motivating example
Xiang et al. [5] examined the effect of co-infection with GBV-C virus on the survival of HIV-infected patients. The data set is doubly censored in that the origin time (HIV infection) is interval censored and the endpoint event (death) is right censored. The date of subjects’ first known positive HIV test is used as the right limit of the interval: the right limit ranges from 1988 to 1999. 1 January 1978 (or date of birth for subjects born after 1 January 1978) is treated as the left limit of the interval, because it is reasonable to assume that no HIV infections occurred prior to 1 January 1978 in this population [25]. The data set has 362 subjects with mean interval width of 11.4 years.
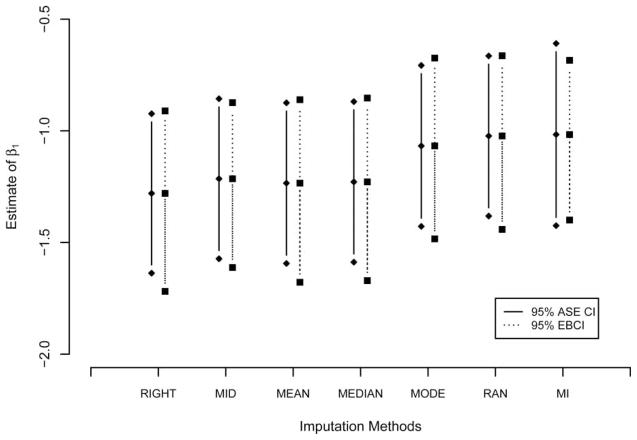
We applied all seven imputation methods described in Section 2. Results are summarized in Figure 1. The estimate of β1, log(hazard ratio) of GBV-C co-infection, varies from − 1.0 to − 1.3 based on different imputation methods. For all but MI the 95% asymptotic standard error (ASE) CI of β1 is the confidence interval based on the ASE of β1 from the Cox model, treating imputed date as if it were known. The 95% ASE CI underestimates the variability of β1 by ignoring the imputation uncertainty and the 95% EBCI is wider than the 95% ASE CI for every imputation method except for MI. In MI, an ASE is computed based on Rubin’s variance formula [17] to attempt to account for imputation uncertainty and only in this case is the 95% ASE CI wider than the 95% CI.
Figure 1.
Estimate of Cox regression coefficient for the Xiang study by seven imputation methods.
To assess the performance of point estimates based on these seven imputation methods and the validity of using bootstrap inference for doubly censored data, simulation studies are implemented. Section 3 describes the design of the simulations, and Section 4 presents the results.
3. Simulation design
Simulation studies are designed: (1) to evaluate the Kaplan–Meier estimator in the one-sample problem, (2) to evaluate the size and power of the logrank test in the two-sample problem and also a new bootstrap-based test which is introduced in Section 4.2, and (3) to evaluate the regression coefficient estimate from the Cox proportional hazards model [26] adjusting for an interval-censored covariate (to be consistent with the motivating example), based on seven imputation methods. We also assess the validity of the bootstrap inference procedure through simulation studies.
Distributions for the infection time X and the subsequent survival time T are the key parts of simulation studies for doubly censored data. Law and Brookmeyer [10] assume a log-logistic distribution for HIV infection time X with restriction X ∈ [1978, 1986] (subjects involved in their study were exposed to HIV from early 1978 to mid-1985) and a Weibull distribution W (2.51, 11.66) for survival time T, to evaluate the effect of midpoint imputation on the Kaplan–Meier estimator and logrank test. In a study to assess the effect of a binary covariate using the Cox model, Goggins et al. [13] adopt a log-normal distribution LN(3.8, 0.3) for X, and simulate survival time T1 for one group from W (2.5, 70.1) and survival time T2 for another group from W (2.5, 60) so that the logarithm of hazard ratio β1 is 0.389. This mimics the haemophilia data described by Kim et al. [2]. To investigate RIGHT, MID, and MEAN imputation methods for Kaplan–Meier estimator, Geskus [18] specifies a shifted log-logistic distribution for the infection time X, X ∈ [1980, 1997] and W (2, 11) for survival time T. Pan [14] uses a similar simulation design to that in Goggins et al. [13].
3.1. Distribution for X and interval censoring
The variable X is simulated from a log-normal distribution as in Goggins et al. [13] and Pan [14], and is truncated with an upper limit of 65 and with parameters chosen so that the simulated data are similar to the real data in Xiang et al. [5]. Specifically, X is distributed as log-normal LN(3.55, 0.24) and truncated to [0, 65]. To mimic screening studies, we simulate a subject’s first visit as a random number from a uniform distribution U (0, 5). After the first visit, each subject is scheduled to have annual follow-ups. Whether or not a subject completes each annual follow-up is modelled as an independent Bernoulli variable. The probability of making an annual visit P can be tuned to result in intervals with specified average censoring width of w years for X. A subject’s HIV infection time Xi is accordingly censored between two consecutive visits, Xi ∈ [Li, Ri], i = 1, …, n. These intervals are then used to obtain an NPMLE Ĝ of G using Turnbull’s self-consistency algorithm. For convenience, we refer to the simulation setting described here as the GA setting.
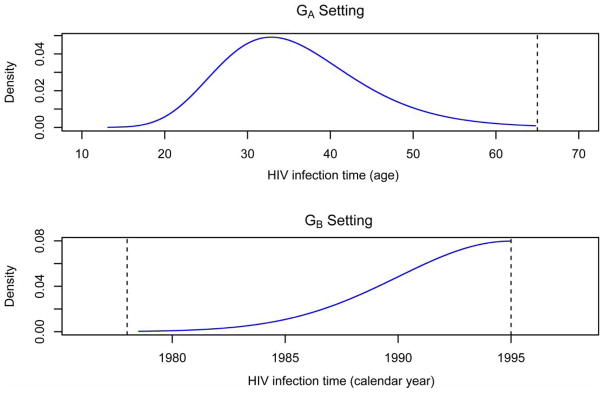
Suppose HIV-positive subjects entered the HIV study before the year 1995. Given HIV infections before 1978 are extremely rare [25] it is reasonable to assume the HIV infection time X is between 1978 and 1995. The HIV infection time X is simulated from a truncated normal distribution N(1995, 5) with upper limit 1995. Assume that for half of the subjects, we are able to establish intervals for HIV infection time based on their annual seronegative tests. For these subjects, censoring intervals are generated using the algorithm described in the above paragraph. The NPMLE Ĝ for the distribution of X is estimated using only these subjects. For the other half of the subjects, it is known that they were HIV-positive only at the time of entry. If an individual was born before 1978, we use 1978 as the left limit, otherwise we use his/her birth year as this person’s left limit. For convenience, we refer the simulation setting described here as the GB setting. Table 1 summarizes both GA and GB settings, and Figure 2 portrays the distribution of X in these two settings.
Table 1.
Two simulation settings: GA and GB.
| Specifications | GA setting | GB setting |
|---|---|---|
| Sample size | n | n |
| Dist. of X | LN(3.55, 0.24) | N(1995, 5) |
| Range of X | X ∈ [0, 65] | X ∈ [1978, 1995] |
| NPMLE Ĝ | based on n intervals | based on n/2 intervals |
| Dist. of T | ||
| One-sample | W (2, 10) | W (2, 10) |
| Two-sample | (1) W (2, 10) vs. W (2, 12.84) | W (2, 10) vs. W (2, 12.84) |
| (2) LN(2.2, 04) vs. LN(2.4, 0.4) | LN(2.2, 04) vs. LN(2.4, 0.4) | |
| Cox model | W (2, 10 · exp(−zβ/2)) | W (2, 10 · exp(−zβ/2)) |
| Right censoring for T | 10% | 10% |
Figure 2.
True distribution for HIV infection time X in two simulation settings.
3.2. Distribution for T and right censoring
For the one-sample problem, the survival time T is simulated from a Weibull distribution W (2, 10). For the two-sample problem, a binary covariate is used to indicate group membership. T is simulated from two different distributions for two groups with equal sample size n/2. Distributions considered for the two groups include Weibull W (2, 10) vs. W (2, 12.84), and log-normal LN(2.2, 0.4) vs. LN(2.4, 0.4). To compare the size of a test resulting from different imputation methods, T is simulated from the same distribution for the two groups with equal sample size n/2, either W (2, 10) or LN(2.2,0.4).
For the Cox regression problem, survival time T is simulated from the distribution W (γ, λ) where λ = λ0 · exp(−zβ/γ), resulting in a proportional hazards model with log (H R) = β. To mimic the study by Xiang et al., we let z = (z1, z2) denote the GBV-C co-infection (yes/no) and age at HIV infection, respectively, with corresponding coefficients β = (β1, β2) = (−0.5, 0.1). We set γ = 2 and λ0 = 10.
In all scenarios, a censoring random variable C is simulated from W (γ, a · λ), where a is a positive coefficient. As in Goggins et al. [13], Sun et al. [4] and Pan [14], the survival time T is subject to right censoring. The coefficient a can be tuned so that Pr(T ≤ C) = 0.9, that is T is subject to 10% random right-censoring.
For each problem, 1000 independent doubly censored data sets are repeatedly simulated. For each simulated data set, 1000 bootstrap data sets are generated to obtain the 95% EBCI for the parameter of interest. Specifically, in the one-sample problem, the pointwise 95% EBCI for the survival function is calculated; in the Cox regression model, the 95% EBCI for β1 is constructed. To facilitate comparison, the results based on the data with exact HIV infection time X and the right-censored survival time T are also included (these data are refered to as exact data). With MI for the missing data problems, Rubin [17, p. 114] shows that in most situations there is little advantage in producing and analysing more than a few imputed data sets, and claims that only 3–10 imputations may be needed. For the doubly censored data, Pan [23] suggests that M = 5 or M = 10 would suffice. We use M = 10 for the MI method. The RAND method corresponds to M = 1.
4. Simulation results
4.1. One-sample problem
The survival probability S(t) at 2.5, 5, 7.5, and 10 years after HIV infection is estimated by the Kaplan–Meier estimator, using different imputation methods to impute the infection time X. The probability of making an annual visit is chosen to be P = 0.3, which results in an average interval width about 5.3 years. Seven imputation methods are compared with respect to bias, mean squared error (MSE) and coverage probability of 95% EBCI.
Table 2 summarizes results for the Kaplan–Meier estimator of survival function for n = 200 in the GA and GB settings, with the results for the GB setting displayed in parentheses. The true values for the S(t) at (2.5, 5, 7.5, 10) years after HIV infection are (0.94, 0.78, 0.57, 0.37), respectively. In the GA setting, all imputation methods give similar bias and MSE except RIGHT for which the bias and MSE is much larger. All imputation methods except the methods RIGHT and MI give acceptable coverage probability of 95% EBCI. Methods MID and MEAN have the smallest biases, followed by MEDIAN. MEAN has smaller bias than MID in the early years, but MID has smaller bias at, and after 7.5 years. MID, MEAN, and MEDIAN have comparable MSEs to that of the exact data. In the GB setting, the probability-based imputation methods perform better than the simple imputation methods for the Kaplan–Meier estimator in terms of bias, MSE, and coverage probability of 95% EBCI. Indeed, both RIGHT and MID work badly for the GB setting.
Table 2.
Comparison of imputation methods for the Kaplan–Meier estimator with heavy interval censoring.
| Imputation | Years after HIV infection |
|||
|---|---|---|---|---|
| 2.5 | 5 | 7.5 | 10 | |
| S(t) = 0.9394 | 0.7788 | 0.5698 | 0.3679 | |
| Bias ×102 | ||||
| EXACT | 0.00 (0.05) | −0.05 (0.05) | 0.01 (−0.02) | −0.16 (−0.07) |
| RIGHT | 18.85 (13.92) | 21.31 (16.85) | 18.97 (15.63) | 14.23 (11.98) |
| MID | −0.22 (−3.30) | 0.26 (−11.55) | −0.65 (−21.11) | −1.59 (−25.46) |
| MEAN | −0.15 (−2.17) | 0.02 (−4.71) | −1.12 (−7.29) | −1.99 (−7.61) |
| MEDIAN | 0.43 (−0.95) | 0.09 (−2.63) | −1.26 (−4.62) | −2.28 (−5.08) |
| MODE | 2.29 (2.75) | 0.89 (1.37) | −1.35 (−1.24) | −2.95 (−3.13) |
| RAND | 2.42 (0.43) | 1.16 (−2.73) | −1.02 (−6.18) | −2.91 (−7.91) |
| MI | 2.44 (0.54) | 1.12 (−2.77) | −1.15 (−6.27) | −2.87 (−8.01) |
| MSE ×102 | ||||
| EXACT | 0.03 (0.03) | 0.09 (0.09) | 0.14 (0.13) | 0.13 (0.12) |
| RIGHT | 3.65 (2.02) | 4.68 (2.97) | 3.73 (2.57) | 2.12 (1.54) |
| MID | 0.03 (0.12) | 0.08 (1.38) | 0.14 (4.52) | 0.16 (6.58) |
| MEAN | 0.03 (0.07) | 0.09 (0.30) | 0.15 (0.67) | 0.18 (0.72) |
| MEDIAN | 0.03 (0.04) | 0.09 (0.17) | 0.15 (0.36) | 0.19 (0.41) |
| MODE | 0.10 (0.15) | 0.12 (0.27) | 0.19 (0.46) | 0.25 (0.62) |
| RAND | 0.10 (0.03) | 0.10 (0.16) | 0.15 (0.51) | 0.22 (0.77) |
| MI | 0.08 (0.02) | 0.08 (0.14) | 0.12 (0.50) | 0.19 (0.75) |
| Coverage probability of 95% EBCI | ||||
| RIGHT | 0.0 (0.0) | 0.0 (0.1) | 0.1 (0.9) | 1.2 (5.1) |
| MID | 91.7 (22.8) | 94.7 (0.1) | 92.3 (0.0) | 90.7 (0.0) |
| MEAN | 97.8 (70.2) | 96.3 (62.3) | 93.6 (47.8) | 90.2 (45.4) |
| MEDIAN | 99.2 (94.6) | 97.6 (86.2) | 94.0 (76.1) | 90.6 (72.0) |
| MODE | 96.3 (99.7) | 99.0 (99.8) | 98.0 (99.2) | 93.3 (98.5) |
| RAND | 89.7 (98.9) | 97.3 (85.2) | 95.3 (56.5) | 88.3 (38.6) |
| MI | 74.3 (93.8) | 94.6 (81.8) | 92.2 (51.4) | 84.4 (33.1) |
Note: Numbers not in parenthesis are based on GA setting; numbers in parenthesis are based on GB setting. Sample size n = 200, 1000 simulated data sets, and 1000 bootstraps per simulated dataset.
4.2. Two-sample problem
Let F1 and F2 be the distribution functions for groups 1 and 2, respectively. For the right-censored data, under the null hypothesis H0: F1 = F2, the logrank test statistic S is asymptotically a standard normal N(0, 1) random variable. There are two tests for which the power and size can be estimated. One is for the regular logrank test by ignoring the fact that the origin time is imputed. The second is a new test that incorporates the double censoring, where the asymptotic distribution of the logrank statistic S is not known. The latter test is based on the bootstrap empirical distribution of the logrank statistic. For convenience, we call the latter the empirical logrank (ELR) test. The power for the ELR test is defined as the probability that the 100(1 − α)% EBCI of S exclude 0, based on the data simulated under an alternative hypothesis H1: F1 ≠ F2. The size for the ELR test is defined as the probability that the 100(1 − α)% EBCI of S exclude 0, based on the data simulated under the null hypothesis H0.
The results for power and size comparison in the GA and GB settings are shown in Table 3. In the GA setting, the probability of making an annual visit P is set as 0.3 for both groups resulting in a mean interval width w = 5 years for each group. For each imputation method, the power of the ELR test and of the regular logrank test is similar. The power based on MODE and RAND tends to be smaller than that from the other imputation methods. Overall, the loss in power using MEAN imputation is negligible compared to the EXACT approach where the original time is known. The size of the logrank test is close to the 5% nominal level. The size of the ELR test is also close to the 5% nominal level, except that the size based on MODE or RAND tends to be lower than nominal. Overall, the size of both tests based on MEDIAN is closer to the 5% nominal level than that of other imputation methods.
Table 3.
Comparison of imputation methods for power and size of two-sample tests with heavy interval censoring.
| Distribution of T | Imputation methods |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Test | EXACT | RIGHT | MID | MEAN | MEDIAN | MODE | RAND | MI | |
| Power | |||||||||
| W (2, 10) vs. W (2, 12.84) | Logrank | 0.895 | 0.872 | 0.871 | 0.868 | 0.865 | 0.851 | 0.845 | 0.877 |
| ELR | – | 0.871 | 0.866 | 0.871 | 0.860 | 0.844 | 0.862 | 0.874 | |
| Logrank | (0.911) | (0.892) | (0.809) | (0.893) | (0.899) | (0.845) | (0.836) | (0.894) | |
| ELR | (−) | (0.892) | (0.814) | (0.892) | (0.894) | (0.858) | (0.856) | (0.892) | |
| LN (2.2, 0.4) vs. LN(2.4, 0.4) | Logrank | 0.882 | 0.800 | 0.828 | 0.833 | 0.823 | 0.776 | 0.778 | 0.821 |
| ELR | – | 0.804 | 0.828 | 0.825 | 0.807 | 0.754 | 0.783 | 0.820 | |
| Logrank | (0.889) | (0.844) | (0.727) | (0.851) | (0.862) | (0.784) | (0.760) | (0.836) | |
| ELR | (−) | (0.849) | (0.722) | (0.850) | (0.847) | (0.791) | (0.784) | (0.847) | |
| Size | |||||||||
| W (2, 10) vs. W (2, 10) | Logrank | 0.048 | 0.043 | 0.054 | 0.054 | 0.054 | 0.061 | 0.050 | 0.055 |
| ELR | – | 0.048 | 0.052 | 0.056 | 0.047 | 0.041 | 0.043 | 0.053 | |
| Logrank | (0.054) | (0.052) | (0.037) | (0.052) | (0.053) | (0.057) | (0.058) | (0.053) | |
| ELR | (−) | (0.050) | (0.036) | (0.054) | (0.052) | (0.041) | (0.035) | (0.056) | |
| LN (2.2, 0.4) vs. LN(2.2, 0.4) | Logrank | 0.043 | 0.056 | 0.055 | 0.056 | 0.051 | 0.054 | 0.057 | 0.052 |
| ELR | – | 0.063 | 0.060 | 0.059 | 0.048 | 0.039 | 0.039 | 0.057 | |
| Logrank | (0.042) | (0.047) | (0.026) | (0.040) | (0.038) | (0.047) | (0.036) | (0.042) | |
| ELR | (−) | (0.050) | (0.023) | (0.036) | (0.031) | (0.029) | (0.023) | (0.037) | |
Note: Numbers not in parenthesis are based on GA setting; numbers in parenthesis are based on GB setting. Sample size n = 200, 1000 simulated data sets, and 1000 bootstraps per simulated dataset.
In the GB setting, the probability of making an annual visit P is again 0.3 but for half of subjects, the other half having the left limit of the interval being 1978. The power of the ELR test is similar to that of the logrank test. Overall, the power based on the MEDIAN and MEAN is greater than the one based on other imputation methods. The MID, MODE, and RAND methods perform worst in terms of power. The size of the logrank test based on the MID method is lower than nominal. The size of the ELR test based on MID, MODE, and RAND methods is also low.
In the case where the mean interval width w is 2.1 years (P = 0.65) for both groups in the GA setting (see Table 4 of the technical report by Zhang et al. [27]), the power of each imputation method is closer to the test with exact data comparing to the scenario where mean interval width is about 5 years. This is reasonable since more information is lost in the case of heavy interval censoring for origin event. The sizes of the ELR test and the regular logrank test are comparable and close to the 5% nominal level.
4.3. Cox regression problem
For doubly censored data with interval-censored HIV infection time X, once X is imputed using the imputation methods described in Section 2, we can make inference based on the methods for the right-censored data. If we regard the date of birth as time 0, then the HIV infection time X can be treated as age at HIV infection. For subject i, let Xi, z2i, and Ti denote the HIV infection time, age at HIV infection, and time from HIV infection to death, respectively, for i = 1, … n. Let X̂i denote the imputed HIV infection time for subject i regardless of the imputation methods. Since the HIV infection time X is interval-censored, so is age at HIV infection z2. Once X is imputed as X̂, z2 is estimated by ẑ2 = X̂. The survival time of interested Ti is then estimated by , where is the time from study entry to event. The performance of β̂1, the estimator of Cox regression coefficient β1 after adjusting for z2, is of interest.
The results for the GA and GB settings with β = (−0.5, 0.1) and heavy interval censoring (mean interval width w = 5.3 years) for HIV infection time are summarized in Table 4. In the GA setting, the estimator of β1 is a little biased (towards 0) for all seven imputation methods with the bias percentage ranging from 1.2–4.3%. The method MID has the smallest bias, followed by the RIGHT, MEAN, MODE, and MEDIAN. The estimator based on RAND and MI has relatively larger bias. These results are based on the same 1000 simulated data sets for each of the seven imputation methods. Treating each simulated data as a block, a two-way ANOVA can be carried out to test for differences among biases of the seven imputation methods. Overall, biases of seven imputation methods differ significantly (F6,6×999 = 37.69, p < 0.001). The bias using the MID method is significantly smaller than the bias based on any other imputation method (p < 0.001). The mean ASE of β̂1 based on the MI is slightly larger than those based on other imputation methods, since it incorporates the between-imputation variability using Rubin’s variance formula [17]. There are some differences (F6,6×999 = 2.61, p = 0.016) among the MSEs of the seven imputation methods but the differences are small. Table 4 gives coverage probability, power, and size for testing β1 = 0 based on two different estimation procedures. The first is based on the asymptotic standard error using the normality assumption of β̂1. The second is based on the 2.5 and 97.5% empirical bootstrap quantiles. All seven imputation methods work reasonably well. For the method MI, the coverage probability of 95% ASE CI is slightly bigger than 0.95; the ASE power is the smallest one; and the ASE size is below the 5% nominal level.
Table 4.
Comparison of imputation methods for β = (−0.5, 0.1) under the Cox model with heavy interval censoring.
| EXACT | Imputation methods |
|||||||
|---|---|---|---|---|---|---|---|---|
| RIGHT | MID | MEAN | MEDIAN | MODE | RAND | MI | ||
| Mean of β̂1 | −0.5036 (−0.5021) | −0.4832 (−0.4897) | −0.4938 (−0.4764) | −0.4829 (−0.4919) | −0.4813 (−0.4920) | −0.4823 (−0.4838) | −0.4792 (−0.4845) | −0.4786 (−0.4849) |
| SD(β̂1) | 0.1569 (0.1606) | 0.1578 (0.1589) | 0.1600 (0.1614) | 0.1569 (0.1596) | 0.1561 (0.1597) | 0.1590 (0.1610) | 0.1585 (0.1619) | 0.1554 (0.1568) |
| Mean ASE(β̂1) | 0.1554 (0.1553) | 0.1552 (0.1552) | 0.1553 (0.1553) | 0.1551 (0.1553) | 0.1551 (0.1553) | 0.1552 (0.1553) | 0.1551 (0.1553) | 0.1585 (0.1605) |
| MSE | 0.0246 (0.0258) | 0.0252 (0.0253) | 0.0256 (0.0266) | 0.0249 (0.0255) | 0.0247 (0.0256) | 0.0256 (0.0262) | 0.0255 (0.0264) | 0.0246 (0.0248) |
| Coverage of 95% CI (ASE) | 0.949 | 0.943 | 0.946 | 0.952 | 0.953 | 0.940 | 0.948 | 0.957 |
| Coverage of 95% EBCI | – | 0.938 | 0.944 | 0.948 | 0.949 | 0.955 | 0.953 | 0.942 |
| Coverage of 95% CI (ASE) | (0.942) | (0.947) | (0.944) | (0.945) | (0.945) | (0.949) | (0.935) | (0.956) |
| Coverage of 95% EBCI | (−) | (0.947) | (0.943) | (0.945) | (0.947) | (0.949) | (0.949) | (0.940) |
| Power (ASE) | 0.904 | 0.881 | 0.882 | 0.876 | 0.878 | 0.882 | 0.864 | 0.861 |
| Power (EBCI) | – | 0.890 | 0.887 | 0.884 | 0.880 | 0.875 | 0.877 | 0.885 |
| Power (ASE) | (0.912) | (0.903) | (0.870) | (0.901) | (0.903) | (0.890) | (0.887) | (0.883) |
| Power (EBCI) | (−) | (0.912) | (0.872) | (0.908) | (0.906) | (0.898) | (0.899) | (0.909) |
| Size (ASE) | 0.044 | 0.048 | 0.052 | 0.052 | 0.052 | 0.056 | 0.048 | 0.040 |
| Size (EBCI) | – | 0.059 | 0.060 | 0.057 | 0.055 | 0.052 | 0.054 | 0.059 |
| Size (ASE) | (0.060) | (0.062) | (0.053) | (0.064) | (0.064) | (0.055) | (0.061) | (0.042) |
| Size (EBCI) | (−) | (0.065) | (0.053) | (0.062) | (0.066) | (0.051) | (0.055) | (0.060) |
Note: Numbers not in parenthesis are from GA setting; numbers in parenthesis are from GB setting. Sample size n = 200, 1000 simulated data sets, and 1000 bootstraps per simulated dataset.
In the GB setting, the estimator of β1 also shows some bias towards 0 for all seven imputation methods. The estimate β̂1 based on any imputation method is small with the bias percentage ranging from 1.6–4.7%. The method MID has the largest bias; the methods MEAN and MEDIAN have the smallest biases. Overall, there are significant differences between the seven imputation methods in biases (F6,6×999 = 41.95, p < 0.001). The bias using the MID method is significantly larger than the bias based on any other imputation method (p < 0.001). Again, the mean ASE of β̂1 based on MI is slightly bigger than the one based on other imputation methods. There are significant differences (F6,6×999 = 4.92, p < 0.001) among the MSEs based on the seven imputation methods. All imputation methods work reasonably well for the coverage probability of 95% CI, power, and size. For the method MI, the coverage probability of 95%ASE CI is slightly bigger than 0.95; the ASE power is the smallest one except MID; and the ASE size is below the 5% nominal level.
In the scenario GA with light interval censoring for HIV infection time (w = 2.1 years, see Table 6 of the technical report by Zhang et al. [27]), the bias of β̂1 shrinks for every imputation method, resulting in bias percentage ranging from 1–2%. In the scenario GB with light interval censoring for HIV infection time (w = 2.1 years, see Table 7 the technical report), the bias of β̂1 also shrinks for every imputation method, resulting in bias percentage ranging from 0.5–4.3%.
5. Discussion
In the one-sample scenario, the method RIGHT does not perform well in terms of estimating the Kaplan–Meier curve. The method MID works very well in the GA setting, but fails in the GB setting, when half of the left limits of the interval correspond to the date 1978. Caution is therefore suggested in using simple imputation methods to impute the actual HIV infection time. The probability-based imputation methods perform well for estimating the Kaplan–Meier curve in both simulation settings. Methods MEDIAN and MODE stand out as preferred ones in estimating the Kaplan–Meier curve.
In the two-sample scenario, the regular logrank test and ELR test perform similarly in terms of power and size regardless of imputation methods. Methods MEAN and MEDIAN are recommended for their robust performance in both simulation settings.
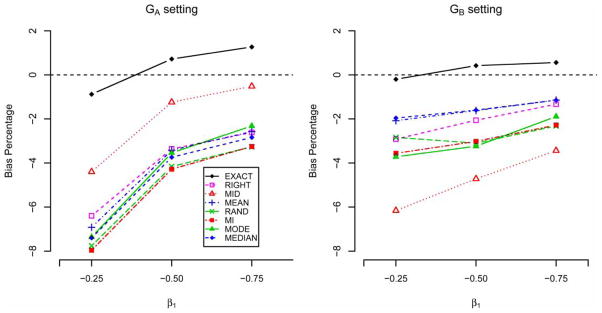
In the Cox model scenario, all seven imputation methods yield acceptable bias in estimating Cox regression coefficient. We also studied the performance of seven imputation methods for different values of the Cox regression coefficient. As shown in Figure 3, all imputation methods tend to yield a downward bias, and the bias’ percentage appears to decrease as the magnitude of the Cox regression coefficient increases. The method MID works well in the GA setting, but fails in the GB setting. Though the method RIGHT works well in both simulation settings, it fails in a scenario when both groups have different interval censoring widths (data not shown). Overall, probability-based imputation methods, especially MEAN and MEDIAN, appear to perform robustly against different simulation settings.
Figure 3.
Bias percentage for the estimator of β1 based on seven imputation methods for data with n = 200 and w = 5.3 years.
To account for the imputation uncertainty, Pan [14] adopts the method MI and makes inference by using the variance formula proposed by Rubin [17] under a Bayesian inference framework. The validity of using Rubin’s formula in a frequentist framework has been discussed by Zhang [28] and Nielsen [29]. Our simulation results show that validity of this procedure may be questionable in the scenarios examined in this paper. For the Cox regression problem in the simulation studies, the comparison between mean ASE(β̂1) and SD(β̂1) implies that using Rubin’s formula may overcorrect the standard error of β̂1 and this is also evident in Figure 2. Zhang [28] presents rules for making MI inferences with missing data. Those rules are not directly applicable for doubly censored data.
In all the problems considered, as the interval width decreases, the performance of each imputation method improves. In the simulation studies note also that, ignoring the uncertainty in the imputed date of origin event, the usual inference based on the ASE performs surprisingly well. The bootstrap inference procedure is recommended, however, since the computational demand with imputation methods is not excessive. Zhang et al. [30] propose a Bayesian approach to analyse doubly censored data by making a parametric assumption for the interval-censored origin and treating it as an unknown quantity. This approach could be used as an alternative to the bootstrap inference procedure.
Acknowledgments
This work was supported in part by a Dissertation Fellowship from University of Iowa Graduate College and an NIH/NIAID grant: R01 058740.
References
- 1.De Gruttola V, Lagakos SW. Analysis of doubly-censored survival data, with application to AIDS. Biometrics. 1989;45:1–11. [PubMed] [Google Scholar]
- 2.Sun J. Statistical analysis of doubly interval-censored failure time data, Advances in Survival Analysis. Handbook of Statistics. 2004;23:105–122. [Google Scholar]
- 3.Kim MY, De Gruttola V, Lagakos SW. Analyzing doubly censored data with covariates, with application to AIDS. Biometrics. 1993;49:13–22. [PubMed] [Google Scholar]
- 4.Sun J, Liao Q, Pagano M. Regression analysis of doubly censored failure time data with applications to AIDS studies. Biometrics. 1999;55:909–924. doi: 10.1111/j.0006-341x.1999.00909.x. [DOI] [PubMed] [Google Scholar]
- 5.Xiang J, Wünschmann S, Diekema DJ, Klinzman D, Patrick KD, George SL, Stapleton JT. Effect of coinfection with GB virus C on survival among patients with HIV infection. N England J Med. 2001;345:707–714. doi: 10.1056/NEJMoa003364. [DOI] [PubMed] [Google Scholar]
- 6.Tillmann HL, Heiken H, Knapir-Botor A, Heringlake S, Ockenga J, Wilber JC, Goergen B, Detmer J, McMorrow M, Stoll M, Schmidt RE, Manns MP. Infection with GB virus C and reduced mortality among HIV-infected patients. N England J Med. 2001;345:715–724. doi: 10.1056/NEJMoa010398. [DOI] [PubMed] [Google Scholar]
- 7.Liu KJ, Darrow WW, Rutherford GW. A model-based estimate of the mean incubation period for AIDS in homosexual men. Science. 1988;240:1333–1335. doi: 10.1126/science.3163848. [DOI] [PubMed] [Google Scholar]
- 8.Mariotto AB, Mariotti S, Pezzotti P, Rezza G, Verdecchia A. Estimation of the acquired-immunodeficiency-syndrome incubation period in intravenous-drug-users – a comparison with male homosexuals. Amer J Epidemiol. 1992;135:428–437. doi: 10.1093/oxfordjournals.aje.a116303. [DOI] [PubMed] [Google Scholar]
- 9.Williams CF, Klinzman D, Yamashita TE, Xiang JH, Polgreen PM, Rinaldo C, Liu CL, Phair J, Margolick JB, Zdunek D, Hess G, Stapleton JT. Persistent GB virus C infection and survival in HIV-infected men. N England J Med. 2004;350:981–990. doi: 10.1056/NEJMoa030107. [DOI] [PubMed] [Google Scholar]
- 10.Law CG, Brookmeyer R. Effects of mid-point imputation on the analysis of doubly censored data. Stat Med. 1992;11:1569–1578. doi: 10.1002/sim.4780111204. [DOI] [PubMed] [Google Scholar]
- 11.Gauvreau K, De Gruttola V, Pagano M. The effect of covariates on the induction time of AIDS using improved imputation of exact seroconversion times. Stat Med. 1994;13:2021–2030. doi: 10.1002/sim.4780131912. [DOI] [PubMed] [Google Scholar]
- 12.Turnbull BW. The empirical distribution function with arbitrarily grouped, censored and truncated data. J Roy Stat Soc Ser B. 1976;38:290–295. [Google Scholar]
- 13.Goggins WB, Finkelstein DM, Zaslavsky AM. Applying the Cox proportional hazards model for analysis of latency data with interval censoring. Stat Med. 1999;18:2737–2747. doi: 10.1002/(sici)1097-0258(19991030)18:20<2737::aid-sim199>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 14.Pan W. A multiple imputation approach to regression analysis for doubly censored data with application to AIDS studies. Biometrics. 2001;57:1245–1250. doi: 10.1111/j.0006-341x.2001.01245.x. [DOI] [PubMed] [Google Scholar]
- 15.Rubin DB. The Bayes bootstrap. Ann Stat. 1981;9:130–134. [Google Scholar]
- 16.Efron B. Missing data, imputation, and the bootstrap. J Amer Stat Assoc. 1994;89:463–475. [Google Scholar]
- 17.Rubin DB. Multiple Imputation for Nonresponse in Surveys. Wiley; New York: 1987. [Google Scholar]
- 18.Geskus RB. Methods for estimating the AIDS incubation time distribution when date of seroconversion is censored. Stat Med. 2001;20:795–812. doi: 10.1002/sim.700. [DOI] [PubMed] [Google Scholar]
- 19.Groeneboom P, Wellner JA. DMV Seminar, Brand 19. Birkhäuser; New York: 1992. Information Bounds and Nonparametric Maximum Likelihood Estimation. [Google Scholar]
- 20.Dempster AP, Laird NM, Rubin DB. Maximum likelihood from incomplete data via the EM algorithm. J Roy Stat Soc Ser B. 1977;39:1–38. [Google Scholar]
- 21.Sun J. The Statistical Analysis of Interval-Censored Failure Time Data. Springer; New York: 2006. [Google Scholar]
- 22.Muñoz A, Xu J. Models for the incubation of AIDS and variations according to age and period. Stat Med. 1996;15:2459–2473. doi: 10.1002/(sici)1097-0258(19961130)15:22<2459::aid-sim464>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 23.Pan W. A multiple imputation approach to Cox regression with interval censored data. Biometrics. 2000;56:199–203. doi: 10.1111/j.0006-341x.2000.00199.x. [DOI] [PubMed] [Google Scholar]
- 24.Little RJA, Robin DB. Statistical Analysis with Missing Data. 2. Wiley; New York: 2002. [Google Scholar]
- 25.Jaffe HW, Darrow WW, Echenberg DF, O’Malley PM, Getchell JP, Kalyanaraman VS, Byers RH, Drennan DP, Braff EH, Curran JW, Francis DP. The acquired immunodeficiency syndrome in a cohort of homosexual men – a six year follow-up-study. Ann Int Med. 1985;103:210–214. doi: 10.7326/0003-4819-103-2-210. [DOI] [PubMed] [Google Scholar]
- 26.Cox DR. Regression models and life-tables (with discussion) J Roy Stat Soc Ser B. 1972;34:187–200. [Google Scholar]
- 27.Zhang W, Zhang Y, Chaloner K, Stapleton JT. Tech Rep 2008-2. Department of Biostatistics, University of Iowa; 2008b. Imputation methods for doubly censored HIV data. available at http://www.public-health.uiowa.edu/biostat/research/reports.html. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang P. Multiple imputation: theory and method. Int Stat Rev. 2003;71:581–592. [Google Scholar]
- 29.Nielsen SF. Proper and improper multiple imputation. Int Stat Rev. 2003;71:593–627. [Google Scholar]
- 30.Zhang W, Chaloner K, Cowles MK, Zhang Y, Stapleton JT. A Bayesian pooled analysis of doubly censored data using a hierarchical Cox model. Stat Med. 2008a;27:529–542. doi: 10.1002/sim.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]