Abstract
The intestinal flora may promote colon tumor formation. Here we explore immunologic mechanisms of colonic carcinogenesis by a human colonic bacterium, enterotoxigenic Bacteroides fragilis (ETBF). ETBF that secretes B. fragilis toxin (BFT) causes human inflammatory diarrhea but also asymptomatically colonizes a proportion of the human population. Our results indicate that whereas both ETBF and nontoxigenic B. fragilis (NTBF) chronically colonize mice, only ETBF triggers colitis and strongly induces colonic tumors in multiple intestinal neoplasia (Min) mice. ETBF induces robust, selective colonic signal transducer and activator of transcription-3 (Stat3) activation with colitis characterized by a selective T helper type 17 (TH17) response distributed between CD4+ T cell receptor-αβ (TCRαβ)+ and CD4−8−TCRγδ+ T cells. Antibody-mediated blockade of interleukin-17 (IL-17) as well as the receptor for IL-23, a key cytokine amplifying TH17 responses, inhibits ETBF-induced colitis, colonic hyperplasia and tumor formation. These results show a Stat3- and TH17-dependent pathway for inflammation-induced cancer by a common human commensal bacterium, providing new mechanistic insight into human colon carcinogenesis.
Infection-associated inflammatory processes are known to enhance carcinogenesis in the affected organs. In humans, for example, chronic hepatitis (hepatitis B virus or hepatitis C virus) leads to liver cancer, and chronic Helicobacter pylori infection leads to gastric cancer in some individuals1,2. Increased cancer incidence is likewise found in experimental mouse models of both infection-induced and noninfectious inflammation. Conditional knockout mice have shown the importance of nuclear factor-κB (NF-κB) signaling not only in the epithelial cells that are the target of transformation but also in myeloid cells that contribute to inflammation3,4. How NF-κB–induced inflammatory processes drive carcinogenesis is unclear, although IL-6 seems to be pivotal5,6. IL-6 induces the procarcinogenic Stat3 pathway and transcriptionally activates proliferative, antiapoptotic and proangiogenic genes involved in cancer growth7. Stat3 signaling organizes the immune microenvironment of tumors to block generation of antitumor immune responses8.
In contrast, little information exists on how adaptive immunity, particularly T cell responses, promote cancer. Given that T cell responses generate antitumor responses and more tumors occur in Rag−/− mice9 and mice with defective interferon signaling10, chronic innate inflammatory responses are postulated to promote carcinogenesis, whereas T cell–dependent responses are postulated to inhibit carcinogenesis. Three effector pathways of T cell differentiation are now defined: TH1 responses promoted by Stat1 and Stat4 signaling, TH2 responses promoted by Stat6 signaling and TH17 responses promoted by Stat3 signaling11,12. TH1 responses, driven by IL-12 and characterized by interferon-γ (IFN-γ) production, are typically anticarcinogenic, whereas little is known about the contribution of TH2 or TH17 responses to cancer10.
The role of infectious and inflammatory processes in colon carcinogenesis is of considerable interest, as ~1 × 1013 commensal bacteria colonize the colon, with inflammation resulting if colonic epithelial homeostasis is disrupted13. Indeed, ulcerative colitis results in predictable development of colon cancer over time. The key role of inflammation in colonic carcinogenesis is emphasized by the diminished tumor formation in multiple intestinal neoplasia (Min) mice (heterozygous for the adenomatous polyposis coli (Apc) gene) when Toll-like receptor signaling is abrogated14,15. Because certain human enteric bacteria cause colitis, there is interest in whether any of them can promote colon cancer, analogous to the H. pylori promotion of stomach cancer1,16. We are studying a molecular subgroup of B. fragilis, ETBF, that produces a metalloprotease toxin termed BFT. ETBF causes acute inflammatory diarrheal disease in children and adults but also asymptomatically colonizes up to 20%–35% of adults17. One study has found higher ETBF colonization in individuals with colon cancer relative to controls without colon cancer18.
Here we explored potential mechanisms of ETBF-induced colitis and carcinogenesis. We show that ETBF persistently colonizes Min mice with a rapid, strong selective activation of Stat3, whereas the non–toxin-producing NTBF strain colonizes but induces neither colitis nor Stat activation. ETBF colitis is characterized by a selective TH17 response with markedly increased colonic tumor formation. The TH17 response directly contributes to ETBF-induced tumorigenesis. These results demonstrate a Stat3- and TH17-dependent pathway for colon carcinogenesis induced by a common human commensal bacterium, thereby defining a distinct role for adaptive immunity in colon cancer pathogenesis.
RESULTS
ETBF stimulates rapid colitis and colon tumors in Min mice
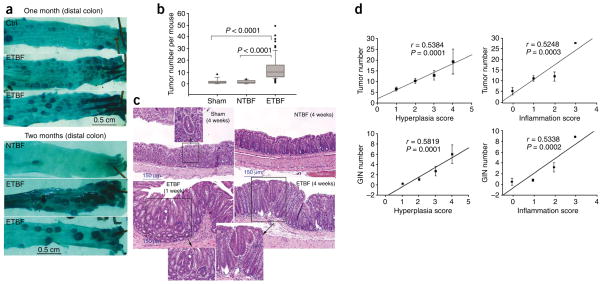
Min mice colonized with ETBF, but not NTBF, usually developed brief diarrhea by 2–3 d, with resolution of the symptoms by 4–5 d after colonization. Asymptomatic high-level colonization (≥1 × 109 colony-forming units per g feces) with NTBF or ETBF occurred by day 3 after infection and persisted. Only ETBF-colonized mice showed a marked increase in colonic thickness, inflammation and visible colonic tumors, especially distally, at 4 weeks or later (Fig. 1a–c and Table 1). Histopathology of ETBF-colonized colons confirmed increases in inflammation, hyperplasia and gastrointestinal intraepithelial neoplasia (GIN) foci compared to sham-treated or NTBF-infected colons (Table 1 and Fig. 1c). Linear regression analysis of inflammation or hyperplasia severity supported an association between ETBF-induced inflammation or hyperplasia with GIN and gross colon tumor detection (Fig. 1d). Furthermore, we detected GIN, inflammation and hyperplasia only in ETBF-colonized colons at 1 week after colonization (Fig. 1c). These data suggest that ETBF induces de novo tumor formation quickly and may enhance tumor growth rates. Tumors in ETBF-colonized mice were typically laden with inflammatory infiltrates comprised of granulocytes and mononuclear cells not seen in tumors in sham-inoculated or NTBF-colonized Min mice (Fig. 1c). We did not observe an increase in the number of small bowel tumors between experimental groups (data not shown), consistent with the known colonic niche for B. fragilis colonization.
Figure 1.
ETBF stimulates colonic inflammation and enhances colonic tumor formation in Min mice. (a) Methylene blue–stained representative samples of distal colons of sham control, NTBF-colonized and ETBF-colonized mice showing thickened mucosal folds and excess tumors, visualized in mice colonized with ETBF for 1–2 months. (b) Distribution of visible tumor numbers detected in sham control, NTBF- or ETBF-colonized mice at 4–6 weeks after inoculation. Tumor distributions are shown as box-and-whisker plots. n = 14, 10 or 75 for sham control, NTBF or ETBF, respectively. (c) Distal colon histopathology of sham control and NTBF-colonized mice at 4 weeks and ETBF-colonized mice at 1 week and 4 weeks after inoculation. Insets show GIN foci in sham and ETBF-colonized mice. (d) Linear regression analysis of histological scores of ETBF-colonized colons for inflammation and hyperplasia versus visible colon tumor formation or GIN foci. Error bars represent means ± s.e.m.
Table 1.
Min mouse colon histological scores 1 week and 4–6 weeks after ETBF or NTBF colonization
| Median (range) |
||||
|---|---|---|---|---|
| Inflammation | Hyperplasia | GIN | Gross tumors | |
| 1 week | ||||
| Sham (n=6) | 0 (0–0) | 1 (0–1) | 0 (0–0) | NA |
| NTBF (n=4) | 0 (0–0) | 0 (0–0) | 0 (0–0) | NA |
| ETBF (n=16) | 2 (0–3)a | 3 (2–4)b | 1.5 (0–4)c | NA |
| 4–6 weeks | ||||
| Sham (n=9) | 0 (0–1) | 0 (0–1) | 0 (0–0) | 2 (0–8) |
| NTBF (n=5) | 0 (0–0) | 0 (0–1) | 0 (0–0) | 3 (2–4) |
| ETBF (n=59) | 1 (0–3)d | 2 (1–4)e | 1 (0–16)f | 9 (2–49)g |
NA, not applicable.
P < 0.015 versus 1 week sham and NTBF; independent comparisons.
P < 0.0004 versus 1 week sham and NTBF; independent comparisons.
P < 0.042 versus 1 week sham and NTBF; independent comparisons.
P < 0.0006 versus 4–6 week sham and NTBF; independent comparisons.
P < 0.0004 versus 4–6 week sham and NTBF; independent comparisons.
P < 0.018 versus 4–6 week sham and P =0.10 versus NTBF; independent comparisons.
P < 0.0005 versus 4–6 week sham and NTBF; independent comparisons.
ETBF selectively activates Stat3 in the colon
To address the mechanisms of ETBF-induced colitis and carcinogenesis, we assessed activation of Stat proteins. Stat proteins are a family of transcription factors activated by cytokine receptor signaling through tyrosine phosphorylation with nuclear translocation and are central to the regulation of immune responses19. Stat1 and Stat4 contribute to TH1-dependent immune responses, whereas Stat6 has a key role in TH2 responses. Stat3 transduces signals from numerous growth factor and cytokine receptors, is constitutively activated in diverse cancers and is absolutely required for TH17 cell generation while simultaneously negatively regulating TH1-mediated inflammation11,20–22.
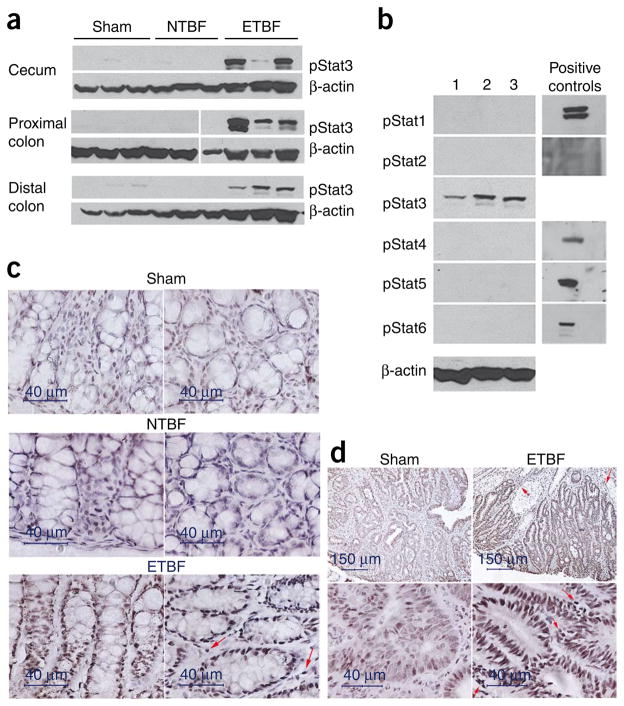
Using antibodies specific for each phosphorylated Stat protein, we found that only phosphorylated Stat3 (pStat3) was abundant in the colonic mucosa of ETBF-colonized Min mice at 2 d after infection (Fig. 2a), whereas we found only faint pStat3 staining in some sham or NTBF-colonized Min mouse colons (Fig. 2a). We did not detect pStat1, pStat2, pStat5 or pStat6 in the colons of any mouse experimental group (Fig. 2b). We did detect very faint pStat4 signals in some ETBF-colonized Min mice (data not shown). The colons of wild-type (WT) C57BL/6 mice revealed identical pStat staining (Supplementary Fig. 1a), indicating that the highly selective activation of predominantly Stat3 by ETBF colonization is independent of the Apc mutation in Min mice.
Figure 2.
ETBF specifically activates Stat3 in the colons of Min mice. (a) Western blot analysis for activated Stat3 (pStat3) in colon samples of sham control Min mice or Min mice colonized with NTBF or ETBF for 2 d. Three individual mice are shown for each experimental condition. β-actin serves as a protein benchmark; protein concentrations per sample were equivalent (4.3–4.9 μg μl−1). The break in the gel (proximal colon) indicates that samples were run on separate gels analyzed in parallel for the same experiment. Data are representative of five sham-inoculated, six NTBF-colonized and six ETBF-colonized Min mice. (b) Western blot analysis for pStat proteins in colons of three ETBF-colonized Min mice. Positive controls for each pStat antibody are shown. β-actin served as a protein loading control. (c) Immunohistochemistry for pStat3 in distal colon of ETBF-colonized mice 4 weeks after inoculation compared to sham or NTBF-colonized mice. Arrows depict a subset of inflammatory cells in the lamina propria of ETBF-colonized mice that show pStat3 staining (see also Supplementary Fig. 1b). Representative of two sham, four NTBF-colonized and seven ETBF-colonized Min mice. (d) Immunohistochemistry for pStat3 in a large colon tumor from an eight-week-old, sham-inoculated Min mouse and a similar-sized colon tumor in a Min mouse colonized with ETBF for 4 weeks. Arrows designate pStat3 staining of inflammatory cells in the interstitium.
We used immunohistochemistry to examine the cellular localization and time course of pStat3 activation in ETBF-colonized mice. Stat3 activation occurred in colonic epithelial cells and a subset of infiltrating immune cells in ETBF-colonized Min mice at 2 d to 4 weeks after colonization compared to pStat3 staining in NTBF- colonized or sham mice at the same time points (Fig. 2c and Supplementary Fig. 1b). In addition to nontumorous epithelium, we found 13 tumors of variable sizes in random colon histopathology sections of seven ETBF-colonized Min mice, and all 13 tumors showed intense epithelial cell pStat3 activation and pStat3 staining in a subset of mucosal immune cells (Fig. 2d). The low frequency of tumors in NTBF-colonized or sham-treated Min mice limited our ability to detect tumors in random colon sections. However, in three tumors that we identified in NTBF-colonized or sham Min mice sections, the pStat3 staining was less consistent and less intense, particularly in the epithelial compartment (Fig. 2d). Thus, beyond inducing tumors in Min colons, ETBF colonization quantitatively alters at least one oncogenic signaling pathway in already established tumors.
ETBF induces dominant colonic TH17 inflammatory infiltrates
Stat3 signaling is absolutely required for the generation of TH17 cells, and pStat3 binds the Il17a and Il17f promoters20–22. We, therefore, wondered whether pStat3 activation by ETBF colonization of Min mice initiates a TH17 mucosal immune response. FACS analysis (n = 8 experiments) of isolated intraepithelial lymphocyte and lamina propria lymphocyte populations showed an approximately four- to fivefold higher number of CD4+ T cells in the lamina propria of ETBF-colonized Min mice after 1 week as compared to NTBF- colonized or sham Min mice (data not shown).
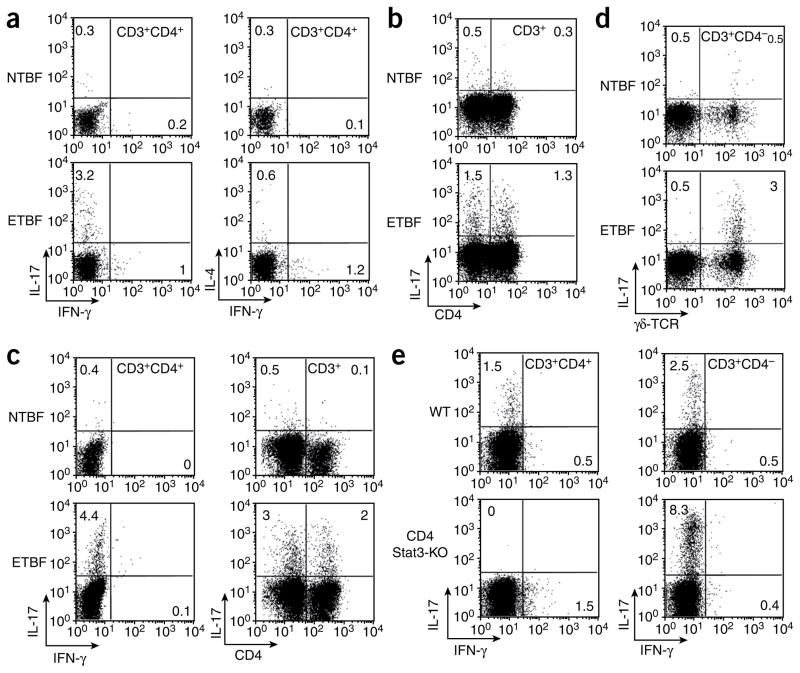
ETBF-colonized Min mice indeed developed a strongly skewed TH17 response characterized by equally contributory IL-17–secreting CD3+CD4+ and CD3+CD4− effector populations in the lamina propria (Fig. 3a,b). We did not find expanded IL-4–producing T cell effector populations, and the modest number of IFN-γ–producing CD3+CD4+ T cells produced low amounts of IFN-γ (Fig. 3a). In contrast to colonic lymphocytes, CD4+TCRαβ+ and CD8+TCRαβ+ splenic cells isolated from ETBF-colonized mice showed enhanced IFN-γ staining with minimal IL-17 production (Supplementary Fig. 2). We obtained similar results for WT mice (Fig. 3c).
Figure 3.
ETBF, but not NTBF, induces IL-17–producing CD3+CD4+ T lymphocytes and γδ T lymphocytes in the colon lamina propria of Min and WT mice 1 week after NTBF or ETBF inoculation. (a) ICS for IL-17, IFN-γ and IL-4 in CD3+CD4+ T lymphocytes of Min mice. Dot plots are derived from the CD3+CD4+ gate. (b) ICS for IL-17 in CD3+CD4+ and CD3+CD4− lymphocytes from the lamina propria of ETBF-colonized Min mice. Dot plots are derived from CD3+ gate. (c) ICS for IL-17 and IFN-γ in CD3+CD4+ and CD3+CD4− T lymphocytes of C57BL/6 mice. Dot plots are derived from CD3+CD4+ and CD3+ gates. (d) ICS for IL-17 in γδ T cells from the lamina propria of ETBF-colonized Min mice. Dot plots are derived from CD3+CD4− gate. (e) ICS staining in CD3+CD4+ and CD3+CD4− lymphocytes from WT and CD4 Stat3-KO C57BL/6 mice. Dot plots are derived from the CD3+ gate. Each panel is representative of at least three independent experiments except e (two independent experiments). The numbers inside the plots indicate the percentage of the cell population showing the quadrant characteristic.
To further identify the IL-17–producing mucosal T cell populations in ETBF-colonized Min mice, we used antibodies to distinguish between classical (TCRαβ-bearing) and nonclassical (TCRγδ-bearing) T cells. IL-17 production by CD3+CD4− T cells in ETBF-colonized Min or WT mice was attributable to CD3+TCRγδ+ lamina propria cells (Fig. 3d). In contrast, neither lamina propria CD3+CD8+ cells nor CD3− cells showed intracellular IL-17 staining in ETBF-colonized Min or WT mice (Supplementary Fig. 3).
Beyond Stat3 activation, induction of a TH17 immune response typically requires IL-6, which, together with transforming growth factor-β (TGF-β) (and augmented by IL-1β), induces TH17 differentiation, whereas expansion of IL-17–producing CD4+ lymphocytes is promoted by IL-23 (ref. 11). Thus, we examined whether Stat3 is required for ETBF-induced IL-17 production by colonic CD3+CD4+ T cells isolated from ETBF-colonized WT mice with functional Stat3 knockout in the CD4 T cell compartment (CD4 Stat3-KO). CD4-targeted Stat3 knockout obliterated ETBF induction of IL-17 in this T cell subset, whereas IL-17 persisted in Stat3-competent CD3+CD4− T cells (Fig. 3e). Histopathology of ETBF-colonized, CD4-targeted, Stat3-knockout mice revealed significant decreases in inflammation and hyperplasia compared to littermate Stat3-sufficient mice, consistent with the contribution of CD4+ cells and Stat3 signaling to ETBF colitis (P ≤ 0.03, Supplementary Table 1).
By quantitative RT-PCR (qRT-PCR), we detected markedly higher levels of IL-17 messenger RNA in the colonic mucosa of ETBF- colonized Min and WT mice relative to NTBF-colonized mice at 1 week after colonization (Supplementary Fig. 4), consistent with our detection of IL-17 protein in CD4+ and CD4− T lymphocytes (Fig. 3b). We also found higher levels of IL-1β, IL-6, IL-23 and TGF-β mRNA 1 week after ETBF colonization, although the differences in TGF-β mRNA were not significant (Supplementary Fig. 4). A major component of TGF-β regulation occurs after transcription; thus, its mRNA levels are less informative than the mRNA levels of the other cytokines. Lastly, we examined sorted CD3+CD4+ lymphocytes from Min mice colonized with ETBF or NTBF by qRT-PCR for induction of the gene encoding the TH17-specific transcription factor, RORγt23, in parallel with the Il17a gene. In CD3+CD4+ T cells isolated from the colons of ETBF-colonized mice, expression of the gene encoding RORγt was tenfold higher (± 2.6, mean ± s.e.m.) and Il17a gene expression was 21-fold higher (± 2.6) compared to NTBF-colonized mice.
Blockade of IL-17 inhibits ETBF-induced colon tumors
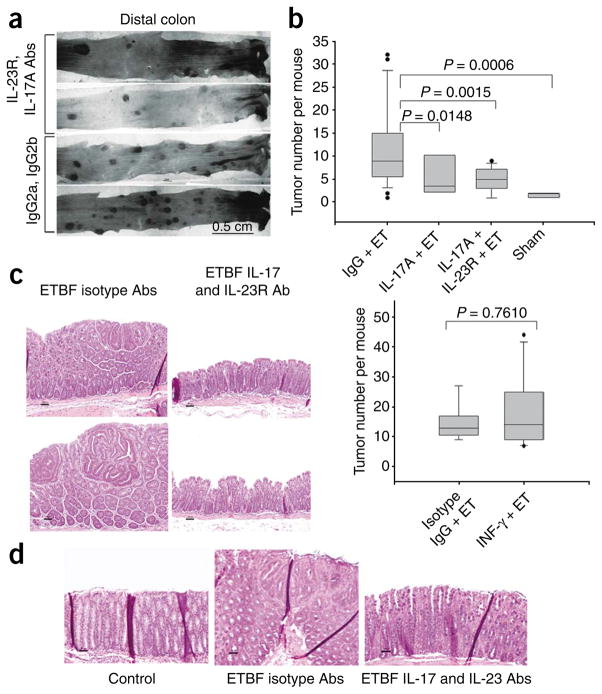
To evaluate the contribution of TH17 inflammatory cells to ETBF-induced tumor formation in Min mice, we conducted experiments with IL-17A–, IL-23 receptor (IL-23R)- or IFN-γ–neutralizing antibodies. Of the six isoforms of IL-17, IL-17A predominates in humans and mice and in the colonic mucosa after 1 week in ETBF-colonized Min or WT mice (Supplementary Fig. 4). Blockade of IL-17A alone or combined blockade with IL-23R significantly inhibited colon tumor formation at 5 weeks after colonization (Fig. 4a,b). The size distribution of the tumors did not differ between the mice treated with IL-17– and IL-23R–neutralizing antibodies mice compared to isotype controls (Supplementary Fig. 5), emphasizing the contribution of the TH17 response in tumor initiation and suggesting a minor role in tumor growth rate. In contrast, IFN-γ blockade did not modify ETBF-induced colon tumorigenesis (Fig. 4b). IL-17A blockade did not detectibly modify pStat3 levels, as determined by western blotting, nor did it affect the cellular distribution of pStat3, as determined by immunohistochemistry, in the colons of ETBF-colonized WT or Min mice, suggesting that Stat3 activation is upstream of IL-17 induction (Supplementary Fig. 6).
Figure 4.
Blockade of IL-17 and IL-23R, but not IFN-γ, inhibits ETBF-induced colonic tumor formation in Min mice. (a) Methylene blue–stained representative samples of distal colons of mice colonized with ETBF for 5 weeks and treated with IL-17 and IL-23R blocking antibodies or isotype control antibodies. (b) Depiction of tumor number distribution by box-and-whisker plots in ETBF-colonized mice treated with isotype-matched antibodies (IgG + ET; experimental positive control) and ETBF-colonized mice treated with IL-17– (IL-17A + ET), IL-17– and IL-23R– (IL-17 + IL-23R + ET) or IFN-γ– (IFN-γ + ET) blocking antibodies after 5 weeks. Sham-inoculated mice served as an experimental negative control. Top, n = 24 for IgG + ET, 8 for IL-17 + ET, 14 for IL-17 + IL-23R + ET and 7 for sham. Bottom, n = 9 for IgG + ET and 11 for IFN-γ + ET. (c) Histopathology of distal colon tumors in Min mice colonized with ETBF for 5 weeks and treated with isotype control antibodies (left) or IL-17– and IL-23R–blocking antibodies (right). Two representative mice of 24 (isotype control) or 14 (IL-17–blocking and IL-23R–blocking antibody treated) per treatment group are shown. (d) Histopathology of distal colon of Min mice colonized with ETBF for 1 week and treated with isotype control antibody (center) or IL-17– and IL-23R–blocking antibodies (right). Left image shows the distal colon of a sham control Min mouse. Micrographs are representative of three sham control, five ETBF and isotype control antibody–treated and four ETBF, IL-17– and IL-23R–neutralizing antibody–treated mice.
Histopathology revealed marked inhibition of colonic mucosal proliferation with fewer infiltrating leukocytes (Fig. 4c) and GIN foci on random colon tissue sections in ETBF-colonized Min mice treated with IL-17A–blocking antibodies or both IL-17A– and IL-23R–blocking antibodies for 5 weeks compared to ETBF-colonized mice treated with isotype control antibodies for 5 weeks (P < 0.02, Supplementary Table 2). We obtained similar results in ETBF-colonized Min mice treated with IL-17A and IL-23R blockade for 1 week (Fig. 4d).
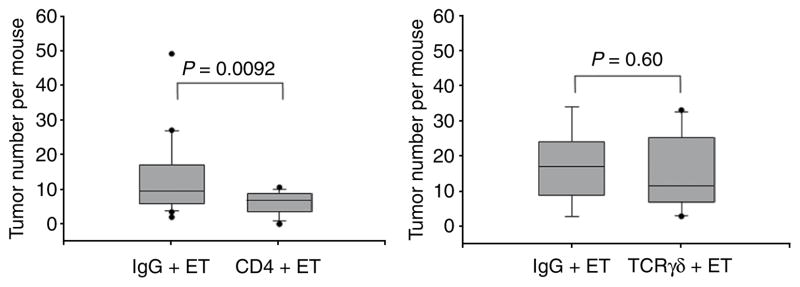
We next examined whether depletion of CD4+ or TCRγδ+ T lymphocytes modifies tumor induction in ETBF-colonized Min mice. Antibody-mediated depletion of CD4+ T lymphocytes significantly inhibited the accelerated tumor formation detected 4–6 weeks after ETBF inoculation when compared to ETBF-colonized mice treated with an isotype control antibody (P = 0.009), whereas TCRγδ+ T cell depletion did not modify ETBF-induced colon tumors (Fig. 5). This result provides a direct example that endogenous CD4+ T responses contribute to infection-induced carcinogenesis.
Figure 5.
CD4+, but not γδ+, T cell depletion inhibits tumor formation in ETBF-colonized Min mice. CD4+ T cells (CD4 + ET) or γδ T cells (TCRγδ + ET) were depleted in ETBF-colonized mice using specific monoclonal antibodies and compared to ETBF-colonized mice treated in parallel with IgG isotype control antibodies (IgG + ET). Distribution of tumor numbers is shown using box-and-whisker plots. Left, n = 22 for IgG + ET and 24 for CD4 + ET. Right, n = 9 for IgG + ET and 11 for TCRγδ + ET.
DISCUSSION
To our knowledge, the results presented here are the first to demonstrate a direct role for endogenous T cell immune responses in infection-induced carcinogenesis. Our intracellular cytokine staining (ICS) and in vivo antibody blockade experiments further implicate a TH17 response, driven by Stat3 activation, as being crucial to the procarcinogenic effect. This newly described mechanism for infection-induced carcinogenesis may be highly relevant for human carcinogenesis, as, in contrast to all other models of murine colitis, our model uses colonization with a human commensal bacterium, ETBF, that has been epidemiologically linked to colon cancer18. In this model, essentially all of the IL-17 production is similarly distributed between two T cell subsets, CD4+TCRαβ+ and CD4−8−TCRγδ+ T cells. Our in vivo depletion experiments emphasize the contribution of classical CD4+ TH17 cells in ETBF-induced colon tumorigenesis, as CD4+, but not TCRγδ+, T cell depletion markedly lowers tumor number. It is still possible that colonic γδ T cells contribute to the overall tumorigenesis process but that IL-17 production by CD4+ TH17 cells is sufficient to induce tumorigenesis in the absence of γδ T cells.
In addition to promoting TH17 development and IL-17 transcription, it has been found that Stat3 activation in the tumor microenvironment inhibits IL-12p35 transcription while enhancing IL-23p19 transcription, thereby shifting the balance from IL-12 to IL-23 (ref. 24). This finding, together with the finding that 9,10-dimethyl-1,2-benzanthracene-induced skin carcinogenesis is diminished in IL-23p19–knockout mice and enhanced in IL-12p35–knockout mice25 as well as the results presented here, suggest that Stat3 potentially promotes a complex procarcinogenic TH17-type immune response. Beyond the immune compartment, Stat3 activation in the intestinal epithelial compartment also contributes to colon carcinogenesis in the axozymethane with dextran sulfate sodium model26,27. Tissue-selective Stat3 knockouts on the Min background will be necessary to define the specific roles of Stat3 activation in the various cell types in ETBF colitis.
Although recent work on inflammation-induced carcinogenesis has focused on innate pathways, particularly the NF-κB and myeloid differentiation factor-88 pathways3,14, little is known about the direct role of adaptive responses in general and T cell responses in particular. In a transgenic model of skin carcinogenesis driven by keratinocyte-specific expression of the human papillomavirus-16 E6 and E7 oncogenes, B lymphocytes proved to be major promoters of tumor formation28. Recent adoptive transfer studies of activated T cells into Rag2−/− (T and B cell deficient) × Min mice29,30 and Ifng−/− TCR transgenic T cells into RIP1-Tag2 (rat insulin promoter driving T antigen expression) transgenic mice that develop islet cell tumors31 demonstrated the potential for T cells to promote tumor development. However, to our knowledge, no previous study has yet documented a direct role for endogenous T cell responses as a mechanism for infection-induced cancer.
In contrast, a number of recent studies in Rag-knockout mice and mice deficient in interferon signaling show a clear role for lymphocytes in inhibiting cancer development and forcing emerging tumors to edit themselves to evade immune elimination10,32. The finding that the procarcinogenic T cell response in our system is TH17 mediated suggests that the role of T cell responses in inhibiting or promoting carcinogenesis may depend on the qualitative response. Stat3-driven TH17 responses, characterized by production of IL-17A in mice and humans and driven by IL-23, are crucial in mucosal inflammatory responses in the lung and gut and are implicated in a number of autoimmune disorders11,33–35.
Although the mechanisms by which ETBF-induced TH17 responses promote colon carcinogenesis remain undefined, two of our notable histopathological findings in ETBF-colonized colons are the marked epithelial hyperproliferative response and the inflammatory infiltrates, both of which were substantially lessened upon in vivo blockade with IL-17– and IL-23R–blocking antibodies. We observed abundant granulocytes in ETBF-colonized colons, consistent with the reported role of TH17 responses in amplifying granulocytic inflammatory responses11. We are currently evaluating whether IL-17 and other TH17 cytokines promote colonic epithelial hyperproliferation and whether specific TH17-induced granulocyte products such as reactive oxygen or nitrogen species contribute to the rapid GIN induction (1 week) by ETBF.
Although the intestinal TH17 mucosal response to ETBF is not unique, we believe the rapid induction of colonic tumors in young Min mice is unique among reported data on enteric pathogens. Two mouse enteric pathogens, Citrobacter rodentium and Helicobacter hepaticus, may induce colonic mucosal TH17 immune responses33–35. However, IL-23 deficiency results in a fatal colitis in mice infected with C. rodentium but diminished colitis in mice infected with H. hepaticus, suggesting pathogen-specific roles for TH17 immunity in colitis. Both of these nonhuman enteric pathogens can induce colonic tumors in Min mice, with C. rodentium inducing modest colonic tumor induction after 5 months36 and H. hepaticus-associated colonic oncogenesis observed only in aged immune-insufficient mice such as Rag2−/− × Min or 129/SvEv Rag2−/− mice or mice with, for example, defective TGF signaling37,38. Salmonella typhimurium induces TH17-associated ileitis in rhesus macaques, and uncharacterized commensal flora in mice induce CD4+ TH17 cells with colitis induction upon adoptive transfer to Rag1−/− mice, but links to colonic tumor pathogenesis have not been reported39,40.
NTBF strains that do not secrete BFT, the only identified ETBF virulence factor, do not stimulate colonic Stat3 activation, TH17 mucosal immune responses nor enhance colonic tumor formation in Min mice, suggesting that BFT has a central role in triggering a procarcinogenic colonic mucosal response. Mechanistic data suggest that BFT acts as an oncogenic bacterial toxin through cleavage of E-cadherin, a tumor suppressor protein, triggering β-catenin nuclear signaling and colonic epithelial cell proliferation41,42. BFT also triggers activation of NF-κB, resulting in colonic epithelial cell secretion of proinflammatory cytokines43. We postulate that ETBF is a human oncogenic bacterium, owing to its production of BFT in vivo43 and its association with colonic inflammation17,44. To date, one report from Turkey supports this hypothesis18. Colonic tumor induction by ETBF in human populations would probably require long-term colonization. Although longitudinal carriage of B. fragilis is poorly characterized, ETBF is prevalent, at least in some locales, with 4–35% of studied populations showing asymptomatic fecal carriage43.
Commensal colonic bacteria are often cited as crucial environmental factors influencing the development of colorectal cancer, but linkages to specific organisms and the mechanisms promoting oncogenesis have been tenuous16. We have demonstrated the oncogenic potential of a human colonic commensal organism, and our data are reminiscent of early studies of H. pylori, an ancient gastric commensal, colonizing more than 50% of the global population, that routinely induces gastritis and, infrequently, also induces gastric cancer1. The mucosal immune response to H. pylori is also TH17 skewed, consistent with our observations linking ETBF-induced colonic mucosal TH17 inflammation to colonic tumor formation45. Together, our observations underpin the necessity of human studies to identify potential links between ETBF colonization, colonic Stat3 activation, colonic TH17 responses and human colorectal cancer.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemedicine/.
Supplementary Material
Acknowledgments
This work was supported by the Crohn’s and Colitis Foundation through a Senior Investigator Award (to C.L.S.) and a Research Fellowship Award (to K.-J.R.), RO1 DK45496 (to C.L.S.), RO1 DK080817 (to C.L.S.), US National Institutes of Health grants (to D.M.P.), Special Projects of Research Excellence grant CA62924, R24 DK64388 (to M. Donowitz, the principal investigator of this grant that provided resource support to this project), RR00171 grant (to D.L.H.), Institutional Training for Pediatricians 5 T32 HD44355 (to G. Dover, the principal investigator of this grant that provided partial salary support to S.R.), Clinical Pharmacology Training Program 2 T32GM066691 (to T. Shapiro, the principal investigator of this grant that provided salary support to F.M.) and F32 DK079509 (to S.R.). This work was also supported by gifts from B. Schwartz, W. and B. Topercer, D. Needle, B. Swartz and the Commonwealth Foundation. D.M.P. is a Januey scholar and holds the Abeloff Chair in Oncology at Johns Hopkins University. We thank J. Wolfe for her assistance with some experiments; L. Myers (formerly Montana State University) for ETBF strain 86-5443-2-2; B. Vogelstein and K. Kinzler (Johns Hopkins University School of Medicine) for Min mice and E. Jaffee (Johns Hopkins University School of Medicine) for GK1.5 antibody.
Footnotes
AUTHOR CONTRIBUTIONS
S.W. and K.-J.R. performed the majority of tumorigenesis experiments. E.A., S.R. and E.W. performed Stat experiments. X.W. did most of the mouse breeding and assisted with experiments. H.-R.Y. assisted with conditional CD4 Stat3-KO mouse experiments. D.L.H. evaluated and interpreted the histopathology. F.L.B. contributed the statistical analyses. F.M. performed qRT-PCR experiments. F.H. provided oversight and strategic planning for colonic immunology analyses. D.M.P. and C.L.S. designed the study, reviewed and discussed experiments and wrote the manuscript with input from co-authors.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
Note: Supplementary information is available on the Nature Medicine website.
References
- 1.Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 2.El Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 3.Greten FR, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 4.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 5.Naugler WE, et al. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- 6.Naugler WE, Karin M. The wolf in sheep’s clothing: the role of interleukin-6 in immunity, inflammation and cancer. Trends Mol Med. 2008;14:109–119. doi: 10.1016/j.molmed.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nat Rev Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 8.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 9.Erdman SE, et al. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am J Pathol. 2003;162:691–702. doi: 10.1016/S0002-9440(10)63863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunn GP, Koebel CM, Schreiber RD. Interferons, immunity and cancer immunoediting. Nat Rev Immunol. 2006;6:836–848. doi: 10.1038/nri1961. [DOI] [PubMed] [Google Scholar]
- 11.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 12.Laurence A, O’Shea JJ. TH-17 differentiation: of mice and men. Nat Immunol. 2007;8:903–905. doi: 10.1038/ni0907-903. [DOI] [PubMed] [Google Scholar]
- 13.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 14.Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317:124–127. doi: 10.1126/science.1140488. [DOI] [PubMed] [Google Scholar]
- 15.Su LK, et al. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science. 1992;256:668–670. doi: 10.1126/science.1350108. [DOI] [PubMed] [Google Scholar]
- 16.Hope ME, Hold GL, Kain R, El Omar EM. Sporadic colorectal cancer–role of the commensal microbiota. FEMS Microbiol Lett. 2005;244:1–7. doi: 10.1016/j.femsle.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 17.Sears CL, et al. Enterotoxigenic Bacteroides fragilis infection is associated with inflammatory diarrhea. Clin Infect Dis. 2008;47:797–803. doi: 10.1086/591130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toprak NU, et al. A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin Microbiol Infect. 2006;12:782–786. doi: 10.1111/j.1469-0691.2006.01494.x. [DOI] [PubMed] [Google Scholar]
- 19.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, et al. Selective regulatory function of Socs3 in the formation of IL-17–secreting T cells. Proc Natl Acad Sci USA. 2006;103:8137–8142. doi: 10.1073/pnas.0600666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laurence A, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Harris TJ, et al. Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J Immunol. 2007;179:4313–4317. doi: 10.4049/jimmunol.179.7.4313. [DOI] [PubMed] [Google Scholar]
- 23.Ivanov II, et al. The orphan nuclear receptor RORγ at directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 24.Kortylewski M, et al. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell. 2009;15:114–123. doi: 10.1016/j.ccr.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langowski JL, et al. IL-23 promotes tumour incidence and growth. Nature. 2006;442:461–465. doi: 10.1038/nature04808. [DOI] [PubMed] [Google Scholar]
- 26.Bollrath J, et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Grivennikov S, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Visser KE, Korets LV, Coussens LM. De novo carcinogenesis promoted by chronic inflammation is B lymphocyte dependent. Cancer Cell. 2005;7:411–423. doi: 10.1016/j.ccr.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 29.Poutahidis T, et al. Rapid reversal of interleukin-6–dependent epithelial invasion in a mouse model of microbially induced colon carcinoma. Carcinogenesis. 2007;28:2614–2623. doi: 10.1093/carcin/bgm180. [DOI] [PubMed] [Google Scholar]
- 30.Rao VP, et al. Proinflammatory CD4+ CD45RBhi lymphocytes promote mammary and intestinal carcinogenesis in ApcMin/+ mice. Cancer Res. 2006;66:57–61. doi: 10.1158/0008-5472.CAN-05-3445. [DOI] [PubMed] [Google Scholar]
- 31.Müller-Hermelink N, et al. TNFR1 signaling and IFN-γ signaling determine whether T cells induce tumor dormancy or promote multistage carcinogenesis. Cancer Cell. 2008;13:507–518. doi: 10.1016/j.ccr.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 32.Koebel CM, et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 33.Mangan PR, et al. Transforming growth factor-β induces development of the TH17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 34.Kullberg MC, et al. IL-23 plays a key role in Helicobacter hepaticus–induced T cell–dependent colitis. J Exp Med. 2006;203:2485–2494. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hue S, et al. Interleukin-23 drives innate and T cell–mediated intestinal inflammation. J Exp Med. 2006;203:2473–2483. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman JV, Kosaka T, Sheppard BJ, Fox JG, Schauer DB. Bacterial infection promotes colon tumorigenesis in ApcMin/+ mice. J Infect Dis. 2001;184:227–230. doi: 10.1086/321998. [DOI] [PubMed] [Google Scholar]
- 37.Nagamine CM, et al. Helicobacter hepaticus infection promotes colon tumorigenesis in the BALB/c-Rag2−/−ApcMin/+ mouse. Infect Immun. 2008;76:2758–2766. doi: 10.1128/IAI.01604-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maggio-Price L, et al. Helicobacter infection is required for inflammation and colon cancer in SMAD3-deficient mice. Cancer Res. 2006;66:828–838. doi: 10.1158/0008-5472.CAN-05-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raffatellu M, et al. Simian immunodeficiency virus–induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niess JH, Leithauser F, Adler G, Reimann J. Commensal gut flora drives the expansion of proinflammatory CD4 T cells in the colonic lamina propria under normal and inflammatory conditions. J Immunol. 2008;180:559–568. doi: 10.4049/jimmunol.180.1.559. [DOI] [PubMed] [Google Scholar]
- 41.Wu S, Lim KC, Huang J, Saidi RF, Sears CL. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc Natl Acad Sci USA. 1998;95:14979–14984. doi: 10.1073/pnas.95.25.14979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu S, Morin PJ, Maouyo D, Sears CL. Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology. 2003;124:392–400. doi: 10.1053/gast.2003.50047. [DOI] [PubMed] [Google Scholar]
- 43.Sears CL. Enterotoxigenic Bacteroides fragilis: a rogue among symbiotes. Clin Microbiol Rev. 2009;22:349–369. doi: 10.1128/CMR.00053-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rhee KJ, et al. Induction of persistent colitis by a human commensal, enterotoxigenic Bacteroides fragilis, in wild-type C57BL/6 mice. Infect Immun. 2009;77:1708–1718. doi: 10.1128/IAI.00814-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caruso R, et al. IL-23–mediated regulation of IL-17 production in Helicobacter pylori–infected gastric mucosa. Eur J Immunol. 2008;38:470–478. doi: 10.1002/eji.200737635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.