Abstract
The human cytomegalovirus DNA polymerase contains a catalytic subunit, UL54, and an accessory protein, UL44. Recent studies suggested that UL54 might interact via its extreme C terminus with UL44 (A. Loregian, R. Rigatti, M. Murphy, E. Schievano, G. Palu', and H. S. Marsden, J. Virol. 77:8336-8344, 2003). To address this hypothesis, we quantitatively measured the binding of peptides corresponding to the extreme C terminus of UL54 to UL44 by using isothermal titration calorimetry. A peptide corresponding to the last 22 residues of UL54 was sufficient to bind specifically to UL44 in a 1:1 complex with a dissociation constant of ca. 0.7 μM. To define individual residues in this segment that are crucial for interacting with UL44, we engineered a series of mutations in the C-terminal region of UL54. The UL54 mutants were tested for their ability to interact with UL44 by glutathione S-transferase pulldown assays, for basal DNA polymerase activity, and for long-chain DNA synthesis in the presence of UL44. We observed that deletion of the C-terminal segment or substitution of alanine for Leu1227 or Phe1231 in UL54 greatly impaired both the UL54-UL44 interaction in pulldown assays and long-chain DNA synthesis without affecting basal polymerase activity, identifying these residues as important for subunit interaction. Thus, like the herpes simplex virus UL30-UL42 interaction, a few specific side chains in the C terminus of UL54 are crucial for UL54-UL44 interaction. However, the UL54 residues important for interaction with UL44 are hydrophobic and not basic. This information might aid in the rational design of new drugs for the treatment of human cytomegalovirus infection.
Most biological processes depend on the coordinated formation of protein-protein interactions. Aside from their importance for viral biology, several interactions between viral proteins have been proposed as attractive targets for antiviral drug discovery, since the exquisite specificity of such cognate interactions affords the possibility of interfering with them in a highly specific and effective manner (for a review, see reference 16). There is a considerable need for new anti-human cytomegalovirus (HCMV) drugs since, although there are agents, most of which target the polymerization activity of the viral DNA polymerase, their use is limited by pharmacokinetic issues, antiviral resistance, and toxicity (4). A potential novel target for anti-HCMV drugs could be the interaction between the two subunits of the viral DNA polymerase.
Analogous to the DNA polymerase of other herpesviruses, the HCMV DNA polymerase is composed of a 1,242-residue catalytic subunit, Pol or UL54 (13, 14), and the 433-residue accessory protein UL44 (9). The UL54 protein, which is the homolog of herpes simplex virus type 1 (HSV-1) UL30, has DNA-dependent DNA polymerase activity and 3′-5′ exonuclease activity (3, 18, 20). The UL44 accessory subunit, which is analogous to the HSV-1 UL42 protein, has been shown to bind double-stranded DNA, to specifically interact with UL54, and to stimulate long-chain DNA synthesis, possibly by increasing the processivity of the polymerase along the DNA template (9, 28). However, the details of the UL44-UL54 interaction and its role in UL44 function have not been elucidated.
The observations that both UL54 and UL44 are essential for HCMV DNA replication (21-23) and that the UL54-UL44 interaction is specific (17) make this interaction an attractive drug target. Recently, it was reported that a peptide corresponding to the C-terminal 22 residues of UL54 could both disrupt the physical interaction between UL54 and UL44 and specifically inhibit the stimulation of UL54 activity by the accessory protein (17). These findings suggested, but did not demonstrate, that UL54 interacts via its extreme C terminus with UL44. They did show that inhibition of HCMV DNA polymerase can indeed be obtained by disruption of the interaction between the two enzyme subunits, suggesting that this could represent a novel strategy for anti-HCMV chemotherapy. However, one obstacle to the inhibition of protein-protein interactions is that these interactions often involve a large surface area and multiple nonspecific hydrophobic contacts (1, 27), making the development of small-molecule inhibitors difficult. This kind of information is currently lacking for the UL54-UL44 interaction, although a previous study suggested that the two cysteines at the extreme C terminus of UL54 might be involved in the interaction with UL44, since a mutant peptide lacking these residues was markedly less effective in disrupting the UL54-UL44 physical interaction than the wild-type peptide (17). However, the precise contribution of these residues to UL44 binding has not been assessed.
To determine whether the C terminus of UL54 is sufficient to bind to UL44 in solution and to examine the contributions to binding energy of the two C-terminal cysteines of UL54, isothermal titration calorimetry (ITC) assays were performed with UL44 and peptides derived from the C terminus of UL54. These assays yielded binding affinities, stoichiometries, and thermodynamic parameters for the UL54-UL44 interaction. Moreover, to define individual residues in UL54 that are crucial for binding to UL44, we engineered a series of substitutions in the C-terminal region of UL54 and tested the effect of the mutations on physical and functional interaction between UL54 and UL44. Our findings define important determinants of the UL54-UL44 interaction, information that might aid in the discovery of new drugs for the treatment of HCMV infection based on disruption of the UL54-UL44 interaction.
MATERIALS AND METHODS
Plasmids.
The pRSET-Pol plasmid, containing the HCMV strain AD169 UL54 gene excised as an EcoRI/HindIII fragment from pUC19-Pol (8) and subcloned in EcoRI/HindIII sites of pRSETC (Invitrogen), was a gift from P. F. Ertl (GlaxoSmithKline, Stevenage, United Kingdom). The pT730 plasmid expressing HSV-1 UL30 under a T7 promoter has been previously described (6). The pDEST15 vector (Invitrogen) was modified to contain an in-frame stop codon and express glutathione S-transferase (GST) alone (pD15-GST). The pD15-UL44 and pD15-UL44ΔC290 plasmids, which express full-length UL44 and the N-terminal 290 residues of UL44 as a GST fusion protein, respectively, were created by utilizing the Gateway lambda recombination system (Invitrogen). PCR primers were designed with 5′ overhangs to amplify the region of UL44 of interest and to add attB recombination sites, a PreScission protease site and an XhoI site immediately upstream of the start codon, an MluI site immediately downstream of the stop codon, and a stop codon located 3′ of nucleotide 890 for pD15-UL44ΔC290. Since this strategy required the addition of over 60 nucleotides located 5′ of UL44, the flanking sites were added with two rounds of PCR. The sequence of the primer pairs used is available online (http://coen.med.harvard.edu) or in printed form from the authors upon request. The PCR products were generated by using pRSET44 (a gift from P. F. Ertl) as a template, gel purified, and recombined with pDONR201 (Invitrogen) to generate entry clones. Entry clones were subsequently recombined into pDEST15 (Invitrogen) and sequenced. The amplification did not introduce any new mutation. To create the UL54 ΔC1212 mutant, an UL54 entry clone, pENT-Pol, was created via subcloning of the EcoRI/HindIII fragment from pRSET-Pol into a modified pENTR1A plasmid (Invitrogen). An entry vector was created to express the UL54 ΔC1212 mutant as follows. A PCR product was amplified from pRSET-Pol from nucleotides 3230 to 3636, which contained a stop codon and a HindIII site located 3′ of nucleotide 3636, and subcloned into pCR-Blunt (Invitrogen). The DNA fragment was excised from this plasmid as a BspEI/HindIII fragment and subcloned into pENT-Pol, where BspEI cleaves at nucleotide 3317 of UL54 and HindIII cleaves downstream of the UL54 gene. The pENT-Pol and pENT-PolΔC1212 plasmids were recombined with pDEST17 (Invitrogen) to generate plasmids pD17-Pol and pD17-PolΔC1212, respectively. All other UL54 mutants were obtained by using the QuikChange mutagenesis kit (Stratagene), amplifying the pRSET-Pol plasmid with primer pairs containing appropriate nucleotide change(s). A list of the mutagenic primers is available at http://coen.med.harvard.edu or in printed form from the authors on request. The mutated region of UL54 gene was sequenced by the Biopolymers Laboratory in the Department of Biological Chemistry and Molecular Pharmacology at Harvard Medical School and shown to contain wild-type sequences except for the engineered mutation. Finally, for each UL54 mutant a BssHII/HindIII fragment containing the mutation was subcloned into BssHII/HindIII sites of pRSET-Pol.
Proteins and protein purification.
Purified baculovirus-expressed HCMV UL54, prepared as described previously (17), was kindly provided by H. S. Marsden (Institute of Virology, Glasgow, United Kingdom). Full-length UL44, purified from insect cells infected with the appropriate recombinant baculovirus as previously reported (17), was also provided by H. S. Marsden. The purification of MBP-UL42ΔC340 fusion protein, which contains the N-terminal 340 residues of UL42, has been previously described (2). Recombinant GST-UL44 and GST-UL44ΔC290 fusion proteins and GST were purified from Escherichia coli BL21(DE3)/pLysS harboring the respective plasmid pD15-UL44, pD15-UL44ΔC290, or pD15-GST. Typically, 2 liters of cells was grown in 2YT medium containing 100 μg of ampicillin/ml. The cells were grown until the A600 was between 0.6 and 0.8 and then induced by the addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside (ICN) for 40 h at 16°C. The pelleted cells were resuspended in 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 10 mM EDTA, 2 mM dithiothreitol (DTT), 20% glycerol, and Complete protease inhibitors (Roche Molecular Biochemicals) and then stored at −80°C until needed. The cells were thawed and lysed by three passes through an Emulsiflex-C5 apparatus (Avestin). The lysate was centrifuged at 43,000 × g for 1 h, applied to a 5-ml glutathione-Sepharose 4 FastFlow column (Amersham Pharmacia Biotech) that had been equilibrated in buffer A (50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 2 mM DTT, 500 mM NaCl, and 20% glycerol), and then washed extensively with buffer A. To remove E. coli DnaK protein (24), the column was equilibrated with buffer B (buffer A minus EDTA) and then washed with buffer B plus 10 mM ATP (Sigma). Finally, GST, GST-UL44, or GST-UL44ΔC290 was eluted with buffer A containing 15 mM glutathione. Purified proteins were stored in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 30% glycerol, 0.1 mM EDTA, and 2 mM DTT. For some experiments, the GST-UL44ΔC290 fusion protein eluted from the glutathione column was cleaved with PreScission protease (Pharmacia) at 4°C for 16 h and then applied to a 5-ml HiTrap Heparin HP column (Amersham Pharmacia Biotech); washed with 50 mM Tris-HCl (pH 7.5), 250 mM NaCl, 10% glycerol, 1 mM EDTA, and 2 mM DTT; and eluted with a linear NaCl gradient from 250 mM to 1 M. The eluted protein was further purified on Sephacryl S-100 HR column (Amersham Pharmacia Biotech) equilibrated in 50 mM Tris-HCl (pH 7.5), 500 mM NaCl, 10% glycerol, 1 mM EDTA, and 2 mM DTT and then eluted with the same buffer. The concentrations of all proteins were determined by using amino acid analysis at the Molecular Biology Core Facility, Dana-Farber Cancer Institute.
Peptides.
Peptide 1 (17), which corresponds to the last 22 residues of UL54, was synthesized by the Biopolymers Laboratory in the Department of Biological Chemistry and Molecular Pharmacology at Harvard Medical School. A variant of peptide 1 that lacks the two C-terminal cysteines (termed peptide 8 in reference 17) and a peptide corresponding to last 10 residues of UL54 (here termed peptide 9) were generously provided by H. S. Marsden. Peptides were dissolved in water, and concentrations were determined by quantitative amino acid analysis performed by the Molecular Biology Core Facility, Dana-Farber Cancer Institute. The peptides were then lyophilized prior to use.
ITC.
ITC experiments were performed as reported elsewhere (2), with some modifications. Briefly, purified GST, GST-UL44, GST-UL44ΔC290, UL44ΔC290, or MBP-UL42ΔC340 protein was dialyzed against buffer containing 50 mM Tris-Cl (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, and decreasing concentrations of DTT up to 0.5 mM immediately before the experiments were performed in order to reduce the concentration of DTT, which otherwise would interfere with calorimetric measurements. Lyophilized synthetic peptides were suspended in the last dialysis buffer of proteins. ITC was performed with a VP-ITC calorimeter (MicroCal, Inc.) and 5 to 10 μM protein concentrations and 150 to 250 μM peptide concentrations. Peptides were titrated into the protein-containing sample cell in 10-μl injections at 25°C with a mixing speed of 270 rpm. The heats of dilution of both protein and ligand were determined and subtracted prior to analysis, and the data were integrated to generate curves in which the areas under the injection peaks were plotted against the ratio of peptide to protein. Analysis of the data was performed as previously described (2).
GST-pulldown assays.
In vitro transcription-translation of wild-type HSV-1 UL30 or wild-type or mutant full-length HCMV UL54 was performed from the appropriate plasmids by using the TNT T7 coupled reticulocyte lysate system from Promega according to the manufacturer's suggestion. The translation products were labeled with [35S]methionine (Amersham Pharmacia Biotech). GST or GST-UL44 fusion protein (0.15 mg) was incubated with 50 μl of in vitro transcription-translation reactions on ice for 2 h in binding buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10% glycerol, 0.1 mM EDTA, 2 mM DTT) containing 5 μl of RNAce-It RNase Cocktail (Stratagene) and 50 U of Benzonase (Sigma) and then loaded onto 0.2-ml glutathione columns. The columns were washed with 5 ml of wash buffer (50 mM Tris-HCl [pH 7.5], 500 mM NaCl, 10% glycerol, 0.1 mM EDTA, 2 mM DTT, 0.5% NP-40, and 0.5% Triton X-100), and bound proteins were eluted with wash buffer containing 15 mM glutathione. The proteins were visualized by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and autoradiography.
DNA polymerase assays.
The basal DNA polymerase activity of UL54 and stimulation of UL54 activity by UL44 were measured by a filter-based assay as previously described (17) but with 5 μl of in vitro-transcribed and -translated wild-type or mutant UL54 in the absence or in the presence of the indicated amounts of purified baculovirus-expressed UL44. Long-chain DNA synthesis by wild-type or mutant UL54 in the presence of UL44 was assayed with 5 μl of in vitro-transcribed and -translated wild-type or mutant UL54 and various amounts of purified baculovirus-expressed UL44 in a reaction mixture containing 75 mM Tris-HCl (pH 8.0), 150 mM KCl, 6.5 mM MgCl2, 1.67 mM β-mercaptoethanol, 400 μg of bovine serum albumin/ml, 10 μg of poly(dA)-oligo(dT)12-18 (Amersham Pharmacia Biotech)/ml, and 50 μM [32P]TTP (5 Ci/mmol; Perkin-Elmer Life Sciences) in a final volume of 25 μl. Reactions were carried out at 37°C for 90 min. Reactions were stopped by placing the mixtures on ice and adding 5 μl of alkaline loading buffer (2 mM EDTA, 50 mM NaOH, 2.5% glycerol, 0.025% bromcresol green) and then loading them onto a 4% alkaline agarose gel. Gels were dried overnight and used to expose film and phosphorescence screens. Newly synthesized DNA larger than 20 bases was defined as long-chain DNA and quantified by using Personal Molecular Imager FX software (Bio-Rad).
RESULTS
A peptide corresponding to the C-terminal 22 residues of UL54 specifically binds UL44 in solution.
Previous studies suggested that the segment corresponding to the C-terminal 22 residues (amino acids 1221 to 1242) of UL54 might be involved in binding to UL44, since a synthetic peptide corresponding to this region could specifically disrupt the UL54-UL44 interaction (17). However, binding of this segment had not been directly assessed, and the thermodynamic parameters of the binding had not been measured. To examine this interaction directly and quantitatively, we used ITC. ITC measures heat generated or absorbed upon binding. Titration curves that are obtained can be analyzed by curve-fitting algorithms to provide values for the stoichiometry, the change in enthalpy (ΔH), and the dissociation constant (Kd) of the interaction as a measure of affinity. The Kd value then allows calculation of the change in free energy (ΔG), which together the ΔH allows the calculation of the entropic term TΔS.
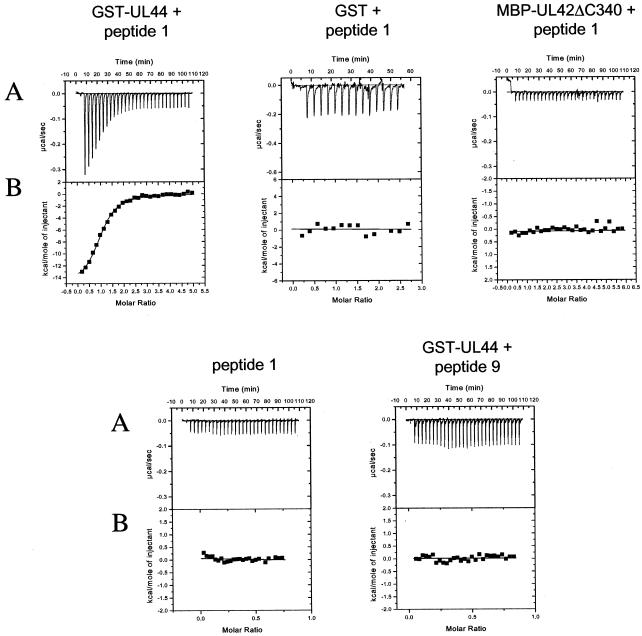
A typical titration experiment for binding of the UL54 C-terminal peptide (here termed peptide 1 as in reference 17) to a purified GST-UL44 fusion protein (Fig. 1) is shown in Fig. 2. The raw data, which are shown in upper panel (Fig. 2A), indicate an exothermic interaction, based on the negative values observed for the peaks, each of which corresponds to a fixed amount of peptide 1 injected into a solution containing the GST-UL44 fusion protein. With each injection of peptide, less and less heat was released until constant values were obtained (corresponding to the heat released due to dilution, see Fig. 2A, peptide 1 alone), reflecting a saturable process. The area under each injection peak was integrated, resulting in the curve shown in each lower panel (Fig. 2B), in which the molar ratio of peptide to protein is plotted against the kilocalories per mole of injected peptide. The parameters for the binding of peptide 1 to GST-UL44 fusion protein are summarized in Table 1. Analysis of the binding data indicated a stoichiometry of about 1 molecule of peptide per molecule of UL44 fusion protein (Table 1) and a Kd of ∼0.7 μM. This Kd value corresponded to a ΔG value of about −8.3 kcal/mol (Table 1). Very similar results (Table 1) were obtained when peptide 1 was titrated with a GST-UL44ΔC290 fusion protein or with cleaved UL44ΔC290 (Fig. 1), a truncated protein that contains the N-terminal 290 residues of UL44 and retains all known biochemical activities of full-length UL44 (28; B. A. Appleton, A. Loregian, D. M. Coen, and J. M. Hogle, unpublished results). Evidence for the specificity of this interaction comes from the fact that no binding of the peptide was detected either with GST or with an MBP-UL42ΔC340 fusion protein, which contains the N-terminal 340 residues of HSV-1 UL42 (Fig. 2).
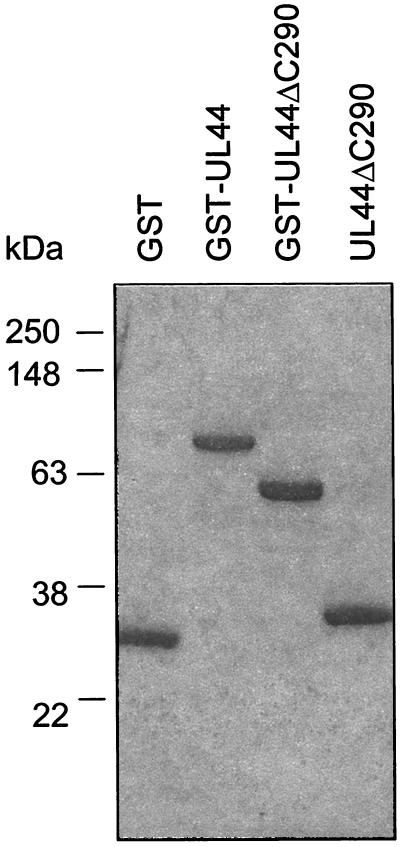
FIG. 1.
Purified GST, GST-UL44, GST-UL44ΔC290, and cleaved UL44ΔC290 preparations. Proteins were expressed in E. coli and purified by column chromatography as described in Materials and Methods. Samples after the final step of purification for each protein were analyzed by SDS-12% PAGE. The molecular masses based on protein markers analyzed on the same gel are indicated on the left.
FIG. 2.
Binding to UL44 of synthetic peptides corresponding to the C-terminal 22 residues (peptide 1) or 10 residues (peptide 9) of UL54 measured by ITC. Titrations were performed with 10-μl injections of peptide into a sample cell containing full-length GST-UL44, GST-UL44ΔC290, cleaved UL44ΔC290, or, as controls, GST or MBP-UL42Δ340. (A) Raw data for the titrations, in which the power output in microcalories per second is measured as a function of time in minutes. (B) The heats of dilution of both protein and ligand in A were subtracted, and the area under each injection curve was integrated to generate the points, which represent heat exchange in kilocalories per mole, which are plotted against the cumulative peptide-to-protein ratio for each injection.
TABLE 1.
Parameters for binding of C-terminal UL54 peptide 1 to UL44a
| Protein | Mean value ± SDb
|
||||
|---|---|---|---|---|---|
| Stoichiometry | ΔH (kcal/mol) | Kd (μM) | ΔG (kcal/mol) | TΔS (kcal/mol) | |
| GST-UL44 | 1.1 ± 0.1 | −16.4 ± 2.1 | 0.67 ± 0.18 | −8.3 ± 1.2 | −8.1 ± 0.9 |
| GST-UL44ΔC290 | 1.0 ± 0.1 | −16.5 ± 1.9 | 0.52 ± 0.11 | −8.6 ± 0.9 | −7.9 ± 0.8 |
| UL44ΔC290 | 1.0 ± 0.1 | −15.3 ± 2.2 | 0.62 ± 0.09 | −8.4 ± 1.3 | −6.9 ± 1.1 |
| GST | -c | - | - | - | - |
| MBP-UL42ΔC340 | - | - | - | - | - |
Titrations were performed with 10-μl peptide injections. Heats of dilution of both peptide and protein were subtracted from the raw data prior to analysis. Nonlinear least-squares analysis of the raw data was performed with the fitting algorithm provided with the calorimeter. Values are means of three to five determinations.
SD, standard deviation.
-, No binding was detected.
No release of heat was detected when GST-UL44 was titrated with a peptide corresponding to the last 10 residues of UL54 (amino acids 1233 to 1242, here termed peptide 9) (Fig. 2), suggesting that this region of UL54 is not sufficient for binding UL44.
The two C-terminal cysteines of UL54 are important but not essential for UL44 binding.
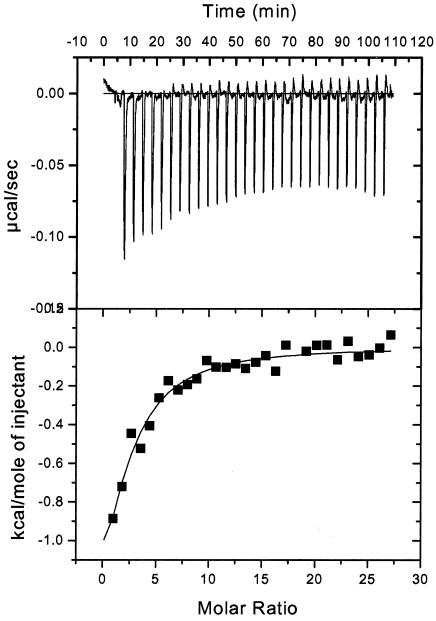
At the very C terminus of UL54 are two cysteines, which are unusual among herpesvirus DNA polymerases. Previous studies suggested that these residues of UL54 might have a role in the interaction with UL44, since the deletion of the two cysteines from the C terminus of peptide 1 greatly impaired the ability of the peptide to interfere with the physical interaction between HCMV UL54 and UL44, while having no discernible effect on peptide structure (17). To examine quantitatively the contribution of these residues of the UL54 C terminus to UL44 binding, ITC experiments were performed by titrating GST-UL44 with a variant of peptide 1 lacking the two C-terminal cysteines (here termed peptide 8, as in reference 17). Titration of binding is shown in Fig. 3. Even though the mutant peptide had lower values for binding energy than peptide 1 and more injections were required to reach saturation, it still measurably bound UL44. Analysis of the binding data indicated that the Kd of this ΔCC variant (peptide 8) was ∼10-fold higher than that of peptide 1 (Kd = 8 μM). Thus, even though the two C-terminal cysteines of UL54 most likely play a role in the interaction with UL44, they are not essential for the binding.
FIG. 3.
Binding to GST-UL44 of a variant of UL54 peptide 1 lacking the two C-terminal cysteines (peptide 8) measured by ITC. Titrations and data analysis were performed as described in the legend to Fig. 2.
Role of the C-terminal region and of the last two residues of UL54 in the binding of full-length protein to UL44.
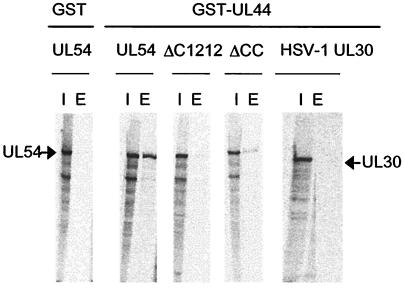
We next investigated the importance of the C-terminal region of UL54 to UL44 binding in the context of full-length protein. In order to detect the physical interaction between full-length UL54 and UL44, a GST-pulldown assay was developed by using an E. coli-expressed GST-UL44 fusion protein and in vitro-expressed and -radiolabeled UL54. Purified GST-UL44 was preincubated with wild-type or mutant in vitro-translated UL54 or, as a negative control, in vitro-expressed HSV-1 UL30 and then was loaded onto glutathione columns. The columns were washed, and bound proteins were eluted with 15 mM glutathione. As expected (9, 17), wild-type UL54 bound UL44 (Fig. 4). Control experiments indicated that the binding was specific, since GST did not bind UL54 and GST-UL44 did not bind HSV-1 UL30. To examine the role of the C-terminal region in UL44 binding, we constructed a mutant, ΔC1212, lacking the C-terminal 30 residues of UL54. No UL44 binding could be detected with this mutant (Fig. 4). Thus, in the context of full-length protein the C terminus of UL54 is necessary for binding to UL44. This also provides further evidence for the specificity of the assay. To examine the role of the two C-terminal cysteines in UL44 binding, we constructed a second deletion mutant, ΔCC, which lacks these two residues. Consistent with ITC data, we observed that deletion of the two C-terminal cysteines of UL54 only partially impaired UL54-UL44 interaction (Fig. 4).
FIG. 4.
Detection of UL54-UL44 interaction by a GST-pulldown assay. Purified GST or GST-UL44 was incubated with in vitro-expressed and -radiolabeled wild-type or mutant UL54 proteins (as indicated at the top of the figure) or, where indicated, HSV-1 UL30 as a control and allowed to bind to glutathione columns. The columns were washed, and the proteins were eluted with 15 mM glutathione. The radiolabeled proteins were visualized by autoradiography after SDS-7.5% PAGE. The position of UL54 and UL30 are marked by arrows. Lanes: I, input; E, eluted by glutathione.
Identification of residues of the UL54 C terminus that are critical for binding to UL44.
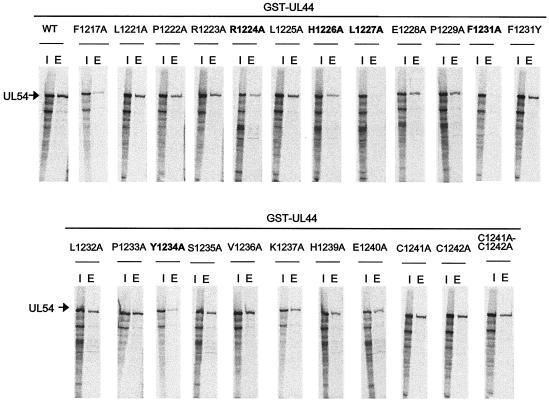
In an attempt to identify single residues of the UL54 C terminus crucial for binding to UL44, “alanine scan” variants of UL54, in which each nonalanine residue in the region corresponding to the last 22 residues of UL54 was individually converted to an alanine (Fig. 5), were created and then tested by GST-pulldown assays for their ability to bind UL44. The results are reported in Fig. 6. We found that substitution of either or both the two C-terminal cysteines with an alanine did not affect binding of UL54 to UL44. In contrast, L1227A and F1231A mutations drastically impaired binding of UL54 to UL44. Mutations at positions R1224, H1226, and Y1234 of UL54 reduced binding to UL44. All other UL54 mutants bound UL44 in a manner similar to that of wild-type UL54 when values were normalized to the input amount of in vitro-transcribed and -translated protein.
FIG. 5.
UL54 mutants. The sequence of the C-terminal 22 residues (amino acids 1221 to 1242) of wild-type UL54 protein, in single-letter code, is reported on the top. A series of mutations were engineered in this region of UL54 and at position 1217 as described in Materials and Methods. For each mutant, the sequence of the region containing the mutation is shown, with the mutated residue(s) indicated by underlined boldface letter(s).
FIG. 6.
Binding of mutant UL54 proteins to GST-UL44. The physical binding of wild-type or the mutant UL54 proteins, as indicated at the tops of the figure panels, to GST-UL44 was tested by GST-pulldown assays as described in legend to Fig. 4. Lanes: I, input; E, eluted by glutathione.
To determine whether an aromatic side chain at position 1231 of UL54 could be important for interaction with UL44, another UL54 point mutant in which the codon for Phe1231 was replaced by one for tyrosine (F1231Y) was constructed and then tested by GST-pulldown assays for the ability to associate with UL44. We observed that this substitution restored binding to UL44 (Fig. 6). We also examined the importance of a UL54 phenylalanine residue, Phe1217, located farther upstream. The F1217A mutant was partially impaired in the ability to bind to UL44 (Fig. 6).
The L1227A and F1231A mutants are highly defective for long-chain DNA synthesis.
To analyze the importance of individual residues of UL54 C terminus in a functional assay of the UL54-UL44 interaction, the UL54 mutants were also tested for their ability to synthesize long-chain DNA products in the presence of UL44 with a poly(dA)-oligo(dT)12-18 template-primer.
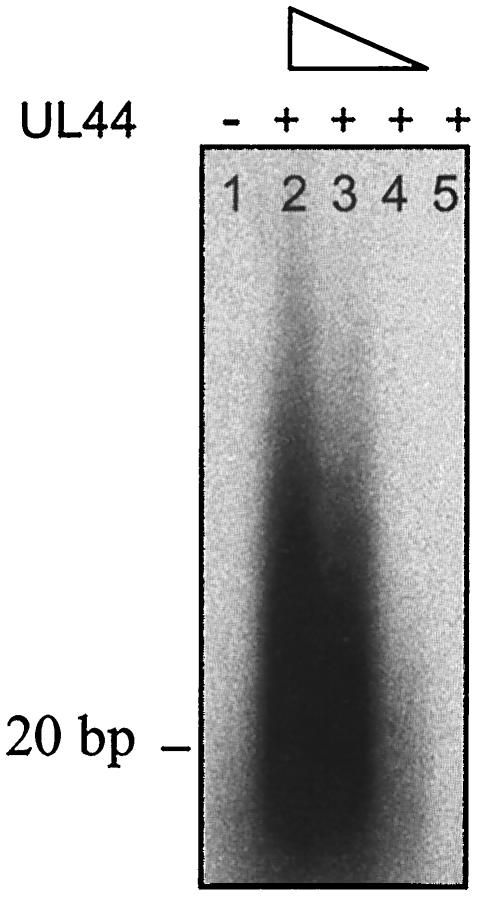
HSV UL30 alone adds only one or two bases on this primer-template but, when UL42 is present, longer DNA products are formed (7, 12). Similarly, no long-chain DNA synthesis was detected in the presence of in vitro-transcribed and -translated UL54 alone (Fig. 7, lane 1) or by purified UL44 alone (lane 5), whereas formation of long DNA products was observed when UL54 was incubated with increasing amounts of purified baculovirus-expressed UL44 (lanes 2, 3, and 4) or of purified E. coli-expressed GST-UL44 (data not shown). We observed that the reaction was slower than that performed by HSV UL30-UL42 complex. In fact, detectable synthesis of long-chain DNA products by UL54-UL44 complex was obtained only after a 30-min incubation, whereas formation of long DNA products by UL30-UL42 is typically detected after 5 to 10 min (data not shown). Very similar results were obtained when long-chain DNA synthesis by purified baculovirus-expressed UL54 in the presence of UL44 was assayed in the same manner (not shown).
FIG. 7.
Long-chain DNA synthesis by UL54 in the presence of UL44. Wild-type UL54 expressed in reticulocyte lysates was assayed for the ability to be stimulated by purified baculovirus-expressed UL44 by measuring the incorporation of labeled TTP on a poly(dA)-oligo(dT) template. The reaction products were visualized by autoradiography after electrophoresis on a 4% alkaline agarose gel. Lane 1 contains UL54 alone; lanes 2, 3, and 4 contain UL54 plus 1,000, 500, or 200 fmol of UL44, respectively; lane 5 contains 1,000 fmol of UL44 alone.
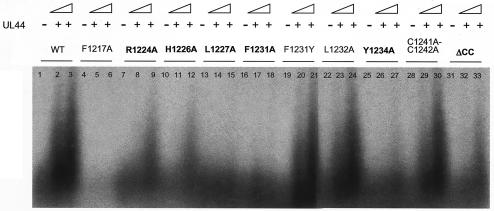
In this assay, UL54 mutants L1227A and F1231A, which were most impaired for interaction with UL44, exhibited no detectable long-chain DNA synthesis (Fig. 8, lanes 13 to 15 and 16 to 18). UL54 mutants R1224A, H1226A, Y1234A, and ΔCC, which exhibited reduced interaction with UL44, showed impaired synthesis of long DNA products (Fig. 8, lanes 7 to 9, 10 to 12, 25 to 27, and 31 to 33) when similar amounts of each UL54 mutant, as assessed by SDS-PAGE and autoradiography (data not shown) were used. Mutant F1231Y exhibited levels of long-chain DNA synthesis similar to those of the wild-type protein (Fig. 8, lanes 19 to 21), a finding in agreement with GST-pulldown data (Fig. 6). The F1217A mutant exhibited no long-chain DNA synthesis in the presence of UL44 (Fig. 8, lanes 5 and 6) but also had very low basal activity in the absence of UL44 (Fig. 8, lane 4; see also below). All other UL54 mutants, including L1232A mutant and the C1241A-C1242A double mutant as representative examples (Fig. 8, lanes 22 to 24 and 28 to 30) showed levels of long-chain DNA synthesis similar to those of wild-type UL54. Similar results were obtained by assaying the rate of incorporation of labeled dTTP into a poly(dA)-oligo(dT) template by using a filter-based assay (data not shown) as previously described (17).
FIG. 8.
Long-chain DNA synthesis by UL54 mutants in the presence of UL44. Wild-type or mutant UL54 proteins expressed in reticulocyte lysates were assayed for their ability to be stimulated by UL44 as described in legend to Fig. 7. Lanes 1, 4, 7, 10, 13, 16, 19, 22, 25, 28, and 31 contain wild-type or mutant UL54 alone; lanes 2, 5, 8, 11, 14, 17, 20, 23, 26, 29, and 32 and lanes 3, 6, 9, 12, 15, 18, 21, 24, 27, 30, and 33 contain wild-type or mutant UL54 plus 400 or 800 fmol of UL44, respectively.
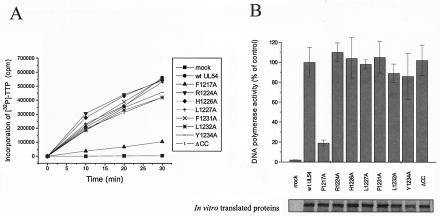
To test whether the inhibitory effects of the R1224A, H1226A, L1227A, F1231A, Y1234A, and ΔCC mutations could be due to impairment of UL54 basal DNA polymerase activity, the activities of wild-type and mutant in vitro-expressed UL54 proteins were tested in the absence of UL44 by using filter-based assays with a poly(dA)-oligo(dT) template-primer (Fig. 9A) or an activated calf thymus DNA template (data not shown) and normalized to the amount of each protein (Fig. 9B). These control experiments demonstrated that all UL54 mutants, apart from F1217A mutant, have basal DNA polymerase activities similar to that of the wild-type protein (Fig. 9 and data not shown).
FIG. 9.
Basal DNA polymerase activity of UL54 mutants. (A) The DNA polymerase activity of wild-type or mutant UL54 proteins expressed in reticulocyte lysates was measured by determining the incorporation of [32P]dTTP into a poly(dA)-oligo(dT) DNA template. Samples were taken after 0, 10, 20, and 30 min of incubation at 37°C and spotted onto DE81 filters. The filters were washed, and the radioactivity was counted. (B) The values were normalized to the amount of in vitro-translated proteins (lower panel), as estimated by SDS-PAGE analysis and phosphorimager quantification. The graph (upper panel) shows the average of data from three independent experiments, together with the standard deviations.
The biochemical properties of selected UL54 mutants are summarized in Table 2. These data show that the phenylalanine residue at position 1231 and the leucine 1227 of UL54 have crucial roles in both physical and functional interaction with UL44.
TABLE 2.
Summary of the biochemical properties of selected UL54 mutants
| Mutation | Activitya
|
||
|---|---|---|---|
| UL44 binding | Basal DNA polymerase activity | Stimulation by UL44 | |
| F1217A | +/−a | +/− | − |
| R1224A | +/− | + | +/− |
| H1226A | +/− | + | +/− |
| L1227A | − | + | − |
| F1231A | − | + | − |
| F1231Y | + | + | + |
| L1232A | + | + | + |
| Y1234A | +/− | + | +/− |
| C1241A | + | + | + |
| C1242A | + | + | + |
| C1241A-C1242A | + | + | + |
| ΔCC | +/− | + | +/− |
+, Wild-type levels of activity; +/−, partially impaired activity; −, no detectable activity. Binding to UL44 was determined by GST-pulldown experiments (Fig. 6), basal DNA polymerase activity was tested in a filter-based assay (Fig. 9), and stimulation of long-chain DNA synthesis in the presence of UL44 was determined by analyzing the products on alkaline agarose gels (Fig. 8).
DISCUSSION
In contrast to our relatively detailed understanding of the HSV-1 UL30-UL42 interaction, the characterization of the interaction between the two subunits of HCMV DNA polymerase, UL54 and UL44, has lagged. Recent data led to the hypothesis that UL54 might interact with the cognate accessory subunit through the C-terminal region (17), as already reported for HSV-1 UL30 (5, 6, 15, 19, 25, 26). The C-terminal region of UL54 is relatively disordered in aqueous solution but in certain solvents can fold into a partially helical structure (17), similar to that of the C terminus of HSV-1 UL30 (7, 29), even though the C termini of these HCMV and HSV proteins share almost no amino acid sequence homology. In addition, UL44 has been predicted to possess a structure with a “processivity fold” similar to that reported for UL42 (29), and preliminary data from the crystal structure of UL44 do in fact indicate that UL44 has an overall fold strikingly similar to that of UL42 (B. A. Appleton, D. J. Filman, D. M. Coen, and J. M. Hogle, unpublished results), again even though the HCMV accessory protein has very little sequence homology to UL42. Taken together, these observations suggest that the two subunits of HCMV DNA polymerase most likely interact in a way that is analogous to that of the two subunits of HSV DNA polymerase.
However, no direct demonstration that the C terminus of UL54 indeed represents the UL44 binding site existed, nor had the exact nature of the HCMV UL54-UL44 interaction been investigated. Thus, the goal of the present study was to dissect such an interaction, investigating whether the C terminus of UL54 is necessary and sufficient to specifically bind to UL44 and identifying discrete elements of UL54 that are important for interacting with UL44, by using both calorimetric and mutational approaches.
UL44 binding site of UL54.
A previous study showed that a peptide corresponding to the C-terminal 22 amino acids of UL54 could selectively inhibit the physical association between UL54 and UL44, suggesting that this region of UL54 might be involved in the interaction with UL44 (17). The data presented here show that such a peptide does specifically bind in solution to UL44 and that the deletion of this segment from UL54 eliminates binding to UL44. Thus, this relatively small region of UL54 is both necessary and sufficient for UL44 binding. Conversely, a shorter peptide, corresponding to the last 10 residues, exhibited no detectable binding to UL44 in solution (Fig. 2) and no detectable inhibitory activity against DNA synthesis by UL54 and UL44 (data not shown). Thus, this shorter region of UL54 is not sufficient for the interaction with UL44. Consistent with this observation, we found that the UL54 residues that are most important for UL54-UL44 interaction lie upstream of this segment. However, although the C-terminal 10-residue segment is not sufficient for UL44 binding, the extreme C terminus of UL54 is involved in the UL54-UL44 interaction, as deletion of the two C-terminal cysteines reduces binding of peptide 1 or UL54 to UL44.
The peptide corresponding to the last 22 amino acids of UL54 binds UL44 with a stoichiometry of one molecule of peptide per molecule of UL44, similar to the stoichiometry of binding of UL42 to peptides derived from HSV-1 UL30 (2). However, the stoichiometry of full-length UL54 and UL44 in the viral holoenzyme could be different from 1:1. In fact, preliminary results suggest that UL44 can form homodimers in solution (Appleton et al., unpublished), unlike UL42, which is a monomer (10, 11, 22a; B. A. Appleton, and D. M. Coen, unpublished results).
For the interaction between the C-terminal UL54 peptide and UL44, we measured a dissociation constant of ∼0.7 μM. Similar Kd (1 to 2 μM) and thus ΔG values (−7.9 to −8.4 kcal/mol) were previously obtained for the binding of C-terminal UL30 peptides to UL42 (2). However, the thermodynamics of the two interactions differ substantially, with much larger ΔH values for the HCMV interaction than for the HSV-1 interaction. This may relate to the more crucial role of hydrophobic versus hydrophilic residues in the two systems.
Contribution of the two C-terminal cysteines of UL54 to binding to UL44.
Our initial studies aimed at identifying residues of UL54 important for the interaction with UL44 focused on the last two residues of UL54, two cysteines, which are not present in any other known herpesvirus DNA polymerase. A previous study showed that a mutant UL54 C-terminal peptide lacking the two carboxy-terminal cysteines was >40-fold less potent than the wild-type peptide for disruption of the physical interaction between UL54 and UL44, suggesting an essential role for these residues in the UL54-UL44 interaction (17).
Here we demonstrate that the two C-terminal cysteines of UL54 most likely play a role in the UL54-UL44 interaction, although not an essential one. In fact, a mutant UL54 peptide lacking the two C-terminal cysteines still bound UL44 in solution, although with an affinity lower than that of the wild-type peptide. Consistent with these data, the deletion of these two residues in full-length UL54 caused only a partial impairment in the ability of UL54 to interact with UL44 in GST-pulldown assays. We also observed that replacement of either or both the C-terminal cysteines by alanine did not cause any significant effect on binding of UL54 to UL44. These findings suggest that interactions between UL44 and the extreme C terminus of UL54 likely involve the main chain of the two C-terminal cysteines rather than their side chains. Similarly, the crystal structure of HSV-1 UL42/UL30 peptide complex revealed that the last two residues of UL30 peptide, Leu1234 and Ala1235, interact with UL42 via hydrogen bonds between the main chain carbonyl oxygens and the amino group of the side chain of Lys289 of UL42 (29). More generally, the results indicate that changes in potency of inhibition do not correspond quantitatively to changes in binding affinity.
Single hydrophobic residues of UL54 are crucial for binding to UL44.
Biophysical studies of the UL30-UL42 binding interface demonstrated that, although numerous hydrophobic interactions are observed in the crystal structure of the UL30 peptide bound to UL42 (29), a few specific hydrogen bonds are crucial determinants of binding energy, and single amino acid changes, i.e., substitutions at positions corresponding to UL30 residues His1228 and Arg1229, can disrupt the UL30-UL42 interaction (2). Similarly, here we show that substitution of alanine for Leu1227 or Phe1231 in UL54 greatly impaired both the UL54-UL44 interaction in pulldown assays and long-chain DNA synthesis. Since the L1227A and F1231A mutants were not impaired in their basal DNA polymerase activity, we conclude that their defect in UL44 binding is specific and not due to global misfolding of the protein. Thus, these observations suggest a converging theme in the interaction between the two subunits of herpesvirus DNA polymerases, in that in both the HSV-1 UL30-UL42 interaction and the HCMV UL54-UL44 interaction a few residues are crucial for the binding of the catalytic subunit with the cognate accessory protein. However, the side chains of the residues that have been identified as important for HSV-1 UL30-UL42 and HCMV UL54-UL44 interaction are remarkably different, being basic in the first case and hydrophobic in the second. In fact, although two basic residues of UL54, Arg1224 and His1226, also seem to participate in UL44 binding, they are not crucial. Since both the Leu1227-to-Ala and the Phe1231-to-Ala change substitutes a larger side chain in a smaller one, maintaining the hydrophobic character of the residue, one may speculate that large side chains at these positions in UL54 are necessary to make intermolecular contacts with UL44. The importance of an aromatic hydrophobic side chain at position 1231 of UL54 for UL44 binding is suggested by the observation that substitution of Phe1231 with a tyrosine restores both binding to UL44 in pulldown assays and long-chain DNA synthesis by UL54 in the presence of UL44.
It is interesting that Leu1227 and Phe1231 are four residues apart. It is tempting to speculate that they might lie on the same face of an α-helix, given the helical propensity of this segment of UL54 (17). However, they are separated by a proline, making this possibility less likely. Alternatively, the presence of the proline leads to the speculation that UL54 makes contacts with two separated surfaces of UL44. Crystallographic studies will be necessary to test these ideas.
Although the data presented here identify the C-terminal residues as being both necessary and sufficient for binding to UL44, they do not exclude possible contributions to UL44 binding by regions upstream of the UL54 C terminus. The only upstream mutation that we tested, F1217A, resulted in decreased UL44 binding but also resulted in almost no basal DNA polymerase activity. Thus, this substitution either independently affects both activities or, more simply, causes a global misfolding of the protein. A previous study showed that peptides spanning the C proximal region of UL54, up to residue 1161, had no effect on UL54-UL44 physical interaction (17). These observations suggest that if there is a contribution of upstream residues of UL54 to UL44 binding, it is small relative to that of the C terminus. However, the role of this region remains to be defined.
Implications for drug discovery.
The importance and specificity of protein associations in viral replication and pathogenesis make protein-protein interactions attractive targets for therapeutic intervention. Compounds capable of selectively interfering with such interactions represent novel potential agents for antiviral therapy. However, although the disruption of specific protein-protein contacts is a promising strategy for drug development, the nature of these interactions can make this goal impractical. Many protein-protein interactions involve large surfaces or multiple contacts, making it unlikely that a single small molecule could interfere with them. Our studies identify aspects of the subunit interface of UL54 that are important for its contact with UL44. Only a relatively small surface of UL54 appears to be necessary and sufficient for UL44 binding, and the substitution of single residue side chains is sufficient to disrupt the HCMV UL54-UL44 interaction and thus to inhibit the holoenzyme activity. These findings herald the prospect that small molecules targeting the relevant side chains could interfere with UL54-UL44 binding. Encouragement for this approach comes from the recent identification of small inhibitory molecules able to block the HSV-1 UL30-UL42 interaction in vitro, as well as virus replication (B. D. Pilger, C. Cui, and D. M. Coen, unpublished data). Since the residues most important for the HSV-1 UL30-UL42 and HCMV UL54-UL44 interactions are not conserved, small molecule inhibitors targeting the residue side chains could be significantly more virus specific than most of the drugs currently licensed for antiherpesvirus chemotherapy.
Acknowledgments
We thank P. F. Ertl for kindly providing the pRSET-Pol and pRSET44 plasmids, H. S. Marsden for purified baculovirus-expressed UL54 and UL44 proteins and for provision of peptide 1 variants, and C. Cui for supplying purified MBP-UL42ΔC340. We also thank J. C. W. Randell for advice on ITC and E. Fontaine for help in constructing some of the UL54 mutants.
This study was supported by NIH grant AI19838.
REFERENCES
- 1.Archakov, A. I., V. M. Govorun, A. V. Dubanov, Y. D. Ivanov, A. V. Veselovsky, P. Lewi, and P. Janssen. 2003. Protein-protein interactions as a target for drugs in proteomics. Proteomics 3:380-391. [DOI] [PubMed] [Google Scholar]
- 2.Bridges, K. G., C. S. Chow, and D. M. Coen. 2001. Identification of crucial hydrogen-bonding residues for the interaction of herpes simplex virus DNA polymerase subunits via peptide display, mutational, and calorimetric approaches. J. Virol. 75:4990-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cihlar, T., M. D. Fuller, and J. M. Cherrington. 1997. Expression of the catalytic subunit (UL54) and the accessory protein (UL44) of human cytomegalovirus DNA polymerase in a coupled in vitro transcription/translation system. Protein Expr. Purif. 11:209-218. [DOI] [PubMed] [Google Scholar]
- 4.Coen, D. M., and P. A. Schaffer. 2003. Antiherpesvirus drugs: a promising spectrum of new drugs and drug targets. Nat. Rev. Drug Discov. 2:278-288. [DOI] [PubMed] [Google Scholar]
- 5.Digard, P., and D. M. Coen. 1990. A novel functional domain of an α-like DNA polymerase. J. Biol. Chem. 265:17393-17396. [PubMed] [Google Scholar]
- 6.Digard, P., W. Bebrin, K. Weisshart, and D. M. Coen. 1993. The extreme C terminus of herpes simplex virus DNA polymerase is crucial for functional interaction with processivity factor UL42 and for viral replication. J. Virol. 67:398-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Digard, P., K. P. William, P. Hensley, I. S. Brooks, C. E. Dahl, and D. M. Coen. 1995. Specific inhibition of herpes simplex virus DNA polymerase by helical peptides corresponding to the subunit interface. Proc. Natl. Acad. Sci. USA 92:1456-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ertl, P. F., M. S. Thomas, and K. L. Powell. 1991. High-level expression of DNA polymerases from herpesviruses. J. Gen. Virol. 72:1729-1734. [DOI] [PubMed] [Google Scholar]
- 9.Ertl, P. F., and K. L. Powell. 1992. Physical and functional interaction of human cytomegalovirus DNA polymerase and its accessory protein (ICP36) expressed in insect cells. J. Virol. 66:4126-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallo, M. L., D. H. Jackwood, M. Murphy, H. S. Marsden, and D. S. Parris. 1988. Purification of the herpes simplex virus type 1 65-kilodalton DNA-binding protein: properties of the protein and evidence of its association with the virus-encoded DNA polymerase. J. Virol. 62:2874-2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb, J., A. I. Marcy, D. M. Coen, and M. D. Challberg. 1990. The herpes simplex virus type 1 UL42 gene product: a subunit of DNA polymerase that functions to increase processivity. J. Virol. 64:5976-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamatake, R. K., M. Bifano, D. J. Tenney, W. W. Hurlburt, and M. G. Cordingley. 1993. The herpes simplex virus type 1 DNA polymerase accessory protein, UL42, contains a functional protease-resistant domain. J. Gen. Virol. 74:2181-2189. [DOI] [PubMed] [Google Scholar]
- 13.Heilbronn, R., G. Jahn, A. Burkle, U. K. Freese, B. Fleckenstein, and H. zur Hausen. 1987. Genomic localization, sequence analysis, and transcription of the putative human cytomegalovirus DNA polymerase gene. J. Virol. 61:119-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouzarides, T., A. T. Bankier, S. C. Satchwell, K. Weston, P. Tomlinson, and B. G. Barrell. 1987. Sequence and transcription analysis of the human cytomegalovirus DNA polymerase gene. J. Virol. 61:125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loregian, A., E. Papini, B. Satin, H. S. Marsden, T. R. Hirst, and G. Palù. 1999. Intranuclear delivery of an antiviral peptide mediated by the B subunit of Escherichia coli heat-labile enterotoxin. Proc. Natl. Acad. Sci. USA 96:5221-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loregian, A., H. S. Marsden, and G. Palù. 2002. Protein-protein interactions as targets for antiviral chemotherapy. Rev. Med. Virol. 12:239-262. [DOI] [PubMed] [Google Scholar]
- 17.Loregian, A., R. Rigatti, M. Murphy, E. Schievano, G. Palù, and H. S. Marsden. 2003. Inhibition of human cytomegalovirus (HCMV) DNA polymerase by C-terminal peptides from the UL54 subunit. J. Virol. 77:8336-8344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mar, E. C., J. F. Chiou, Y. C. Cheng, and E. S. Huang. 1985. Human cytomegalovirus-induced DNA polymerase and its interaction with the triphosphates of 1-(2′-deoxy-2′-fluoro-β-d-arabinofuranosyl)-5-methyluracil, -5-iodocytosine, and -5-methylcytosine. J. Virol. 56:846-851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsden, H. S., M. Murphy, G. L. McVey, K. A. MacEachran, A. M. Owsianka, and N. D. Stow. 1994. Role of the carboxy terminus of herpes simplex virus type 1 DNA polymerase in its interaction with UL42. J. Gen. Virol. 75:3127-3135. [DOI] [PubMed] [Google Scholar]
- 20.Nishiyama, Y., K. Maeno, and S. Yoshida. 1983. Characterization of human cytomegalovirus-induced DNA polymerase and the associated 3′-to-5′ exonuclease. Virology 124:221-231. [DOI] [PubMed] [Google Scholar]
- 21.Pari, G. S., M. A. Kacica, and D. G. Anders. 1993. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 67:2575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Randall, J. C. W., and D. M. Coen. The herpes simplex virus processivity factor, UL42, binds DNA as a monomer. J. Mol. Biol., in press. [DOI] [PubMed]
- 23.Ripalti, A., M. C. Boccuni, F. Campanini, and M. P. Landini. 1995. Cytomegalovirus-mediated induction of antisense mRNA expression to UL44 inhibits virus replication in astrocytoma cell line: identification of an essential gene. J. Virol. 69:2047-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva, N. L., R. S. Haworth, D. Singh, and L. Fliegel. 1995. The carboxyl-terminal region of the Na+/H+ exchanger interacts with mammalian heat shock protein. Biochemistry 34:10412-10420. [DOI] [PubMed] [Google Scholar]
- 25.Stow, N. D. 1993. Sequences at the C terminus of the herpes simplex virus type 1 UL30 protein are dispensable for DNA polymerase activity but not for viral origin-dependent DNA replication. Nucleic Acids Res. 21:87-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tenney, D. J., P. A. Micheletti, J. T. Stevens, R. K. Hamatake, J. T. Matthews, A. R. Sanchez, W. W. Hurlburt, M. Bifano, and M. G. Cordingley. 1993. Mutations in the C terminus of herpes simplex virus type 1 DNA polymerase can affect binding and stimulation by its accessory protein UL42 without affecting basal polymerase activity. J. Virol. 67:543-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsai, C. J., S. L. Lin, H. J. Wolfson, and R. Nussinov. 1997. Studies of protein-protein interfaces: a statistical analysis of the hydrophobic effect. Protein Sci. 6:53-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiland, K. L., N. L. Oien, F. Homa, and M. W. Wathen. 1994. Functional analysis of human cytomegalovirus polymerase accessory protein. Virus Res. 34:191-206. [DOI] [PubMed] [Google Scholar]
- 29.Zuccola, H. J., D. J. Filman, D. M. Coen, and J. M. Hogle. 2000. The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase. Mol. Cell 5:267-278. [DOI] [PubMed] [Google Scholar]