Abstract
The herpes simplex virus type 1 (HSV-1) US3 kinase is likely important for primary envelopment of progeny nucleocapsids since it localizes to the nuclear envelope of infected cells and largely determines the phosphorylation state and localization of the necessary primary envelopment factor, the UL34 protein. In HEp-2 cells, the production of infectious US3 null progeny is delayed and decreased relative to that of the parental strain, HSV-1(F). Furthermore, the US3 kinase affects the morphology of primary envelopment such that in its absence, UL34 protein-containing enveloped virions accumulate within membrane-bound vesicles. These vesicles are most often found along the interior periphery of the nucleus and may be derived from the inner nuclear membrane. Since the US3 and UL34 proteins comprise a kinase-substrate pair, a reasonable hypothesis is that the US3 kinase influences these replication parameters by direct phosphorylation of the UL34 protein. For this report, recombinant viruses were constructed to determine the significance of UL34 protein phosphorylation and US3 catalytic activity on UL34 protein localization, single-step growth, and envelopment morphology in both HEp-2 and Vero cells. The data presented suggest that the significance of UL34 phosphorylation is cell type dependent and that efficient viral morphogenesis requires US3-mediated phosphorylation of an infected cell protein other than UL34.
All known herpesviruses assemble progeny nucleocapsids within the nucleus of the host cell and therefore must have a mechanism to transport nucleocapsids across the nuclear envelope. Among members of the herpesvirus family, nuclear egress has been most extensively studied with herpes simplex viruses types 1 and 2 (HSV-1 and -2) and pseudorabies virus (PRV). Data indicate that progeny nucleocapsids can exit the nucleus of the host cell through envelopment at the inner nuclear membrane and subsequent deenvelopment at the outer nuclear membrane (13, 41). Both envelopment and deenvelopment appear to be facilitated by a specific set of virus-encoded (and probably host-encoded) proteins. Gene deletion studies indicate that the HSV-1 envelopment-deenvelopment process involves the UL34, UL31, UL20, UL11, and US3 proteins, although the specific functions of these proteins are unknown (2-4, 11, 15, 16, 34, 35, 37). The envelopment-deenvelopment machinery may include other virus-encoded proteins, and no host-encoded factors have been identified at the time of this report.
The UL34 gene of HSV-1 (and PRV) encodes a phosphoprotein that is primarily localized to the nuclear envelope of infected cells and is a necessary component of an envelopment complex that also includes the UL31 protein (34, 35, 37). Localization data and sequence analysis suggest that the UL34 protein is anchored within the inner nuclear membrane by a C-terminal hydrophobic domain, leaving the bulk of the protein exposed to the interior of the nucleus (34, 35). There are at least three non-mutually exclusive ways by which UL34 protein could facilitate nuclear egress. First, UL34 could directly mediate envelopment through bridging interactions between the nucleocapsid and the inner nuclear membrane. Second, the UL34 protein might direct the localization of other important nuclear egress factors. Finally, the nuclear lamina, which lines the interior face of the nuclear envelope, may represent a significant physical barrier to nuclear egress and herpesvirus infection alters this structure (39). Therefore, UL34 may affect the architecture of the nuclear lamina to allow nucleocapsids to access the envelopment machinery. Data supporting this function have been reported for the UL34 homologue of murine cytomegalovirus (23).
The US3 gene of HSV-1 encodes a serine/threonine kinase whose functions in virus replication remain enigmatic. In addition to protecting infected cells from apoptosis (1, 12, 19, 22), there is evidence that the US3 kinase facilitates nuclear egress of progeny nucleocapsids (15, 35, 42). Consistent with this hypothesis, the HSV-1 US3 protein is localized to the nuclear envelope and is believed to phosphorylate the envelopment factor encoded by the UL34 gene (31, 32, 35).
Evidence for a catalytic relationship between the HSV-1 US3 and UL34 proteins is limited but highly suggestive. Biochemical studies have elucidated the optimal target sequence for US3-directed phosphorylation (sometimes referred to as the US3 consensus sequence) and the predicted sequence of the UL34 protein contains a threonine and serine within this context (threonine 195 and serine 198) (18, 29). Also, in HSV-1-infected BHK cells, the UL34 protein was found to be phosphorylated, but when the US3 gene was deleted or the US3 consensus site within the UL34 protein was mutated, UL34 phosphorylation was not detected (31, 32). These data indicate that, in BHK cells, the phosphorylation state of the UL34 protein is dependent on US3 and strongly suggest direct phosphorylation of UL34 by US3. Unresolved issues include (i) what effect the US3 protein has on UL34 protein phosphorylation in other cell types, (ii) what effect other infected-cell kinases have on phosphorylation of UL34 protein, and (iii) what effect phosphorylation of UL34 or other proteins has on nuclear egress. These are important considerations because HSV-1 replicates in a wide variety of cell types and the necessity of at least one other nuclear egress factor is cell type dependent (4). Furthermore, phosphorylation of UL34 protein may modulate any of the potential functions noted above.
Given the likely catalytic relationship between the UL34 and US3 proteins and their apparent involvement in nuclear egress, a reasonable hypothesis is that phosphorylation of the UL34 protein (directly or indirectly) by the US3 kinase facilitates the envelopment-deenvelopment process. In addressing this hypothesis, we present data indicating that the importance of the UL34-US3 catalytic relationship for viral replication is cell type dependent. Furthermore, data presented here suggest that in some cell types, efficient viral replication requires US3-mediated phosphorylation of an infected cell protein(s) other than the UL34 protein.
MATERIALS AND METHODS
Cells and viruses.
HEp-2 and Vero cells (American Type Culture Collection) were grown in Dulbecco's modified Eagle’s medium supplemented with 5% fetal calf serum. The construction and maintenance of the UL34 complementing cell line 143/1099E were described previously (37). The wild-type virus HSV-1(F) was described previously (10). The construction and growth characteristics of the UL34 null mutant HSV-1(F), vRR1072, were also previously described (37). Dilution of virus stocks and incubation of virus-infected cells were done in V medium (Dulbecco's modified Eagle’s medium supplemented with 1% heat-inactivated calf serum).
Antibodies.
Generation and characterization of the anti-UL34 and anti-US3 antibodies were described previously (22, 34, 37).
Plasmids.
Plasmid pRR1098 contains the HSV-1(F) US3 open reading frame (ORF) under the transcriptional control of the human cytomegalovirus major immediate-early promoter (hCMV-MIEP) and was described previously (34). Plasmid pRR1072rep, which contains the UL34 ORF within a SacI-HindIII fragment of the HSV-1(F) genome, was also previously described (6). Plasmid pRR1201, which contains the complete HSV-1(F) US2, US3, and US4 ORFs as well as portions of the US1 and US5 ORFs, was constructed by ligation of the HindIII-SacI fragment of the HSV-1(F) genome into the HindIII-SacI site of pGEM-3Z (Promega). Plasmid pRR1202 was constructed by ligation of the NsiI-MfeI Klenow fragment of pDsRed1-N1 (Clontech), containing the hCMV-MIEP and the red fluorescent protein (RFP) ORF, into the PstI-BamHI Klenow site of pRR1201. Plasmid pRR1204 contains the coding sequence for the K220A mutant form of the US3 protein as well as the genomic flanking sequence and was constructed by ligation of the PstI-BamHI fragment of pRR1203 (see below) into the PstI-BamHI site of pRR1201. The construction of plasmid pRR1205 is described below.
PCR-based site-directed mutagenesis.
To generate plasmid pRR1203, which encodes a mutant US3 in which lysine 220 is mutated to alanine (K220A), two independent PCRs were performed, using HSV-1(F) DNA as template. Reaction 1 used primers BR21 (5′-TTCCTCATGGCCCCTTTTAT-3′) and BR24 (5′-CACCCAGCTGCCACGATTAC-3′) and gave a 576-nucleotide (nt) product, while reaction 2 used primers BR23 (5′-GTAATCGTGGCAGCTGGGTG-3′) and BR22 (5′-CTGGGCCTGTCGGATGAT-3′) and gave a 569-nt product. Both products were gel purified and used in an equal molar ratio as template in a third PCR using primers BR21 and BR22. The 1,126-nt product of reaction 3 was gel purified, digested with restriction enzymes BspEI and PmlI, and ligated into the BspEI-PmlI site of pRR1098.
Plasmid pRR1205 encodes a mutant form of the HSV-1(F) UL34 protein in which the predicted target residues of the US3 kinase (threonine 195 and serine 198) are both mutated to alanine (T195A/S198A). To generate this plasmid, two independent PCRs were performed, using plasmid pRR1072rep as template. Reaction 1 used primers BR1 (5′-AACCCTTTGGTGGGTTTACG-3′) and BR38 (5′-CGTACGCCTCCCGGGCCCGCCGAGCTCGGCGGCGACGGGT-3′) and gave a 656-nt product, and reaction 2 used primers BR37 (5′-ACCCGTCGCCGCCGAGCTCGGCGGGCCCGGGAGGCGTACG-3′) and BR7 (5′-AACATAGGCTGCGGGGTAA-3′) and gave a 601-nt product. Both products were gel purified and used in an equal molar ratio as template in a third PCR using primers BR1 and BR7. The 1,218-nt product of reaction 3 was gel purified, digested with restriction enzymes NcoI and BspEI, and ligated into the NcoI-BspEI site of pRR1072.
Purification of viral genomic DNA.
Viral genomic DNA was purified from infected cells for Southern blot analysis and construction of recombinant viruses as previously described (33). Vero cells were used for HSV-1(F), vRR1202, vRR1202rep, vRR1204 #1, and vRR1204 #2, while 143/1099E cells were used for vRR1072, vRR1205 #1, and vRR1205 #2.
Construction of recombinant viruses.
All recombinant viruses were generated by a method in which 4.5-cm2 cultures of Vero or 143/1099E cells at approximately 50% confluence were cotransfected with 200 ng of viral genomic DNA and 100 ng of a linearized plasmid carrying the mutated gene of interest and viral genomic flanking sequence. Cotransfection was performed with Lipofectamine reagent according to the manufacturer's suggested protocol (Invitrogen). Cultures were then incubated at 37°C until cytopathic effects were evident, at which time cultures were frozen at −80°C and then thawed. Cotransfection stocks were then prepared by diluting cultures 1:1 with autoclaved nonfat dry milk and sonicating them for 30 s at power level 0 with a Fisher Scientific sonic dismembrator. Recombinant viruses, which result from homologous recombination between the viral genome and linearized plasmid, were then isolated by at least three successive rounds of plaque purification on either Vero or 143/1099E cells as follows. Serial 10-fold dilutions of each cotransfection stock were plated on confluent 55-cm2 cultures and incubated in the presence of neutralizing antibody (pooled human immunoglobulin) until plaques were evident. Plaques resulting from infection with recombinant viruses were identified visually by virtue of expected RFP or green fluorescent protein (GFP) expression, picked, and diluted into 500 μl of V medium. Picked plaques were frozen at −80°C, thawed, sonicated as described above, and passed through a 0.45-μm-pore-size syringe filter, and serial 10-fold dilutions were plated onto 9.6-cm2 cultures.
The US3 null virus, vRR1202, was derived from cotransfection of Vero cells with HSV-1(F) genomic DNA and HindIII-treated plasmid pRR1202. Recombinant viruses vRR1202rep, vRR1204 #1, and vRR1204 #2 were derived from independent cotransfections of Vero cells with vRR1202 genomic DNA and HindIII-treated plasmids pRR1201, pRR1204, and pRR1204, respectively. vRR1202 encodes RFP, while vRR1202rep, vRR1204 #1, and vRR1204 #2 do not; all four recombinant viruses were plaque purified on Vero cells.
The UL34 mutant viruses vRR1205 #1 and vRR1205 #2 were derived from independent cotransfections of 143/1099E cells with vRR1072 genomic DNA with HindIII-treated plasmid pRR1205 and by plaque purification on 143/1099E cells. vRR1072 encodes GFP, while vRR1205 #1 and vRR1205 #2 do not.
Southern blot analysis.
For genome structure analysis of vRR1202, vRR1202rep, vRR1204 #1, and vRR1204 #2, the BlpI-NsiI fragment of pRR1201 was radiolabeled with [32P]CTP by use of Ready-To-Go DNA labeling beads according to the supplied protocol (Amersham Pharmacia). The same method was used to label the NcoI-PmlI fragment of pRR1072rep for genome structure analysis of vRR1205 #1 and vRR1205 #2.
Purified, restriction enzyme-digested, viral genomic DNA was separated by electrophoresis on 0.8% agarose, transferred to a nylon membrane, and analyzed with the appropriate probe by standard methods (21).
HSV-1(F) US3 ORF sequencing.
Automated sequencing was performed by the University of Iowa DNA Sequencing Facility, using plasmid pRR1201 as template. Sense and antisense strands were sequenced by use of primers BR26 (5′-GGGTCTTTTTGTGCCAACC-3′), BR27 (5′-ATCCACCGCGACATTAAGAC-3′), BR28 (5′-GTAGGGGAAGGGGCTTGAT-3′), BR29 (5′-ATTTACCGCGGAAGAGCTG-3′), BR30 (5′-GCTCCGTGGATCGTAAAGC-3′), and BR31 (5′-CTCGTTGGTTGGCACTCAC-3′).
Generation of 32P-labeled infected cell lysates.
Confluent 9.6-cm2 cultures of HEp-2 or Vero cells were infected at a multiplicity of infection (MOI) of 5 with the indicated viruses. At 12 hours postinfection (h.p.i.), infected cells were washed twice with methionine-free minimum essential Eagle medium (Sigma) supplemented with 2% dialyzed horse serum (Sigma) (−Met/MEM) and then incubated for 1 h in 2 ml of −Met/MEM. Ten microcuries of 32P as inorganic phosphate (Amersham Pharmacia) was then added to each culture. At 17 h.p.i., infected cell lysates were generated as follows. All manipulations were performed on ice. Cells were washed twice with phosphate-buffered saline, scraped into 1 ml of phosphate-buffered saline, and pelleted by centrifugation at 1,200 × g for 5 min. Pelleted cells were resuspended in RIPA buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10 mM EDTA, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1.0% Triton X-100) supplemented with 2 mM sodium vanadate, 5 mM NaF, 10 mM 2-mercaptoethanol, and 40 μg of phenylmethylsulfonyl fluoride per ml. Cell lysates were centrifuged at 10,000 × g for 15 min and pellets were discarded. Total protein was determined by use of Bio-Rad protein assay reagent (Bio-Rad) and lysates were either stored at −80°C or analyzed by Western blotting or immunoprecipitation.
Western blot analysis.
For Western blotting, cell lysates were prepared as described above, and 50 μg of protein from each lysate was separated on SDS-polyacrylamide gels and transferred to nitrocellulose membranes by standard methods (21). Blots were probed with anti-UL34 antibody diluted 1:1,000 or anti-US3 antibody diluted 1:2,000 and detected with anti-chicken alkaline phosphatase conjugate (Aves Labs Inc.) diluted 1:1,500 or anti-rabbit alkaline phosphatase conjugate (Sigma) diluted 1:5,000 as described previously (36).
Immunoprecipitation.
For immunoprecipitation, cell lysates were prepared as described above, and 200 μg of protein from each lysate was diluted in RIPA buffer to a total volume of 300 μl. To preclear lysates, chicken preimmune serum (1:1,000) (Aves Labs Inc.) and 40 μl of anti-chicken agarose conjugate (Promega) were added and each lysate was incubated for 2 h at 4°C while rocking gently. Anti-chicken agarose conjugate and associated proteins were removed by centrifugation at 1,600 × g for 1 min. Anti-UL34 antibody was then added to a dilution of 1:1,000 and each lysate was incubated overnight at 4°C while rocking gently. Forty microliters of anti-chicken agarose was then added and each lysate was incubated for 2 h as described above. Precipitated proteins were then collected by centrifugation as described above, washed three times in RIPA buffer, and separated in an SDS-12% polyacrylamide gel electrophoresis (PAGE) gel. The gel was dried and analyzed by autoradiography.
Confocal microscopy.
Confocal microscopy was performed as previously described (34). Briefly, HEp-2 or Vero cells were infected at an MOI of 5. At 12 h.p.i., cells were formaldehyde fixed and immunostained with anti-UL34 antibody, detected with anti-chicken fluorescein isothiocyanate conjugate (Aves Labs), and anti-US3 antibody, detected with anti-rabbit Texas red conjugate (Molecular Probes). To reduce the background associated with the anti-US3 antibody, a blocking procedure using pooled human immunoglobulin was performed as described previously (35).
Growth analysis of recombinant viruses.
Single-step growth analysis was performed as previously described (35, 37). Briefly, replicate cultures of HEp-2 or Vero cells were infected at an MOI of 5, and residual virus was removed or inactivated with a low-pH-buffer wash. At the indicated times, virus was harvested and total culture PFU were calculated by titration on 143/1099E cells.
Electron microscopy.
For electron microscopy, HEp-2 or Vero cells were infected with the indicated viruses at an MOI of 5. At 20 h.p.i., cells were fixed, processed, and analyzed by electron microscopy as described previously (37).
RESULTS
Construction of recombinant HSV-1 strains encoding mutant UL34 and US3 proteins.
Previously published evidence suggests that the US3 and UL34 proteins comprise a kinase-substrate pair (31, 32). To examine the significance of this catalytic relationship on the viral life cycle, we engineered recombinant HSV-1 strains to encode either a mutant, inactive form of US3 protein or a mutant UL34 protein in which the predicted target serine and threonine residues within the US3 consensus phosphorylation site (threonine 195 and serine 197) had both been changed to alanine. Purves et al. constructed recombinant viruses in which threonine 195 and serine 197 of UL34 had been independently mutated to alanine (31, 32). However, we found these viruses to be syncytial (data not shown), and therefore, it seemed likely that they contained secondary mutations. To avoid potential interpretation difficulties due to secondary mutations, we chose to construct two independent recombinant viruses encoding a UL34 protein in which both potential phosphotargets were mutated.
To generate recombinant HSV encoding mutant US3, a suitable parental strain was required. For this purpose, a US3 null HSV was generated by replacing most of the coding sequence of US3 with an expression cassette encoding RFP under the transcriptional control of the hCMV-MIEP. This sequence substitution preserved the promoter elements for the US2 ORF and would not be expected to interfere with expression from this locus.
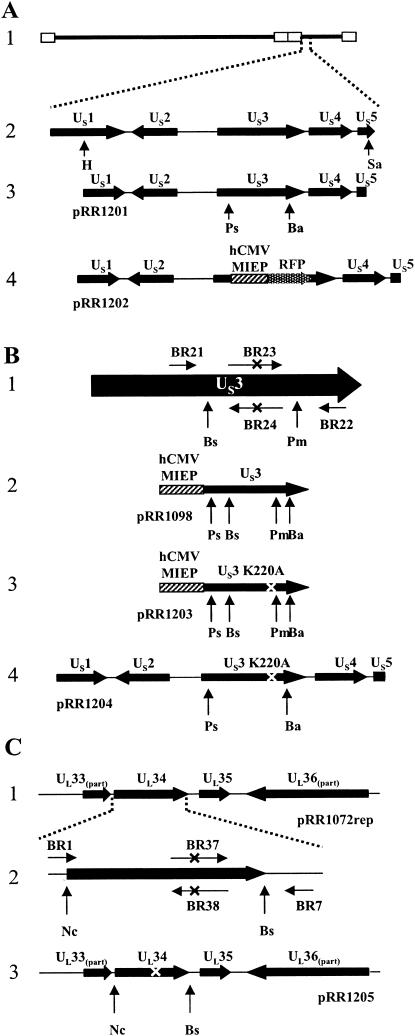
As described in Materials and Methods and shown in Fig. 1A, the RFP expression cassette of pDsRed-N1 was ligated into plasmid pRR1201, removing a large portion of the US3 ORF. Cotransfection of the resulting plasmid, designated pRR1202, with purified HSV-1(F) genomic DNA into Vero cells yielded progeny (designated vRR1202) that produced red fluorescent plaques (not shown). A homologous repair virus, designated vRR1202rep, was then constructed by cotransfecting purified vRR1202 genomic DNA with plasmid pRR1201.
FIG. 1.
Construction of US3 and UL34 mutant plasmids. (A) Structure 1, schematic representation of the HSV-1(F) genome. Open rectangles indicate inverted repeats. Structure 2, HSV-1(F) genome expanded to show the arrangement of the US1 to US5 ORFs. Restriction sites used for construction of plasmid pRR1201 are indicated. Structure 3, arrangement of HSV-1 ORFs contained on plasmid pRR1201, showing restriction sites used for deletion of the US3 ORF. Structure 4, gene arrangement of US3 deletion plasmid pRR1202. US3 sequences were replaced with an hCMV-MIEP-RFP expression cassette. (B) PCR-based mutagenesis of the US3 protein. Structure 1,schematic representation of the US3 ORF (horizontal arrows represent PCR primers); structure 2, diagram of the US3 expression cassette of plasmid pRR1098; structure 3, diagram of the K220A US3 expression cassette of plasmid pRR1203; structure 4, diagram of the HSV-1 sequences of plasmid pR1204. For all structures, “X” indicates the position of the K220A mutation and generated PvuII restriction site and vertical arrows represent restriction sites used for cloning. (C) PCR-based mutagenesis of the UL34 protein. Structure 1, arrangement of HSV-1 ORFs contained on plasmid pRR1072rep. Structure 2, expanded diagram of the UL34 ORF. Horizontal arrows represent PCR primers and “X” indicates the position of the T195A/S198A mutation and generated SacI restriction site. Also shown are restriction sites used for cloning of the mutation-containing PCR product. Structure 3, arrangement of the HSV-1 ORFs contained on plasmid pRR1205. Restriction sites used for cloning are indicated and “X” indicates the position of the introduced mutation. Restriction enzyme abbreviations: Ha, HindII; Sa, SacI; Ps, PstI; Ba, BamHI; Bs, BspEI; Pm, PmlI; Nc, NcoI.
A conserved feature of serine/threonine kinases, such as the US3 protein, is a lysine residue within the active site that may be involved in positioning the γ-phosphate of ATP, thus facilitating phosphotransfer (14). Substitution of alanine for the invariant lysine is expected to significantly reduce the activity of the kinase. Sequence analysis indicated that the invariant lysine residue of the US3 protein is at position 220 (14).
A plasmid designated pRR1204, in which the codon specifying the invariant lysine residue was mutated to an alanine codon, was constructed as described in Materials and Methods and depicted in Fig. 1B. Two recombinant viruses were derived from independent cotransfections of vRR1202 genomic DNA with plasmid pRR1204. These viruses, designated vRR1204 #1 and vRR1204 #2, encode the K220A mutant form of the US3 protein.
The US3 consensus phosphorylation site within the UL34 protein contains two potential target residues, threonine 195 and serine 198 (18, 29). To create recombinant HSV strains that encode a UL34 protein with an inactive US3 phosphorylation site, a plasmid designated pRR1205 was constructed as described in Materials and Methods and depicted in Fig. 1C. Two recombinant HSV strains encoding the mutant UL34 protein were generated by independent cotransfection of 143/1099E cells with plasmid pRR1205 and genomic DNA derived from the UL34 null HSV strain, vRR1072. The resulting progeny were designated vRR1205 #1 and vRR1205 #2.
The US3, UL34, and RFP or GFP genotypes of all viruses used are listed in Table 1.
TABLE 1.
Recombinant virus genotypes
| Virus | Genotype or mutation
|
Presence of RFP or GFP | |
|---|---|---|---|
| US3 | UL34 | ||
| HSV-1(F) | WTa | WT | No |
| vRR1202 | Null | UL34 | RFP |
| vRR1202rep | WT | WT | No |
| vRR1204 #1 | K220A | WT | No |
| vRR1204 #2 | K220A | WT | No |
| vRR1205 #1 | WT | T195A, S198A | No |
| vRR1205 #2 | WT | T195A, S198A | No |
| vRR1072 | WT | Null | GFP |
WT, wild type.
The predicted amino acid sequence of HSV-1(F) US3 protein differs from that of HSV-1 strain 17.
During the construction of the US3 mutant viruses, the sequence of the HSV-1(F) US3 gene was determined. When it was compared to the US3 sequence shown in GenBank (accession no. NC_001806), derived from strain 17, three codon deviations were discovered. These strain differences are summarized in Table 2.
TABLE 2.
Strain-specific US3 sequence
| Amino acid no. | US3 sequence flanking and including indicated amino acida
|
|
|---|---|---|
| HSV-1(17) | HSV-1(F) | |
| 62 | ...cccAgcgag... | ...cccGgcgag... |
| 63 | ...agcgaGgcc... | ...agcgaTgcc... |
| 122 | ...gcgAccggt... | ...gcgCccggt... |
The amino acid of concern is shown in bold.
Southern blot analysis of recombinant virus genomes.
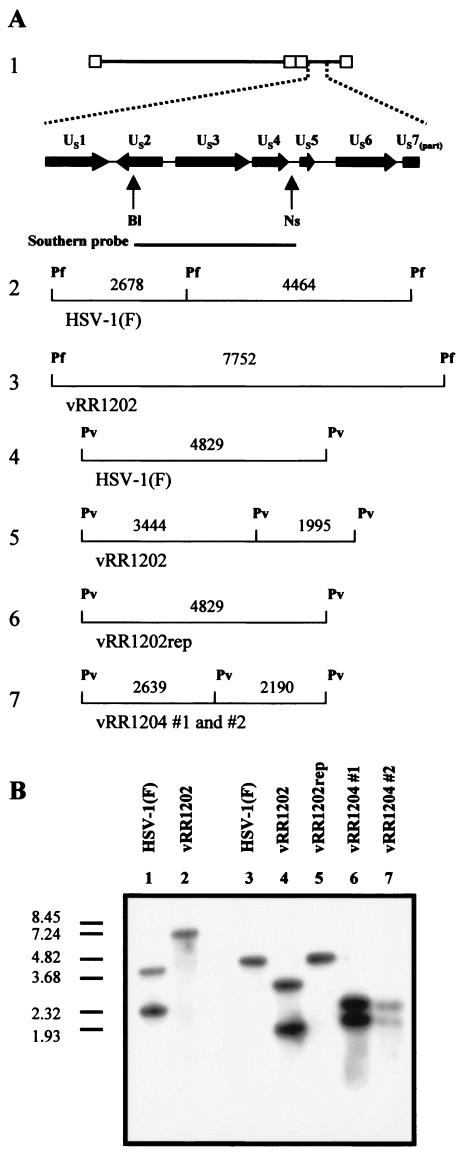
For verification of the genome structure of the US3 null virus (vRR1202), genomic DNA derived from HSV-1(F) and vRR1202 was digested with PflFI, electrophoretically separated, and analyzed by Southern blotting with a probe designed to hybridize with the sequence spanning the US2 to US4 ORFs (Fig. 2A). As expected, the probe hybridized with two fragments of digested HSV-1(F) genomic DNA (4,464 and 2,678 bp) (Fig. 2B, lane 1). Since replacement of the US3 sequence with the RFP expression cassette eliminated a PflFI site and added 610 bp to the resulting fragment, the Southern probe hybridized to a single 7,752-bp fragment of digested vRR1201 genomic DNA (Fig. 2B, lane 2).
FIG. 2.
Verification of US3 mutant recombinant virus genomic structure by Southern blotting. (A) Structure 1, schematic representation of the HSV-1(F) genome expanded to show the US1 ORF through part of the US7 ORF. The probe used recognized the sequence between the indicated restriction sites. Structures 2 and 3, probe-specific fragments expected for PflFI-digested HSV-1(F) and vRR1202 genomic DNA. Structures 4, 5, 6, and 7, probe-specific fragments expected for PvuII-digested genomic DNA from HSV-1(F), vRR1202, vRR1202rep, vRR1204 #1, and vRR1204 #2, respectively. Restriction site abbreviations: Bl, BlpI; Ns, NsiI; Pf, PflFI; Pv, PvuII. (B) Southern blot analysis of PflFI-digested genomic DNA derivedfrom HSV-1(F) (lane 1) and vRR1202 (lane 2) or PvuII-digested genomic DNA derived from HSV-1(F) (lane 3), vRR1202 (lane 4), vRR1204rep (lane 5), vRR1204 #1 (lane 6), and vRR1204 #2 (lane 7). The DNA size marker indicates sizes in kilobase pairs.
For verification of the genome structures of vRR1204 #1, vRR1204 #2, and vRR1202rep, genomic DNA derived from these viruses and from HSV-1(F) was digested with PvuII, separated electrophoretically, and analyzed by Southern blotting with the probe mentioned above (Fig. 2A). Again as expected, the probe hybridized with a single 4,829-bp fragment of digested HSV-1(F) genomic DNA and vRR1202rep (Fig. 2B, lanes 3 and 5, respectively). A silent mutation that created a PvuII site was engineered as part of the K220A mutation, and therefore PvuII digestion of the vRR1204 #1 and vRR1204 #2 genomes was expected to yield 2,639- and 2,190-bp fragments (Fig. 2A, structure 7). This pattern was observed and is shown in Fig. 2B, lanes 6 and 7. vRR1202 genomic DNA was also included in this analysis. Replacement of the US3 sequence with the RFP expression cassette added 610 bp and introduced a novel PvuII site, yielding two fragments (3,444 and 1,995 bp) that hybridized with the probe (Fig. 2B, lane 4).
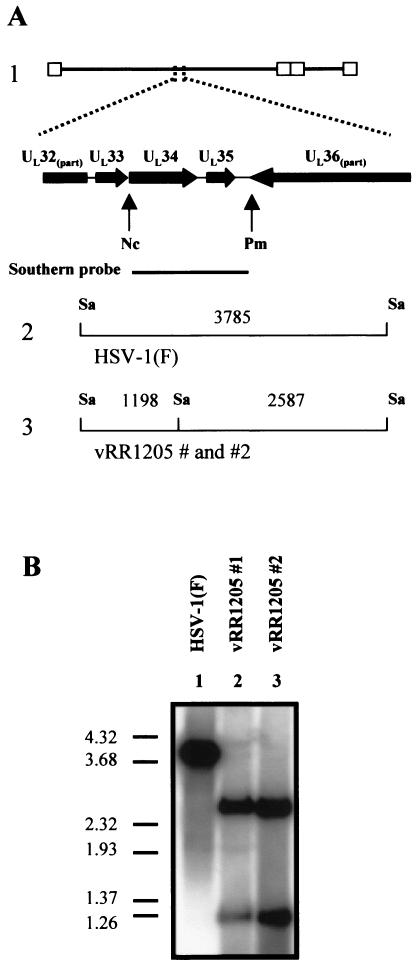
A similar analysis was performed to verify the genomic structure of the UL34 mutant viruses vRR1205 #1 and vRR1205 #2. As shown in Fig. 3A, the UL34 ORF of HSV-1(F) is contained within a 3,785-bp SacI fragment. Mutation of the US3 consensus phosphorylation site within the UL34 sequence of vRR1205 #1 and vRR1205 #2 was designed to generate a SacI site. Genomic DNA was digested with SacI and analyzed by Southern blotting with a probe that spanned the mutated sequences. As expected, this probe hybridized with a single 3,785-bp fragment of HSV-1(F) DNA and with two fragments (2,587 and 1,198 bp) of genomic DNA derived from vRR1205 #1 and vRR1205 #2 (Fig. 3B).
FIG. 3.
Verification of UL34 mutant recombinant virus genomic structure by Southern blotting. (A) Structure 1, schematic representation of the HSV-1(F) genome expanded to show a portion of the UL32 ORF through a portion of the UL36 ORF. The probe used recognized the sequence between the indicated restriction sites. Structures 2 and 3, probe-specific fragments expected for SacI-digested HSV-1(F) and for vRR1205 #1 and vRR1205 #2 genomic DNA, respectively. Restriction site abbreviations: Nc, NcoI; Pm, PmlI; Sa, SacI. (B) Southern blot analysis of SacI-digested genomic DNA derived from HSV-1(F) (lane 1), vRR1205 #1 (lane 2), and vRR1205 #2 (lane 3). The DNA marker indicates sizes in kilobase pairs.
Mutations do not affect UL34 or US3 protein accumulation but do disrupt their catalytic relationship.
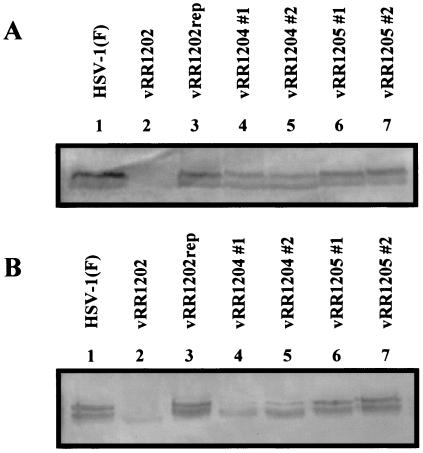
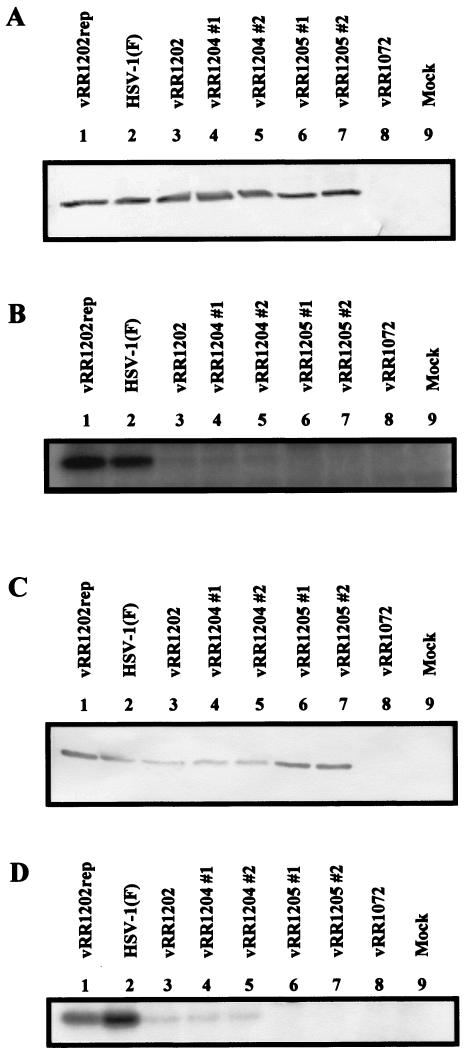
Any observed phenotype associated with the UL34 or US3 mutations might be due to disruption of the catalytic relationship or to a lack of US3 protein accumulation. To rule out the latter possibility, replicate cultures of HEp-2 and Vero cells were infected with HSV-1(F) or each of the recombinant viruses and cell lysates were analyzed by Western blotting for the accumulation of the US3 protein (Fig. 4). In both cell types, US3 protein was detected in all infected cell lysates except those derived from vRR1202 (US3 null). Consistent with a previous report, the HSV-1(F) US3 protein was resolved as doublet bands, of approximately 68 and 69 kDa, with the slower migrating form predominating (22). The ratio between these two species in vRR1204 #1- and vRR1204 #2-infected cells was more equal than in HSV-1(F)-infected cells. Furthermore, lysates of cells infected with viruses harboring the US3 K220A mutation contained slightly less US3 protein.
FIG. 4.
Expression of US3 protein in recombinant virus-infected cells. HEp-2 cells (A) or Vero cells (B) were infected at an MOI of 5 for 12 h with HSV-1(F) (lane 1), vRR1202 (lane 2), vRR1202rep (lane 3), vRR1204 #1 (lane 4), vRR1204 #2 (lane5), vRR1205 #1 (lane 6), or vRR1205 #2 (lane 7). Equal amounts of total infected cell proteins were separated by SDS-PAGE (8%), transferred to a nitrocellulose membrane, and probed with an anti-US3 antibody (conjugated with alkaline phosphatase).
For verification that the introduced mutations disrupted the catalytic relationship between the US3 and UL34 proteins, replicate cultures of HEp-2 and Vero cells were mock infected or infected with HSV-1(F) or each of the recombinant viruses. Cultures were labeled with 32P and cell lysates were prepared. Equal amounts of total cell proteins were either separated by SDS-PAGE, transferred to nitrocellulose, and analyzed by Western blotting for the accumulation of UL34 protein (Fig. 5A and C) or immunoprecipitated with anti-UL34 antibody, separated by SDS-PAGE, and analyzed by autoradiography (Fig. 5B and D).
FIG. 5.
Expression and phosphorylation of UL34 protein in recombinant virus-infected cells. HEp-2 cells (A and B) or Vero cells (C and D) were infected at an MOI of 5 with vRR1202rep (lane 1), HSV-1(F) (lane 2), vRR1202 (lane 3), vRR1204 #1 (lane 4), vRR1204 #2 (lane 5), vRR1205 #1 (lane 6), vRR1205 #2 (lane 7), or vRR1072 (lane 8) or mock infected (lane 9) and were then labeled with 32P as inorganic phosphate. Equal amounts of total infected cell proteins were either separated by SDS-PAGE (12%), transferred to a nitrocellulose membrane, and probed with an anti-UL34 antibody (conjugated with alkaline phosphatase) (A and C) or immunoprecipitated with an anti-UL34 antibody, separated by SDS-PAGE, and analyzed by autoradiography (B and D).
Lysates of all infected cells except those infected with vRR1072 (UL34 null) contained similar amounts of UL34 protein, indicating that the introduced mutations did not significantly affect the expression or stability of the UL34 protein (Fig. 5A and C). As shown in Fig. 5B and D, when lysates of cells infected with HSV-1(F) or vRR1202rep were immunoprecipitated with the anti-UL34 antibody, strong phosphoprotein bands with an Mr of 30,000 were detected. Very faint phosphoprotein bands of similar mobility were detected in lysates of vRR1202-, vRR1204 #1-, and vRR1204 #2-infected cells, while no 30-kDa phosphoproteins were detected in lysates of cells infected with vRR1205 #1 or vRR1205 #2.
Purves et al. reported that when the UL34 protein is unphosphorylated in infected BHK cells, either as the result of US3 deletion or UL34 mutation, four phosphoproteins are detected in place of the UL34 protein (32). We did not observe similar phenomena in HEp-2 or Vero cells (data not shown).
These results demonstrate several properties of the recombinant viruses and the relationship between the UL34 and US3 proteins. First, since deletion of the US3 ORF significantly reduced the amount of phosphorylated UL34 protein and repair of the US3 gene restored UL34 phosphorylation to wild-type levels, the phosphorylation state of UL34 protein in both HEp-2 and Vero cells is largely determined by the US3 protein. Second, the observation that lysates of cells infected with viruses encoding the K220A variant of US3 contained no more phosphorylated UL34 protein than lysates of cells infected with US3 null virus strongly suggests that mutation of lysine 220 to alanine efficiently inactivated the kinase activity of the US3 protein. Third, since low-level phosphorylation of UL34 protein was observed in cells infected with US3 null (or mutant) HSV, other infected cell kinases must be capable of phosphorylating UL34 protein. Finally, since phosphorylation of the mutant UL34 protein that lacks a US3 consensus phosphorylation site was undetectable, it is unlikely that the UL34 protein contains other significant phosphorylation sites for HEp-2 or Vero cell kinases.
Localization of UL34 and US3 proteins in cells infected with recombinant HSV strains.
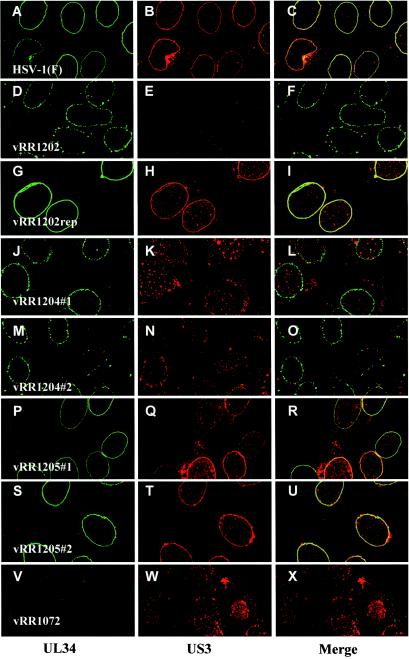
We and members of our laboratory have previously shown that in HEp-2 cells, the US3 kinase localizes to the nuclear envelope and influences the localization of the UL34 protein such that in US3 null virus-infected cells, the UL34 protein is mislocalized to punctate structures at the nuclear envelope (34, 35). These structures appeared to be accumulations of enveloped virions within vesicles that line the interior periphery of the nucleus (35). Given the apparent US3-UL34 catalytic relationship, a reasonable hypothesis was that the US3 kinase influences UL34 localization by directing the phosphorylation of the UL34 protein. To test this possibility, replicate monolayers of HEp-2 and Vero cells were infected with HSV-1(F) or each of the recombinant viruses and the localization of US3 and UL34 proteins under each condition was analyzed by confocal microscopy. Similar results were obtained with both cell types, and representative images of infected Vero cells are shown in Fig. 6.
FIG.6.
Proper localization of the UL34 and US3 proteins requires US3-directed phosphorylation of an infected cell protein other than UL34. Vero cells were infected at an MOI of 5 with HSV-1(F) (A to C), vRR1202 (D to F), vRR1202rep (G to I), vRR1204 #1 (J to L), vRR1204 #2 (M to O), vRR1205 #1 (P to R), vRR1205 #2 (S to U), or vRR1072 (V to X). At 12 h.p.i., infected cells were formaldehyde fixed, immunostained with anti-UL34 (conjugated with fluorescein isothiocyanate) and anti-US3 (conjugated with Texas red), and analyzed by confocal microscopy. Representative confocal Z sections are shown. Original magnification, ×1,000.
Consistent with previous results, the UL34 and US3 proteins in HSV-1(F)-infected cells localized to the nuclear envelope in a uniform distribution (Fig. 6A to C). In vRR1202 (US3 null)-infected cells, UL34 protein was detected in punctate structures at the nuclear envelope (Fig. 6D). As expected, repair of the US3 deletion (vRR1202rep) yielded UL34 and US3 localization that was indistinguishable from that of HSV-1(F), confirming that the observed clustering of UL34 protein was specifically due to deletion of the US3 ORF (Fig. 6, compare panels A to C with panels G to I).
In cells infected with either vRR1204 #1 or vRR1204 #2, both of which encode the K220A mutant form of US3 protein, UL34 protein localization was similar to that observed for US3 null virus-infected cells (Fig. 6, compare panel D with panels J and M). Interestingly, the mutant US3 protein itself was aberrantly localized to large, punctate structures throughout the cytoplasm and nucleus (Fig. 6K and N). In contrast, when cells were infected with vRR1205 #1 or vRR1205 #2, both of which encode the phosphorylation-defective mutant UL34 protein, neither UL34 nor US3 localization differed significantly from that of HSV-1(F)-infected cells (Fig. 6, compare panels A to C with panels P to R and S to U). These results indicate that the catalytic function of the US3 protein is necessary for proper localization of both itself and the UL34 protein but that phosphorylation of the UL34 protein is not critical.
Additionally, in cells infected with UL34 null HSV (vRR1072), US3 protein failed to localize to the nuclear envelope as it did in HSV-1(F)-infected cells (Fig. 6W). This indicates that the US3 protein is recruited to the nuclear envelope by the UL34 protein.
Growth analysis of recombinant viruses.
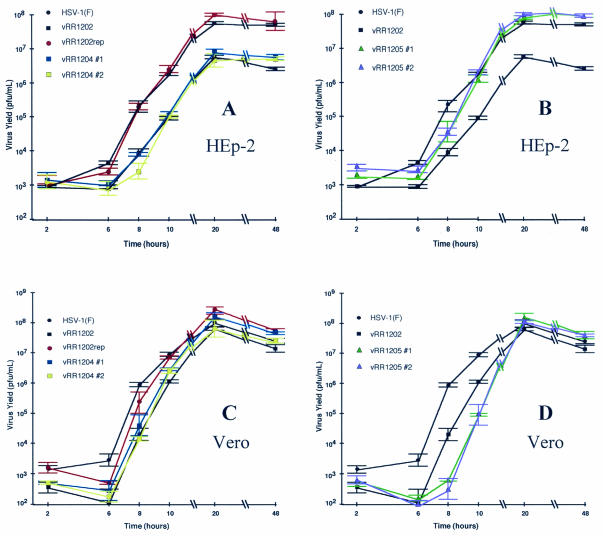
Published data concerning the effect of US3 mutation on HSV-1 replication are limited and somewhat conflicting. We previously reported that recombinant HSV-1 strains that fail to express the US3 protein are impaired for growth on HEp-2 cells, while others have observed that US3 null viruses grow to plateau titers equivalent to those for HSV-1(F) in Vero cells (20, 30, 35). To test the hypothesis that US3 facilitates HSV replication in HEp-2 cells by directing the phosphorylation of the UL34 protein and to characterize the significance of the US3-UL34 catalytic relationship for growth in Vero cells, we conducted a single-step growth experiment. Replicate cultures of HEp-2 and Vero cells were infected, as described in Materials and Methods, with HSV-1(F), vRR1202, vRR1202rep, vRR1204 #1, vRR1204 #2, vRR1205 #1, or vRR1205 #2. At the indicated time postinfection, the virus yield was determined, as plotted in Fig. 7. For ease of comparison, the growth curves of HSV-1(F) and the US3 null virus (vRR1202) are shown in both Fig. 7A and B and in both Fig. 7C and D for HEp-2 and Vero cells, respectively.
FIG. 7.
The significance of the US3-UL34 catalytic relationship to virus replication is cell type dependent. Replicate cultures of HEp-2 cells (A and B) or Vero cells (C and D) were infected an at MOI of 5 with HSV-1(F) (A to D), vRR1202 (A to D), vRR1202rep (A and C), vRR1204 #1 (A and C), vRR1204 #2 (A and C), vRR1205 #1 (B and D), or vRR1205 #2 (B and D). Residual virus was removed or inactivated with a low-pH wash, and at the indicated times total culture virus was titrated on 143/1099E cells. Virus yields are expressed as PFU per milliliter. Each datum point represents the mean of three experiments. Error bars indicate the sample standard deviations. For ease of comparison with recombinant viruses, the growth data presented in panels A and C for HSV-1(F) and vRR1202 are duplicated in panels B and D.
In HEp-2 cells (Fig. 7A and B), HSV-1(F) replication entered the productive phase by 6 h.p.i. and reached a plateau titer of approximately 7.7 × 107 PFU per ml between 20 and 48 h.p.i. Conversely, replication of the US3 null virus (vRR1202) did not enter the productive phase until between 6 and 8 h.p.i. and reached a plateau titer of <7 × 106 PFU/ml between 20 and 48 h.p.i. These results are consistent with our previous report, which utilized independent US3 null viruses (35). Repair of the US3 locus (vRR1202rep) restored replication to HSV-1(F)-like kinetics. This result confirms that the observed growth defect is specifically due to the loss of US3 expression.
Recombinant viruses encoding the K220A mutant form of US3 protein (vRR1204 #1 and vRR1204 #2) displayed growth characteristics similar to that of the US3 null virus (Fig. 7A), while recombinant viruses encoding the phosphorylation-defective mutant UL34 protein (vRR1205 #1 and vRR1205 #2) replicated to levels similar to those of HSV-1(F) (Fig. 7B). These data suggest that efficient replication of HSV-1 in HEp-2 cells requires a US3-directed phosphorylation event but that the relevant substrate is not the UL34 protein.
In Vero cells (Fig. 7C and D), HSV-1(F) replication entered the productive phase between 6 and 8 h.p.i. and reached a plateau titer of approximately 1.5 × 108 to 2.5 × 107 PFU/ml between 20 and 48 h.p.i. The onset of replication for the US3 null virus (vRR1202) and for both of the US3 K220A recombinants (vRR1204 #1 and #2) was slightly delayed compared to that of HSV-1(F). However, since an equal delay was observed for the US3 repair virus (vRR1202rep), this cannot be attributed to the US3 protein (Fig. 7C). US3 null, US3 K220A, and US3 repair viruses all grew to final titers that were similar to that of HSV-1(F). These results are consistent with previous reports (20, 30).
Interestingly, replication of recombinant viruses encoding the phosphorylation-defective mutant UL34 protein (vRR1205 #1 and vRR1205 #2) did not enter the productive phase until after 8 h.p.i., but reached plateau titers similar to that of HSV-1(F) (Fig. 7D).
Ultrastructural morphology of recombinant virus-infected cells.
We and members of our laboratory previously reported that loss of US3 protein expression results in an accumulation of enveloped virions within intranuclear vesicles (35). However, the goal of those previous experiments was to determine the ultrastructural localization of the UL34 and UL31 proteins, and consequently, the morphology of these structures was unclear. Here we present an ultrastructural analysis of infected cells with an emphasis on morphological characteristics.
HEp-2 and Vero cells were infected with HSV-1(F), vRR1202, vRR1202rep, vRR1204 #1, vRR1204 #2, vRR1205 #1, or vRR1205 #2 for 20 h, fixed, and processed for electron microscopy. Similar results were obtained with both cell types and representative images of infected Vero cells are shown in Fig. 8.
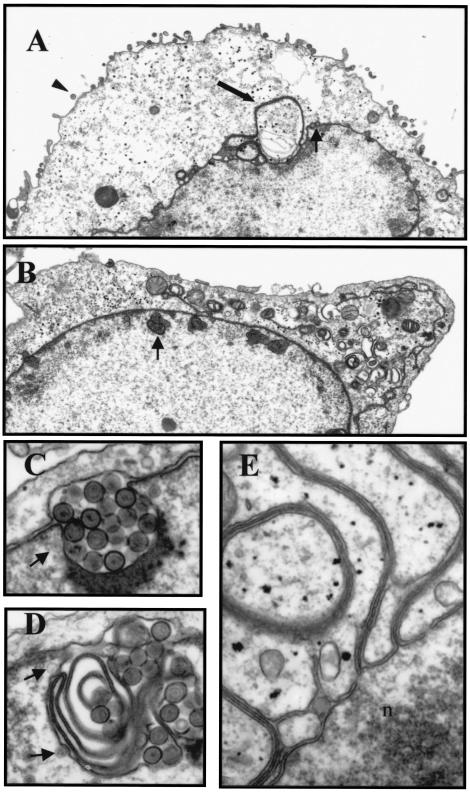
FIG. 8.
Ultrastructural analysis of recombinant virus-infected cells. Vero cells were infected at an MOI of 5 with HSV-1(F) (A and E) or the US3 null virus vRR1202 (B, C, and D). At 20 h.p.i., cells were analyzed by transmission electron microscopy. (A) The arrowhead indicates an extracellular virion, the thick arrow points to four-membrane-bound vesicle, and the thin arrow shows a perinuclear virion. (B) The arrow indicates an example of an intranuclear vesicle containing enveloped virions observed in US3 null virus-infected cells. (C) High magnification of a US3 null virus-associated intranuclear vesicle, like that indicated in panel B, appearing as expanded perinuclear space. (D) High magnification of a US3 null virus-associated intranuclear vesicle, like that indicated in panel B, appearing as concentric lamellae. (E) High magnification of four-membrane-bound vesicle at the junction with the nuclear envelope. n, interior of the nucleus. Original magnification: ×7,000 (A), ×8,000 (B), ×60,000 (C, D, and E).
Cells infected with HSV-1(F) (Fig. 8A) had numerous extracellular virions attached to their surfaces as well as many primary enveloped virions within the perinuclear space. Primary enveloped virions were generally isolated or in small, linear groups. Unenveloped nucleocapsids within the cytoplasm were also observed, as were enveloped virions within cytoplasmic membrane compartments. These observations are consistent with those previously reported (13).
HSV-1(F)-infected cells also contained large vesicle-like structures that either appeared free in the cytoplasm or as projections of the nuclear envelope. These structures varied in relative size from about 1/64 to 1/8 the size of the nucleus. At a low magnification, they appeared to contain nucleoplasm (including viral capsids) and their membranes were thicker and more heavily poststained than would be expected for a single or double lipid bilayer (Fig. 8A). Higher magnification analysis of these membrane structures demonstrated that they are derived from the nuclear envelope and that they consist of four lipid bilayers (Fig. 8E). Since the cells were thin-sectioned prior to electron microscopy analysis, it is possible that those structures that appeared free within the cytoplasm were actually connected to the nuclear envelope either above or below the thin-section plane. Similar structures have been reported, but how they might form and what significance they might have on virus replication are not at all understood (5, 38).
Consistent with previous results, infection of cells with a US3 null virus (vRR1202) yielded very different results (Fig. 8B) (15, 35). The surfaces of these cells were noticeably devoid of enveloped virions, and enveloped virions accumulated within vesicles that lined the interior periphery of the nuclei. Intranuclear vesicles appeared either as obvious expansions of the perinuclear space (Fig. 8C) or as convoluted membrane structures (Fig. 8D). Additionally, these structures had a disproportionate number of what appeared to be envelopment events associated with them, suggesting that they represent preferred sites of envelopment (Fig. 8C and D, arrows).
Interestingly, US3 null virus-infected cells did not contain membrane structures similar to the four-membrane-bound vesicles observed in HSV-1(F)-infected cells. Instead, the cytoplasm of US3 null-infected cells was heavily vesiculated (Fig. 8B).
Cells infected with recombinant viruses carrying the K220A US3 mutation (vRR1204 #1 and vRR1204 #2) appeared similar to those infected with the US3 null virus (not shown). In contrast, cells infected with either the US3 repair virus (vRR1202rep) or recombinants carrying the mutation of the UL34 phosphorylation site (vRR1205 #1 and vRR1205 #2) appeared similar to HSV-1(F)-infected cells, including the presence of the described four-membrane-bound structures (not shown).
Quantitative ultrastructural analysis of recombinant virus-infected cells.
Given the effect that US3 mutation or deletion had on ultrastructural morphology of the infected cell, it was of interest to quantitate the differences in distribution of viral particles in recombinant virus-infected cells. Low-magnification electron micrographs, similar to those shown in Fig. 8A and B, were examined and enveloped virus particles were counted and classified according to the morphological characteristics described above, i.e., in what type of cellular compartment they were found. These data are presented in Table 3 as percentages of total particles counted that were observed in each classification.
TABLE 3.
Enveloped virus particles observed in electronmicroscopy thin sections
| Virusa | % of enveloped virus particles in compartment
|
Total counted (particles/cells) | |||
|---|---|---|---|---|---|
| Perinuclear area | Intranuclear vesiclesb | Extracellular | Otherc | ||
| HSV-1(F) | 35.3 | 0.0 | 38.4 | 26.3 | 365/9 |
| vRR1202rep | 24.2 | 0.0 | 57.0 | 18.8 | 293/9 |
| vRR1202 | 3.2 | 73.1 | 6.7 | 17.0 | 505/9 |
| vRR1204 #1 | 9.6 | 75.0 | 6.6 | 8.8 | 604/8 |
| vRR1204 #2 | 5.0 | 76.8 | 4.3 | 14.0 | 564/9 |
| vRR1205 #1 | 49.0 | 0.0 | 23.0 | 23.2 | 296/10 |
| vRR1205 #2 | 34.9 | 0.0 | 18.2 | 46.9 | 418/8 |
Vero cells were infected with the indicated virus at an MOI of 5 for 20 h.
As shown in Fig. 7.
Golgi complex, endoplasmic reticulum, or other vesicular compartment.
In cells infected with HSV-1(F), 35.3% of the total particles counted were enveloped and within the perinuclear space. The corresponding values for viruses vRR1202rep, vRR1205 #1, and vRR1205 #2 were similar. However, in cells infected with US3 mutant viruses (vRR1202, vRR1204 #1, and vRR1204 #2), the frequency of isolated perinuclear, enveloped particles was reduced approximately 3.5- to 11-fold.
Enveloped particles were commonly observed on the surfaces of HSV-1(F)-infected cells and represented 38.4% of the total particles counted. In cells infected with US3 mutant viruses, the frequency of enveloped extracellular particles was reduced approximately six- to ninefold. Interestingly, the frequency of extracellular virions observed on the surfaces of cells infected with UL34 phosphorylation-site mutant viruses was reduced about twofold compared to HSV-1(F)-infected cells. Significantly, in US3 mutant virus-infected cells, approximately 75% of observed particles were enveloped and contained within the intranuclear vesicles described above, which were absent from cells infected with viruses encoding wild-type US3 protein.
DISCUSSION
The US3 gene of HSV-1 encodes a serine/threonine kinase whose functions in virus replication remain enigmatic. In addition to protecting infected cells from apoptosis (12, 19, 22), data presented in this report and elsewhere indicate that the US3 kinase determines the phosphorylation state of the UL34 protein and may be a component of the envelopment-deenvelopment machinery (31, 32, 35). This report describes experiments aimed at better understanding the mechanism by which US3 facilitates nuclear egress and the effect(s) that phosphorylation might have on UL34 protein function.
Phosphorylation of the UL34 protein by the US3 gene-encoded kinase.
To examine the significance of the US3-UL34 catalytic relationship for primary envelopment and viral replication, we constructed recombinant viruses harboring mutations in either the UL34 or US3 gene. The UL34 protein was immunoprecipitated from 32P-labeled infected HEp-2 or Vero cell lysates and analyzed by SDS-PAGE (Fig. 5). Deletion or mutational inactivation of the US3 kinase dramatically reduced, but did not eliminate, phosphorylation of the UL34 protein. Conversely, when the US3 consensus phosphorylation site within the UL34 protein was inactivated by mutation, phosphorylation of the UL34 protein was undetectable.
Several conclusions may be drawn from these results. First, the phosphorylation state of the UL34 protein is largely determined by the US3 kinase. Second, at least one other infected cell kinase can phosphorylate the UL34 protein, albeit with less efficiency than the US3 protein. Finally, phosphorylation of the UL34 protein by any kinase in HEp-2 or Vero cells is limited to threonine 195 and/or serine 198 (the potential targets within the US3 consensus phosphorylation site). It is unknown whether either of these two residues is the preferred target of phosphorylation or if they are used equally.
Purves et al. reported that, in BHK cells, the UL34 protein is exclusively phosphorylated by the US3 protein (31, 32). The results presented here are similar inasmuch as in both cases, the US3 protein mostly determines the phosphorylation state of the UL34 protein. One discrepancy, however, is that Purves et al. observed that when UL34 is unphosphorylated, because of either US3 deletion or UL34 mutation, four other phosphoproteins can be coimmunoprecipitated with the UL34 protein (32). Since we did not observe a similar phenomenon with HEp-2 or Vero cells, this may represent a peculiarity of BHK cells.
Also relevant to this analysis is the fact the phosphorylation state of the UL34 homologue encoded by PRV does not appear to be affected by the US3 protein in infected RK-13 cells (15). However, the phenotypes of UL34 and US3 deletion mutants of PRV are similar to those of the corresponding recombinant HSV-1 mutants (15, 16, 34, 35, 37).
The significance of the US3 catalytic activity on HSV replication.
Data presented here and elsewhere indicate that the US3 protein facilitates nuclear egress of HSV-1 (15, 35, 42). It is likely that the mechanism involves US3-directed phosphorylation of one or more infected cell proteins since mutational inactivation of the US3 protein and deletion of the US3 ORF yield similar phenotypes. Specifically, in the absence of functional US3 protein, the HSV-1 envelopment factor, the UL34 protein, is mislocalized into punctate structures at the nuclear envelope. At the ultrastructural level, these punctate structures appear to be vesicular accumulations of enveloped virus particles at the interior perimeter of the nucleus (35). Electron microscopic analysis indicates that these vesicles are derived from the perinuclear space (Fig. 8) and may represent a significant block to viral morphogenesis (Table 3).
The causal relationship between mislocalization of the UL34 protein into punctate structures and the observed intranuclear vesicles is unclear. It is possible that mislocalization of the UL34 protein into clusters results in the formation of intranuclear vesicles by directing envelopment at discrete positions along the inner nuclear membrane. Alternatively, UL34 protein clustering might be a secondary effect of intranuclear vesicle formation.
In HEp-2 cells, US3 deletion or mutation also results in a significant growth defect. Production of infectious US3 mutant (or null) progeny is both delayed and decreased compared to that for HSV-1(F) (Fig. 7). It is tempting to speculate that the growth defects associated with US3 mutation are directly related to the observed alterations in virion morphogenesis. However, this hypothesis is complicated by the fact that US3 mutation yields a similar morphological phenotype in Vero cells without apparent growth defects.
The discrepancy between HEp-2 and Vero cells in the ability to support the replication of viruses that fail to express functional US3 is likely a complex issue since there is not only a decrease, but also a delay in onset, of replication in HEp-2 cells. While many possible hypotheses exist to explain the differences between cell types, two seem particularly attractive.
The first hypothesis is based on the fact that the growth assay used measures both cell-free and cell-associated virus. Some data have suggested that primary enveloped virions may possess some infectivity (4). However, they are likely less infectious than mature virions. It is possible that the difference in specific infectivity between perinuclear and mature virions may be greater in HEp-2 cells than in Vero cells. If the accumulation of perinuclear virions in US3 mutant virus-infected cells represents a significant block to virus morphogenesis, then most of the infectivity produced should be due to perinuclear virions. In contrast, most of the infectivity produced in wild-type virus-infected cells should be derived from mature virions. Therefore, if the specific infectivity of perinuclear virions in Vero cells is near that of mature virions, then the growth assay used may not detect the difference between the US3 mutant and wild-type HSV-1.
Alternatively, since US3 has some ability to prevent apoptosis in infected cells (12, 19, 22), it is possible that HEp-2 cells are more sensitive to virus-induced apoptosis than are Vero cells. If so, then US3 mutant HSV-infected HEp-2 cells might die by apoptosis before the virus can replicate to high titers. However, this seems unlikely to account for the observed delay in the onset of replication. The effects of apoptosis on single-step growth of HSV-1 are currently being investigated.
It has been suggested that the reduced pathogenicity of US3 null HSV in mice is due to the lack of US3 antiapoptotic activity (1, 17, 28). The data presented here and elsewhere suggest that the egress defects associated with US3 deletion may also contribute to the reduced pathogenicity (15, 35, 42).
The significance of UL34 phosphorylation on HSV replication.
Phosphorylation is a common mechanism for regulating the function of proteins. As described here and elsewhere, the HSV-1 UL34 protein is phosphorylated principally by the US3 kinase (31, 32). However, the significance of UL34 phosphorylation is unknown. A reasonable hypothesis was that the effect of US3 deletion or mutation on viral replication is mediated by the loss of UL34 protein phosphorylation, suggesting that UL34 function is regulated by US3-directed phosphorylation. As was the case for US3 catalytic activity, the importance of UL34 protein phosphorylation differed between the cell lines tested.
In HEp-2 cells, recombinant HSV-1 strains that encoded a nonphosphorylated mutant UL34 protein did not differ significantly from HSV-1(F) in any parameter tested. Therefore, since US3 catalytic activity is necessary for proper virion morphogenesis (see above), there likely exists an unidentified US3 substrate that is important for HSV-1 replication.
In Vero cells, the same recombinant viruses displayed a significant delay in the onset of infectious progeny production (Fig. 7). Unlike the situation with US3 catalytic activity, however, the growth defect observed for these viruses was not associated with any obvious morphological abnormality or mislocalization of the UL34 protein. Since no growth defect was observed when US3 was deleted or mutated (see above), it may be that low-level phosphorylation of the UL34 protein by a cellular kinase facilitates an interaction with a cellular factor(s) that is important in Vero cells but dispensable in HEp-2 cells. Alternatively, mutation of the US3 consensus phosphorylation site of UL34 may destroy an important interaction site independent of phosphorylation.
Potential substrates of the US3 kinase.
The data presented in this report indicate that the importance of the catalytic relationship between the HSV-1 US3 kinase and the UL34 protein is cell type dependent. Additionally, it seems likely that an infected cell protein other than UL34 exists that must be phosphorylated to allow efficient virion morphogenesis. In HEp-2 cells, HSV-1 apparently uses the US3 protein to accomplish this, while Vero cells seem to possess an endogenous kinase(s) that can suffice. This is consistent with results obtained with PRV. As mentioned above, deletion of the US3 ORF from either PRV or HSV-1 results in similar phenotypes in spite of the fact that in PRV-infected cells, the US3 and UL34 proteins do not appear catalytically related (15). However, those experiments were only performed with one cell type and it would be interesting to see if there is a similar cell type dependence in PRV as described in this report for HSV-1.
Physiologically relevant substrates for the US3 protein may be either virus or host encoded. The HSV-2 US3 kinase has been reported to affect the phosphorylation of the viral US9 and UL12 proteins and the cellular protein cytokeratin 17 (9, 24). Given the information on US9 and UL12 function, they seem unlikely to account for the US3-dependent effects described in this report and the effect of US3 on cytokeratin 17 phosphorylation was only observed under conditions of induced overexpression of US3 in a stably transfected cell line (7, 24, 27, 40). Therefore, the significance of these potential catalytic relationships in viral replication is unclear.
The following observations suggest that a relevant US3 substrate may be a cellular nuclear envelope protein: (i) the necessity of US3 catalytic activity is a cell type-dependent phenomenon, (ii) the US3 protein is localized to the nuclear envelope in a UL34 protein-dependent manner, and (iii) US3 deletion appears to affect nuclear envelope architecture.
Previous data indicated that the nuclear lamina represents a significant physical barrier to nuclear egress and that herpesviruses encode proteins that can disrupt the integrity of the lamina, thereby allowing envelopment and deenvelopment (23, 39). Specifically, the UL34 homologue of murine cytomegalovirus was shown to recruit protein kinase C to the nuclear envelope, which led to breakdown of the nuclear lamina (23).
The reported consensus target sequence of the PRV US3 protein is (R)nX(S/T)YY, where n ≥ 3; X = R, A, V, P, or S; Y = X ≠ P or acidic residues; and S/T is the phosphorylated residue (18, 29). Of the nuclear envelope proteins whose functions are likely relevant to HSV nuclear egress, lamin B receptor contains this consensus sequence near the N terminus. Hyperphosphorylation of lamin B receptor by cellular kinases near the N terminus is involved in dismantling the nuclear lamina during mitosis (8, 25, 26).
Acknowledgments
We thank Bernard Roizman of the University of Chicago for the US3 antisera and Jean Ross of the University of Iowa Central Microscopy Research Facility for expert technical assistance. We also thank Stanley Perlman of the University of Iowa for critical manuscript review.
This work was supported by the University of Iowa and Public Health Service Award AI41478 and NIH training grant AI07533 to the University of Iowa (B.J.R.).
REFERENCES
- 1.Asano, S., T. Honda, F. Goshima, D. Watanabe, Y. Miyake, Y. Sugiura, and Y. Nishiyama. 1999. US3 protein kinase of herpes simplex virus type 2 plays a role in protecting corneal epithelial cells from apoptosis in infected mice. J. Gen. Virol. 80:51-56. [DOI] [PubMed] [Google Scholar]
- 2.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines, J. D., and B. Roizman. 1992. The UL11 gene of herpes simplex virus 1 encodes a function that facilitates nucleocapsid envelopment and egress from cells. J. Virol. 66:5168-5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baines, J. D., P. L. Ward, G. Campadelli-Fiume, and B. Roizman. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65:6414-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibor-Hardy, V., M. Suh, M. Pouchelet, and R. Simard. 1982. Modifications of the nuclear envelope of BHK cells after infection with herpes simplex virus type 1. J. Gen. Virol. 63:81-94. [DOI] [PubMed] [Google Scholar]
- 6.Bjerke, S. L., J. M. Cowan, J. K. Kerr, A. E. Reynolds, J. D. Baines, and R. J. Roller. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J. Virol. 77:7601-7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandimarti, R., and B. Roizman. 1997. US9, a stable lysine-less herpes simplex virus 1 protein, is ubiquitinated before packaging into virions and associates with proteasomes. Proc. Natl. Acad. Sci. USA 94:13973-13978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courvalin, J. C., N. Segil, G. Blobel, and H. J. Worman. 1992. The lamin B receptor of the inner nuclear membrane undergoes mitosis specific phosphorylation and is a substrate for the p34cdc2-type protein kinase. J. Biol. Chem. 267:19035-19038. [PubMed] [Google Scholar]
- 9.Daikoku, T., R. Kurachi, T. Tsurumi, and Y. Nishiyama. 1994. Identification of a target protein of US3 protein kinase of herpes simplex virus type 2. J. Gen. Virol. 75:2065-2068. [DOI] [PubMed] [Google Scholar]
- 10.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characteristics of herpes simplex virus strains differing in their effect on social behavior of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs, W., B. G. Klupp, H. Granzow, and T. C. Mettenleiter. 1997. The UL20 gene product of pseudorabies virus functions in virus egress. J. Virol. 71:5639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galvan, V., and B. Roizman. 1998. Herpes simplex virus 1 induces and blocks apoptosis at multiple steps during infection and protects cells from exogenous inducers in a cell-type-dependent manner. Proc. Natl. Acad. Sci. USA 95:3931-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanks, S. K., A. M. Quinn, and T. Hunter. 1988. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241:42-52. [DOI] [PubMed] [Google Scholar]
- 15.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363-2371. [DOI] [PubMed] [Google Scholar]
- 16.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurachi, R., T. Daikoku, T. Tsurumi, K. Maeno, Y. Nishiyama, and T. Kurata. 1993. The pathogenicity of a US3 protein kinase-deficient mutant of herpes simplex virus type 2 in mice. Arch. Virol. 133:259-273. [DOI] [PubMed] [Google Scholar]
- 18.Leader, D. P., A. D. Deana, F. Marchiori, F. C. Purves, and L. A. Pinna. 1991. Further definition of the substrate specificity of the alpha-herpesvirus protein kinase and comparison with protein kinases a and c. Biochim. Biophys. Acta 1091:426-431. [DOI] [PubMed] [Google Scholar]
- 19.Leopardi, R., C. Van Sant, and B. Roizman. 1997. The herpes simplex virus 1 protein kinase US3 is required for protection from apoptosis induced by the virus. Proc. Natl. Acad. Sci. USA 94:7891-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longnecker, R., and B. Roizman. 1987. Clustering of genes dispensable for growth in culture in the s component of the HSV-1 genome. Science 236:573-576. [DOI] [PubMed] [Google Scholar]
- 21.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 22.Munger, J., A. V. Chee, and B. Roizman. 2001. The US3 protein kinase blocks apoptosis induced by the d120 mutant of herpes simplex virus 1 at a premitochondrial stage. J. Virol. 75:5491-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muranyi, W., J. Haas, M. Wagner, and U. H. Koszinowski. 2002. Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 287:854-856. [DOI] [PubMed] [Google Scholar]
- 24.Murata, T., F. Goshima, Y. Nishizawa, T. Daikoku, H. Takakuwa, K. Ohtsuka, T. Yoshikawa, and Y. Nishiyama. 2002. Phosphorylation of cytokeratin 17 by herpes simplex virus type 2 US3 protein kinase. Microbiol. Immunol. 46:707-719. [DOI] [PubMed] [Google Scholar]
- 25.Nikolakaki, E., J. Meier, G. Simos, S. D. Georgatos, and T. Giannakouros. 1997. Mitotic phosphorylation of the lamin B receptor by a serine/arginine kinase and p34cdc2. J. Biol. Chem. 272:6208-6213. [DOI] [PubMed] [Google Scholar]
- 26.Nikolakaki, E., G. Simos, S. D. Georgatos, and T. Giannakouros. 1996. A nuclear envelope-associated kinase phosphorylates arginine-serine motifs and modulates interactions between the lamin B receptor and other nuclear proteins. J. Biol. Chem. 271:8365-8372. [DOI] [PubMed] [Google Scholar]
- 27.Nishiyama, Y., R. Kurachi, T. Daikoku, and K. Umene. 1993. The US9, 10, 11, and 12 genes of herpes simplex virus type 1 are of no importance for its neurovirulence and latency in mice. Virology 194:419-423. [DOI] [PubMed] [Google Scholar]
- 28.Nishiyama, Y., Y. Yamada, R. Kurachi, and T. Daikoku. 1992. Construction of a US3 lacZ insertion mutant of herpes simplex virus type 2 and characterization of its phenotype in vitro and in vivo. Virology 190:256-268. [DOI] [PubMed] [Google Scholar]
- 29.Purves, F. C., A. D. Deana, F. Marchiori, D. P. Leader, and L. A. Pinna. 1986. The substrate specificity of the protein kinase induced in cells infected with herpesviruses: studies with synthetic substrates indicate structural requirements distinct from other protein kinases. Biochim. Biophys. Acta 889:208-215. [DOI] [PubMed] [Google Scholar]
- 30.Purves, F. C., R. M. Longnecker, D. P. Leader, and B. Roizman. 1987. Herpes simplex virus 1 protein kinase is encoded by open reading frame US3 which is not essential for virus growth in cell culture. J. Virol. 61:2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purves, F. C., D. Spector, and B. Roizman. 1991. The herpes simplex virus 1 protein kinase encoded by the US3 gene mediates posttranslational modification of the phosphoprotein encoded by the UL34 gene. J. Virol. 65:5757-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purves, F. C., D. Spector, and B. Roizman. 1992. UL34, the target of the herpes simplex virus US3 protein kinase, is a membrane protein which in its unphosphorylated state associates with novel phosphoproteins. J. Virol. 66:4295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rauch, D. A., N. Rodriguez, and R. J. Roller. 2000. Mutations in herpes simplex virus glycoprotein D distinguish entry of free virus from cell-cell spread. J. Virol. 74:11437-11446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reynolds, A. E., B. J. Ryckman, J. D. Baines, Y. Zhou, L. Liang, and R. J. Roller. 2001. UL31 and UL34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803-8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reynolds, A. E., E. G. Wills, R. J. Roller, B. J. Ryckman, and J. D. Baines. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939-8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roller, R. J., and B. Roizman. 1992. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunits. J. Virol. 66:3624-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roller, R. J., Y. Zhou, R. Schnetzer, J. Ferguson, and D. DeSalvo. 2000. Herpes simplex virus type 1 UL34 gene product is required for viral envelopment. J. Virol. 74:117-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlehofer, J. R., H. Hampl, and K. O. Habermehl. 1979. Differences in the morphology of herpes simplex virus infected cells. I. Comparative scanning and transmission electron microscopic studies on HSV-1 infected Hep-2 and chick embryo fibroblast cells. J. Gen. Virol. 44:433-442. [DOI] [PubMed] [Google Scholar]
- 39.Scott, E., and P. O'Hare. 2001. Fate of the inner nuclear membrane protein lamin B receptor and nuclear lamins in herpes simplex virus type 1 infection. J. Virol. 75:8818-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao, L., L. M. Rapp, and S. K. Weller. 1993. Herpes simplex virus 1 alkaline nuclease is required for efficient egress of capsids from the nucleus. Virology 196:146-162. [DOI] [PubMed] [Google Scholar]
- 41.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment-deenvelopment-reenvelopment pathway. J. Virol. 75:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagenaar, F., J. M. Pol, B. Peeters, A. L. Gielkens, N. de Wind, and T. G. Kimman. 1995. The US3-encoded protein kinase from pseudorabies virus affects egress of virions from the nucleus. J. Gen. Virol. 76:1851-1859. [DOI] [PubMed] [Google Scholar]